Abstract
Simple Summary
We investigated the influence of GWAS-identified variants for T2D in modulating prostate cancer (PCa) risk through a meta-analysis of our data with those from the UKBiobank and FinnGEn cohorts and four large European cohorts. We found that genetic variants within the FTO, HNF1B, and JAZF1 loci were associated with PCa risk. Our results also suggested, for the first time, a potentially interesting association of SNPs within NOTCH2 and RBMS1 genes that need to be further explored and validated. This study also shed some light onto the functional mechanisms behind the observed associations, and demonstrated that the HNF1Brs7501939 polymorphism correlated with lower levels of SULT1A1, an enzyme responsible for the sulfate conjugation of multiple endogenous and exogenous compounds. Furthermore, we found that SNPs within the HFN1B, NOTCH2, and RBMS1 genes impacted PCa risk through the modulation of mRNA gene expression levels of their respective genes. However, given the healthy nature of the subjects included in the cohort used for functional experiments, the link between the HNF1B locus and SULT1A1 should be considered still speculative and, therefore, requires further validation.
Abstract
In this study, we have evaluated whether 57 genome-wide association studies (GWAS)-identified common variants for type 2 diabetes (T2D) influence the risk of developing prostate cancer (PCa) in a population of 304 Caucasian PCa patients and 686 controls. The association of selected single nucleotide polymorphisms (SNPs) with the risk of PCa was validated through meta-analysis of our data with those from the UKBiobank and FinnGen cohorts, but also previously published genetic studies. We also evaluated whether T2D SNPs associated with PCa risk could influence host immune responses by analysing their correlation with absolute numbers of 91 blood-derived cell populations and circulating levels of 103 immunological proteins and 7 steroid hormones. We also investigated the correlation of the most interesting SNPs with cytokine levels after in vitro stimulation of whole blood, peripheral mononuclear cells (PBMCs), and monocyte-derived macrophages with LPS, PHA, Pam3Cys, and Staphylococcus Aureus. The meta-analysis of our data with those from six large cohorts confirmed that each copy of the FTOrs9939609A, HNF1Brs7501939T, HNF1Brs757210T, HNF1Brs4430796G, and JAZF1rs10486567A alleles significantly decreased risk of developing PCa (p = 3.70 × 10−5, p = 9.39 × 10−54, p = 5.04 × 10−54, p = 1.19 × 10−71, and p = 1.66 × 10−18, respectively). Although it was not statistically significant after correction for multiple testing, we also found that the NOTCH2rs10923931T and RBMS1rs7593730 SNPs associated with the risk of developing PCa (p = 8.49 × 10−4 and 0.004). Interestingly, we found that the protective effect attributed to the HFN1B locus could be mediated by the SULT1A1 protein (p = 0.00030), an arylsulfotransferase that catalyzes the sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds. In addition to these results, eQTL analysis revealed that the HNF1Brs7501939, HNF1Brs757210, HNF1Brs4430796, NOTCH2rs10923931, and RBMS1rs7593730 SNPs influence the risk of PCa through the modulation of mRNA levels of their respective genes in whole blood and/or liver. These results confirm that functional TD2-related variants influence the risk of developing PCa, but also highlight the need of additional experiments to validate our functional results in a tumoral tissue context.
Keywords: prostate cancer, genetic susceptibility, type 2 diabetes-related variants
1. Introduction
Prostate cancer (PCa) is the second most common cancer worldwide and one of the first leading causes of cancer-related deaths in men in developed countries [1]. It accounts for 7.3% of all cancers, with an incidence of 37.5 per 100,000 individuals [2]. Despite the refinements in prevention strategies, a total of 1.4 million new cases were diagnosed in 2020 [2], and the incidence of the disease is increasing over the world, likely due to the interaction of both inherited and modifiable factors [3,4].
Although PCa has a high prevalence among males, only age, family history, and ethnicity have been established as major risk factors for the disease, with an attributable effect ranging from 5 to 9% of the cases [5]. In addition, rare highly penetrant mutations in specific genes, high levels of endogenous androgens, smoking, alcohol consumption, exposure to chemical compounds, sexually transmitted infections, diet, obesity, insulin-like growth factors, and type 2-diabetes (T2D) have been suggested as important modulators of the prostatic tumorigenesis [6]. Among the modifiable risk factors, T2D has attracted significant attention, since it has been consistently identified as a protective factor for PCa development [7,8,9], but it seems to induce disease progression. Several studies have suggested that the use of anti-diabetic drugs such as metformin might account for this protective effect of T2D on PCa risk [10,11], but other studies were not able to confirm these results in larger cohorts [12], which suggested that the protective effect attributed to T2D on PCa depend on common molecular pathways between these traits rather than the use of anti-diabetic drugs.
Although research on the specific pathways interfering in the development of T2D and PCa traits is still under way, a mounting body of evidence suggests that these diseases might share a genetic component [13,14,15,16]. Recent studies have reported that common genetic polymorphisms within T2D-related genes have an important role in modulating the risk of many cancers [17,18,19], but, so far, only a few studies have investigated the impact of diabetogenic variants on PCa risk, showing controversial results [20,21,22,23]. Considering this background, we decided to conduct a population-based case-control study including 994 subjects (304 PCa patients and 686 controls) to evaluate whether 57 diabetogenic variants identified through genome-wide association studies (GWAS) are associated with the risk of developing Pca. In order to validate the association of T2D-related markers with Pca risk, when possible, we performed a meta-analysis with data from previous genetic studies. Finally, we analyzed whether the most interesting markers correlated with absolute numbers of 91 blood-derived cell populations, 106 immunological serum proteins, 7 steroid hormones, and 9 cytokines (IFNγ, IL1β, IL1Ra, IL6, IL8, IL10, IL17, IL22, and TNFα) after stimulation of whole blood, peripheral blood mononuclear cells (PBMCs), and monocyte-derived macrophages with LPS, PHA, Pam3Cys, and Staphylococcus aureus.
2. Materials and Methods
2.1. Study Population
The study cohort consisted of 304 Caucasian PCa patients and 686 male healthy controls recruited in the Virgen de las Nieves University hospital (Granada, Spain) and the Complejo Hospitalario de Cáceres (Cáceres, Spain). Only patients without any prior history of malignancy, and who were not treated before blood withdrawal, were enrolled in this study. Patient characteristics are included in Table 1. The diagnosis of PCa was assigned by physicians, and fulfilled the international criteria [24]. Male controls with a mean age of 58.92 were blood donors from the Centro Regional de Transfusiones Sanguíneas de Granada (CRTS) and were selected from the same geographical region of the cases. In accordance with the Declaration of Helsinki, all participants gave their written informed consent to participate in the study and the ethical committees of the participant institutions approved the study.
Table 1.
Demographic and clinical characteristics of PCa patients.
| Demographic Characteristics | Study Population (n = 990) | |
|---|---|---|
| Age (years) | 62.35 ± 11.51 | |
| Clinical assessment | ||
| PSA | PSA (4–10) | 137 (46.13) |
| PSA (10–20) | 68 (22.90) | |
| PSA (>20) | 92 (30.97) | |
| Gleason | Gleason (≤7) | 220 (73.58) |
| Gleason (8–10) | 79 (26.42) | |
| TNM Staging system | T1–T2 | 209 (76.28) |
| T3–T4 | 65 (23.72) | |
| Risk | High | 63 (26.58) |
| Intermediate | 79 (33.33) | |
| Low | 95 (40.09) | |
2.2. SNP Selection and Genotyping
An extensive literature search concerning the mechanism of action of T2D-related genes was performed to select candidate genes that might affect the risk of developing PCa. SNPs were assessed on the basis of NCBI data, and were selected according to their known or putative functional consequences, i.e., their modifying influence on the structure of proteins, transcription level, or alternative splicing mechanisms. In total, 57 SNPs in 49 genes were selected for this study (Table 2).
Table 2.
Selected type 2 diabetes-related SNPs.
| Gene Name | dbSNP rs# | Nucleotide Substitution | GWAS-Identified Risk Allele for T2D | Location/Aa Substitution | References |
|---|---|---|---|---|---|
| ADAM30 | rs2641348 ɱ | T/C | C | L359P | [25,26] |
| ADAMTS9 | rs4607103 | T/C | C | Near gene | [26,27,28] |
| ADCY5 | rs11708067 | T/C | T | Intronic | [29,30] |
| ADRA2A | rs10885122 | G/T | G | Near ADRA2A | [29] |
| ARAPI, CENTD2 | rs1552224 | C/A | A | Near gene | [31,32] |
| CDC123 | rs12779790 | A/G | G | Near gene | [26,27,28] |
| CDKAL1 | rs7754840 | C/G | C | Intronic | [33,34,35] |
| CDKN2A-2B | rs10811661 | T/C | T | Near gene | [26,27,28,33,35,36,37] |
| COL5A1 | rs4240702 | C/T | n/s | Intronic | [38] |
| CRY2 | rs11605924 | A/C | A | Intronic | [29] |
| DCD | rs1153188 | A/T | A | Near gene | [26] |
| EXT2 | rs1113132 | C/G | C | Intronic | [34,39] |
| FADS1 | rs174550 | C/T | T | Intronic | [29] |
| FAM148B | rs11071657 | A/G | A | Near gene | [29,40] |
| FLJ39370 | rs17044137 | A/T | A | Near gene | [33] |
| FTO | rs9939609 | A/C | A | Intronic | [27,41,42] |
| G6PC2 | rs560887 | G/A | G | Intronic | [29,38,43,44,45] |
| GCK | rs1799884 | G/A | A | Near gene | [29,38,43,44,45] |
| GCKR | rs1260326 | A/G | A | Leu446Pro | [46,47,48,49] |
| HHEX | rs1111875 | G/A | C | Near gene | [27,33,34,35,39,41,42] |
| HMGA2 | rs1531343 | C/G | C | Near gene | [31,32] |
| HNF1A, TCF1 | rs7957197 | A/T | T | Intronic | [31,32] |
| HNF1B, TCF2 | rs7501939 ʯ | C/T | T | Intronic | [14,50] |
| HNF1B, TCF2 | rs757210 | C/T | T | Intronic | [14,31] |
| HNF1B, TCF2 | rs4430796 | G/A | G | Intronic | [14] |
| IGF1 | rs35767 | C/T | C | Near gene | [29,51] |
| IGF2BP2 | rs4402960 | G/T | T | Intronic | [27,33,34,35,42,52] |
| IL13 | rs20541 | C/T | T | R144Q | [33] |
| IRS1 | rs2943641 | C/T | C | Near gene | [31,52,53] |
| JAZF1 | rs864745 | T/C | T | Intronic | [26,28] |
| JAZF1 | rs10486567 | A/G | A | Intronic | [26,28] |
| KCNJ11 | rs5215 | T/C | C | V337I | [27,33,35,41,42,54,55] |
| KCNJ11 | rs5219 ʚ | C/T | T | K23E | |
| KCNQ1 | rs2237897 ʠ | C/T | C | Intronic | [36,56,57,58] |
| KCNQ1 | rs2074196 | G/T | G | Intronic | |
| KCNQ1 | rs2237892 | C/T | C | Intronic | |
| KCNQ1 | rs2237895 | A/C | C | Intronic | |
| KCNQ1OT1 | rs231362 | C/T | G | Intronic | [31,32,57] |
| LTA | rs1041981 | A/C | A | T60N | [59] |
| MADD | rs7944584 | A/T | A | Intronic | [29] |
| MCR4 | rs12970134 | A/G | A | Near gene | [40] |
| MTNR1B | rs1387153 | C/T | T | Near gene | [31,38,45] |
| NOTCH2 | rs10923931 | G/T | T | Intronic | [26,27] |
| PKN2 | rs6698181 | C/T | T | Intergenic | [33] |
| PPARG | rs1801282 | C/G | C | P12A | [26,27,33,35,41,42,54,60] |
| PRC1 | rs8042680 | A/C | A | Intronic | [31,32] |
| PROX1 | rs340874 | A/G | G | Promoter | [29] |
| RBMS1 | rs7593730 | T/C | T | Intronic | [61,62] |
| SLC2A2 | rs11920090 | A/T | T | Intronic | [29] |
| SLC30A8 | rs13266634 | C/T | C | R325W | [27,28,29,33,34,35,39,41,42,63] |
| TCF7L2 | rs7903146 ʞ | C/T | T | Intronic | [27,29,30,33,34,35,39,41,42,63,64,65] |
| TCF7L2 | rs12255372 | G/T | T | Intronic | [66] |
| THADA | rs7578597 | T/C | C | Thr1187Ala | [26,67] |
| TP53INP1 | rs896854 | T/C | A | Intronic | [31,47,67,68] |
| TSPAN8, LGR5 | rs7961581 | C/T | C | Near gene | [69] |
| VEGFA | rs9472138 | C/T | T | Near gene | [26] |
| WFS1 | rs10010131 | A/G | G | Intronic | [50] |
n/s, not specified; Aa, amino acid; GWAS, genome-wide association studies. ɱ That SNP rs2641348 is in complete linkage disequilibrium with the rs10923931, r2 = 1.00. ʞ That SNP rs7903146 is in strong linkage disequilibrium with the rs12255372, r2 = 0.72. ʠ That SNP rs2237897 is in strong linkage disequilibrium with the rs2237892, r2 = 0.79. ʚ That SNP rs5219 is in complete linkage disequilibrium with the rs5215, r2 = 1.00. ʯ That SNP rs7501939 is in strong linkage disequilibrium with the rs4430796, r2 = 0.77.
Selected variants for T2D were genotyped using KASPar® assays (LGC Genomics, London, UK) according to the manufacturer’s instructions. For internal quality control, 5% of samples were randomly included as duplicates. Concordance between the original and the duplicate samples for the 57 SNPs was ≥99.0%. Call rates for all SNPs were ≥90.0%.
2.3. Statistical Analysis
The Hardy–Weinberg Equilibrium (HWE) tests were performed in the control group by a standard observed–expected chi-squared (χ2) test. Logistic regression analyses were used to assess the effects of the genetic polymorphisms on PCa risk using dominant, recessive, and log-additive models of inheritance. Overall analyses were adjusted for age, and conducted using Stata (v12.1). Statistical power was calculated using the Quanto software (vs. 12.4; log-additive model).
In order to account for multiple testing, we calculated an adjusted significance level using data from the SNPclip Tool (https://ldlink.nci.nih.gov/?tab=snpclip, accessed on 8 May 2020), which consider the number of independent marker loci (n = 52). Given the high correlation between the log-additive and dominant models of inheritance, we corrected by log-additive and recessive models, resulting in a significant threshold for the main effect analysis of 0.00048 (0.05/52 SNPs/2 inheritance models). Since a study-wide significance threshold considering all these factors is generally perceived as a too conservative test, we also assessed the magnitude of observed associations between selected SNPs and risk of PCa through a quantile–quantile (QQ) plot generated from the results of the study population. The observed association p-values were ranked in order from smallest to largest on the y-axis and plotted against the expected results from a theoretical ~χ2-distribution under the null hypothesis of no association on the x-axis.
2.4. Meta-Analysis
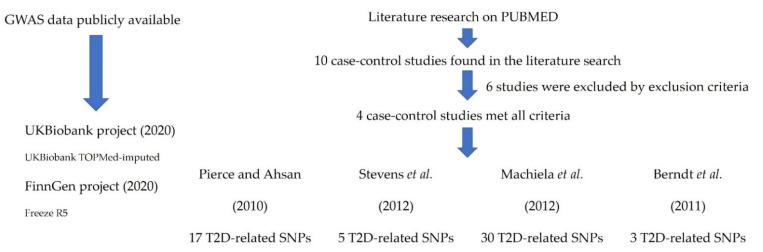
With the aim of assessing the consistency of the association between T2D-related SNPs and the risk of developing PCa, we performed a meta-analysis of our data with those from publicly available GWAS. We downloaded association estimates from the PheWeb site (https://pheweb.sph.umich.edu/, accessed on 11 May 2020) for 6311 PCa cases; 74,685 controls from the FinnGen research project; and 5993 PCa cases and 168,999 controls from the UK Biobank project (UKBiobank TOPMed-imputed). Details on genome-wide associations have been previously reported [70]. Briefly, analyses on binary outcomes were conducted using the SAIGE generalized mixed logistic regression model, adjusting for genetic relatedness, sex, birth year, and the first four principal components. For White British participants of the UK Biobank, endpoint definitions were generated from electronic health-records-derived ICD billing codes, and endpoint definitions for the FinnGen data can be found at risteys.finngen.fi (Risteys = intersection in Finnish). We also validated the association of genetic markers using data from previously published studies that were selected according to the following criteria: (1) GWAS or candidate-gene association studies found in PUBMED (https://www.ncbi.nlm.nih.gov/pubmed, accessed on 13 May 2020) using the following key words: prostate cancer, case-control association study, type 2 diabetes, genetic polymorphisms; (2) Studies using Caucasian populations; (3) Availability of association estimates according to a log-additive model of inheritance; (4) Hardy–Weinberg equilibrium in the control group; and (5) Written in English (Figure 1). We pooled the Odds Ratios (ORs) using a fixed-effect model. Coefficients with a p-value ≤ 0.05 were considered significant. I2 statistic was used to assess heterogeneity between studies. All statistics were calculated using STATA (v. 12).
Figure 1.
2.5. cQTL Analysis of the T2D-Related Variants
Cytokine stimulation experiments were conducted in the 500 Functional Genomics (500FG) cohort from the Human Functional Genomics Project (HFGP; http://www.humanfunctionalgenomics.org/, accessed on 7 July 2020). The HFGP study was approved by the Arnhem-Nijmegen Ethical Committee (no. 42561.091.12), and biological specimens were collected after informed consent was obtained. We investigated whether any of the 57 T2D-related SNPs correlated with cytokine levels (IFNγ, IL1β, IL1Ra, IL6, IL8, IL10, IL17, IL22, and TNFα) after the stimulation of peripheral blood mononuclear cells (PBMCs), macrophages, or whole blood from 172 healthy men with LPS (1 or 100 ng/mL), PHA (10 μg/mL), Pam3Cys (10 μg/mL), and Staphylococcus aureus. After log transformation, linear regression analyses adjusted for age were used to determine the correlation of selected SNPs with cytokine expression quantitative trait loci (cQTLs). All analyses were performed using R software (http://www.r-project.org/, accessed on 8 May 2020). In order to account for multiple comparisons, we used a significant threshold of 0.000106 (0.05/52 independent SNPs × 9 cytokines).
Details on PBMCs isolation, macrophage differentiation, and stimulation assays have been reported elsewhere [72,73,74]. Briefly, PBMCs were washed twice in saline, and suspended in medium (RPMI 1640) supplemented with gentamicin (10 mg/mL), L-glutamine (10 mM), and pyruvate (10 mM). PBMC stimulations were performed with 5 × 105 cells/well in round-bottom 96-well plates (Greiner) for 24 h in the presence of 10% human pool serum at 37 °C and 5% CO2. Supernatants were collected and stored in −20 °C until used for ELISA. LPS (100 ng/mL), PHA (10 μg/mL), and Pam3Cys (10 μg/mL) were used as stimulators for 24 or 48 h. Whole blood stimulation experiments were conducted using 100 μL of heparin blood that was added to a 48-well plate and subsequently stimulated with 400 μL of LPS and PHA (final volume 500 μL) for 48 h at 37 °C and 5% CO2. Supernatants were collected and stored in −20 °C until used for ELISA. Concentrations of human cytokines were determined using specific commercial ELISA kits (PeliKine Compact, Amsterdam, The Netherlands or R&D Systems, Minneapolis, MN, USA), following the manufacturer’s instructions.
2.6. Correlation between T2D-Related Polymorphisms and Cell Counts of 91 Blood-Derived Immune Cell Populations and 103 Serum/Plasmatic Immunological Proteins
We also investigated whether selected polymorphisms had an impact on blood cell counts by analyzing a set of 91 manually annotated immune cell populations and genotype data from the 500 FG cohort that consisted of 172 healthy men (Supplementary Table S1). Cell populations were measured by 10-color flow cytometry (Navios flow cytometer, Beckman Coulter) after blood sampling (2–3 h), and cell count analysis was performed using the Kaluza software (Beckman Coulter, v. 1.3). In order to reduce inter-experimental noise and increase statistical power, cell count analysis was performed by calculating parental and grandparental percentages, which were defined as the percentage of a certain cell type within the cell populations one or two levels higher in the hierarchical definitions of cell sub-populations [75]. Detailed laboratory protocols for cell isolation, reagents, gating, and flow cytometry analysis have been reported elsewhere [76], and the accession number for the raw flow cytometry data and analyzed data files are available upon request to the authors (http://hfgp.bbmri.nl, accessed on 8 May 2020). A proteomic analysis was also performed in serum and plasma samples from the 500 FG cohort. Circulating proteins were measured using the commercially available Olink® Inflammation panel (Olink, Sweden), which resulted in the measurement of 103 different biomarkers (Supplementary Materials Table S2). Proteins levels were expressed on a log2-scale as normalized protein expression values, and normalized using bridging samples to correct for batch variation. Considering the number of proteins (n = 103) and cell populations (n = 91) tested, p-values of 9.33 × 10−6 and 1.05 × 10−5 were set as significant thresholds for the proteomic and cell-level variation analysis, respectively.
2.7. Correlation between Steroid Hormone Levels and T2D-Related SNPs
We also measured serum levels of seven steroid hormones (androstenedione, cortisol, 11-deoxy-cortisol, 17-hydroxy progesterone, progesterone, testosterone, and 25 hydroxy vitamin D3) in the 500 FG cohort. Complete protocol details of steroid hormone measurements have been reported elsewhere [74]. Hormone levels and genotyping data were available for a total of 167 subjects. After log-transform, correlation between steroid hormone levels and T2D-related SNPs was evaluated by linear regression analysis adjusted for age. In order to avoid a possible bias, we excluded from the analysis those subjects that were using oral contraceptives, or those subjects in which this information was not available. The significance threshold was set to 0.000137 considering the number of independent SNPs tested (n = 52) and the number of hormones determined (n = 7).
2.8. In Silico Functional Analysis
Once we assessed the correlation of T2D-related SNPs with cytokine and steroid hormone levels, we used the HaploReg SNP annotation tool to further investigate the functional consequences of each specific variant (http://www.broadinstitute.org/mammals/haploreg/haploreg.php, accessed on 8 July 2020). We also assessed whether any of the potentially interesting markers correlated with mRNA expression levels of their respective genes using data from public eQTL browsers (GTex portal; www.gtexportal.org/home/, accessed on 8 July 2020; https://genenetwork.nl/bloodeqtlbrowser/, accessed on 8 July 2020) [77].
3. Results
3.1. Overall Associations of Selected SNPs with PCa Risk
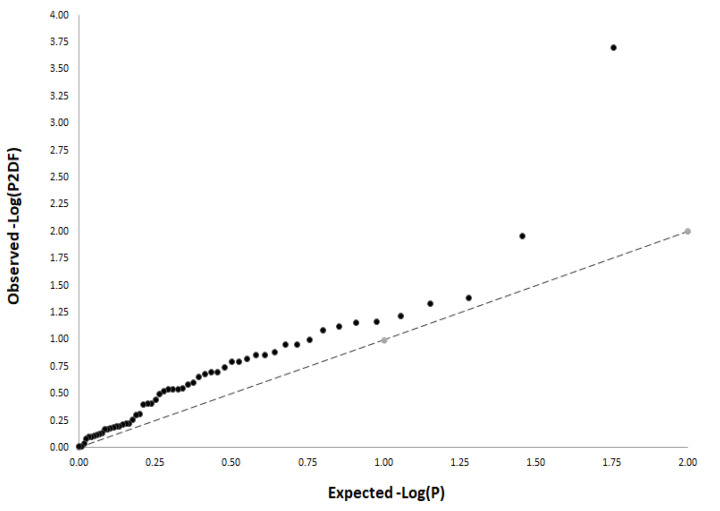
All SNPs were in HWE in the control group (p > 0.001). Logistic regression analysis adjusted for age showed that carriers of the IGF2BP2rs4402960T/T, TCF7L2rs12255372T/T, and TSPAN8|LGR5rs7961581C/C genotypes had an increased risk of PCa (p = 0.037; 0.005 and 0.024), whereas those carrying the CDKAL1rs7754840C, FLJ39370rs17044137A, FTOrs9939609A, HNF1Brs7501939T, HNF1Brs757210T, JAZF1rs10486567A, KCNQ1rs2237897C, and KCNQ1rs2237892C alleles showed a decreased risk of developing the disease (p = 0.022, 0.021, 0.046, 0.030, 0.024, 0.011, 0.041, and 0.0002; Table 3). Although none of the reported associations remained statistically significant after a stringent correction for multiple testing (p = 0.00048), the QQ plot showed a pronounced and early deviation of identity line, which confirmed that the effect attributed to SNPs in T2D-related loci was more than expected under the null hypothesis and, therefore, might represent true associations (Figure 2).
Table 3.
Association of T2D-related variants and risk of developing PCa in the discovery population.
| Variant_dbSNP | Gene | Nucleotide Substitution | Risk Allele | OR (95% CI) † | p |
|---|---|---|---|---|---|
| rs2641348 | ADAM30 | T/C | C | 0.93 (0.66–1.29) | 0.66 |
| rs4607103 | ADAMTS9 | T/C | C | 1.06 (0.83–1.37) | 0.63 |
| rs11708067 | ADCY5 | A/G | G | 1.08 (0.80–1.48) | 0.60 |
| rs10885122 | ADRA2A | G/T | T | 1.12 (0.81–1.55) | 0.49 |
| rs1552224 | ARAPI, CENTD2 | C/A | A | 1.04 (0.76–1.41) | 0.82 |
| rs12779790 | CDC123, CAMK1D | A/G | G | 1.05 (0.80–1.38) | 0.73 |
| rs7754840 | CDKAL1 | C/G | C | 0.69 (0.51–0.95) ¥ | 0.022 |
| rs10811661 | CDKN2A-2B | T/C | T | 0.84 (0.64–1.10) | 0.22 |
| rs4240702 | COL5A1 | C/T | T | 0.82 (0.66–1.02) | 0.082 |
| rs11605924 | CRY2 | A/C | A | 1.03 (0.83–1.28) | 0.79 |
| rs1153188 | DCD | A/T | T | 0.93 (0.73–1.18) | 0.55 |
| rs1113132 | EXT2 | C/G | C | 1.02 (0.80–1.30) | 0.13 |
| rs174550 | FADS1 | C/T | C | 0.96 (0.76–1.22) | 0.75 |
| rs11071657 | FAM148B | A/G | G | 1.16 (0.94–1.44) | 0.16 |
| rs17044137 | FLJ39370 | A/T | A | 0.68 (0.49–0.94) ¥ | 0.021 |
| rs9939609 | FTO | A/C | A | 0.80 (0.63–0.99) | 0.046 |
| rs560887 | G6PC2 | G/A | G | 1.15 (0.90–1.46) | 0.28 |
| rs1799884 | GCK | G/A | A | 1.07 (0.80–1.44) | 0.65 |
| rs1260326 | GCKR | C/T | T | 0.93 (0.73–1.20) | 0.60 |
| rs1111875 | HHEX | C/T | C | 0.90 (0.72–1.13) | 0.36 |
| rs1531343 | HMGA2 | C/G | C | 0.74 (0.53–1.02) | 0.068 |
| rs7957197 | HNF1A (TCF1) | A/T | T | 0.82 (0.63–1.07) | 0.16 |
| rs7501939 | HNF1B (TCF2) | C/T | T | 0.70 (0.50–0.96) ¥ | 0.030 |
| rs757210 | HNF1B (TCF2) | C/T | T | 0.67 (0.48–0.95) ¥ | 0.024 |
| rs4430796 | HNF1B (TCF2) | G/A | G | 0.73 (0.50–1.06) ¥ | 0.10 |
| rs35767 | IGF1 | C/T | C | 0.87 (0.66–1.14) | 0.30 |
| rs4402960 | IGF2BP2 | G/T | T | 1.66 (1.03–2.68) § | 0.037 |
| rs20541 | IL13 | C/T | T | 0.82 (0.60–1.11) | 0.20 |
| rs2943641 | IRS1 | C/T | C | 0.97 (0.77–1.21) | 0.80 |
| rs864745 | JAZF1 | T/C | T | 1.05 (0.84–1.30) | 0.67 |
| rs10486567 | JAZF1 | A/G | A | 0.69 (0.52–0.91) | 0.011 |
| rs5215 | KCNJ11 | T/C | C | 0.87 (0.70–1.08) | 0.21 |
| rs5219 | KCNJ11 | C/T | T | 0.89 (0.71–1.11) | 0.29 |
| rs2237897 | KCNQ1 | C/T | C | 0.66 (0.44–0.98) | 0.041 |
| rs2074196 | KCNQ1 | G/T | T | 0.99 (0.53–1.84) | 0.97 |
| rs2237892 | KCNQ1 | C/T | C | 0.41 (0.26–0.66) | 0.0002 |
| rs2237895 | KCNQ1 | A/C | C | 0.92 (0.73–1.17) | 0.50 |
| rs231362 | KCNQ1OT1 | C/T | C | 0.94 (0.75–1.18) | 0.61 |
| rs1041981 | LTA | A/C | A | 0.87 (0.68–1.12) | 0.29 |
| rs7944584 | MADD | A/T | T | 1.16 (0.93–1.46) | 0.18 |
| rs12970134 | MCR4 | A/G | A | 0.85 (0.66–1.11) | 0.25 |
| rs1387153 | MTNR1B | C/T | T | 0.81 (0.63–1.04) | 0.10 |
| rs10923931 | NOTCH2 | G/T | T | 0.92 (0.66–1.28) | 0.63 |
| rs6698181 | PKN2 | C/T | T | 0.90 (0.72–1.13) | 0.39 |
| rs1801282 | PPARG | C/G | C | 0.99 (0.70–1.42) | 0.98 |
| rs8042680 | PRC1 | A/C | A | 1.10 (0.87–1.37) | 0.40 |
| rs340874 | PROX1 | A/G | G | 0.89 (0.72–1.10) | 0.29 |
| rs7593730 | RBMS1 | C/T | T | 0.77 (0.59–1.02) | 0.070 |
| rs11920090 | SLC2A2 | A/T | T | 0.81 (0.59–1.12) | 0.20 |
| rs13266634 | SLC30A8 | C/T | C | 0.83 (0.65–1.05) | 0.11 |
| rs7903146 | TCF7L2 | C/T | T | 1.01 (0.80–1.29) | 0.91 |
| rs12255372 | TCF7L2 | G/T | T | 1.85 (1.20–2.86) § | 0.005 |
| rs7578597 | THADA | T/C | C | 0.93 (0.58–1.49) | 0.76 |
| rs896854 | TP53INP1 | G/A | A | 0.73 (0.52–1.03) ¥ | 0.070 |
| rs7961581 | TSPAN8, LGR5 | C/T | C | 1.72 (1.07–2.76) § | 0.024 |
| rs9472138 | VEGFA | C/T | T | 1.04 (0.81–1.32) | 0.78 |
| rs10010131 | WFS1 | A/G | G | 0.90 (0.72–1.13) | 0.39 |
Abbreviations: OR, odds ratio; CI, confidence interval. Estimates were adjusted for age. p < 0.05 in bold. † Estimates calculated according to a log-additive model of inheritance and adjusted for age. ¥ Estimates calculated according to a dominant model of inheritance and adjusted for age. § Estimates calculated according to a recessive model of inheritance and adjusted for age.
Figure 2.
QQ plot showing early deviation of the identity line.
The identity line represents the null hypothesis (no significant association between T2D-related SNPs and PCa risk). Early deviation of the identity line might represent true associations.
3.2. Meta-Analysis
In order to confirm these potentially interesting associations, we conducted a meta-analysis with GWAS data from two large cohorts (UKBiobank and FinnGen) and previously published genetic association studies. After filtering all studies found in the literature according to selected key words, we found that four case-control studies met the eligibility criteria [20,21,22,71]. The meta-analysis of our data with those from all these studies confirmed that carriers of the FTOrs9939609A, HNF1Brs7501939T, HNF1Brs757210T, HNF1Brs4430796G, and JAZF1rs10486567A alleles had a decreased risk of developing PCa (p = 3.70 × 10−5, 9.39 × 10−54, 5.04 × 10−54, 1.19 × 10−71, and 1.66 × 10−18; Table 4). Although the effect of the FTO and HNF1B loci on PCA risk has been consistently validated in previous studies, this is the first validation study confirming the association of the JAZF1 variant with the risk of developing the disease. In addition, although the association did not remain significant after correction for multiple testing, the meta-analysis suggested modest associations with the risk of developing the disease for SNPs within the NOTCH2 and RBMS1 loci (p = 8.49 × 10−4 and 0.004; Table 4). These associations are potentially interesting and need to be further investigated.
Table 4.
Meta-analysis of association estimates with previous candidate gene association studies according to a log-additive model of inheritance.
| Study Population (304 PCa Cases and 686 Controls) |
UKBiobank (2020) (5993 PCa Cases and 168,999 Controls) |
FinnGen (2020) (6311 PCa Cases and 74,685 Controls) |
Machiela et al. (2012) (2782 PCa Cases and 4458 Controls) |
Pierce and Ahsan (2010) (1230 PCa Cases and 1160 Controls) |
Stevens et al. (2010) (2935 PCa Cases and 2932 Controls) |
Berndt et al. (2011) (10,272 PCa Cases and 9123 Controls) |
Meta-Analysis (29,827 PCa Cases and 262,042 Controls) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene_SNP | Risk Allele | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | OR (95% CI) a | p Value | PHet |
| rs2641348 | ADAM30 | C | 0.93 (0.66–1.29) | 0.95 (0.89–1.01) | 0.96 (0.90–1.02) | - | 0.87 (0.71–1.05) | - | - | 0.95 (0.91–0.99) | 0.020 | 0.826 |
| rs4607103 | ADAMTS9 | C | 1.07 (0.83–1.37) | 1.02 (0.97–1.07) | 0.97 (0.93–1.02) | 0.99 (0.91–1.08) | 0.98 (0.85–1.12) η | - | - | 0.99 (0.96–1.02) | 0.660 | 0.641 |
| rs11708067 | ADCY5 | G | 1.09 (0.80–1.48) | 1.00 (0.96–1.05) | 0.99 (0.94–1.05) | 0.91 (0.84–0.99) | - | - | - | 0.98 (0.94–1.02) | 0.307 | 0.217 |
| rs10885122 | ADRA2A | T | 1.12 (0.81–1.54) | 0.99 (0.94–1.05) | 1.00 (0.94–1.06) | - | - | - | - | 1.00 (0.96–1.04) | 0.863 | 0.750 |
| rs1552224 | ARAPI | A | 1.04 (0.76–1.41) | 1.00 (0.95–1.06) | 1.00 (0.95–1.05) | 1.00 (0.91–1.10) | - | - | - | 1.00 (0.96–1.03) | 0.948 | 0.951 |
| rs12779790 | CDC123 | G | 1.05 (0.80–1.37) | 1.04 (0.99–1.09) | 0.98 (0.93–1.03) | 1.06 (0.97–1.16) | 1.03 (0.89–1.19) ξ | - | - | 1.02 (0.99–1.05) | 0.251 | 0.441 |
| rs7754840 | CDKAL1 | C | 0.80 (0.63–1.02) | 1.00 (0.96–1.05) | 1.03 (0.98–1.07) | 1.04 (0.97–1.13) | 1.00 (0.88–1.14) # | - | - | 1.01 (0.98–1.05) | 0.316 | 0.283 |
| rs10811661 | CDKN2A-2B | T | 0.84 (0.64–1.10) | 1.02 (0.97–1.07) | 0.95 (0.90–1.01) | 0.91 (0.83–1.00) | - | - | - | 0.98 (0.94–1.01) | 0.168 | 0.063 |
| rs4240702 | COL5A1 | T | 0.83 (0.67–1.03) | 1.00 (0.97–1.04) | 1.01 (0.97–1.05) | - | - | - | - | 1.00 (0.97–1.03) | 0.907 | 0.211 |
| rs11605924 | CRY2 | A | 1.03 (0.82–1.28) | 1.01 (0.97–1.04) | 1.00 (0.95–1.04) | - | - | - | - | 1.01 (0.98–1.03) | 0.637 | 0.924 |
| rs1153188 | DCD | T | 0.93 (0.73–1.18) | 0.97 (0.93–1.02) | 0.98 (0.93–1.03) | - | - | - | - | 0.97 (0.94–1.01) | 0.122 | 0.892 |
| rs1113132 | EXT2 | C | 0.93 (0.73–1.19) | 0.99 (0.95–1.02) | 1.01 (0.97–1.06) | - | - | - | - | 1.00 (0.97–1.02) | 0.890 | 0.646 |
| rs174550 | FADS1 | C | 0.96 (0.76–1.21) | 0.98 (0.94–1.02) | 0.98 (0.94–1.03) | - | - | - | - | 0.98 (0.95–1.01) | 0.182 | 0.985 |
| rs11071657 | FAM148B | G | 1.16 (0.94–1.44) | 1.02 (0.98–1.06) | 1.01 (0.96–1.05) | - | - | - | - | 1.02 (0.99–1.05) | 0.227 | 0.456 |
| rs17044137 | FLJ39370 | A | 0.79 (0.61–1.03) | 1.02 (0.98–1.07) | 0.94 (0.90–0.99) | - | - | - | - | 0.98 (0.95–1.01) | 0.199 | 0.013 |
| rs9939609 | FTO | A | 0.80 (0.63–0.99) | 0.96 (0.92–1.00) | 0.96 (0.92–1.00) | 0.93 (0.86–1.00) | 0.87 (0.77–0.98) δ | 0.93 (0.85–1.02) ς | - | 0.95 (0.92–0.97) | 3.70 × 10−5 | 0.388 |
| rs560887 | G6PC2 | G | 1.15 (0.90–1.46) | 1.02 (0.98–1.07) | 0.96 (0.92–1.01) | - | - | - | - | 1.00 (0.94–1.06) | 0.705 | 0.088 |
| rs1799884 | GCK | A | 1.07 (0.80–1.44) | 1.03 (0.98–1.08) | 0.99 (0.93–1.06) | 1.06 (0.96–1.16) ∂ | - | - | - | 1.02 (0.99–1.06) | 0.220 | 0.643 |
| rs1260326 | GCKR | C | 1.07 (0.83–1.37) | 0.99 (0.96–1.03) | 0.98 (0.94–1.02) | 0.98 (0.91–1.05) ∏ | - | - | - | 0.99 (0.97–1.00) | 0.170 | 0.889 |
| rs1111875 | HHEX | C | 0.90 (0.72–1.13) | 1.01 (0.97–1.05) | 1.01 (0.97–1.05) | 1.01 (0.94–1.09) | 0.98 (0.87–1.10) | - | - | 1.01 (0.98–1.03) | 0.586 | 0.876 |
| rs1531343 | HMGA2 | C | 0.74 (0.53–1.20) | 0.98 (0.92–1.04) | 1.01 (0.97–1.05) | 0.98 (0.88–1.10) | - | - | - | 0.99 (0.94–1.03) | 0.534 | 0.512 |
| rs7957197 | HNF1A | T | 0.82 (0.63–1.07) | 1.01 (0.97–1.06) | 0.99 (0.94–1.04) | 0.96 (0.88–1.05) | - | - | - | 0.99 (0.96–1.02) | 0.673 | 0.370 |
| rs7501939 | HNF1B | T | 0.84 (0.67–1.05) | 0.83 (0.80–0.86) | 0.83 (0.79–0.87) | - | - | 0.87 (0.80–0.94) | 0.84 (0.80–0.87) | 0.84 (0.82–0.86) | 9.39 × 10−54 | 0.873 |
| rs757210 | HNF1B | T | 0.84 (0.67–1.04) | 0.84 (0.81–0.88) | 0.82 (0.79–0.86) | 0.85 (0.79–0.92) | - | 0.85 (0.79–0.92) | 0.84 (0.80–0.88) | 0.84 (0.82–0.86) | 5.04 × 10−54 | 0.902 |
| rs4430796 | HNF1B | G | 0.89 (0.71–1.12) | 0.81 (0.79–0.85) | 0.82 (0.78–0.85) | - | 0.87 (0.77–0.97) | 0.85 (0.79–0.92) | 0.81 (0.77–0.84) | 0.82 (0.80–0.84) | 1.19 × 10−71 | 0.688 |
| rs35767 | IGF1 | C | 0.87 (0.66–1.13) | 0.99 (0.94–1.04) | 1.01 (0.96–1.06) | - | - | - | - | 1.00 (0.96–1.03) | 0.901 | 0.516 |
| rs4402960 | IGF2BP2 | T | 1.05 (0.83–1.32) | 0.99 (0.95–1.03) | 1.00 (0.95–1.04) | 1.03 (0.95–1.11) | 0.91 (0.81–1.04) | - | - | 0.99 (0.97–1.02) | 0.733 | 0.552 |
| rs20541 | IL13 | T | 0.82 (0.60–1.11) | 0.97 (0.93–1.02) | 1.04 (0.99–1.08) | - | - | - | - | 1.00 (0.93–1.06) | 0.788 | 0.042 |
| rs2943641 | IRS1 | C | 0.97 (0.77–1.21) | 1.01 (0.97–1.05) | 1.02 (0.98–1.06) | 0.95 (0.88–1.02) | - | - | - | 1.01 (0.98–1.03) | 0.641 | 0.403 |
| rs864745 | JAZF1 | T | 1.05 (0.84–1.30) | 1.02 (0.98–1.06) | 0.99 (0.95–1.03) | 1.08 (1.01–1.16) | 0.98 (0.87–1.10) ℵ | - | - | 1.02 (0.99–1.05) | 0.269 | 0.283 |
| rs10486567 | JAZF1 | A | 0.69 (0.52–0.91) | 0.87 (0.83–0.91) | 0.86 (0.82–0.91) | - | - | 0.86 (0.73–0.94) | - | 0.86 (0.83–0.89) | 1.66 × 10−18 | 0.459 |
| rs5215 | KCNJ11 | C | 0.87 (0.70–1.08) | 1.02 (0.98–1.06) | 0.99 (0.95–1.03) | 1.01 (0.94–1.09) | 0.89 (0.78–1.00) | - | - | 0.99 (0.96–1.03) | 0.921 | 0.182 |
| rs5219 | KCNJ11 | T | 0.89 (0.71–1.11) | 1.02 (0.98–1.06) | 0.99 (0.95–1.04) | - | - | - | - | 1.00 (0.97–1.04) | 0.746 | 0.349 |
| rs2237897 | KCNQ1 | C | 0.66 (0.44–0.98) | 0.94 (0.86–1.04) | 0.98 (0.91–1.06) | - | - | - | - | 0.94 (0.86–1.04) | 0.136 | 0.148 |
| rs2074196 | KCNQ1 | T | 0.99 (0.53–1.84) | 1.03 (0.94–1.14) | 0.97 (0.88–1.07) | - | - | - | - | 1.00 (0.93–1.07) | 0.996 | 0.693 |
| rs2237892 | KCNQ1 | C | 0.41 (0.26–0.66) | 0.98 (0.91–1.06) | 1.02 (0.93–1.12) | 0.85 (0.74–0.98) | 0.88 (0.69–1.12) | - | - | 0.89 (0.78–1.02) | 0.105 | 0.001 |
| rs2237895 | KCNQ1 | C | 0.92 (0.73–1.16) | 0.99 (0.95–1.03) | 0.96 (0.92–1.00) | - | - | - | - | 0.97 (0.95–1.00) | 0.078 | 0.517 |
| rs231362 | KCNQ1OT1 | C | 0.94 (0.75–1.18) | 0.99 (0.96–1.03) | 1.03 (0.99–1.08) | 0.92 (0.86–0.98) | - | - | - | 0.99 (0.94–1.03) | 0.515 | 0.042 |
| rs1041981 | LTA | A | 0.88 (0.69–1.13) | 0.95 (0.91–0.99) | 0.99 (0.94–1.04) | - | - | - | - | 0.96 (0.93–1.00) | 0.028 | 0.359 |
| rs7944584 | MADD | T | 1.16 (0.93–1.46) | 1.03 (0.99–1.07) | 1.04 (0.99–1.10) | - | - | - | - | 1.04 (1.00–1.07) | 0.026 | 0.585 |
| rs12970134 | MCR4 | A | 0.85 (0.65–1.11) | 0.99 (0.95–1.04) | 0.99 (0.94–1.04) | - | - | - | - | 0.99 (0.95–1.02) | 0.466 | 0.541 |
| rs1387153 | MTNR1B | T | 0.81 (0.63–1.04) | 1.02 (0.97–1.06) | 0.98 (0.94–1.03) | 1.10 (1.01–1.19) ς | - | - | - | 1.01 (0.96–1.08) | 0.517 | 0.029 |
| rs10923931 | NOTCH2 | T | 0.92 (0.66–1.28) | 0.95 (0.90–1.01) | 0.95 (0.89–1.01) | 0.86 (0.76–0.96) | 0.87 (0.71–1.05) * | - | - | 0.94 (0.90–0.97) | 8.49 × 10−4 | 0.552 |
| rs6698181 | PKN2 | T | 0.90 (0.72–1.13) | 0.99 (0.96–1.03) | 1.01 (0.96–1.06) | - | - | - | - | 0.99 (0.97–1.02) | 0.732 | 0.551 |
| rs1801282 | PPARG | C | 1.00 (0.70–1.42) | 1.01 (0.95–1.06) | 1.00 (0.94–1.05) | 0.96 (0.87–1.07) | 0.88 (0.74–1.04) | - | - | 0.99 (0.96–1.03) | 0.733 | 0.596 |
| rs8042680 | PRC1 | A | 1.10 (0.87–1.37) | 0.99 (0.95–1.03) | 0.96 (0.91–0.99) | 1.04 (0.97–1.12) | - | - | - | 0.99 (0.95–1.03) | 0.300 | 0.204 |
| rs340874 | PROX1 | G | 0.89 (0.72–1.10) | 1.01 (0.97–1.05) | 1.00 (0.96–1.05) | 1.01 (0.94–1.08) | - | - | - | 1.00 (0.98–1.03) | 0.758 | 0.708 |
| rs7593730 | RBMS1 | T | 0.77 (0.59–1.02) | 1.07 (1.03–1.12) | 1.03 (0.98–1.09) | - | - | - | - | 1.03 (0.96–1.11) | 0.004 | 0.045 |
| rs11920090 | SLC2A2 | T | 0.82 (0.59–1.12) | 0.94 (0.89–0.99) | 0.99 (0.93-1.06) | - | - | - | - | 0.96 (0.92–1.00) | 0.036 | 0.307 |
| rs13266634 | SLC30A8 | C | 0.83 (0.65–1.05) | 0.99 (0.95–1.03) | 1.00 (0.96–1.05) | 1.00 (0.93–1.08) | 0.97 (0.86–1.11) | - | - | 0.99 (0.96–1.02) | 0.551 | 0.659 |
| rs7903146 | TCF7L2 | T | 1.01 (0.80–1.29) | 1.04 (1.00–1.08) | 0.99 (0.94–1.04) | 0.90 (0.83–0.97) | 0.97 (0.85–1.10) | - | - | 0.98 (0.93–1.03) | 0.872 | 0.047 |
| rs12255372 | TCF7L2 | T | 1.17 (0.94–1.46) | 1.02 (0.98–1.06) | 0.97 (0.92–1.02) | - | - | - | - | 1.00 (0.96–1.06) | 0.778 | 0.123 |
| rs7578597 | THADA | T | 1.08 (0.67–1.72) | 1.05 (0.98–1.11) | 1.04 (0.95–1.14) | 1.03 (0.91–1.16) | 1.10 (0.92–1.32) Ϯ | - | - | 1.05 (1.00–1.10) | 0.044 | 0.982 |
| rs896854 | TP53INP1 | T | 0.88 (0.71–1.10) | 0.99 (0.95–1.02) | 0.99 (0.94–1.03) | 1.02 (0.95–1.09) | - | - | - | 0.99 (0.97–1.02) | 0.570 | 0.615 |
| rs7961581 | TSPAN8 | C | 1.19 (0.94–1.51) | 0.98 (0.94–1.02) | 1.00 (0.95–1.05) | 1.05 (0.97–1.13) | 1.04 (0.92–1.19) τ | - | - | 1.00 (0.97–1.04) | 0.924 | 0.295 |
| rs9472138 | VEGFA | T | 1.04 (0.82–1.33) | 1.01 (0.97–1.05) | 0.98 (0.94–1.03) | - | - | - | - | 1.00 (0.97–1.03) | 0.877 | 0.586 |
| rs10010131 | WFS1 | G | 0.90 (0.72–1.13) | 0.99 (0.95–1.02) | 1.01 (0.97–1.05) | 1.00 (0.93–1.07) | - | - | - | 1.00 (0.97–1.02) | 0.859 | 0.716 |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; C, cytosine; T, thymine; A, adenine; G, guanosine; n/s, not specified. a Estimates calculated according to a log-additive model of inheritance and adjusted for age. Meta-analysis was performed assuming a fixed-effect model. p < 0.05 in bold. η Authors report the effect found for the rs4411878 (a SNP in complete linkage disequilibrium with the rs4607103, r2 = 0.97). ξ Authors report the effect found for the rs11257655 (a SNP in strong linkage disequilibrium with the rs12779790, r2 = 0.82). # Authors report the effect found for the rs7756992 (a SNP in strong linkage disequilibrium with the rs7754840, r2 = 0.75). δ Authors report the effect found for the rs8050136 (a SNP in complete linkage disequilibrium with the rs9969309, r2 = 0.98). ℵAuthors report the effect found for the rs1635852 (a SNP in complete linkage disequilibrium with the rs864745, r2 = 0.98). * Authors report the effect found for the rs2641348 (a SNP in complete linkage disequilibrium with the rs10923931, r2 = 1.00). ∂ Authors report the effect found for the rs4607517 (a SNP in complete linkage disequilibrium with the rs1799884, r2 = 1.00). ∏ Authors report the effect found for the rs780094 (a SNP in complete linkage disequilibrium with the rs1260326, r2 = 0.92). Ϯ Authors report the effect found for the rs13414140 (a SNP in complete linkage disequilibrium with the rs7578597, r2 = 1.00). τ Authors report the effect found for the rs1353362 (a SNP in strong linkage disequilibrium with the rs7961581, r2 = 0.92). ς Authors report the effect found for the rs10830963 (a SNP in moderate to high linkage disequilibrium with the rs1387153, r2 = 0.67). ς Results from Lewis et al. Plos One 2010; 5: e13485 [78].
3.3. Functional Characterization of T2D-Related Variants in the HFGP Cohort
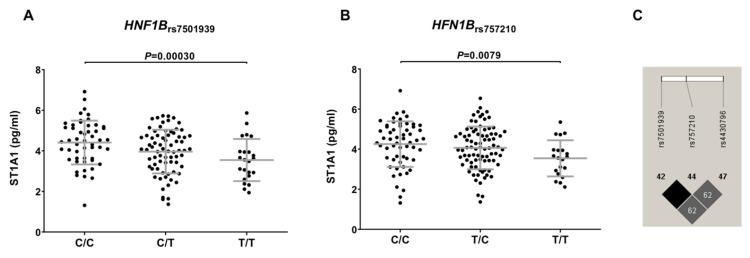
In order to test the possible functional relevance of the most interesting SNPs, we analyzed data from the HFGP cohort. The proteomic analysis of the immunological serum proteins showed that carriers of the HNF1Brs7501939T allele had decreased circulating levels of ST1A1 protein (p = 0.00030; Figure 3). Although this correlation did not survive multiple testing correction, this result supported the implication of the HNF1B locus in modulating PCa risk, likely by the regulation of the sulfatation of multiple compounds in the liver. In addition, given the healthy nature of the subjects included in the HFGP cohort, this result is still speculative and needs to be further confirmed in tumor samples of PCa patients. No significant correlation was found between HFN1B, FTO, JAZF1, and NOTCH2 SNPs and blood-derived cells populations, steroid hormones, or cQTL data, which suggested that these SNPs might affect PCa risk likely through the regulation of mRNA levels of their respective genes.
Figure 3.
Correlation of the HFN1Brs7501939 (A) and HFN1Brs757210 (B) polymorphisms with reduced levels of ST1A1 protein and linkage disequilibrium values among the HFN1B SNPs included in the study (C). C/C, cytosine/cytosine; C/T, cytosine/thymine; T/T, thymine/thymine; T/C, thymine/cytosine.
Next, we assessed functional information from the HaploReg SNP annotation tool, and we also assessed whether any of the potentially interesting markers correlated with mRNA expression levels of their respective genes using data from public eQTL browsers. We found that, according to Haploreg data, the HNF1Brs7501939, HNF1Brs757210, and HNF1Brs4430796 SNPs were modestly associated with mRNA HNF1B expression levels in peripheral whole blood (p = 9.23 × 10−4, 1.00 × 10−3, and 2.00 × 10−3) [77], and that they mapped among histone marks (H3K4me1, H3K4me3) in several tissues and changed motifs for CEBPB, DMRT5, p300, HES1, and Maf (Supplementary Table S3). Similarly, we found that the NOTCH2rs10923931 and RBMS1rs7593730 SNPs also correlated with NOTCH2 and RBMS1 mRNA expression levels in peripheral whole blood (p = 7.3 × 10−6 and 3.31 × 10−7, respectively) [77]. On the other hand, we found that the FTOrs9939609 and JAZF1rs10486567 SNPs mapped among histone marks in multiple tissues and several immune cells, and changed regulatory motifs for multiple regulatory transcription factors (Supplementary Table S3). In addition, GTEx portal data suggested that the NOTCH2rs10923931 SNP is an eQTL in the pancreas and liver.
4. Discussion
T2D has been consistently identified as protective factor for PCa development and disease progression [7,8,9]. Several studies have also suggested that both diseases might share a genetic component [13,14,15,16], and some others have attempted to demonstrate the impact of diabetogenic variants on PCa risk, showing controversial results [20,21,22,23]. With this background, we decided to further investigate the association of diabetogenic variants identified through GWAS with the risk to PCa, and attempted to identify the biological mechanisms underlying the most interesting associations through the analysis of functional data from the HFGP cohort and eQTL browsers.
The meta-analysis of the Spanish cohort with those from the UKBiobank, FinnGen, and previously published studies [20,21,22] confirmed that carriers of the FTOrs9939609A, HNF1Brs7501939T, HNF1Brs757210T, HNF1Brs4430796G, and JAZF1rs10486567A alleles showed a decreased risk of developing the disease. Although the association did not reach the stringent significance threshold, we also found that the NOTCH2rs10923931 and RBMS1rs7593730 SNPs associated with the risk of developing the disease. The strongest effect on PCa risk was observed for SNPs within the HNF1B locus (rs757210, rs7501939, and rs4430796), which showed a similar direction across all study populations. The HNF1B (TCF2) gene is located at chromosome 17q12, and it encodes for a transcription factor implicated in the control of regulatory networks related to pancreas and kidney development. It has been reported that the HNF1B locus participates not only in the generation of endocrine precursors, but also in the modulation of acinar cell identity and duct morphogenesis. In addition to these functions, it has been consistently reported that the HNF1B locus plays a key role in modulating tumorigenesis in solid [79] and hematological cancers [80], and that its methylation or mRNA expression levels can be used for patient stratification [81] and prediction of disease outcome [82]. The association of the HNF1Brs7501939, HNF1Brs757210, and HNF1Brs4430796 polymorphisms with PCa risk was in agreement with results recently reported in GWAS for PCa [14,83,84], whereas large-scale fine mapping studies have even found additional polymorphisms that might contribute to the development of PCa [71]. These findings, together with our functional results reporting that carriers of the HNF1Brs7501939T and HNF1Brs757210T alleles showed decreased levels of SULT1A1 protein (also known as ST1A1), suggest that the effect of the HFN1B locus on PCa risk might be mediated through the regulation of SULT1A1 expression levels. This protein is an enzyme that catalyzes the sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds, among other compounds. It also has been demonstrated that SULT1A1 regulates the metabolic activation of carcinogenic N-hydroxyarylamines, leading to highly reactive intermediates capable of forming DNA adducts, which could result in mutagenesis [85]. In support of the hypothesis of a tumorigenic effect of the SULT1A1, several studies have shown that an increased expression of the SULT1A1 mRNA expression levels contributes to PCa development [86,87]. Although our functional experiments were conducted in a cohort of healthy donors and, therefore, cannot be directly translated to a disease context, our experimental data are in agreement with previous studies, and suggest that the protective effect attributed to the HNF1Brs7501939 and HNF1Brs757210 SNPs could be mediated by a reduction in the expression of the SULT1A1 protein. Furthermore, it has been reported that the HNF1Brs7501939, HNF1Brs757210, and HNF1Brs4430796 SNPs are modestly associated with mRNA HNF1B expression levels in peripheral blood, which might help to explain how these genetic variants may influence the risk of developing PCa [77]. However, despite these interesting data, we think that the biological link between the HNF1B locus and SULT1A1 is still speculative and needs to be further explored and validated, since, if confirmed, it might represent a potentially interesting therapeutic target. An option to confirm this hypothesis would be to measure SULT1A1 levels in tumoral tissues.
Besides these results, this study also confirmed the association of the FTO locus with PCa risk. The FTO gene is located on chromosome 16q12.2, and it has been implicated in determining not only obesity, but also other symptoms of the metabolic syndrome. In addition, it has been reported that the FTO gene acts as a tumor suppressor gene by regulating the proliferation, migration, and invasion of PCa cells, and the FTO expression level had a relevance with the development of PCa and the prognosis of PCa patients [88]. Although the association of the FTOrs9939609 polymorphism with PCa risk was weak in all previous studies, and might depend on different confounding factors, the meta-analysis performed in this study confirmed a strong and consistent association of this intronic variant with a decreased risk of PCa. A recent study also demonstrated that the association of the FTOrs9939609 SNP with a decreased risk of PCa was found in non-European populations, and that the presence of this genetic marker tended to be associated with disease severity in patients that were overweighted [89]. These findings, together with the lack of significant results in our functional studies, suggest that the role of the FTO locus in determining PCa might be mediated by complex obesogenic and/or diabetogenic mechanisms. In support of this hypothesis, we found that the association of the FTOrs9939609 SNP with a decreased risk of PCa showed a similar direction to the one observed in the GWAS for T2D.
Similarly, we also found that the presence of the JAZF1rs10486567 SNP was inversely associated with the risk of developing PCa. These results were again in concordance with previous GWAS that have consistently reported that JAZF1 is a susceptibility locus for PCa [84,90,91,92,93]. The JAZF1 gene is located at 7p15, and it encodes for a zinc finger protein that is overexpressed in the human prostate tissue where it induces cell proliferation, migration, and invasion, and tumor development [94]. Even though there is not much evidence about the functional role of the JAZF1rs10486567 SNP in PCa, it has been demonstrated that the deletion of the JAZF1 locus is associated with reduced levels of IGF-1 and insulin resistance in mice [95], which suggests that the presence of functional polymorphisms within this locus might act to promote PCa development through diabetogenic mechanisms.
Finally, although the association was not statistically significant after correction for multiple testing, it seems to be reasonable to suggest that genetic variants within the NOTCH2 and RBMS1 genes could weakly influence the risk of developing PCa. In this regard, it has been reported that genes of the NOTCH family play a relevant role in multiple cancers, including PCa, and that their deregulation may be a key event in tumor onset and disease progression [96,97,98,99,100,101,102]. The human NOTCH2 locus is located in the chromosomal region 1p11.2, and it plays an important role in modulating prostate development and homeostasis, and its deregulation induces proliferation and expansion of both basal and luminal cells in the prostate [97]. In addition, it has been reported that NOTCH activity promotes prostate cancer cell migration [97], invasion [96,97], aggressiveness [98], and metastasis [99], and that its silencing induces apoptosis and increases the chemosensitivity of PCa cells [100]. However, a tumor suppressive role of the NOTCH pathway has also been suggested in the literature [103,104], which points towards the need of additional studies to elucidate the role of these genes in the etiopathogenesis of PCa. On the other hand, the meta-analysis of all study cohorts suggested that carriers of the RBMS1rs7593730T allele had an increased risk of developing PCa. The RBMS1 gene is located at chromosome 2q24.2, and it encodes for a protein that binds single stranded DNA/RNA and plays an important role in DNA replication, cell cycle progression, gene transcription, and apoptosis. A recent study demonstrated that the RBMS1 locus acts by regulating the expression of the miR-106b [105], which has been found overexpressed in hepatocellular carcinoma [106], cervical cancer [107], renal carcinoma [108], and gastric cancer [109]. At the same time, Dankert and collaborators have also demostrated that miR-106b can regulate endogenous RBMS1 expression in PCa cell lines and, thereby, act as a tumor suppressor gene with inhibitory effects on colony formation and cell growth [105]. Despite the lack of evidence linking RBMS1 SNPs and risk of PCa, it seems to be plausible to suggest that the presence of genetic variants in the RBMS1 locus might control miR-106b levels and, therefore, favors tumorigenesis. In support of this hypothesis, haploreg data showed that the RBMS1rs7593730 SNP is associated with different mRNA RBMS1 expression levels in several tissues and cells [77], and that it maps among histone marks in multiple tissues and several immune cells, and changed regulatory motifs for multiple regulatory transcription factors. Nonetheless, although interesting, the effect of NOTCH2 and RBMS1 SNPs on PCa risk must be considered as preliminary and, therefore, needs to be further confirmed in independent cohorts.
This study has both strengths and weaknesses. The major strength of this study is the large number of genetic markers analyzed that allowed us to perform a well-powered meta-analysis of our data with those from previous studies. The meta-analysis of all study cohorts allowed us to not only confirm previous associations between T2D-related polymorphisms and PCa risk, but also to identify potentially interesting genetic markers for the disease. Although the discovery population was relatively small and the influence of diabetogenic variants on the risk of the disease was expected to be modest, our study was sufficiently powered to detect such small effects. Based on the genotype frequencies observed in our study cohort, we had 80% of power (log-additive model) to detect an odds ratio of 1.59 at alpha = 0.00048 (multiple testing threshold) for a polymorphism with a minor allele frequency of 0.25. Likewise, we comprehensively analyzed the impact of T2D-related SNPs in modulating immune responses, blood cell counts, steroid hormones, and serum and plasma metabolites in a relatively large cohort of healthy subjects. However, we also have important limitations. One of them was the fact that functional characterization of the most interesting SNPs was conducted in a healthy control cohort rather than in PCa patients. In addition, we did not have access to medication history, T2D status, and BMI for a substantial number of PCa cases included in the meta-analyses, which did not allow us to adjust our analyses for these confounding variables and, consequently, to rule out the possibility that some of the reported associations could arise as a result of a different distributions of diabetic and/or obese subjects between PCa cases and controls, or because of the effect of diabetes medication rather than the diabetes condition itself. Nonetheless, previous studies have reported that the effect of T2D-related variants on the risk of PCa was independent of T2D status and BMI.
5. Conclusions
In conclusion, our study indicates that T2D-related variants within the HNF1B, FTO, and JAZF1 genes influence the risk of PCa likely through the modulation of diabetogenic pathways, and suggests, for the first time, an association of SNPs within the NOTCH2 and RBMS1 loci that need to be validated in independent cohorts. This study also suggests that the effect of the HFN1B SNPs on PCa risk might be mediated by not only the ST1A1 protein, but also HFN1B mRNA expression levels, whereas the effect of the FTO, JAZF1, NOTCH2, and RBMS1 SNPs on PCa risk seem to be involved in the regulation of mRNA expression levels of their respective genes.
Acknowledgments
We kindly thank all individuals who agreed to participate in the study, as well as all cooperating physicians and students.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14102376/s1. Table S1: Cell types analyzed either in whole blood or peripheral mononuclear blood cells; Table S2: Serum and plasma metabolites measured in the HFGP cohort; Table S3: Summary of functional and regulatory annotation of the most significant SNPs.
Author Contributions
J.S. and M.J.Á.-C. conceived the study and participated in its design and coordination. R.C., A.J.C.-S., F.J.G.-V., F.G.-M., J.A.L.L., I.R.-F., V.A.-R., B.C.-G., P.S.-R., M.I.B.-F., J.O.-R., M.Á.L.-N., L.F.-P., J.M.C.-O., M.J., J.A.L. and M.J.Á.-C. coordinated patient recruitment and provided the clinical data. J.M.S.-M. performed the genetic experiments and R.T.H., O.B., M.G.N. and Y.L. provided functional data. J.S. and A.J.C.-S. performed all statistical analyses and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Boards of all institutions participating in patient recruitment (Virgen de las Nieves University Hospital, 18012, Granada, Spain; Hospital de San Pedro Alcántara, 10003, Cáceres, Spain).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The genotype data used in the present study are available from the corresponding authors upon reasonable request. Functional data used in this project have been meticulously catalogued and archived in the BBMRI-NL data infrastructure (https://hfgp.bbmri.nl/, accessed on 7 July 2020) using the MOLGENIS open-source platform for scientific data [52]. This allows flexible data querying and download, including sufficiently rich metadata and interfaces for machine processing (R statistics, REST API), and using FAIR principles to optimize findability, accessibility, interoperability, and reusability [53,54].
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by grants from the FIBAO foundation (Granada, Spain) and from the Instituto de Salud Carlos III (PI12/02688, PI17/02256 and PI20/01845; Madrid, Spain).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Fedewa S.A., Miller K.D., Goding-Sauer A., Pinheiro P.S., Martinez-Tyson D., Jemal A. Cancer statistics for Hispanics/Latinos, 2015. CA Cancer J. Clin. 2015;65:457–480. doi: 10.3322/caac.21314. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer. J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Weiner A.B., Matulewicz R.S., Eggener S.E., Schaeffer E.M. Increasing incidence of metastatic prostate cancer in the United States (2004–2013) Prostate Cancer Prostatic Dis. 2016;19:395–397. doi: 10.1038/pcan.2016.30. [DOI] [PubMed] [Google Scholar]
- 4.Wong M.C., Goggins W.B., Wang H.H., Fung F.D., Leung C., Wong S.Y., Ng C.F., Sung J.J. Global Incidence and Mortality for Prostate Cancer: Analysis of Temporal Patterns and Trends in 36 Countries. Eur. Urol. 2016;70:862–874. doi: 10.1016/j.eururo.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Hodson R. Prostate cancer: 4 big questions. Nature. 2015;528:S137. doi: 10.1038/528S137a. [DOI] [PubMed] [Google Scholar]
- 6.Eeles R., Goh C., Castro E., Bancroft E., Guy M., Al Olama A.A., Easton D., Kote-Jarai Z. The genetic epidemiology of prostate cancer and its clinical implications. Nat. Rev. Urol. 2014;11:18–31. doi: 10.1038/nrurol.2013.266. [DOI] [PubMed] [Google Scholar]
- 7.Khan A.E., Gallo V., Linseisen J., Kaaks R., Rohrmann S., Raaschou-Nielsen O., Tjonneland A., Johnsen H.E., Overvad K., Bergmann M.M., et al. Diabetes and the risk of non-Hodgkin’s lymphoma and multiple myeloma in the European Prospective Investigation into Cancer and Nutrition. Haematologica. 2008;93:842–850. doi: 10.3324/haematol.12297. [DOI] [PubMed] [Google Scholar]
- 8.Richardson P.G., Sonneveld P., Schuster M.W., Stadtmauer E.A., Facon T., Harousseau J.L., Ben-Yehuda D., Lonial S., Goldschmidt H., Reece D., et al. Reversibility of symptomatic peripheral neuropathy with bortezomib in the phase III APEX trial in relapsed multiple myeloma: Impact of a dose-modification guideline. Br. J. Haematol. 2009;144:895–903. doi: 10.1111/j.1365-2141.2008.07573.x. [DOI] [PubMed] [Google Scholar]
- 9.Castillo J.J., Mull N., Reagan J.L., Nemr S., Mitri J. Increased incidence of non-Hodgkin lymphoma, leukemia, and myeloma in patients with diabetes mellitus type 2: A meta-analysis of observational studies. Blood. 2012;119:4845–4850. doi: 10.1182/blood-2011-06-362830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haring A., Murtola T.J., Talala K., Taari K., Tammela T.L., Auvinen A. Antidiabetic drug use and prostate cancer risk in the Finnish Randomized Study of Screening for Prostate Cancer. Scand. J. Urol. 2017;51:5–12. doi: 10.1080/21681805.2016.1271353. [DOI] [PubMed] [Google Scholar]
- 11.Murtola T.J., Tammela T.L., Lahtela J., Auvinen A. Antidiabetic medication and prostate cancer risk: A population-based case-control study. Am. J. Epidemiol. 2008;168:925–931. doi: 10.1093/aje/kwn190. [DOI] [PubMed] [Google Scholar]
- 12.Haggstrom C., Van Hemelrijck M., Zethelius B., Robinson D., Grundmark B., Holmberg L., Gudbjornsdottir S., Garmo H., Stattin P. Prospective study of Type 2 diabetes mellitus, anti-diabetic drugs and risk of prostate cancer. Int. J. Cancer. 2017;140:611–617. doi: 10.1002/ijc.30480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frayling T.M., Colhoun H., Florez J.C. A genetic link between type 2 diabetes and prostate cancer. Diabetologia. 2008;51:1757–1760. doi: 10.1007/s00125-008-1114-9. [DOI] [PubMed] [Google Scholar]
- 14.Gudmundsson J., Sulem P., Steinthorsdottir V., Bergthorsson J.T., Thorleifsson G., Manolescu A., Rafnar T., Gudbjartsson D., Agnarsson B.A., Baker A., et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 15.Winckler W., Weedon M.N., Graham R.R., McCarroll S.A., Purcell S., Almgren P., Tuomi T., Gaudet D., Bostrom K.B., Walker M., et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 16.Horikawa Y., Iwasaki N., Hara M., Furuta H., Hinokio Y., Cockburn B.N., Lindner T., Yamagata K., Ogata M., Tomonaga O., et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat. Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 17.Rios R., Lupianez C.B., Campa D., Martino A., Martinez-Lopez J., Martinez-Bueno M., Varkonyi J., Garcia-Sanz R., Jamroziak K., Dumontet C., et al. Type 2 diabetes-related variants influence the risk of developing multiple myeloma: Results from the IMMEnSE consortium. Endocr.-Relat. Cancer. 2015;22:545–559. doi: 10.1530/ERC-15-0029. [DOI] [PubMed] [Google Scholar]
- 18.Sainz J., Rudolph A., Hoffmeister M., Frank B., Brenner H., Chang-Claude J., Hemminki K., Forsti A. Effect of type 2 diabetes predisposing genetic variants on colorectal cancer risk. J. Clin. Endocrinol. Metab. 2012;97:E845–E851. doi: 10.1210/jc.2011-2565. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z., Wen W., Michailidou K., Bolla M.K., Wang Q., Zhang B., Long J., Shu X.O., Schmidt M.K., Milne R.L., et al. Association of genetic susceptibility variants for type 2 diabetes with breast cancer risk in women of European ancestry. Cancer Causes Control. 2016;27:679–693. doi: 10.1007/s10552-016-0741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce B.L., Ahsan H. Genetic susceptibility to type 2 diabetes is associated with reduced prostate cancer risk. Hum. Hered. 2010;69:193–201. doi: 10.1159/000289594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens V.L., Ahn J., Sun J., Jacobs E.J., Moore S.C., Patel A.V., Berndt S.I., Albanes D., Hayes R.B. HNF1B and JAZF1 genes, diabetes, and prostate cancer risk. Prostate. 2010;70:601–607. doi: 10.1002/pros.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machiela M.J., Lindstrom S., Allen N.E., Haiman C.A., Albanes D., Barricarte A., Berndt S.I., Bueno-de-Mesquita H.B., Chanock S., Gaziano J.M., et al. Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium. Am. J. Epidemiol. 2012;176:1121–1129. doi: 10.1093/aje/kws191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters K.M., Wilkens L.R., Monroe K.R., Stram D.O., Kolonel L.N., Henderson B.E., Le Marchand L., Haiman C.A. No association of type 2 diabetes risk variants and prostate cancer risk: The multiethnic cohort and PAGE. Cancer Epidemiol. Biomark. Prev. 2011;20:1979–1981. doi: 10.1158/1055-9965.EPI-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mottet N., Bellmunt J., Bolla M., Briers E., Cumberbatch M.G., De Santis M., Fossati N., Gross T., Henry A.M., Joniau S., et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Lyssenko V., Nagorny C.L., Erdos M.R., Wierup N., Jonsson A., Spegel P., Bugliani M., Saxena R., Fex M., Pulizzi N., et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohlke K.L., Boehnke M., Abecasis G.R. Metabolic and cardiovascular traits: An abundance of recently identified common genetic variants. Hum. Mol. Genet. 2008;17:R102–R108. doi: 10.1093/hmg/ddn275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu X.O., Long J., Cai Q., Qi L., Xiang Y.B., Cho Y.S., Tai E.S., Li X., Lin X., Chow W.H., et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen T., Sparso T., Grarup N., Jorgensen T., Pisinger C., Witte D.R., Diabetes Genetics R., Meta-analysis C., Hansen T., Pedersen O. Type 2 diabetes risk allele near CENTD2 is associated with decreased glucose-stimulated insulin release. Diabetologia. 2011;54:1052–1056. doi: 10.1007/s00125-011-2054-3. [DOI] [PubMed] [Google Scholar]
- 33.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 34.Florez J.C., Manning A.K., Dupuis J., McAteer J., Irenze K., Gianniny L., Mirel D.B., Fox C.S., Cupples L.A., Meigs J.B. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: Replication and integration with other genome-wide datasets. Diabetes. 2007;56:3063–3074. doi: 10.2337/db07-0451. [DOI] [PubMed] [Google Scholar]
- 35.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamauchi T., Hara K., Maeda S., Yasuda K., Takahashi A., Horikoshi M., Nakamura M., Fujita H., Grarup N., Cauchi S., et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat. Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi F., Serizawa M., Yamamoto K., Fujisawa T., Nakashima E., Ohnaka K., Ikegami H., Sugiyama T., Katsuya T., Miyagishi M., et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–1699. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouatia-Naji N., Bonnefond A., Cavalcanti-Proenca C., Sparso T., Holmkvist J., Marchand M., Delplanque J., Lobbens S., Rocheleau G., Durand E., et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat. Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 39.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 40.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 41.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouatia-Naji N., Rocheleau G., Van Lommel L., Lemaire K., Schuit F., Cavalcanti-Proenca C., Marchand M., Hartikainen A.L., Sovio U., De Graeve F., et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 44.Chen W.M., Erdos M.R., Jackson A.U., Saxena R., Sanna S., Silver K.D., Timpson N.J., Hansen T., Orru M., Grazia Piras M., et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J. Clin. Investig. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prokopenko I., Langenberg C., Florez J.C., Saxena R., Soranzo N., Thorleifsson G., Loos R.J., Manning A.K., Jackson A.U., Aulchenko Y., et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vujkovic M., Keaton J.M., Lynch J.A., Miller D.R., Zhou J., Tcheandjieu C., Huffman J.E., Assimes T.L., Lorenz K., Zhu X., et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kichaev G., Bhatia G., Loh P.R., Gazal S., Burch K., Freund M.K., Schoech A., Pasaniuc B., Price A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan A., Wessel J., Willems S.M., Zhao W., Robertson N.R., Chu A.Y., Gan W., Kitajima H., Taliun D., Rayner N.W., et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandhu M.S., Weedon M.N., Fawcett K.A., Wasson J., Debenham S.L., Daly A., Lango H., Frayling T.M., Neumann R.J., Sherva R., et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat. Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pechlivanis S., Wagner K., Chang-Claude J., Hoffmeister M., Brenner H., Forsti A. Polymorphisms in the insulin like growth factor 1 and IGF binding protein 3 genes and risk of colorectal cancer. Cancer Detect. Prev. 2007;31:408–416. doi: 10.1016/j.cdp.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Rung J., Cauchi S., Albrechtsen A., Shen L., Rocheleau G., Cavalcanti-Proenca C., Bacot F., Balkau B., Belisle A., Borch-Johnsen K., et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat. Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 53.Tang Y., Han X., Sun X., Lv C., Zhang X., Guo W., Ren Q., Luo Y., Zhang X., Zhou X., et al. Association study of a common variant near IRS1 with type 2 diabetes mellitus in Chinese Han population. Endocrine. 2013;43:84–91. doi: 10.1007/s12020-012-9693-0. [DOI] [PubMed] [Google Scholar]
- 54.Willer C.J., Bonnycastle L.L., Conneely K.N., Duren W.L., Jackson A.U., Scott L.J., Narisu N., Chines P.S., Skol A., Stringham H.M., et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes. 2007;56:256–264. doi: 10.2337/db06-0461. [DOI] [PubMed] [Google Scholar]
- 55.Gloyn A.L., Weedon M.N., Owen K.R., Turner M.J., Knight B.A., Hitman G., Walker M., Levy J.C., Sampson M., Halford S., et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 56.Unoki H., Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., Ng D.P., Holmkvist J., Borch-Johnsen K., Jorgensen T., et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 57.Tsai F.J., Yang C.F., Chen C.C., Chuang L.M., Lu C.H., Chang C.T., Wang T.Y., Chen R.H., Shiu C.F., Liu Y.M., et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda K., Miyake K., Horikawa Y., Hara K., Osawa H., Furuta H., Hirota Y., Mori H., Jonsson A., Sato Y., et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 59.Hamid Y.H., Urhammer S.A., Glumer C., Borch-Johnsen K., Jorgensen T., Hansen T., Pedersen O. The common T60N polymorphism of the lymphotoxin-alpha gene is associated with type 2 diabetes and other phenotypes of the metabolic syndrome. Diabetologia. 2005;48:445–451. doi: 10.1007/s00125-004-1659-1. [DOI] [PubMed] [Google Scholar]
- 60.Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 61.Qi L., Cornelis M.C., Kraft P., Stanya K.J., Linda Kao W.H., Pankow J.S., Dupuis J., Florez J.C., Fox C.S., Pare G., et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 64.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 65.Scott L.J., Bonnycastle L.L., Willer C.J., Sprau A.G., Jackson A.U., Narisu N., Duren W.L., Chines P.S., Stringham H.M., Erdos M.R., et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55:2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 66.Peng S., Zhu Y., Lu B., Xu F., Li X., Lai M. TCF7L2 gene polymorphisms and type 2 diabetes risk: A comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis. 2013;28:25–37. doi: 10.1093/mutage/ges048. [DOI] [PubMed] [Google Scholar]
- 67.Zhao W., Rasheed A., Tikkanen E., Lee J.J., Butterworth A.S., Howson J.M.M., Assimes T.L., Chowdhury R., Orho-Melander M., Damrauer S., et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J., Spracklen C.N., Marenne G., Varshney A., Corbin L.J., Luan J., Willems S.M., Wu Y., Zhang X., Horikoshi M., et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021;53:840–860. doi: 10.1038/s41588-021-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grarup N., Andersen G., Krarup N.T., Albrechtsen A., Schmitz O., Jorgensen T., Borch-Johnsen K., Hansen T., Pedersen O. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57:2534–2540. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gagliano Taliun S.A., VandeHaar P., Boughton A.P., Welch R.P., Taliun D., Schmidt E.M., Zhou W., Nielsen J.B., Willer C.J., Lee S., et al. Exploring and visualizing large-scale genetic associations by using PheWeb. Nat. Genet. 2020;52:550–552. doi: 10.1038/s41588-020-0622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berndt S.I., Sampson J., Yeager M., Jacobs K.B., Wang Z., Hutchinson A., Chung C., Orr N., Wacholder S., Chatterjee N., et al. Large-scale fine mapping of the HNF1B locus and prostate cancer risk. Hum. Mol. Genet. 2011;20:3322–3329. doi: 10.1093/hmg/ddr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y., Oosting M., Smeekens S.P., Jaeger M., Aguirre-Gamboa R., Le K.T.T., Deelen P., Ricano-Ponce I., Schoffelen T., Jansen A.F.M., et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e1014. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 73.Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Horst R.T., Jansen T., Jacobs L., Bonder M.J., et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167:1897.e113. doi: 10.1016/j.cell.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 74.Ter Horst R., Jaeger M., Smeekens S.P., Oosting M., Swertz M.A., Li Y., Kumar V., Diavatopoulos D.A., Jansen A.F.M., Lemmers H., et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell. 2016;167:1111–1124. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orru V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A., et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aguirre-Gamboa R., Joosten I., Urbano P.C.M., van der Molen R.G., van Rijssen E., van Cranenbroek B., Oosting M., Smeekens S., Jaeger M., Zorro M., et al. Differential Effects of Environmental and Genetic Factors on T and B Cell Immune Traits. Cell Rep. 2016;17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis S.J., Murad A., Chen L., Davey Smith G., Donovan J., Palmer T., Hamdy F., Neal D., Lane J.A., Davis M., et al. Associations between an obesity related genetic variant (FTO rs9939609) and prostate cancer risk. PLoS ONE. 2010;5:e13485. doi: 10.1371/journal.pone.0013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu D.D., Guo S.W., Jing Y.Y., Dong Y.L., Wei L.X. A review on hepatocyte nuclear factor-1beta and tumor. Cell Biosci. 2015;5:58. doi: 10.1186/s13578-015-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rios-Tamayo R., Lupianez C.B., Campa D., Hielscher T., Weinhold N., Martinez-Lopez J., Jerez A., Landi S., Jamroziak K., Dumontet C., et al. A common variant within the HNF1B gene is associated with overall survival of multiple myeloma patients: Results from the IMMEnSE consortium and meta-analysis. Oncotarget. 2016;7:59029–59048. doi: 10.18632/oncotarget.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silva T.D., Vidigal V.M., Felipe A.V., JM D.E.L., Neto R.A., Saad S.S., Forones N.M. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol. Lett. 2013;6:1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim L., Liao J., Zhang M., Talamonti M., Bentrem D., Rao S., Yang G.Y. Clear cell carcinoma of the pancreas: Histopathologic features and a unique biomarker: Hepatocyte nuclear factor-1beta. Mod. Pathol. 2008;21:1075–1083. doi: 10.1038/modpathol.2008.95. [DOI] [PubMed] [Google Scholar]
- 83.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 84.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 85.Chou H.C., Lang N.P., Kadlubar F.F. Metabolic activation of N-hydroxy arylamines and N-hydroxy heterocyclic amines by human sulfotransferase(s) Cancer Res. 1995;55:525–529. [PubMed] [Google Scholar]
- 86.Nowell S., Ratnasinghe D.L., Ambrosone C.B., Williams S., Teague-Ross T., Trimble L., Runnels G., Carrol A., Green B., Stone A., et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol. Biomark. Prev. 2004;13:270–276. doi: 10.1158/1055-9965.EPI-03-0047. [DOI] [PubMed] [Google Scholar]
- 87.Al-Buheissi S.Z., Patel H.R., Meinl W., Hewer A., Bryan R.L., Glatt H., Miller R.A., Phillips D.H. N-Acetyltransferase and sulfotransferase activity in human prostate: Potential for carcinogen activation. Pharmacogenet. Genom. 2006;16:391–399. doi: 10.1097/01.fpc.0000204998.22301.09. [DOI] [PubMed] [Google Scholar]
- 88.Zhu K., Li Y., Xu Y. The FTO m(6)A demethylase inhibits the invasion and migration of prostate cancer cells by regulating total m(6)A levels. Life Sci. 2021;271:119180. doi: 10.1016/j.lfs.2021.119180. [DOI] [PubMed] [Google Scholar]
- 89.Salgado-Montilla J.L., Rodriguez-Caban J.L., Sanchez-Garcia J., Sanchez-Ortiz R., Irizarry-Ramirez M. Impact of FTO SNPs rs9930506 and rs9939609 in Prostate Cancer Severity in a Cohort of Puerto Rican Men. Arch. Cancer Res. 2017;5:148. doi: 10.21767/2254-6081.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoffmann T.J., Van Den Eeden S.K., Sakoda L.C., Jorgenson E., Habel L.A., Graff R.E., Passarelli M.N., Cario C.L., Emami N.C., Chao C.R., et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015;5:878–891. doi: 10.1158/2159-8290.CD-15-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoffmann T.J., Passarelli M.N., Graff R.E., Emami N.C., Sakoda L.C., Jorgenson E., Habel L.A., Shan J., Ranatunga D.K., Quesenberry C.P., et al. Genome-wide association study of prostate-specific antigen levels identifies novel loci independent of prostate cancer. Nat. Commun. 2017;8:14248. doi: 10.1038/ncomms14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Conti D.V., Darst B.F., Moss L.C., Saunders E.J., Sheng X., Chou A., Schumacher F.R., Olama A.A.A., Benlloch S., Dadaev T., et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sung Y., Park S., Park S.J., Jeong J., Choi M., Lee J., Kwon W., Jang S., Lee M.H., Kim D.J., et al. Jazf1 promotes prostate cancer progression by activating JNK/Slug. Oncotarget. 2018;9:755–765. doi: 10.18632/oncotarget.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee H.J.D. Early growth retardation and insulin resistance in JAZF1 KO Mice. Diabetes. 2011;60:A103. [Google Scholar]
- 96.Bin Hafeez B., Adhami V.M., Asim M., Siddiqui I.A., Bhat K.M., Zhong W., Saleem M., Din M., Setaluri V., Mukhtar H. Targeted knockdown of Notch1 inhibits invasion of human prostate cancer cells concomitant with inhibition of matrix metalloproteinase-9 and urokinase plasminogen activator. Clin. Cancer Res. 2009;15:452–459. doi: 10.1158/1078-0432.CCR-08-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Z., Li Y., Banerjee S., Kong D., Ahmad A., Nogueira V., Hay N., Sarkar F.H. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J. Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 98.Kashat M., Azzouz L., Sarkar S.H., Kong D., Li Y., Sarkar F.H. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu H., Zhou X., Redfield S., Lewin J., Miele L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. Am. J. Transl. Res. 2013;5:368–378. [PMC free article] [PubMed] [Google Scholar]
- 100.Ye Q.F., Zhang Y.C., Peng X.Q., Long Z., Ming Y.Z., He L.Y. Silencing Notch-1 induces apoptosis and increases the chemosensitivity of prostate cancer cells to docetaxel through Bcl-2 and Bax. Oncol. Lett. 2012;3:879–884. doi: 10.3892/ol.2012.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Than B.L., Goos J.A., Sarver A.L., O’Sullivan M.G., Rod A., Starr T.K., Fijneman R.J., Meijer G.A., Zhao L., Zhang Y., et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene. 2014;33:3861–3868. doi: 10.1038/onc.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jacobs D.I., Mao Y., Fu A., Kelly W.K., Zhu Y. Dysregulated methylation at imprinted genes in prostate tumor tissue detected by methylation microarray. BMC Urol. 2013;13:37. doi: 10.1186/1471-2490-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shou J., Ross S., Koeppen H., de Sauvage F.J., Gao W.Q. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291–7297. [PubMed] [Google Scholar]
- 104.Whelan J.T., Kellogg A., Shewchuk B.M., Hewan-Lowe K., Bertrand F.E. Notch-1 signaling is lost in prostate adenocarcinoma and promotes PTEN gene expression. J. Cell. Biochem. 2009;107:992–1001. doi: 10.1002/jcb.22199. [DOI] [PubMed] [Google Scholar]
- 105.Dankert J.T., Wiesehofer M., Wach S., Czyrnik E.D., Wennemuth G. Loss of RBMS1 as a regulatory target of miR-106b influences cell growth, gap closing and colony forming in prostate carcinoma. Sci. Rep. 2020;10:18022. doi: 10.1038/s41598-020-75083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gu H., Gu S., Zhang X., Zhang S., Zhang D., Lin J., Hasengbayi S., Han W. miR-106b-5p promotes aggressive progression of hepatocellular carcinoma via targeting RUNX3. Cancer Med. 2019;8:6756–6767. doi: 10.1002/cam4.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zong S., Liu X., Zhou N., Yue Y. E2F7, EREG, miR-451a and miR-106b-5p are associated with the cervical cancer development. Arch. Gynecol. Obstet. 2019;299:1089–1098. doi: 10.1007/s00404-018-5007-y. [DOI] [PubMed] [Google Scholar]
- 108.Miao L.J., Yan S., Zhuang Q.F., Mao Q.Y., Xue D., He X.Z., Chen J.P. miR-106b promotes proliferation and invasion by targeting Capicua through MAPK signaling in renal carcinoma cancer. OncoTargets Ther. 2019;12:3595–3607. doi: 10.2147/OTT.S184674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuan C., Zhang Y., Tu W., Guo Y. Integrated miRNA profiling and bioinformatics analyses reveal upregulated miRNAs in gastric cancer. Oncol. Lett. 2019;18:1979–1988. doi: 10.3892/ol.2019.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotype data used in the present study are available from the corresponding authors upon reasonable request. Functional data used in this project have been meticulously catalogued and archived in the BBMRI-NL data infrastructure (https://hfgp.bbmri.nl/, accessed on 7 July 2020) using the MOLGENIS open-source platform for scientific data [52]. This allows flexible data querying and download, including sufficiently rich metadata and interfaces for machine processing (R statistics, REST API), and using FAIR principles to optimize findability, accessibility, interoperability, and reusability [53,54].