Abstract
We developed a technique to map the availability of sugars and amino acids along live roots in an intact soil-root matrix with native microbial soil flora and fauna present. It will allow us to study interactions between root exudates and soil microorganisms at the fine spatial scale necessary to evaluate mechanisms of nitrogen cycling in the rhizosphere. Erwinia herbicola 299R harboring a promoterless ice nucleation reporter gene, driven by either of two nutrient-responsive promoters, was used as a biosensor. Strain 299RTice exhibits tryptophan-dependent ice nucleation activity, while strain 299R(p61RYice) expresses ice nucleation activity proportional to sucrose concentration in its environment. Both biosensors exhibited up to 100-fold differences in ice nucleation activity in response to varying substrate abundance in culture. The biosensors were introduced into the rhizosphere of the annual grass Avena barbata and, as a control, into bulk soil. Neither strain exhibited significant ice nucleation activity in the bulk soil. Both tryptophan and sucrose were detected in the rhizosphere, but they showed different spatial patterns. Tryptophan was apparently most abundant in soil around roots 12 to 16 cm from the tip, while sucrose was most abundant in soil near the root tip. The largest numbers of bacteria (determined by acridine orange staining and direct microscopy) occurred near root sections with the highest apparent sucrose or tryptophan exudation. High sucrose availability at the root tip is consistent with leakage of photosynthate from immature, rapidly growing root tissues, while tryptophan loss from older root sections may result from lateral root perforation of the root epidermis.
Interactions between roots and microorganisms in distinct spatial and temporal patterns in soil surrounding roots probably influence nitrogen mineralization and cycling. To develop models of this process, information is needed on (i) the spatial and temporal pattern of exudation of sugars and amino acids from roots into the rhizosphere and (ii) the effect of these exudates on numbers of native bacteria present in the rhizosphere. Among the compounds commonly found in exudates, sugars are important because they are found in the largest quantity (14) and constitute a very labile source of C for microorganisms. Amino acids are commonly the second-largest component of exudates (14) and can provide both carbon skeletons and amino N to microorganisms. The proportion of sugars to amino acids in exudates is important because it affects the relative amounts of C and N available for microbial growth.
Most knowledge about root exudation stems from three experimental approaches (8). In the first approach, plants are grown in sterile hydroponic culture and exudates are collected from the solution bathing the roots. The solution is then analyzed for its chemical constituents. The second approach utilizes plants grown in a solid matrix, such as sand, soil, or simulated soil, from which exudates are leached and chemically analyzed. In the third general class of experiments, plants are grown in soil, isotopic tracers are administered via the leaves, and labeled exudates are detected in leachate. While these methods have provided much useful information on the process of root exudation, they do not provide sufficient spatial detail to evaluate fine scale root-microbe interactions that have been hypothesized to accelerate N cycling in the rhizosphere.
An alternative approach utilizes 14CO2 labeling of plants in microcosms in natural soil to detect exudation from roots. This method can provide information on the rate of C flow through the plant into the soil as well as the quantity of C flow into the soil (23). When this method is linked with autoradiography, general patterns of exudation can be observed, but these patterns will be partially obscured by diffusion of labeled CO2 from both root respiration and microbial metabolism of root exudates. This method does not provide information on the composition of exudates.
We used a promoterless ice nucleation gene, inaZ, from the bacterium Pseudomonas syringae as a reporter gene to detect the availability of exudate compounds in soil with native flora and fauna present. When inaZ is expressed, proteins with high ice nucleation activity are produced and incorporated into the outer membrane of bacteria, enabling the cells to catalyze ice formation at relatively warm temperatures (>−10°C). Bacterial ice nuclei are easily quantified by a simple droplet-freezing assay (16, 18, 19).
The objectives of this study were threefold: (i) to demonstrate the utility of bacterial biosensors incorporating inaZ for evaluation of the availability of two substrates (sugars and amino acids); (ii) to map the availability of these substrates along a live, growing root; and (iii) to relate patterns of substrate availability to the anatomy of the root and to population densities of native bacteria around the root. Our ultimate goal is to apply the approach described here to a study of the mechanism by which plant roots may stimulate N cycling in the rhizosphere by the effect of their exudates on soil bacteria.
MATERIALS AND METHODS
Bacterial strains.
Two variants of the rifampin-resistant Erwinia herbicola strain 299R (1) were used as biological sensors. E. herbicola 299RTice (hereafter referred to as 299R-tryp for clarity) produces ice nuclei when transcription of the aatl gene, encoding a tryptophan aminotransferase, is induced in response to the presence of tryptophan in the external environment of the bacterium (5, 6). Tn3-Spice was used to produce fusions of inaZ within aatl when contained within clones in pLAFR3 (27) by methods similar to that used by Lindgren et al. (15). Fusions in the same orientation as the direction of transcription of aatl had high ice nucleation activity only when cells were grown in the presence of tryptophan. The chromosomal aatl gene was replaced with an aatl::Tn3-Spice fusion gene by marker exchange mutagenesis, performed similarly to mutagenesis employed in other studies (2). Marker exchange mutant 299RTice was selected as a rifampin- and streptomycin-resistant colony as described previously (2).
E. herbicola 299R(p61RYice) (hereafter referred to as 299R-sucr) expresses ice nucleation activity when sucrose or fructose is present in its external environment (17, 22). This strain is responsive to concentrations of sucrose that are about 100-fold lower than that of fructose (17, 22). The p61RYice construct consists of the scrY promoter region and the scrR repressor gene from the Salmonella typhimurium plasmid pUR400 (7, 26) cloned into the stable plasmid pVSP61 (18), which confers kanamycin resistance. In this construct, the scrY promoter is cloned immediately 5′ to a promoterless inaZ gene such that expression of ice nucleation is under the control of PscrY and scrR. Cells of 299R-tryp were recovered on culture medium containing both rifampin (100 μg · ml−1) and streptomycin (20 μg · ml−1), while strain 299R-sucr cells were recovered on medium containing both rifampin (100 μg · ml−1) and kanamycin (50 μg · ml−1).
Culturing of bacteria.
Each strain was grown at 24°C and shaken at 200 rpm in minimal A medium (21) containing the appropriate antibiotics. When the cultures were in log phase, tryptophan was added to the 299R-tryp strain at concentrations ranging from 10−8 to 10−2 M. The cultures were then incubated for 6 h. Strain 299R-sucr was exposed to concentrations of sucrose or fructose ranging from 10−6 to 10−2 M.
Bacteria for inoculation into soil were cultured as described above in the absence of added tryptophan or sucrose. Cell concentration was quantified by determining the turbidity of the culture at 600 nm. Cells were removed from the growth medium by centrifugation, washed, and resuspended in Tris NaCl buffer (pH 7.2, 0.2 M). The amount of buffer for resuspension was calculated based on the initial number of cells per milliliter (determined from the optical density [OD]), such that the addition of the resuspended bacteria to soil resulted in the addition of 107 cells per gram of soil and a moisture content of approximately 25%.
Measurement of ice nucleation activity.
The number of ice nuclei per milliliter was estimated by a droplet-freezing assay similar to that employed in other studies (18). Cell suspensions or soil slurries were serially diluted, and 40 10-μl droplets of each sample dilution were placed on a paraffin-coated aluminum foil sheet. The sheet was floated on a refrigerated ethanol bath maintained at −7°C. The fraction of droplets that froze within 5 min was used to calculate the number of ice nuclei in the original sample (18). In all cases, the number of viable cells of either 299R-tryp or 299R-sucr was determined by plating appropriate dilutions on Luria agar containing appropriate antibiotics and enumerating colonies after growth for 2 days at 26°C. The estimated number of ice nuclei in a given sample was normalized for the number of cells of the biosensor strains as in other studies (18).
Plants and soils.
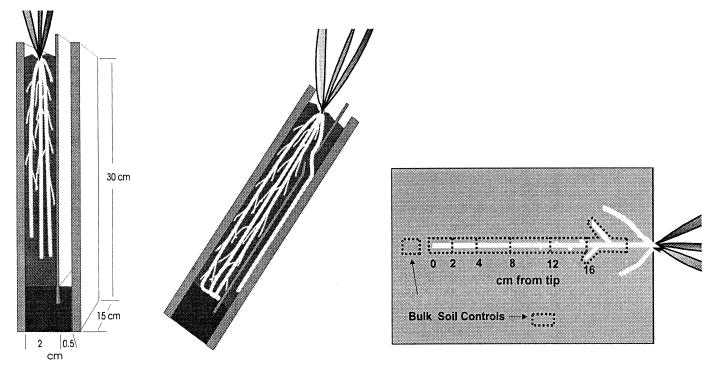
Studies were conducted with the annual graminoid Avena barbata, cultivated from seed collected at the Jasper Ridge Biological Preserve, Palo Alto, Calif., in microcosms in growth chambers. A. barbata has a growth pattern and root anatomy that facilitate microcosm studies. The microcosms (Fig. 1), modified from the design of James et al. (13), can be divided into a large chamber and a small chamber by a thin Plexiglas divider. Plants were grown in the large chamber, which was filled with a mixture of 1:1 Sierra Foothills soil and fine quartz sand (to improve drainage). The soil was a Mollic Haploxeralf from the Argonaut series, collected at the Sierra Foothills Range Experiment Station, Marysville, Calif. The thin sections of the microcosms were filled later in the experimental course with a 1:1 mixture of sand and Yolo silt loam from the Plant Pathology Farm at University of California—Davis. This substitution was necessitated by high background levels of ice nucleation activity and antibiotic-resistant native bacteria in the original soil. An A. barbata seedling was planted into the large chamber of each of five microcosms and then placed in growth chambers with day- and nighttime temperatures of 25 and 15°C, respectively, and a 12-h photoperiod. Plants were watered with tap water twice a week and with a dilute solution of a balanced plant nutrient solution (Peter’s Professional 20-20-20 with micronutrients; Grace-Sierra Horticultural Products Inc., Milpitas, Calif.) once a week. The microcosms were wrapped in aluminum foil to prevent light from reaching the roots.
FIG. 1.
Plant microcosms. Plants were grown in the main section of the microcosm (left). A root growing through a thin section during an experiment is shown in the center. On the right the soil harvest pattern along a root in the thin section of the microcosm is shown. Five bulk soil control samples were taken from each microcosm (only two are shown).
Experimental approach.
When the plants reached peak vegetative growth (maximum aboveground biomass prior to visible onset of flowering) and were producing large numbers of rapidly growing primary roots from their stem bases (usually 6 weeks after germination), the solid divider separating the two sections of the microcosm was replaced by a divider with a 5-mm-high horizontal slot located approximately 3 cm below the soil line (Fig. 1). The small chamber was filled with a soil-and-sand mixture. The microcosms were placed back in the growth chambers and held in racks at a 45° angle. Under the influence of gravity, new roots initiated from the stem base of the plants grew along the divider, entered the thin section of the microcosm through the slot in the divider, and grew through the soil in the thin section, along the face of the microcosm. The progress of the roots was monitored daily to determine root growth rate. When new roots were within 3 cm of the bottom of the thin section, we removed the face of each microcosm and sprayed the exposed soil face with a suspension of the biosensor bacteria. The microcosms were resealed, placed back in the growth chambers for 24 h, and then harvested. The microcosms were not watered during the 24-h period of incubation to prevent redistribution of the biosensors by mass flow of water.
The covers over the thin section were removed, exposing the soil surface of the thin section. Roots were severed from the main plant where they passed through the slot in the divider. Entire roots were excised from the open face of the soil so that an approximately 2-mm-thick layer of rhizosphere remained on the root. The root was placed on a clean surface and cut into sections 2 cm in length for the youngest 4-cm-long segment of the root and into 4-cm lengths for the remainder of the root (Fig. 1). Each root section was placed in a preweighed tube containing buffer, cleansed of soil by gentle agitation, and then removed. The buffer-containing tube was reweighed to determine the weight of soil from the root sample. The procedure yielded at least 0.5 g of wet soil per sample. Bulk soil control samples were taken at a distance of greater than 1.5 cm from any root. The soil-buffer samples were stored at 5°C. A large (∼12 g) soil sample was composited from five different areas in each microcosm, weighed, and dried at 100°C overnight. The next day, the dried soil samples were reweighed and the gravimetric water content was calculated for the soil from each microcosm. Each soil-buffer sample was diluted individually with the amount of dilutant needed to yield a 1:100 dilution; 10-fold dilution series were then generated to 1:10−5 and used for the assay.
To evaluate the possible effects of root exudates on numbers of native soil bacteria, an experiment was conducted under conditions identical to those used in the above-described experiments but without the addition of biosensor bacteria. The thin sections of the microcosms were filled with the Yolo silt loam and sand mixture as described above. Roots were allowed to grow through the thin section for 9 days, and soils were harvested from around specific root sections as noted above except that the soil from the first 4-cm length of root was integrated into one sample. Soil samples were diluted to 1:1,000, stained with acridine orange, filtered onto black polycarbonate filter disks, and examined by epifluorescence microscopy to determine total bacterial numbers per gram of soil (24).
Root sections from each experiment were preserved in an aqueous 50% ethanol solution, and five replicate roots were examined under a dissecting microscope for gross anatomical features, including the distance from the tip at which root hairs appeared and the distance at which lateral roots were initiated.
Statistical tests.
The numbers of ice nuclei per cell and the direct bacterial count data were log transformed to meet the assumption of normality. Means were compared by one-way analysis of variance and Duncan’s multiple range test (JMP statistical software; SAS Institute, Cary, N.C.).
RESULTS
Biosensor strains.
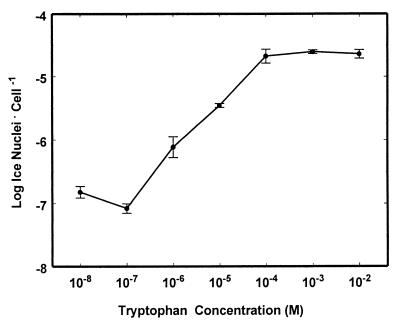
E. herbicola 299R-tryp exhibited a wide range of ice nucleation activity that was dependent on the concentration of tryptophan present in the culture medium (Fig. 2). When 10−8 M tryptophan was added to a minimal medium, 299R-tryp exhibited ice nucleation activity of only 3 × 10−7 ice nuclei per cell. Ice nucleation activity increased with increasing tryptophan concentrations within the range of 10−7 to 5 × 10−5 M. At a tryptophan concentration of 10−4 M, the number of ice nuclei was about 100-fold larger than at 10−7 M tryptophan (Fig. 2). E. herbicola 299R-sucr also exhibited an over-100-fold variation in ice nucleation that was dependent on sucrose concentration in the range of about 10−5 to 10−4 M (22). The ice nucleation activity of strain 299R-sucr was not responsive to any of 23 other sugars and carbohydrates tested except fructose and sorbose (22).
FIG. 2.
Tryptophan-dependent ice nucleation activity in E. herbicola 299R-tryp. Bacteria were grown in flask culture in the absence of tryptophan. The different concentrations of tryptophan shown on the abscissa were added while bacteria were in log phase growth. Each error bar indicates the standard error of the mean for three cultures.
Soil experiment.
The A. barbata roots grew at an average rate of about 2 cm per day. The roots began to show root hairs at an average distance of 1.2 ± 0.15 cm from the tip (Fig. 3A). The root hairs persisted until they were at least 16 cm from the tip. Lateral root primordia first emerged through the root epidermis at an average distance of 9.2 ± 0.50 cm from the tip. Lateral roots reached a length of 3 mm by the time they were 11.9 ± 0.48 cm from the tip.
FIG. 3.
(A) Spatial pattern of root anatomy showing the onset of root hair formation, lateral root primordia, and lateral roots. (B) Ice nucleation activity associated with sucrose biosensors in specific zones of the root. (C) Ice nucleation activity associated with tryptophan biosensors in specific zones of the root. Vertical dotted lines relate the section of the root shown in panel A to levels shown in panels B and C. Data with the same lowercase letter are not significantly different.
The ice nucleation activities of inocula of both 299R-tryp and 299R-sucr were less than 10−6 ice nuclei · cell−1 and were undetectable after introduction into the soil. The sucrose sensor (strain 299R-sucr) exhibited ice nucleation activity of only 2 × 10−4 ice nuclei per cell in bulk soil controls (Fig. 3B). All rhizosphere samples had a higher ice nucleation activity than bulk soil samples. The highest ice nucleation activity in 299R-sucr occurred in soil adjacent to the tip section of the roots (0 to 2 cm); this activity was almost 100-fold higher than that in bulk soil samples. The ice nucleation activity of 299R-sucr decreased progressively to section 4 (8 to 12 cm), where it was about sixfold lower than at the root tip.
The ice nucleation activity of strain 299R-tryp was significantly higher around all sections of the root than that in bulk soil controls (where it was undetectable) (Fig. 3C). Rhizosphere samples from the first two root sections (0 to 2 and 2 to 4 cm from the tip) showed low but detectable induced ice nucleation activity. At the third section (4 to 8 cm), ice nucleation activity increased slightly but was not significantly higher than that at the second or first section. Compared to that at the first section, the ice nucleation activity was increased more than fivefold at the fourth section (8 to 12 cm) and reached its highest value, almost 10-fold higher than that at the first section, at the fifth section (12 to 16 cm from the tip). The response patterns for 299R-suc and 299R-tryp in this experiment were consistent with patterns obtained from two previous replicate experiments using 299R-suc and 299R-tryp.
At harvest time, 299R-tryp cells numbered 6.6 × 106 CFU · g of soil−1 in the bulk soil and averaged slightly fewer (4.9 × 106 CFU · g of soil−1) in the rhizosphere. 299R-sucr cell numbers were 2.9 × 106 CFU · g of soil−1 in the bulk soil and were greater (average, 4.2 × 107 CFU · g of soil−1) in the rhizosphere. Neither strain showed a significant pattern of variation in sensor numbers along the root.
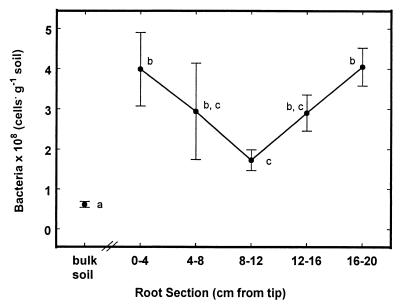
Total native bacterial numbers in the rhizosphere were always greater than those in the bulk soil control samples (6.7 × 107 cells · g of soil−1) (Fig. 4). The first (0 to 4 cm from the tip) and last (16 to 20 cm) root sections showed the largest bacterial numbers (4.0 × 108 cells per g of soil). Numbers declined to a minimum of 1.7 × 108 cells per g of soil adjacent to the third section (8 to 12 cm from the tip). Bacterial numbers then increased in the fourth section, and in the fifth section they reached the same high value as at the tip (4.0 × 108 cells · g of soil−1 at 16 to 20 cm from the tip).
FIG. 4.
Spatial pattern of bacterial population sizes along the root, determined by acridine orange staining and direct counting by epifluorescence microscopy. The first root section corresponds to the first two root sections (0 to 2 and 2 to 4 cm from the tip) for the root for which data are shown in Fig. 3. Data with the same lowercase letter are not significantly different.
DISCUSSION
The tryptophan and sucrose biosensor strains both indicated the presence of considerably higher levels of their respective substrates in rhizosphere compared to those in bulk soil controls. However, the patterns of tryptophan availability and sucrose availability were very different. Apparent rhizosphere tryptophan availability was relatively low within about 8 cm from the tip; tryptophan was detected at much higher levels more than 8 cm from the root tip. In this and other replicate experiments the level of tryptophan detection in the region of the root where laterals had begun to emerge (8 to 12 cm from the tip in this case) was always increased markedly compared to those in younger root sections. Conversely, sucrose availability was highest in the tip section of the root and decreased in progressively older sections, where it reached a minimum at about 8 to 12 cm from the tip.
The data show that different exudate compounds have different patterns of availability along a root. The fact that sucrose was detected most strongly near the tip is consistent with the hypothesized movement of sucrose from the protophloem in the zone of root elongation to the meristem by apoplastic diffusion (3). A portion of the apoplastic sucrose may diffuse out of the root along the concentration gradient that exists between the inside and the outside of the root. Sucrose does not leak from mature sections of the root, where the phloem is fully developed, and diffusion between the root stele and the exterior is blocked by a suberized layer of endodermal cells (11).
The pattern of tryptophan leakage may pertain to all amino acids in the root or could be specific to tryptophan. There may, in general, be higher internal amino acid concentrations in mature sections of the root than in the tip. Perforation of the epidermis by lateral roots in older sections may allow release of a portion of these amino acids. In one of the few studies conducted on longitudinal zonation of N assimilation enzymes within roots, nitrate reductase activity was found to be higher in the 2-cm section beginning at the tip than in the mature section of corn seedling roots, but glutamate dehydrogenase (which is involved in assimilation of NH4+ into amino acids) activity was much higher in mature sections of the root (29). It is possible that quantitatively more amino acid synthesis occurs in mature portions of the root than in the tip and/or that amino acids synthesized in the tip are immediately incorporated into new tissue, while amino acids generated in fully expanded mature roots persist longer and are subject to leakage through lateral root channels.
The presence of tryptophan near roots with laterals also could relate to the role of indoleacetic acid (IAA) in lateral root initiation (4). Tryptophan is a precursor for IAA synthesis, which occurs primarily in the shoot (25). A mechanism for the conversion of IAA to tryptophan in the root or soil surrounding emerging laterals seems to be required for this explanation to apply. An alternative explanation is that tryptophan is metabolically linked to synthesis of compounds affording defense against microbial pathogens. The first root section where increased levels of tryptophan were detected (section 4, 8 to 12 cm from the tip) was also the section where lateral root primordia were expanding out through the root epidermis. Puncturing the epidermis could provide an avenue for pathogen entry, and this tissue disruption may lead to synthesis of plant defense compounds. In corn, the cyclic hydroxamic acids 2,4-dihydroxy-1,4-benzoxazin-3-one and 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one are synthesized via a branch of the tryptophan synthetic pathway and confer resistance to pathogens (12). The tryptophan synthetic pathway is involved in the synthesis of phenolic compounds, including chlorogenic acid, which has been shown to protect potato tubers from fungal pathogens (30).
The pattern of rhizosphere bacterial population sizes along the root (Fig. 4) appears to reflect the pattern of exudation from the root. While all rhizosphere samples had greater numbers of indigenous bacteria than nearby bulk soils, within the rhizosphere, the greatest numbers of bacteria occurred adjacent to the tip sections (0 to 4 and 4 to 8 cm from the tip) where the highest levels of sucrose were detected and in the oldest sections (12 to 16 and 16 to 20 cm from the tip), which showed the highest levels of tryptophan. The smallest populations occurred about 8 to 12 cm from the tip. This section exhibited the lowest level of sucrose exudation and an intermediate level of tryptophan exudation. This pattern could result from bacterial growth supported by large amounts of sugars present at the tip and amino acids (indicated by tryptophan) present around older sections. Total native bacterial numbers may correlate with overall root exudation but they cannot reveal differences in patterns of exudate components. The capability to discern the different patterns may prove useful in understanding root anatomy and physiology, as well as in understanding the spatial distribution of microbial activities in the rhizosphere.
Because the roots were growing during the 24-h period that the biosensor strains were in the soil, the biosensor data point for each root section may include some contribution from the root section previously monitored by the biosensor (except for the root tip data point). It seems unlikely at the resolution reported here (2 to 4 cm) that biosensor motility affects the resolution of the technique over 24 h.
Populations of sensor bacteria did not show a significant pattern of variation along the root. This is probably because the sensor bacteria encountered exudates from the root for only 24 h while the native bacteria were in contact with the root for up to 9 days.
Our results demonstrate that it is possible to determine patterns of exudation from roots growing through soil and subject to the effects of abrasion by soil particles and to the effects of native microbial populations. The method allows us to evaluate exudation while controlling or manipulating factors such as soil moisture, soil nutrient availability, and leaf level environmental factors. Our approach minimizes disruption of the root-soil matrix and of the native microbial community, especially in comparison to hydroponic or nonsoil solid medium culture. At the end of the experiment there was no evidence of root death in the main section resulting from disruption during switching to the slotted barrier. Roots in the thin section showed no sign of damage resulting from removal of the faceplate for spraying when they were harvested 24 h later. Opening the microcosm probably increases the O2 concentration around the roots temporarily, but this should not constitute a major shock to the roots or native microorganisms because the soils are well drained and presumably well aerated. Soils in the microcosms have never shown signs of being under anaerobic conditions.
Other reporter systems have been used to evaluate rhizosphere bacteria. de Weger et al. (9) used a lux reporter to detect Pseudomonas species in the rhizosphere, while van Overbeek and van Elsas (28) used lacZ to observe the response of promoter activity in Pseudomonas fluorescens to root exudates. de Weger et al. (10) used lacZ to evaluate phosphate limitation in rhizosphere bacteria. In all these cases, the best results were achieved in nonsoil media. The challenge of ensuring sufficient metabolic energy for lux constructs in soil and the attenuation of light by soil reduce the utility of this reporter for work in soil. lacZ also gives better results in nonsoil media, in part due to the high background levels of β-galactosidase activity in soils. The inaZ gene appears to be ideally suited for use in soils. It can be easily measured in “dirty” biological systems and is also more sensitive than other genes, such as lacZ, lux, or gus (16, 18, 19). Similar constructs in P. fluorescens have been used to successfully map Fe availability around roots in soil in microcosms (20).
At present we have biosensors that detect only a fraction of the microbially relevant components of root exudation; additional biosensors could be developed for other components of root exudates, such as other amino acids, microbially active secondary plant compounds, or important mineral nutrients such as NH4+ or NO3−. Despite the limited availability of biosensors, our present method has allowed us to delineate patterns of root exudation in more detail and under more-natural conditions than has been possible until now. We have been able to show that different exudate components have distinctly different patterns of availability around roots.
ACKNOWLEDGMENTS
This work was supported by NSF Postdoctoral Fellowship grant DEB 9303165, California AES project 6117-H, and USDA award 96-35107-2780.
We thank Kristine Johnson and Farhad Ghodroti for technical assistance.
REFERENCES
- 1.Brandl M, Clark E M, Lindow S E. Characterization of the indole-3-acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can J Microbiol. 1996;42:586–592. [Google Scholar]
- 2.Brandl M T, Lindow S E. Environmental signals modulate the expression of an indole-3-acetic acid biosynthetic gene in Erwinia herbicola. Mol Plant-Microbe Interact. 1997;10:499–505. [Google Scholar]
- 3.Bret-Harte M S, Silk W E. Nonvascular, symplastic diffusion of sucrose cannot satisfy the carbon demands of growth in the primary root tip of Zea mays L. Plant Physiol (Rockville) 1994;105:19–33. doi: 10.1104/pp.105.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celenza J L, Jr, Grisafi P L, Fink G R. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- 5.Clark E, Brandl M, Lindow S E. Aromatic aminotransferase genes from an indoleacetic acid-producing Erwinia herbicola strain. Phytopathology. 1992;82:1100. [Google Scholar]
- 6.Clark E M, Lindow S E. Proceedings, Seventh International Symposium on Molecular Plant-Microbe Interactions, Edinburgh, Scotland. 1994. A tryptophan aminotransferase gene from an indole-3-acetic acid-producing Erwinia herbicola strain; p. 177. [Google Scholar]
- 7.Cowan P J, Nagesha H, Leonard L, Howard J L, Pittard A J. Characterization of the major promoter for the plasmid-encoded sucrose genes scrY, scrA, and scrB. J Bacteriol. 1991;173:7464–7470. doi: 10.1128/jb.173.23.7464-7470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curl E A, Truelove B. The rhizosphere. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 9.de Weger L A, Dunbar P, Mahafee W F, Lugtenberg B J J, Sayler G S. Use of bioluminescence markers to detect Pseudomonas spp. in the rhizosphere. Appl Environ Microbiol. 1991;57:3641–3644. doi: 10.1128/aem.57.12.3641-3644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Weger L A, Dekkers L C, Van Der Bij A J, Lugtenberg B J J. Use of phosphate-reporter bacteria to study phosphate limitation in the rhizosphere and in bulk soil. Mol Plant-Microbe Interact. 1994;7:32–38. [Google Scholar]
- 11.Esau K. Vascular differentiation in plants. New York, N.Y: Holt, Rinehart and Winston; 1965. [Google Scholar]
- 12.Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 13.James B R, Bartlett R J, Aamadon J F. A root observation and sampling chamber (rhizotron) for pot studies. Plant Soil. 1985;85:291–293. [Google Scholar]
- 14.Juma N G, McGill W B. Decomposition and nutrient cycling in agro-ecosystems. In: Mitchell M J, Nakas J P, editors. Microfloral and faunal interactions in natural and agro-ecosystems. Boston, Mass: Martinus-Nijhoff/DR W. Junk Publishers; 1986. pp. 74–137. [Google Scholar]
- 15.Lindgren P B, Frederick R, Govindaragan A G, Panopoulos N J, Staskawics B J, Lindow S E. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 1989;8:1291–1301. doi: 10.1002/j.1460-2075.1989.tb03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindow S E. The use of reporter genes in the study of microbial ecology. Mol Ecol. 1995;4:555–566. [Google Scholar]
- 17.Lindow S E. Molecular genetic approaches to assessing bacterial habitat composition, modification, and interactions on leaves. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society of Molecular Plant-Microbe Interactions; 1997. pp. 487–492. [Google Scholar]
- 18.Loper J E, Lindow S E. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol. 1994;60:1934–1941. doi: 10.1128/aem.60.6.1934-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loper J E, Lindow S E. Reporter gene systems useful in evaluating in situ gene expression by soil- and plant-associated bacteria. In: Hurst C J, editor. Manual of environmental microbiology. Washington, D.C: American Society for Microbiology; 1996. pp. 482–491. [Google Scholar]
- 20.Marschner P, Crowlely D E. Iron stress and pyoverdin production by a fluorescent pseudomonad in the rhizosphere of white lupine (Lupinus albus L.) and barley (Hordeum vulgare L.) Appl Environ Microbiol. 1997;63:277–281. doi: 10.1128/aem.63.1.277-281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Miller, W. G., M. T. Brandl, and S. E. Lindow. Relative sensitivities of various reporter genes using a whole-cell biosensor for sucrose. Submitted for publication.
- 23.Norton J M, Smith J L, Firestone M K. Carbon flow in the rhizosphere of ponderosa pine seedlings. Soil Biol Biochem. 1990;22:449–455. [Google Scholar]
- 24.Olsen R A, Bakken L R. Viability of soil bacteria: optimization of plate-counting technique and comparison between total counts and plate counts within different size groups. Microb Ecol. 1987;13:59–74. doi: 10.1007/BF02014963. [DOI] [PubMed] [Google Scholar]
- 25.Salisbury F B, Ross C W. Plant physiology. Belmont, Calif: Wadsworth Publishing Co.; 1992. [Google Scholar]
- 26.Schmid K, Ebner R, Altenbuchner J, Schmitt R, Lengeler J W. Plasmid-mediated sucrose metabolism in Escherichia coli K12: mapping of the scr genes of pUR400. Mol Microbiol. 1988;2:1–8. doi: 10.1111/j.1365-2958.1988.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 27.Staskawicz B J, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Overbeek L S, van Elsas J D. Root exudate-induced promoter activity in Pseudomonas fluorescens mutants in the wheat rhizosphere. Appl Environ Microbiol. 1995;61:890–898. doi: 10.1128/aem.61.3.890-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace W. The distribution and characteristics of nitrate reductase and glutamate dehydrogenase in the maize seedling. Plant Physiol. 1973;52:191–196. doi: 10.1104/pp.52.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao K, De Luca V, Brisson N. Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell. 1995;7:1787–1799. doi: 10.1105/tpc.7.11.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]