Abstract
We used randomly amplified polymorphic DNA (RAPD)-PCR to estimate genetic variation among isolates of Trichoderma associated with green mold on the cultivated mushroom Agaricus bisporus. Of 83 isolates examined, 66 were sampled during the recent green mold epidemic, while the remaining 17 isolates were collected just prior to the epidemic and date back to the 1950s. Trichoderma harzianum biotype 4 was identified by RAPD analysis as the cause of almost 90% of the epidemic-related episodes of green mold occurring in the major commercial mushroom-growing region in North America. Biotype 4 was more closely allied to T. harzianum biotype 2, the predominant pathogenic genotype in Europe, than to the less pathogenic biotype 1 and Trichoderma atroviride (formerly T. harzianum biotype 3). No variation in the RAPD patterns was observed among the isolates within biotype 2 or 4, suggesting that the two pathogenic biotypes were populations containing single clones. Considerable genetic variation, however, was noted among isolates of biotype 1 and T. atroviride from Europe. Biotype 4 was not represented by the preepidemic isolates of Trichoderma as determined by RAPD markers and PCR amplification of an arbitrary DNA sequence unique to the genomes of biotypes 2 and 4. Our findings suggest that the onset of the green mold epidemic in North America resulted from the recent introduction of a highly virulent genotype of the pathogen into cultivated mushrooms.
Sinden and Hauser in 1953 (19) formally recognized the importance of Trichoderma spp. in limiting commercial production of the button mushroom, Agaricus bisporus Imbach (Lange). Trichoderma disease, commonly referred to as green mold, was previously considered a minor problem in mushroom production, because it typically occurred episodically in association with low-quality compost or poor hygiene (6, 7). Therefore, the disease could be effectively managed by modifying the composting process, improving sanitation, or chemical intervention.
Severe outbreaks of green mold occurred in Northern Ireland in 1985 and, in the ensuing years, in England, Scotland, Canada, and the United States (1, 9, 15, 16). These episodes of the disease were more severe and difficult to control than those of the past. In the early stages of the disease, Trichoderma flourishes in the composted mushroom substrate as white mycelia and eventually develops a dark green color after sporulation. The mechanism of pathogenesis is not understood, but a cessation in the formation of mushrooms occurs in areas of the substrate colonized by Trichoderma.
The dramatic increase in the incidence and severity of green mold in mushrooms reflects the emergence of highly virulent forms of the pathogen. Historically, Trichoderma viride and Trichoderma koningii caused green mold in mushrooms (19); however, these species are not responsible for the recent escalation of the disease to epidemic proportions. Since the onset of the epidemic in Ireland, Trichoderma harzianum biotypes 1, 2, 3, and 4 have been described from mushroom cultures (15–17). The biotypes were differentiated by mycelial growth rate and colony appearance, as well as microscopic morphological features, including phialides and phialospores (17). The biotypes can also be distinguished by randomly amplified polymorphic DNA (RAPD)-PCR, restriction fragment length polymorphisms in mitochondrial DNA and ribosomal DNA, and sequence analysis of ribosomal DNA (1, 10–13). Biotypes 2 and 4 are the most prevalent and highly virulent biotypes on mushrooms in Europe and North America, respectively, whereas biotypes 1 and 3 may infest mushroom compost but rarely cause crop loss. More recent molecular evidence indicates that biotype 3 is identical to Trichoderma atroviride, which is a part of the T. viride-T. atroviride complex (1, 5, 12).
Our objective was to determine if biotype 4 existed in cultivated mushrooms before the epidemic or rather had emerged coincidentally with the onset of the epidemic. This objective was approached experimentally by using a combination of RAPD-PCR and PCR specific for biotypes 2 and 4 to profile epidemic and preepidemic collections of Trichoderma from cultivated mushrooms.
MATERIALS AND METHODS
Fungal cultures.
Sixty-six isolates of Trichoderma were collected during the epidemic between 1994 and 1996 from visibly infested areas of the compost and casing of 49 mushroom crops grown at 38 different commercial operations in southeastern Pennsylvania. Cultures of Trichoderma initially were isolated from the mushroom substrates on potato dextrose-yeast agar (24 g of potato dextrose broth powder, 1.5 g of yeast extract [Difco Laboratories, Detroit, Mich.], 20 g of flake agar/liter of water) containing 17.5 μg of tetracycline per ml and maintained thereafter on the same medium without the antibiotic. The epidemic-related isolates were assigned Pennsylvania State University (PSU) accession numbers as follows: 2, 3, 6 to 13, 23, 24, 31 to 35, 66 to 81, 83 to 87, 89, 96, 105 to 109, 111 to 114, 116 to 124, 140, 145, 146, 169 to 172, and 178. A description of tester isolates of T. harzianum biotypes 1, 2, and 4 and T. atroviride (formerly T. harzianum biotype 3) is presented in Table 1. Sources of the preepidemic isolates of Trichoderma obtained from cultivated mushrooms primarily in southeastern Pennsylvania between the 1950s and 1990 appear in Table 2.
TABLE 1.
Sources of the biotypes of T. harzianum and T. atroviride from mushroom cultures
| T. harzianum biotype | PSUc accession no. | Source | Isolate designation | Geographic origin |
|---|---|---|---|---|
| 1 | 64 | HRIa | TH1 T28TF | England |
| 65 | HRI | TH1 TD13 | England | |
| 149 | NAVBCb | MRG 1 | Ireland | |
| 150 | NAVBC | MRG 2 | Ireland | |
| 151 | NAVBC | MRG 3 | Ireland | |
| 152 | NAVBC | MRG 4 | Ireland | |
| 153 | NAVBC | MRG 5 | Ireland | |
| 154 | NAVBC | MRG 6 | Ireland | |
| 2 | 62 | HRI | TH2 KPNT | England |
| 63 | HRI | T43 (5) | England | |
| 155 | NAVBC | MRG 1 | Ireland | |
| 156 | NAVBC | MRG 2 | Ireland | |
| 157 | NAVBC | MRG 3 | Ireland | |
| 158 | NAVBC | MRG 4 | Ireland | |
| 159 | NAVBC | MRG 5 | Ireland | |
| 160 | NAVBC | MRG 6 | Ireland | |
| 3 (T. atroviride) | 60 | HRI | TH3 TD6 | England |
| 61 | HRI | TH3 TD7 | England | |
| 161 | NAVBC | MRG 1 | Ireland | |
| 162 | NAVBC | MRG 2 | Ireland | |
| 163 | NAVBC | MRG 3 | Ireland | |
| 164 | NAVBC | MRG 4 | Ireland | |
| 165 | NAVBC | MRG 5 | Ireland | |
| 166 | NAVBC | MRG 6 | Ireland | |
| 4 | 90 | UGd | TH4-RM10 | Canada |
| 91 | UG | TH4-BE | Canada | |
| 92 | UG | TH4-Rinle | Canada |
Peter R. Mills, Department of Plant Pathology and Microbiology, Horticultural Research International, Wellesbourne, Warwick, United Kingdom.
Owen P. E. Doyle, National Agricultural and Veterinary Biotechnology Centre, University College, Belfield, Dublin, Ireland.
Department of Plant Pathology, The Pennsylvania State University, University Park, Pa.
Danny L. Rinker, University of Guelph, Horticultural Research Institute of Ontario, Vineland Station, Ontario, Canada.
TABLE 2.
Description of the preepidemic isolates of Trichoderma from mushroom cultures
| PSU accession no. | Isolate designationa | Yr collected |
|---|---|---|
| 14 | DC149 | 1981 |
| 15 | DC209 | 1990 |
| 125 | DC39 | 1950s |
| 126 | DC40 | 1950s |
| 127 | DC98 | 1970s |
| 128 | DC99 | 1970s |
| 129 | DC100 | 1970s |
| 130 | DC101 | 1970s |
| 131 | DC102 | 1970s |
| 132 | DC104 | 1970s |
| 133 | DC105 | 1970s |
| 134 | DC106 | 1982 |
| 135 | DC179 | 1988 |
| 136 | DC205 | 1988 |
| 137 | DC206 | 1988 |
| 138 | DC207 | 1990 |
| 139 | DC208 | 1990 |
All isolates were obtained from The Pennsylvania State University Mushroom Culture Collection, Department of Plant Pathology, The Pennsylvania State University, University Park, Pa. All isolates were collected in Pennsylvania except isolate 135, which was collected in Washington state.
Isolation of DNA.
Fungal isolates were grown in 50 ml of potato dextrose-yeast broth for 2 days at room temperature with rotary shaking at 150 rpm. Mycelia were harvested by filtration through a piece of filter paper and washed with distilled water. One hundred milligrams of fresh mycelia was homogenized with a micropestle in a 1.5-ml microcentrifuge tube containing 400 μl of a solution containing 20 mM Tris-HCl (pH 7.8), 100 mM LiCl, 10 mM EDTA, and 0.5% sodium dodecyl sulfate. Five hundred microliters of a 25:24:1 (vol/vol/vol) mixture of phenol-chloroform-pentanol was added to the tube, and the tube was vortexed for 30 s and centrifuged at 14,000 × g for 10 min. The upper portion of the aqueous phase (∼250 μl) was recovered, and DNA was precipitated by the addition of 0.5 ml of cold absolute ethanol with incubation at −20°C for 10 min. DNA was collected by centrifugation at 14,000 × g for 10 min, dried, and resuspended in 20 μl of water. DNA preparations were diluted 1/500 with sterile water and used as the template for PCR amplification.
RAPD-PCR.
Amplification was carried out in a volume of 25 μl containing AmpliTaq buffer, a 100 μM concentration of each deoxynucleoside triphosphate, 1.25 mM MgCl2, a 0.2 μM concentration of an oligonucleotide primer, 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.), and 5 μl of the DNA template (20, 21). The reaction mixture was overlaid with 35 μl of mineral oil. Primers 203 (5′-CACGGCGAGT-3′), 211 (5′-GAAGCGCGAT-3′), 220 (5′-GTCGATGTCG-3′), 230 (5′-CGTCGCCCAT-3′), 232 (5′-CGGTGACATC-3′), 238 (5′-CTGTCCAGCA-3′), and 241 (5′-GCCCGACGCG-3′) were obtained from the University of British Columbia, Vancouver, Canada. Primer OPA13 (5′-CAGCACCCAC-3′) was obtained from Operon Technologies, Alameda, Calif. Each amplification included a negative control in which 5 μl of sterile water was substituted for the DNA template. Amplification was carried out with a Perkin-Elmer Gene Amp 480 system with the following program: one cycle at 94°C for 2 min and 35 cycles at 94°C for 1 min, 37°C for 2 min, and 72°C for 2 min. We have not repeated these experiments with another thermocycler.
PCR specific for biotypes 2 and 4.
Primers Th-F (5′-CGGTGACATCTGAAAAGTCGTG-3′) and Th-R (5′-TGTCACCCGTTCGGATCATCCG-3′) targeting an arbitrary sequence in the genomes of biotypes 2 and 4 were designed as described previously (2). PCR was carried out in a volume of 25 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, a 160 μM concentration of each deoxynucleoside triphosphate, 1 U of AmpliTaq, 5 μl of the DNA template, and a 0.5 μM concentration of the Th-F and Th-R primers. The reaction mixture was overlaid with 35 μl of mineral oil. Each experiment included a negative control in which 5 μl of sterile water was substituted for the DNA template. Amplification was carried out in a Perkin-Elmer 480 thermal cycler or a Stratagene RoboCycler GRADIENT 96 as follows: 1 cycle of 94°C for 2 min, 35 cycles at 94°C for 15 s (1 min for the RoboCycler) and 60°C for 1 min, and one cycle at 70°C for 7 min.
Electrophoresis.
Twenty microliters of the PCR product were mixed with 5 μl of a mixture of 1 M sucrose and 2 mM bromophenol blue and subjected to electrophoresis at 70 V for 3 h in a 2% agarose gel. The gel and electrophoresis buffer was 40 mM Tris-acetate–1 mM EDTA, pH 8.4, containing 50 ng of ethidium bromide per ml. A 100-bp DNA ladder (Life Technologies, Inc., Gaithersburg, Md.) was used as a size standard. DNA was visualized with UV transillumination and photographed with type 55 Polaroid film (Polaroid Corp., Cambridge, Mass.).
Genetic distance determination.
Each fungal isolate was scored for the presence or absence of 75 DNA products (markers) generated with the eight RAPD primers. Genetic distances were represented by euclidean metric distances (E), equivalent to the number of observed marker differences among all pairwise combinations of different isolates (4, 8). A dendrogram was constructed from euclidean metric distances by the unweighted pair group analysis of arithmetic means by using numerical taxonomy and multivariate analysis with NTSYS-PC software, version 1.5 (14).
RESULTS
RAPD-PCR.
Eight of the 20 RAPD primers that were initially screened were chosen for this study based on the capacity to reveal polymorphisms among the three biotypes of T. harzianum and T. atroviride. Primers typically generated gel electrophoretic DNA patterns having from one to eight major products ranging from 300 bp to more than 1,500 bp. The eight primers were used to profile 66 epidemic and 17 preepidemic isolates of Trichoderma from cultivated mushrooms and 27 tester isolates of T. harzianum biotypes 1, 2, and 4 and T. atroviride from Europe and Canada. RAPD patterns generated by the eight primers for the various fungal isolates were found to be highly reproducible in two or three separate trials.
RAPD-PCR analysis of epidemic isolates.
Fifty-eight of 66 isolates (88%) of Trichoderma collected at commercial sites following the onset of the epidemic shared an identical, unique RAPD pattern generated by each of the eight primers. The remaining eight isolates (12%) fell into seven genotypic classes, defined by RAPD patterns that were readily distinguishable from each other and from the predominant genotype.
Identification of the predominant epidemic-related genotype.
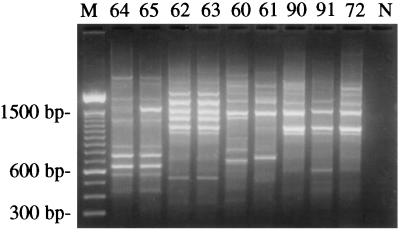
RAPD analysis was carried out on a representative isolate (PSU 72) of the predominant genotype of Trichoderma from Pennsylvania and biotypes 1, 2, and 4 of T. harzianum as well as T. atroviride on mushrooms. The prevalent genotype in Pennsylvania was identical to isolates of biotype 4 originating in Canada and clearly distinct from isolates of biotypes 1 and 2 and T. atroviride from Europe (Fig. 1).
FIG. 1.
Identification of the predominant epidemic-associated genotype of Trichoderma by RAPD-PCR with primer 230. Shown are the gel electrophoretic profiles of isolates 64 and 65 (biotype 1), isolates 62 and 63 (biotype 2), isolates 60 and 61 (T. atroviride), isolates 90 and 91 (biotype 4), and isolate 72 (representing the predominant genotype in Pennsylvania). M, 100-bp DNA ladder. N, negative control in which sterile water was substituted for the DNA template.
Determination of genetic distances.
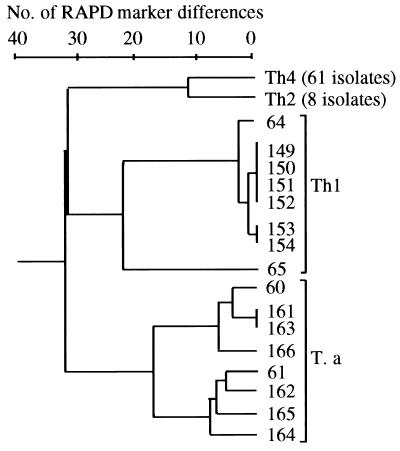
A dendrogram was constructed based on the variation in 75 RAPD markers in order to reveal the genetic variation and relationships among the three biotypes of T. harzianum and T. atroviride (Fig. 2). Cluster analysis showed a distinct separation of the isolates according to biotype and species. Clusters representing biotypes 1 and 2 and T. atroviride each contained the eight tester isolates, whereas the biotype 4 cluster contained 61 isolates, including the 58 isolates of the predominant genotype from Pennsylvania and the three tester isolates from Canada. The cophenetic correlation coefficient for the dendrogram (r = 0.97) indicated an excellent fit between the data and the actual clustering.
FIG. 2.
Dendrogram based on the number of differences in a total of 75 scored RAPD markers among the three biotypes of T. harzianum and T. atroviride. Numbers on the dendrogram refer to PSU accession numbers for the isolates. No genetic variation was observed among isolates within the biotype 2 and 4 clusters (Th2 and Th4). The biotype 1 and 2 and T. atroviride (T. a) clusters each contained eight isolates from Europe, whereas the biotype 4 cluster contained 58 isolates from Pennsylvania and three tester isolates from Canada.
The highly virulent biotypes (2 and 4) from Europe and North America, respectively, were more closely related to each other than to the largely benign biotype 1 and T. atroviride. Biotypes 2 and 4 differed at 12 of the 75 RAPD bands, compared to a difference of 32 to 33 bands between the biotype 1-T. atroviride and biotype 2-biotype 4 clusters. No genetic variation was observed within biotype 2 or 4, indicating that these two populations of pathogenic biotypes were highly homogenous (i.e., represent clones). In contrast, considerable variation was noted among isolates within the biotype 1 and T. atroviride clusters. Within the biotype 1 cluster, isolate 65 was most distantly related to the other isolates.
Characteristics of the specific PCR.
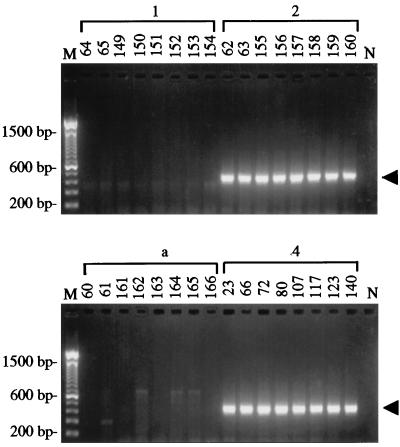
The primer set Th-F and Th-R, which targeted an arbitrary and unique sequence in biotype 4, amplified the expected ∼450-bp PCR product in each of eight different isolates of biotypes 4 and 2 (Fig. 3). In contrast, no major products were generated with isolates of biotype 1 and T. atroviride. The diagnostic amplicons generated from biotypes 2 and 4 each comprised 444 bp and had a 99% sequence similarity (data not shown). The specificity of the primer pair for biotypes 2 and 4 also was demonstrated in other experiments in which no major PCR products were observed following the amplification of genomic DNA templates representing 16 species of Trichoderma, Gliocladium, and Hypocrea or 15 other genera of fungi (2).
FIG. 3.
Specificity of the PCR for biotypes 2 and 4 of T. harzianum from cultivated mushrooms. Shown are the amplification products resolved by agarose gel electrophoresis that were generated with primer pair Th-F and Th-R by using genomic DNA templates of eight isolates of T. harzianum biotypes 1, 2, and 4 and T. atroviride (a). Numbers above the lanes refer to PSU accession numbers for the isolates. The arrowheads indicate the position of the predicted 444-bp amplicon. M, 100-bp DNA ladder. N, negative control in which sterile water was substituted for the DNA template.
PCR analysis of preepidemic isolates.
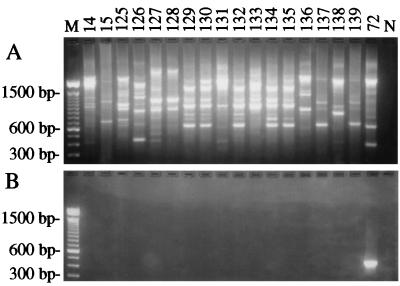
All 17 isolates of Trichoderma that were collected before the epidemic had RAPD electrophoretic patterns that were distinctly different from those of biotype 4. With the exception of one isolate from Washington state (PSU 135), all of these isolates were collected from the same geographic region as the epidemic isolates. Several of the preepidemic isolates (127 and 128; 129, 130, 132, 134, and 135; 137 and 139) had similar or identical profiles (Fig. 4A). In contrast, we estimated that the isolates most closely related to biotype 4 shared only a 15% similarity in RAPD markers. Likewise, all of the preepidemic isolates failed to generate the ∼450-bp amplicon diagnostic for biotypes 2 and 4 in amplifications with the Th-F and Th-R primer pair (Fig. 4B).
FIG. 4.
PCR analysis of 17 isolates of Trichoderma collected from mushroom cultures prior to the epidemic between the 1950s and 1990. Shown are the amplification products resolved by gel electrophoresis after RAPD-PCR with primer OPA13 (A) and PCR with the Th-F and Th-R primer set targeting a unique genomic sequence in biotypes 2 and 4 (B). Numbers above the lanes refer to the isolates by their PSU accession numbers. M, 100-bp DNA ladder. Isolates 14, 15, and 125 to 139 are preepidemic isolates. Isolate 72 is a positive control for the DNA template of biotype 4. N, negative control in which sterile water was substituted for the DNA template.
DISCUSSION
The escalation of green mold disease on cultivated Agaricus mushrooms in North America during the 1990s has been associated with T. harzianum biotype 4 (1, 5, 17). Castle and coworkers (1) surveyed commercial mushroom operations for green mold disease and found biotype 4 to occur at an approximately 40% incidence and to be specifically associated with farms experiencing major crop losses. One of the many questions about this disease that remains unanswered is whether biotype 4 was newly introduced to cultivated mushrooms at the onset of the epidemic or preexisted in cultivated mushrooms in a benign form or as an unrecognized pathogen.
In the present study, we compared the genetic variation in a collection of Trichoderma spp. associated with the green mold epidemic on cultivated mushrooms with that in a collection sampled prior to the onset of the epidemic. We focused on commercial operations located in southeastern Pennsylvania, a region that accounts for approximately 45% of the total U.S. mushroom crop. We found that while biotype 4 was associated with almost 90% of the outbreaks of green mold occurring during the height of the epidemic from 1994 to 1996, we could find no evidence for its earlier existence in mushroom cultivation. PCR analysis with primers specifically targeting biotypes 4 and 2 confirmed and extended the results of our RAPD analysis, indicating that biotype 4 was not represented among the preepidemic isolates. Muthumeenakshi and coworkers (10) alluded to the fact that biotype 2 was not similar to any isolates of T. harzianum deposited in international culture collections. We do not know if our collection of preepidemic isolates is representative of the spectrum of Trichoderma spp. associated with cultivated mushrooms. We suspect that some isolates are largely benign, having been isolated from wooden surfaces, compost, and symptomless mushrooms, whereas others are associated with outbreaks of green mold disease. We conclude that the green mold epidemic in cultivated A. bisporus in North America probably resulted from the recent introduction of a different and highly virulent genotype of the pathogen.
Eight of the 66 epidemic-associated isolates (12%) and all 17 of the preepidemic isolates of Trichoderma did not have any identity with biotype 2 or 4 by RAPD-PCR or PCR specific for biotypes 2 and 4. Aside from the two pathogenic biotypes, several less-pathogenic biotypes and species of Trichoderma are commonplace in mushroom cultivation. For example, biotype 1, T. atroviride (biotype 3), T. viride, T. aureoviride, T. koningii, T. pseudokoningii, T. hamatum, T. citrinoviride, T. longibrachiatum, T. crassum, and T. spirale have been isolated from the raw ingredients of mushroom compost, or Agaricus-colonized compost and casing, but seldom cause significant crop loss (1, 15, 16, 18). DNA sequence analysis of a β-tubulin gene now under way suggests that the epidemic and preepidemic isolates, which were not of biotype 2 or 4, described herein include many of the same species (3).
The findings of our RAPD analysis concur with those of earlier reports (1, 10, 12), indicating a high genetic similarity between biotypes 2 and 4. Further, the highly conserved RAPD profiles displayed by all isolates of biotypes 2 and 4 characterized herein and previously (1, 10) support the clonal nature of the two pathogen populations and suggest that the recent epidemics of green mold in Europe and North America occurred independently and that they originated from single sources. The lack of extensive genetic variation within each biotype makes it unlikely that either biotype 2 or biotype 4 was derived from the other within cultivated mushrooms but rather that they represent two similar clones.
We considered the possibility that biotype 4 existed in cultivated mushrooms in a benign fashion and at a low population level and then flourished to epidemic proportions owing to changes in cultural practices. However, such a disease induction hypothesis would involve some degree of genetic variation among the collected pathogenic isolates as evidenced by polymorphic RAPD markers, which is not the case. Rather, the lack of variation within the pathogen population further supports the hypothesis of a more recent introduction of biotype 4 into cultivated mushrooms.
ACKNOWLEDGMENTS
We thank Vija Wilkinson for technical assistance.
This work was supported by grants from the Pennsylvania Department of Agriculture (ME 445101) and the Mushroom Industry Farmer-based Applied Research Program.
REFERENCES
- 1.Castle A, Speranzini D, Rghel N, Alm G, Rinker D, Bissett J. Morphological and molecular identification of Trichoderma isolates on North American farms. Appl Environ Microbiol. 1998;61:133–137. doi: 10.1128/aem.64.1.133-137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Romaine C P, Ospina-Giraldo M D, Royse D J. A PCR-based test for the identification of Trichoderma harzianum biotypes 2 and 4 inciting the world-wide green mold epidemic in cultivated Agaricus bisporus. 1999. Appl. Microbiol. Biotechnol., in press. [Google Scholar]
- 3.Chen, X., C. P. Romaine, M. D. Ospina-Giraldo, and D. J. Royse. Unpublished data.
- 4.Excoffier L, Smouse P E, Quattro J M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gams W, Meyer W. What exactly is Trichoderma harzianum? Mycologia. 1998;90:904–915. [Google Scholar]
- 6.Geels F, van de Geijin J, Rutjens A. Pests and diseases. In: van Griensven L J L D, editor. The cultivation of mushrooms. East Grinstead, Sussex, England: Interlingua; 1988. pp. 361–422. [Google Scholar]
- 7.Harvey C L, Wuest P J, Schisler L C. Diseases, weed molds, indicator molds, and abnormalities of the commercial mushroom. In: Wuest P J, Bengston G D, editors. Penn State handbook for commercial mushroom growers. University Park, Pa: The Pennsylvania State University; 1982. pp. 19–33. [Google Scholar]
- 8.Huff D R, Peakall R, Smouse P E. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloe dactyloides (Nut) Engelm.] Theor Appl Genet. 1993;86:927–934. doi: 10.1007/BF00211043. [DOI] [PubMed] [Google Scholar]
- 9.Morris E, Doyle O, Clancy K. A profile of Trichoderma species. I. Mushroom compost production. In: Elliot T, editor. Science and cultivation of edible fungi. Rotterdam, The Netherlands: Balkema; 1995. pp. 611–625. [Google Scholar]
- 10.Muthumeenakshi S, Mills P R, Brown A E, Seaby D A. Intraspecific molecular variation among Trichoderma harzianum isolates colonizing mushroom compost in the British Isles. Microbiology. 1994;140:769–777. doi: 10.1099/00221287-140-4-769. [DOI] [PubMed] [Google Scholar]
- 11.Muthumeenakshi S, Mills P R. Detection and differentiation of fungal pathogens of Agaricus bisporus. Mushroom Sci. 1995;14:603–610. [Google Scholar]
- 12.Ospina-Giraldo M D, Royse D J, Thon M R, Chen X, Romaine C P. Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources. Mycologia. 1998;90:76–81. [Google Scholar]
- 13.Ospina-Giraldo M D, Royse D J, Chen X, Romaine C P. Molecular phylogenetic analysis of biological control strains of Trichoderma harzianum and other biotypes of Trichoderma spp. associated with mushroom green mold. Phytopathology. 1999;89:308–313. doi: 10.1094/PHYTO.1999.89.4.308. [DOI] [PubMed] [Google Scholar]
- 14.Rohlf F J. NTYSYS-pc: numerical taxonomy and multivariate analysis system. Setauket, N.Y: Exeter Publishers; 1989. [Google Scholar]
- 15.Seaby D A. Infection of mushroom compost by Trichoderma species. Mushroom J. 1987;179:355–361. [Google Scholar]
- 16.Seaby D A. Further observation on Trichoderma. Mushroom J. 1989;197:147–151. [Google Scholar]
- 17.Seaby D A. Differentiation of Trichoderma taxa associated with mushroom production. Plant Pathol. 1996;45:905–912. [Google Scholar]
- 18.Seaby D A. Investigation of the epidemiology of green mould of mushroom (Agaricus bisporus) compost caused by Trichoderma harzianum. Plant Pathol. 1996;45:913–923. [Google Scholar]
- 19.Sinden J, Hauser E. Nature and control of three mildew diseases of mushrooms in America. Mushroom Sci. 1953;2:177–180. [Google Scholar]
- 20.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]