Abstract
A self-cloning system for Actinomadura verrucosospora, a producer of the angucyclic antibiotic pradimicin A (PRM A), has been developed. The system is based on reproducible and reliable protoplasting and regeneration conditions for A. verrucosospora and a novel plasmid vector that consists of a replicon from a newly found Actinomadura plasmid and a selectable marker cloned from the Actinomadura strain. The system has an efficiency of more than 105 CFU/microgram of DNA. Using this system, we have cloned and identified the polyketide synthase (PKS) genes essential for PRM A biosynthesis from A. verrucosospora. Nucleotide sequence analysis of the 3.5-kb SalI-SphI fragment showed that ketosynthase subunits (open reading frame 1 [ORF1] and ORF2) of the essential PKS genes have strong similarities (59 to 89%) to those for angucyclic antibiotic biosynthesis.
More than 70% of antibiotics (including not only antibacterial agents but also bioactive microbial compounds) have been reported to be produced by actinomycetes, which are composed of the genus Streptomyces (68%) and the rare actinomycetes (32%) (19). Among the rare actinomycetes, the genus Actinomadura is reported to be the most predominant (19). This fact shows that this genus is one of the most important targets in screening programs for pharmacologically active compounds.
Despite these important attributes of the genus Actinomadura, to the best of our knowledge, with the exception of beta-lactamase of Actinomadura sp. strain R39 (10, 16), there is no information available on the basic aspects of gene expression in this genus due to the lack of versatile gene manipulation systems in the genus. In the case of Actinomadura sp. strain R39, a vector was introduced by electrotransformation into the original strain. However, only one transformant was obtained in several assays (10), showing that this method is not reproducible and reliable.
We are interested in the biosynthetic genes and enzymes of pradimicin A (PRM A), which has a unique dihydrobenzo[a]naphthacenequinone aglycone substituted with d-alanine and two sugars and is a potent antifungal agent (21, 23). Actinomadura hibisca and Actinomadura verrucosospora subsp. neohibisca are representatives of PRM A producers (21, 23). We have cloned the putative essential polyketide synthase (PKS) genes (pms open reading frame 1 [ORF1], ORF2, and ORF3) for PRM A biosynthesis from A. hibisca by using oligonucleotide probes based on the conserved amino acid sequences of other PKSs (5). The essential PKS gene consists of the β-ketoacyl synthase units KSα and KSβ (chain-length determination factor) and an acyl carrier protein, which is an enzyme complex which makes the polyketide backbone by repetitive condensation of two-carbon units (13). A homology search of the pms ORF1, ORF2, and ORF3 showed that these ORFs have a high similarity to those of type II essential PKS genes for aromatic antibiotic biosynthesis. Moreover, specific DNA regions homologous to pms genes were found with genomic Southern hybridization in all the PRM A producers examined but not in PRM A nonproducers (5). These results suggested that the pms ORF1, ORF2, and ORF3 would be essential PKS genes for PRM A biosynthesis, though we had no direct evidence for this hypothesis. Therefore, we decided to develop a self-cloning system for Actinomadura strains to confirm this probability by complementation or a gene inactivation technique. In this report, we describe a newly developed self-cloning system for A. verrucosospora and the cloning and identification of the essential PKS genes for PRM A biosynthesis from A. verrucosospora subsp. neohibisca.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. verrucosospora subsp. neohibisca E40 (9) and its derivative JN51 (a PKS gene-deficient mutant) (9) were kindly provided by the Bristol-Myers Squibb Pharmaceutical Research Institute (Wallingford, Conn.). Actinomadura echinospora IFO 14042 was obtained from the Institute for Fermentation (Osaka, Japan). Actinomadura sp. strain TPA0016, Actinomadura sp. strain TPA0019, and Actinomadura sp. strain TPA0123 were isolated from soil in the course of the secondary metabolite screening from actinomycetes (4). Streptomyces lividans TK23 (14) was used as a host for plasmid construction. Plasmid pIJ702 (14) and its derivative plasmid pSE101 (a shuttle vector for Streptomyces strains and Escherichia coli) (3), which is usually used as a vector for the Streptomyces strain, was used for the integration experiment. E. coli XL1-Blue MRF′ (recA1 thi endA1 supE44 gyrA46 relA1 hsdR17 lac/F′ [proAB+ lacIq lacZΔM15::Tn10{Tetr}]) (Toyobo, Osaka, Japan) and cosmid pWE15 (Toyobo) were used for preparation of a genomic library. E. coli XL1-Blue MRF′ and plasmids pBluescriptSK(+), pBluescriptKS(+), pUC18, and pUC19 were used for the subcloning experiment and sequencing analysis.
Media.
All actinomycetes were maintained on modified American Type Culture Collection (ATCC) no. 5 plates (6). V15 (9) and FR-18 (9) were used with all of the actinomycetes as the seed and production media, respectively. Mycelia which had been stocked in 20% glycerol at −80°C were inoculated in 4 ml of V15 medium and were grown at 30°C with shaking for 3 days. Next, 1.5 ml of the seed culture was transferred into 30 ml of FR-18 medium in 300-ml Erlenmeyer flasks and grown at 30°C for 3 to 5 days on a rotary shaker. All transformed actinomycetes were cultivated in the presence of thiostrepton (10 μg/ml; Sigma). Growth conditions for and manipulations of E. coli were as described by Maniatis et al. (17).
Analysis of PRM A.
For analysis of PRM A, Actinomadura strains were cultivated in liquid medium and the products were analyzed by high-pressure liquid chromatography (HPLC). The conditions of the HPLC analysis were described previously (9).
DNA isolation and manipulation.
Genomic DNA and plasmid isolations from streptomycetes were done by the method of Hopwood et al. (14). Plasmids from E. coli were prepared by using the Qiagen Plasmid Kit (QIAGEN, Inc., Valencia, Calif.). All restriction enzymes, T4 ligase, and calf intestinal alkaline phosphatase were obtained from Toyobo and used according to the manufacturer’s protocols. Transformation of E. coli with plasmid DNA by electroporation was performed under standard conditions by using a BTX ECM 600 electroporation system (Biotechnologies and Experimental Research, Inc., San Diego, Calif.). The procedure for cosmid library construction has been described previously (7).
Screening of Actinomadura plasmids.
Cryptic plasmids pAE042, pTPA0016, pTPA0019, and pTPA0123 were isolated from A. echinospora IFO 14042, Actinomadura sp. strain TPA0016, Actinomadura sp. strain TPA0019, and Actinomadura sp. strain TPA0123, respectively.
Sequence analysis.
The 3.5-kb SalI-SphI fragment prepared from cosmid pPRM30 was cloned into the same sites of pUC18 and pUC19. After construction of a series of plasmids subcloned from these plasmids, sequencing was done by the dideoxy chain termination method of Sanger et al. (24) with an automatic DNA sequencer (model 4000L; LI-COR).
Hybridization.
The conditions employed for colony hybridization with the 1.7-kb BglII fragment as a probe, which contains a putative essential PKS gene cloned from A. hibisca (5) and which was 32P labeled (2 × 108 cpm/μg) with a nick translation kit (Takara Syuzo, Kyoto, Japan), were as follows. A nylon membrane (GeneScreen Plus; Dupont, Boston, Mass.) with immobilized DNA was prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer containing 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate (SDS), and 100 μg of heat-denatured salmon sperm DNA/ml at 65°C for 4 h. For overnight hybridization, the same buffer and temperature conditions were used. The filter was washed twice with 0.3× SSC and 0.1% SDS buffer at 65°C for 1 h.
Preparation and transformation of protoplasts of strain E40 and its derivatives. (i) Protoplast preparation.
Frozen stock mycelia in 20% glycerol were inoculated into 4 ml of V15 medium and cultivated to the stationary phase (60 h). Portions (0.5 ml) of this culture were transferred to 30 ml of V15-P medium, consisting of sucrose (20% [wt/vol]), glucose (3%), Bacto soyton (1.5%; Difco), glycine (0.3%), CaCl2 · 2H2O (0.04%), and MgCl2 · 6H2O (0.1%) (pH 7.2) and grown to the early stationary phase. Mycelia were collected by centrifugation (5,000 × g, 10 min), washed with modified P medium (the composition of which was the same as that described by Hopwood et al. [14] except for the sucrose concentration [20%]), and then incubated in 10 ml of the same medium with lysozyme (2.5 mg/ml) and N-acetylmuramidase (0.05 mg/ml; Seikagaku Kogyo, Tokyo, Japan) at 32°C for 3 h. Protoplast formation was monitored microscopically. Protoplasts were recovered by centrifugation at 1,800 × g and 4°C for 10 min and washed twice with ice-cold modified P medium. They were gently resuspended in 5 ml of the same medium and stored at −80°C.
(ii) Transformation.
For transformation, the following regeneration medium (RAM-SM medium) has been developed. RAM-SM medium was prepared by mixing the following four separately autoclaved solutions: 983 ml of nutrient solution (200 g of sucrose, 10 g of glucose, 2 g of glycerol, 7.5 g of yeast extract [Difco], Casamino Acids [Difco], 1.5 g of l-asparagine, 5.2 g of MOPS [morpholinepropanesulfonic acid], 20 g of Bacto agar [Difco] [pH 7.2]), 6 ml of 5 M CaCl2 · 2H2O, 1 ml of 0.5% KH2PO4, and the supernatant of the sonicate of strain E40 prepared from a 50-ml culture. The supernatant of the sonicate was prepared by sonication (in 10 ml of cold water) of the mycelia grown in 50 ml of V15 medium with a pencil-type automatically tuned ultrasonic disruptor (model UD200; output, 5 to 6; TOMY SEIKO Co., Ltd., Tokyo, Japan) at 30-s intervals for a total of 3 min and then centrifugation at 10,000 × g and 4°C for 10 min. Plates were dried under laminar flow for 30 min and were immediately used. Transformation was done with plasmid DNA in 10 μl of Tris-EDTA (TE) buffer to which a 100-μl suspension of 109 to 1010 protoplasts was added. After the mixture was left for 1 min at room temperature, 440 μl of 25% (vol/vol) polyethylene glycol (PEG) 4000 with T medium was added and mixed gently by pipetting. The mixture was diluted with modified P medium, and 100-μl samples were plated on RAM-SM medium. These plates were incubated at 30°C for 3 days and were then overlaid with 2.5 ml of nutrient soft agar containing thiostrepton (final concentration, 10 μg/ml).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB019690.
RESULTS AND DISCUSSION
Protoplasting and regeneration of Actinomadura strains.
Previously, we isolated five Actinomadura strains producing PRMs (4). We chose A. verrucosospora subsp. neohibisca E40 as the host for DNA manipulations in this study because it does not harbor any detectable cryptic plasmids and is the original strain of the previously isolated PRM-nonproducing mutants (9). Moreover, because the strain was sensitive to many antibiotics, including thiostrepton, streptomycin, erythromycin, neomycin, and kanamycin, resistance genes to these antibiotics were thought to be proper selectable markers of the vectors for the strain. Recently, we have developed the protocols for protoplast formation and regeneration of the protoplast of Actinomadura strains (4), almost all of which were extraordinarily resistant to lysozyme. In the case of strain E40, we found that mycelia grown in the presence of sucrose (30%), the addition of which in preculture was reported to be effective for protoplast formation of some Streptomyces strains (15), became sensitive to lysozyme. The addition of sucrose plus glycine (0.3%) in the medium was most effective (4). As for lytic enzymes, the use of both lysozyme and N-acetylmuramidase (1 mg/ml) was even more effective than the use of lysozyme alone (4). However, we could obtain no transformants of strain E40 with ligation mixtures that were prepared by ligating a replicon from a newly found Actinomadura plasmid and a selectable marker, probably because of low regeneration efficiency of the protoplast (2.7%) on RAM-S medium (a previously described regeneration medium) (4). Therefore, several parameters were examined to try to improve the ratio.
With regard to the preparation of protoplasts, substances which accelerate the protoplast formation when added to the culture medium were investigated. Though we examined dozens of compounds, including antibiotics inhibiting cell wall biosynthesis, detergents, amino acids and sugar alcohols, we were not able to find any effective substances other than sucrose as described previously (4). However, we found that the regeneration ratio of protoplasts, which were prepared from mycelia grown in the presence of 20% sucrose, is higher than that of protoplasts prepared from mycelia grown in the presence of 30% sucrose, though prolonged incubation (more than 3 h) with lytic enzymes was needed to complete protoplasting. Next, we examined the condition of treatment of lytic enzyme. When a high concentration (1 mg/ml) of N-acetylmuramidase was used for preparation of the protoplasts, the regeneration was found to be slightly inhibited, though the protoplasts were washed five times with modified P medium to remove N-acetylmuramidase. The optimal concentration of N-acetylmuramidase was determined to be 0.05 mg/ml. The concentration of lysozyme had no effect on regeneration of protoplasts in the range between 0.5 and 10 mg/ml. The suitable temperature of treatment with lytic enzymes was determined to be 32°C. These modified protocols provided us with a regeneration rate of at least 15% on RAM-S medium by several trials.
To increase the regeneration ratio of the protoplasts of strain E40, several parameters of the regeneration medium were also modified. It was reported that the addition of cell extracts of Micromonospora strains to the regeneration medium was effective to increase the regeneration ratio of the protoplasts of these strains (12). This was also the case for Actinomadura strains, and the regeneration ratio was increased by 30% when cell extracts of strain E40 were added to the regeneration medium. Though we do not know what materials in the cell extracts increased the regeneration ratio of the protoplasts, some high-molecular-weight materials were thought to be candidates because the cell extracts after filtration with CENTRIPREP-10 (Amicon Inc., Beverly, Mass.), which cut off those with molecular weights less than 10,000, were still effective. Brief drying of about 30 min in laminar flow (about 3% reduction of medium weight) was also effective in increasing the regeneration ratio. The most important parameter in the regeneration medium was the type of buffer used for pH stabilization. When MOPS was used as a pH stabilizer instead of TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], which is usually used as a pH stabilizer of the regeneration medium of Streptomyces strains (14), the regeneration ratio was increased threefold. Finally, more than 60% of the protoplasts prepared under the optimized conditions could be regenerated on a modified regeneration medium (RAM-SM).
Construction of Actinomadura vectors and transformation of strain E40.
In our preliminary experiment, strain E40 could not be transformed by pIJ61 (14), pGM9 (20), and pIJ702 (14), the typical cloning vectors for Streptomyces strains, with the optimized protoplasting and regeneration conditions as described above and with the use of PEG by the method used for S. lividans (14). This led us to construct vectors of a new type harboring replicons of Actinomadura strains. Cryptic plasmids were screened from our culture stocks and fresh soil isolates of Actinomadura strains. We found four new plasmids (pAE042, pTPA0016, pTPA0019, and pTPA0123) by analysis using agarose gel electrophoresis. Judging from the physical maps of these plasmids, they were thought to be different from one another.
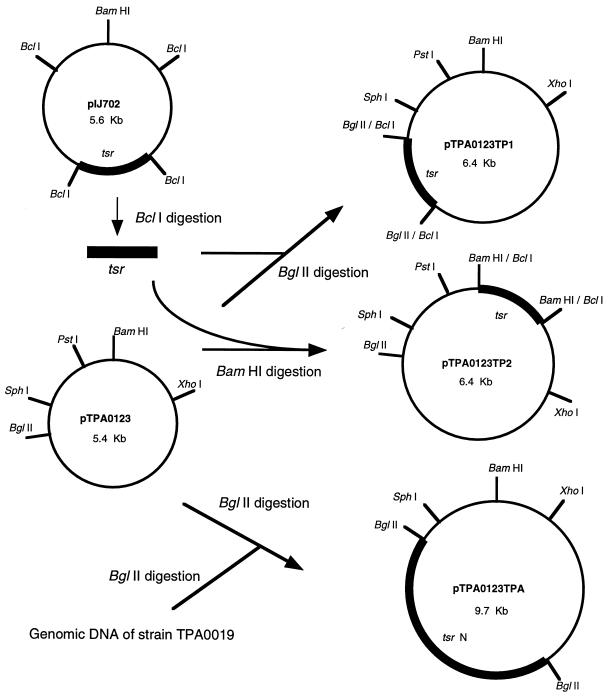
Strain E40 was sensitive to thiostrepton, and we therefore first tried to construct cloning vectors that had the thiostrepton resistance gene (tsr) prepared from pIJ702 as a selectable marker. To make cloning vectors harboring tsr, we completely digested the newly found four plasmids with BglII or BamHI. The 1.0-kb BclI fragment carrying the tsr gene prepared from pIJ702 was ligated with these digested plasmids and introduced into the protoplasts of strain E40 with the assistance of PEG by the method used for the S. lividans system (14). Ten to 50 thiostrepton-resistant transformants were obtained in a single experiment only when BglII or BamHI digests of pTPA0123 (one of the cryptic plasmids; Fig. 1) were used as replicons. The plasmids isolated from thiostrepton-resistant colonies had the structures we had expected (pTPA0123TP1 and pTPA0123TP2; Fig. 1). Next, we wanted a marker originating from Actinomadura strains to produce a cloning system of generically homologous backgrounds. Because Actinomadura sp. strain TPA0019 was moderately resistant to thiostrepton (MIC, 50 μg/ml), the genomic DNA of this strain was used as a source of the thiostrepton resistance gene. The total genomic DNA of strain TPA0019 was digested with BamHI, BclI, and BglII; ligated with BglII-digested pTPA0123; and used to transform strain E40. Thiostrepton-resistant transformants were obtained only when the BglII-digested genomic DNA of strain TPA0019 was used. All transformants examined had plasmids of the same size (9.7 kb). One of these plasmids was selected and named pTPA0123TPA (Fig. 1).
FIG. 1.
Construction of plasmid vectors for A. verrucosospora E40 by using the cryptic plasmid of Actinomadura sp. strain TPA0123 and the thiostrepton resistance genes (tsr). A cryptic plasmid, pTPA0123, digested with BglII or BamHI was treated with calf intestinal alkaline phosphatase, ligated with a 1.0-kb BclI fragment of pIJ702 containing tsr, and introduced into the protoplasts of A. verrucosospora E40. Thiostrepton-resistant transformants were found to harbor pTPA0123TP1 and pTPA0123TP2. For construction of pTPA0123TPA, the genomic DNA of Actinomadura sp. strain TPA0019 digested with BglII was ligated with BglII-digested pTPA0123 and used to transform A. verrucosospora E40. All thiostrepton-resistant transformants examined harbored 9.7-kb plasmids.
The experiments for the construction of vectors described above indicated that the PEG-assisted protocol for the Streptomyces strain worked in strain E40. We optimized the transformation conditions with pTPA0123TP1 and pTPA0123TPA. By tuning the type and the concentration of PEG and optimizing the timing of antibiotic selection, we could obtain 105 transformants/μg of DNA with both pTPA0123TP1 and pTPA0123TPA, which were prepared from strain E40. The strains harboring pTPA0123TP1 or pTPA0123TPA were passed through three cycles of sporulation and germination on modified ATCC no. 5 plates in the presence or absence of thiostrepton. At each cycle, spores were collected and checked for the presence of plasmids by plating them on modified ATCC no. 5 plates with or without thiostrepton. The plasmids were almost perfectly maintained in the presence of the antibiotic. The loss of the plasmid in the absence of the antibiotic is less than 30% even after three cycles.
Cloning of the essential PKS genes for PRM A biosynthesis.
Recently, we cloned the putative essential PKS genes for PRM A biosynthesis from A. hibisca by using oligonucleotide probes based on the conserved amino acid sequences of other PKS genes and found by genomic Southern hybridization that there are specific DNA regions homologous to pms genes in the genome of strain E40 (5). Therefore, we attempted to clone the PKS genes for PRM A biosynthesis from strain E40 by using the DNA fragment containing the putative essential PKS genes from A. hibisca as a probe. Subsequently, we tried to obtain the direct evidence by complementation and gene inactivation. To clone the hybridized fragment and its flanking region simultaneously, a cosmid library in E. coli of strain E40 DNA partially digested with Sau3AI was screened by colony hybridization. Among them, pPRM30 was selected for further analysis. The 7.5-kb PstI fragment that hybridized to the probe was subcloned into pBluescriptSK(+) and used for further analysis.
Nucleotide sequence of the DNA fragment hybridized to the probe.
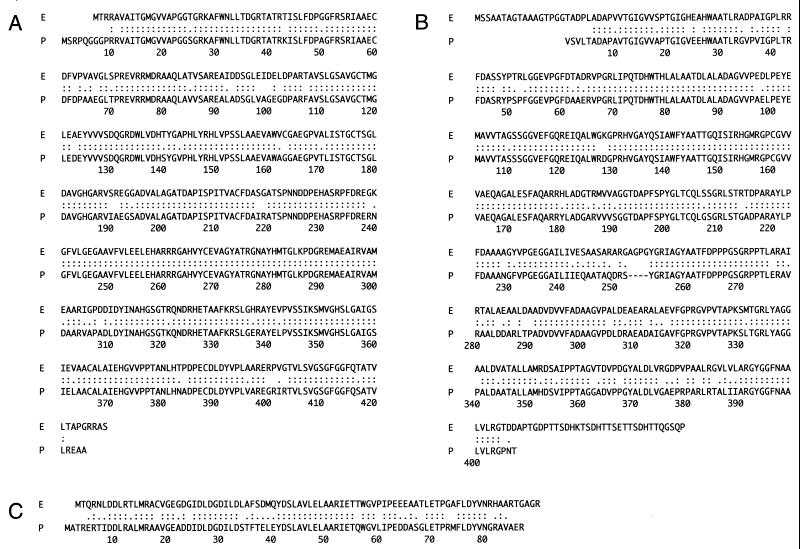
To examine whether the DNA fragment hybridized to the probes carries the PKS gene for biosynthesis of PRM A, the nucleotide sequence of the 3.5-kb SalI-SphI fragment containing the hybridized region (Fig. 2) was determined. Computer analysis of the DNA sequence, using Frame Analysis (2), revealed at least three complete ORFs (ORF1 to -3), which were all oriented in the same direction. We searched the databases with their translated products by means of the sequence similarity search programs BLAST (22) and FASTA (1). As we had expected, ORF1, ORF2, and ORF3 products show strong similarities (89, 82, and 71% identity, respectively; Fig. 3) with those of the putative essential PKS genes for PRM A biosynthesis, which were previously cloned from A. hibisca (5) and used as a probe in this study. Moreover, ORF1 and ORF2 products were found to be significantly similar to ketosynthase-α and -β responsible for urdamycin A (73 and 59% identity, respectively) (8) and jadomycin B (69 and 59% identity, respectively) (11) biosynthesis. On the other hand, ORF3, encoding an acyl carrier protein, is less similar to urdamycin A and jadomycin B biosynthetic genes (40 and 41% identity). Urdamycin A and jadomycin B also have angucyclic structures, and the sequence similarities of ketosynthases might therefore reflect the features of biosynthetic genes for angular-type antibiotics.
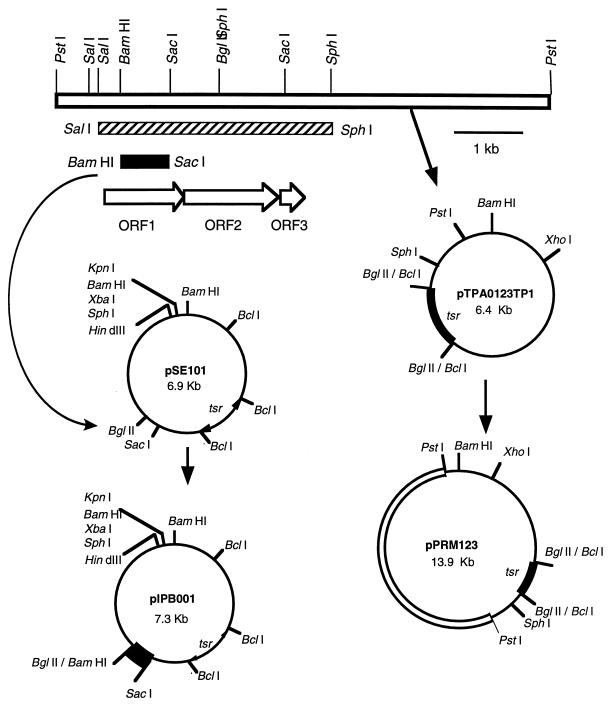
FIG. 2.
Construction of plasmids used for complementation and gene inactivation experiments. The 7.5-kb PstI fragment (open box) prepared from cosmid pPRM30 containing essential PKS genes for PRM A biosynthesis was ligated with pTPA0123TP1, which was digested with PstI and treated with calf intestinal alkaline phosphatase. S. lividans TK23 (14) was transformed with the ligation mixture. A recombinant plasmid (pPRM123) was screened among the thiostrepton-resistant transformants and used to transform a PRM A-nonproducing mutant, JN51. For gene inactivation experiments, pSE101 (a shuttle vector constructed with pIJ702 and pUC19) (3) was digested with BglII and SacI, and the resulting large fragment (6.6 kb) was ligated with the 0.7-kb BamHI-SacI fragment (black box) carrying the internal region of ORF1. The ligation mixture was used to transform S. lividans TK23. A recombinant plasmid (pIPB001) was isolated from the transformant and used to transform A. verrucosospora E40. The 3.5-kb SalI-SphI fragment (hatched box) used for sequencing analysis is also shown.
FIG. 3.
Alignment of the amino acid sequences of the essential PKS gene products from A. verrucosospora E40 with those from A. hibisca P-157-2. Deduced amino acid sequences of ORF1 (A), ORF2 (B), and ORF3 (C) of strain E40 (lines labeled E) and of strain P-157-2 (lines labeled P) are shown. The colons and periods indicate identical and similar residues, respectively.
Complementation and gene inactivation of PKS genes.
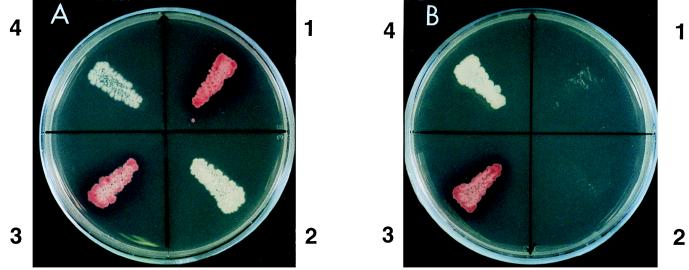
To explore whether the DNA fragment cloned in this study would contain the essential PKS genes for PRM A biosynthesis, the 7.5-kb PstI fragment that hybridized to the probe and contained the essential PKS genes was cloned into the same site of pTPA0123TP1 to give pPRM123 (Fig. 2) and was introduced into mutant JN51, which was derived from strain E40 by N-methyl-N′-nitro-N-nitrosoguanidine treatment (9). Mutant JN51 produced no intermediates as judged by thin-layer chromatography and HPLC analysis and always acted as a converter during the cosynthesis study (9), suggesting that this strain is not a mutant defective in the regulatory gene but rather a mutant deficient in the essential PKS genes. We constructed pPRM123 in S. lividans and then introduced it into strain JN51, because S. lividans was found to be able to maintain pTPA0123TP1 and was tolerant of accepting the DNA fragments prepared from E. coli. All transformants of mutant JN51 harboring the thus-constructed pPRM123 produced PRM A, which has a red color, on the modified ATCC no. 5 plate (Fig. 4), in contrast to those harboring a vector (pTPA0123TP1) which produced no pigments. The transformants harboring pPRM123 were cultivated in liquid medium, and the red pigment was confirmed to be PRM A by HPLC analysis.
FIG. 4.
Colony colors of A. verrucosospora E40 (1), JN51 (2), JN51 harboring pPRM123 (3), and JN51 harboring a vector (pTPA0123TP1) (4). Panels A and B show the modified ATCC no. 5 plate and the modified ATCC no. 5 plate containing thiostrepton (10 μg/ml), respectively.
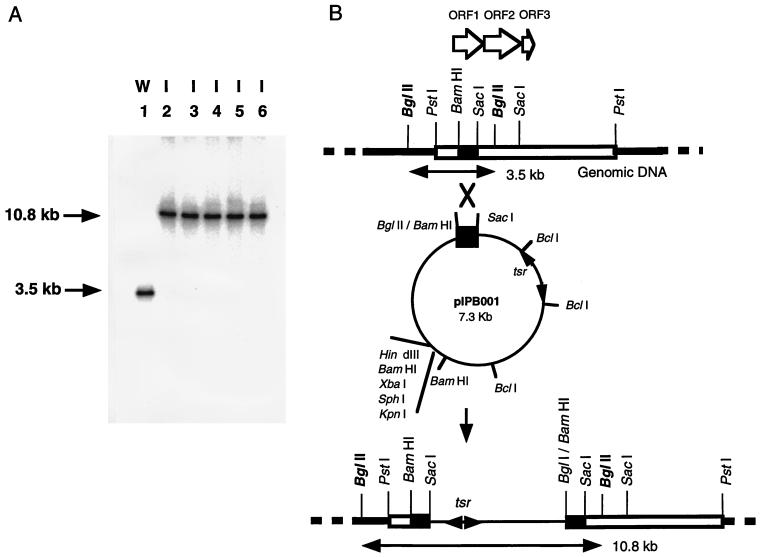
We also confirmed that the essential PKS genes cloned in this study indeed are the genes for PRM A biosynthesis by a gene disruption experiment utilizing plasmid integration. As described above, strain E40 could not be transformed with pIJ702 though the tsr gene of pIJ702 could function in strain E40, suggesting that pIJ702 is useful for an integration vector in strain E40. The 0.7-kb BamHI-SacI fragment carrying the internal sequences of ORF1 was subcloned into the BglII-SacI sites of pSE101 (a shuttle vector for Streptomyces strains and E. coli) (3) to give pIPB001 (Fig. 2). Strain E40 was transformed with pIPB001 prepared from S. lividans. About 10 thiostrepton-resistant colonies were obtained with 1 μg of plasmid. After single colony isolation in the presence of thiostrepton (10 μg/ml), Southern blot hybridization was performed to confirm the integration of pIPB001 (Fig. 5). These pIPB001-integrated strains did not produce any PRM A either on the plate or in liquid cultivation, showing that the essential PKS genes cloned in this study indeed are the genes for PRM A biosynthesis.
FIG. 5.
Southern blot analysis of the genomic DNA of ORF1-inactivated strains. (A) BglII digests of DNA from A. verrucosospora E40 (lane 1) and from ORF1-inactivated strains (lanes 2 to 6) were subjected to Southern blot hybridization with the 32P-labeled 0.7-kb BamHI-SacI fragment as a probe. Arrows indicate the hybridized fragments. (B) The hybridized fragments shown schematically.
In this work, we developed a practical gene cloning system for A. verrucosospora and cloned the essential PKS genes for PRM A biosynthesis. Over the last few years, many type II PKS recombinant gene cassettes have been constructed (13), and a set of predictive design rules for the generation of polyketides was proposed on the basis of these studies (13). However, it is not fully understood how angucyclic antibiotics represented by PRM A and jadomycin B are synthesized (18). Therefore, we are now trying to examine whether the PKS system for angucyclic antibiotics would follow predictive design rules by constructing a series of expression cassettes containing the essential PKS genes from a PRM A producer. These results will be published in the near future.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 3.Dairi T, Aisaka K, Katsumata R, Hasegawa M. A self-defense gene homologous to tetracycline effluxing gene essential for antibiotic production in Streptomyces aureofaciens. Biosci Biotechnol Biochem. 1995;59:1835–1841. doi: 10.1271/bbb.59.1835. [DOI] [PubMed] [Google Scholar]
- 4.Dairi T, Hamano Y, Igarashi Y, Furumai T, Oki T. Protoplasting and regeneration of strains belonging to the genus Actinomadura. Actinomycetologica. 1997;11:1–5. [Google Scholar]
- 5.Dairi T, Hamano Y, Igarashi Y, Furumai T, Oki T. Cloning and nucleotide sequence of the putative polyketide synthase genes for pradimicin biosynthesis from Actinomadura hibisca. Biosci Biotechnol Biochem. 1997;61:1445–1453. doi: 10.1271/bbb.61.1445. [DOI] [PubMed] [Google Scholar]
- 6.Dairi T, Ohta T, Hashimoto E, Hasegawa M. Self-cloning in Micromonospora olivasterospora of fms genes for fortimicin A (astromicin) biosynthesis. Mol Gen Genet. 1992;232:262–270. doi: 10.1007/BF00280005. [DOI] [PubMed] [Google Scholar]
- 7.Dairi T, Ohta T, Hashimoto E, Hasegawa M. Organization and nature of fortimicin A (astromicin) biosynthetic genes studied by a cosmid library of Micromonospora olivasterospora DNA. Mol Gen Genet. 1992;236:39–48. doi: 10.1007/BF00279641. [DOI] [PubMed] [Google Scholar]
- 8.Decker H, Haag S. Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tu2717, which carries the genes for biosynthesis of the angucyclic antibiotic urdamycin A and a gene probably involved in its oxygenation. J Bacteriol. 1995;177:6126–6136. doi: 10.1128/jb.177.21.6126-6136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furumai T, Kakinuma S, Yamamoto H, Komiyama N, Suzuki K, Saitoh K, Oki T. Biosynthesis of the pradimicin family of antibiotics. J Antibiot. 1993;46:412–419. doi: 10.7164/antibiotics.46.412. [DOI] [PubMed] [Google Scholar]
- 10.Granier B, Duez C, Lepage S, Englebert S, Dusart J, Dideberg O, van Beeumen J, Frere J M, Ghuysen J M. Primary and predicted secondary structures of the Actinomadura R39 extracellular DD-peptidase, a penicillin-binding protein (PBP) related to the Escherichia coli PBP4. Biochem J. 1992;282:781–788. doi: 10.1042/bj2820781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Yang K, Ramalingam E, Mosher R H, Vining L C. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuela ISP5230. Microbiology. 1994;140:3379–3389. doi: 10.1099/13500872-140-12-3379. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa M, Dairi T, Ohta T, Hashimoto E. A novel, highly efficient gene-cloning system for Micromonospora strains. J Bacteriol. 1991;173:7004–7011. doi: 10.1128/jb.173.21.7004-7011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopwood D A. Genetic contribution to understanding polyketide synthases. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Gene manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 15.Hopwood D A, Chater K F, Dowding J E, Vivian A. Advances in Streptomyces coelicolor genetics. Bacteriol Rev. 1973;37:371–405. doi: 10.1128/br.37.3.371-405.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houba S, Willem S, Duez C, Moliter C, Dusart J, Frere J M, Ghuysen J M. Nucleotide sequence of the gene encoding the active-site serine beta-lactamase from Actinomadura R39. FEMS Microbiol Lett. 1989;53:241–246. doi: 10.1016/0378-1097(89)90224-3. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 18.Meurer G, Gerlitz M, Wendt-Pienkowski E, Vining L C, Rohr J, Hutchinson C R. Iterative type II polyketide synthases, cyclases and ketoreductases exhibit context-dependent behavior in the biosynthesis of linear and angular decapolyketides. Chem Biol. 1997;4:433–443. doi: 10.1016/s1074-5521(97)90195-2. [DOI] [PubMed] [Google Scholar]
- 19.Miyadoh S. Research on antibiotic screening in Japan over the last decade: a producing microorganism approach. Actinomycetologica. 1993;7:100–106. [Google Scholar]
- 20.Muth G, Nußbaumer B, Wohlleben W, Puhler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 21.Oki T, Konishi M, Tomatsu K, Tomita K, Saitoh K, Tsunakawa M, Nishio M, Miyaki T, Kawaguchi H. Pradimicin, a novel class of potent antifungal antibiotics. J Antibiot. 1988;41:1701–1704. doi: 10.7164/antibiotics.41.1701. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitoh K, Sawada Y, Tomita K, Tsuno T, Hator M, Oki T. New pradimicin congeners from Actinomadura verrucosospora subsp. neohibisca. J Antibiot. 1993;46:387–397. doi: 10.7164/antibiotics.46.387. [DOI] [PubMed] [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]