Abstract
The quiescent-cell expression system is a radical alternative to conventional fermentation for protein overproduction in Escherichia coli. It is dependent on the controlled overexpression of a small RNA called Rcd in hns mutant strains to generate nongrowing, quiescent cells which are not nutrient limited. Quiescent cells no longer produce biomass and have their metabolic resources channelled toward the expression of plasmid-based genes. The biosynthetic capacity of the system is demonstrated by its ability to express chloramphenicol acetyltransferase to more than 40% of total cell protein. Quiescent cells may provide an ideal environment for the expression of toxic as well as benign proteins.
Escherichia coli continues to be a favored host for large-scale expression of heterologous proteins (12). Developments to promote high-level protein expression in E. coli continue at a rapid pace, although a general cloning strategy for the overexpression of heterologous proteins remains elusive. The use of high-biomass fermentations for high-level protein expression leads to cells becoming stressed and starved and may trigger the synthesis of stress-specific proteins. Furthermore, the expression of a foreign protein is often detrimental to the cell’s integrity, severely reducing the long-term protein synthetic capacity of the culture (36). Recent attempts to address these problems include strain development and coexpression of chaperone and suppressor proteins (1, 20, 23). Despite these advances, neoteric approaches to protein expression are still required, particularly when attempting to express and retain the functionality of polytopic prokaryotic and eukaryotic membrane proteins (9).
A fundamental problem associated with the expression of recombinant proteins in growing cells is that energy and nutritional resources are channelled toward biomass production. Expression of the “product gene” is simultaneous with the expression of hundreds of host genes which compete for the transcription-translation machinery and metabolic resources. Furthermore, the metabolic stress imposed by the expression of a recombinant gene from a multicopy plasmid reduces the growth rate and viability of the host cell (16). Cells that have lost the cloning vector or have mutated or deleted the cloned gene will almost invariably outgrow the original cell type, reducing the yield and purity of the product (26). The desirability of uncoupling biomass production from the expression of cloned genes has stimulated interest in the basis of bacterial dormancy, stress response, and entry into stationary phase with the aim of placing genes under the control of starvation-inducible promoters (10, 13, 17, 31).
This study concerns the development of a system for gene expression in a nongrowing but metabolically active (quiescent) cell culture. It offers a radical, alternative solution to the much-debated problem of sustaining protein synthesis in the absence of rapid cell division (8). The quiescent-cell expression system is a “spin-off” from our studies on plasmid stability in E. coli. It is well known that plasmid dimers, arising initially by recombination and proliferating by over-replication, are a major cause of segregational instability among cloning vectors (25, 29). Dimers and higher multimers of ColE1 and other natural plasmids are resolved to monomers by unidirectional, site-specific recombination requiring a 250-bp region of ColE1 (the cer site [28]) and at least four proteins (XerC, XerD, ArgR, and PepA) encoded by the host bacterium (5). The presence of multimers also triggers the expression of a small RNA called Rcd (regulator of cell division) from its promoter (Pcer) within the ColE1 cer site (21). Transient expression of the Rcd transcript is proposed to delay cell division, allowing time for plasmid multimers to be removed and avoiding the production of plasmid-free offspring (27). Cells in which Rcd is overexpressed for an extended period have a distinct cell cycle arrest phenotype. The majority of cells are of uniform size (two to four cell lengths) with correctly partitioned, condensed nucleoids, but they remain undivided since no cell septum is formed. As distinct from other situations in which cell division is blocked, Rcd overexpression does not lead to cell filamentation. We describe here how these cells can be exploited as factories for the high-level expression of plasmid-borne genes.
MATERIALS AND METHODS
Strains and plasmids.
DS941 and DS903 are derivatives of E. coli K-12 strain AB1157 (3). DS941 is AB1157 recF lacIq lacZΔM15 (lacY). DS903 is AB1157 recF and carries wild-type lacI and lacZ genes. The hns-205::Tn10 derivatives of DS941 and DS903 were constructed by P1 transduction from strain GM230 (11) selecting for mucoid, tetracycline-resistant colonies.
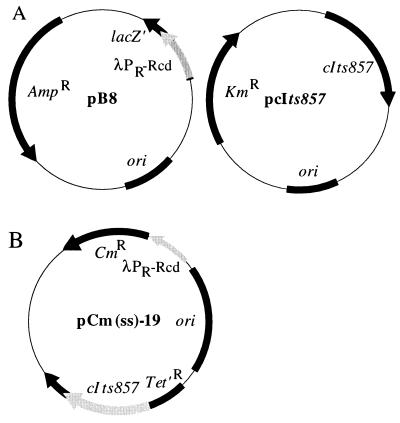
PCR was used to generate DNA fragments where Rcd was placed under the control of the λPR promoter by using the oligonucleotide primers 5′-ATGCATATGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTGATAATGGTTGCAGGCGCGATCGCGGCAG-3′ and 5′-ATGCATATGATTTACCATAATCCC-3′ and pKS490 (21) as template. PCR-generated DNA fragments were cloned into the SmaI site of pUC18 (35) to generate plasmid B8. Our original vector system for generating a quiescent state used two plasmids with compatible origins: plasmid B8 (a 2,915-bp pUC18-based vector containing a 179-bp PCR-generated fragment with rcd under control of the λPR promoter) and pcIts857 (22). The single-shot vector pCm(ss)-19 (5,177 bp) was constructed by ligating the 1,220-bp BglI-BglII fragment of pcIts857 containing the cIts857 temperature-sensitive repressor allele with the 3,456-bp BamHI-BglI fragment of pACYC184 (4) to create plasmid pACYC/cIts857-68 (4,676 bp). Plasmid B8 was cut with PvuII, and the 501-bp fragment containing the λPR-rcd fusion cloned into the Bst1107I site of pACYC/cIts857-68 to create pCm(ss)-19 (5,177 bp). Plasmid pSC1 contains the hns gene under the control of the Ptac promoter (18).
Plasmids were introduced by electroporation with a Gene Pulser (Bio-Rad Laboratories, Ltd., Hemel Hempstead, England) according to the manufacturer’s specifications. Transformants were selected on Iso-Sensitest broth agar (which is used for antimicrobial susceptibility testing; Oxoid/UniPath, Basingstoke, England) at 30°C so that Rcd expression was repressed by the cIts857 protein, allowing colonies to form. Where appropriate, media were supplemented with kanamycin (50 μg/ml), ampicillin (100 μg/ml), or chloramphenicol (30 μg/ml).
Cell culture and quiescence.
Cells were grown in L broth (14) in 50-ml conical shake flasks containing antibiotics, where appropriate, for plasmid selection. A single colony of the plasmid-containing strains was picked from a selective plate and cultured overnight in L broth at 30°C. Portions (0.2 to 1 ml) of the overnight culture were used to inoculate 20 to 30 ml of L broth prewarmed to 30°C. After the cells had undergone several generations in exponential phase, reaching an optical density at 600 nm (OD600) of between 0.2 and 0.3, 1 to 4 ml of culture was withdrawn and used to inoculate 17 to 21 ml of L broth prewarmed to 42°C. The shift in growth temperature induces Rcd expression from the λPR promoter, since the cIts857 protein does not function as a transcriptional repressor at 42°C. An Rcd-induced quiescent state was reached within 3 h. Then, 20 μl of carbenicillin (50 μg/ml) was added each hour to maintain selection for pB8. The viability of cells in quiescent culture was assessed by colony formation (measured as CFU per milliliter) on selective Iso-Sensitest agar plates at 30°C (repressing conditions for Rcd expression).
DAPI and viability staining.
For staining with DAPI (4′,6-diamidino-2-phenylindole) cells from broth culture were heat fixed onto Poly-Prep slides (Sigma-Aldrich, Poole, England) coated with poly-l-lysine. Cells were permeabilized with 4% para-formaldehyde for 1 min and stained with DAPI (Sigma-Aldrich) (0.1 μg/ml) for 2 min. The excess stain was removed, and cells were mounted by using 70% glycerol and sealed under a coverslip. Cells in broth culture were tested for viability by use of the Live/Dead Baclight bacterial viability kit (Molecular Probes, Leiden, The Netherlands), which utilizes the fluorescent nucleic acid stains SYTO9 and propidium iodide. Cells were stained in suspension according to the manufacturer’s instructions and examined by fluorescent light microscopy. Live cells are stained by SYTO9 and fluoresce green, while dead cells are stained by propidium iodide and fluoresce red.
Measurements of β-galactosidase expression.
Expression of the lacZ gene was monitored by assaying β-galactosidase activity by the method of Miller (19). β-Galactosidase activity was expressed in Miller units, which are equivalent to the increase in o-nitrophenol concentration per minute per bacterium. A 1 mM concentration of isopropyl-β-d-thiogalactopyranoside (IPTG) was used to induce lacZ expression.
[35S]methionine incorporation and protein sequencing.
Samples (10 ml) of the DS941hns-205 pCm(ss)-19 culture were labelled for 1 h with 20 μCi of protein labelling mix ([l-35S]methionine, 1,175 Ci/mmol; NEN Life Science Products, Hounslow, England). Total protein was prepared from 10 ml of culture. Cells were centrifuged for 6 min at 4,000 rpm at room temperature; they were then washed twice with 5 ml of phosphate buffer (pH 7.0) and suspended in 100 μl of phosphate buffer and an equivalent volume of 2× sodium dodecyl sulfate (SDS) Laemmli sample buffer (Sigma-Aldrich). Then 1 μl of protein sample was loaded onto a SDS–4 to 15% polyacrylamide gel electrophoresis (PAGE) homogeneous Phast-Gel (Amersham Pharmacia Biotech, Little Chalfont, England) and run for 40 min at 250 V against Rainbow 14C-methylated protein molecular weight markers (Amersham Pharmacia Biotech). Proteins were fixed for 30 min in isopropanol-water-acetic acid (25:65:10), and the gel was treated with Amplify (Amersham Pharmacia Biotech) for 20 min, followed by soaking in 1% glycerol for 30 min. The gel was dried at 65°C overnight before autoradiography. Two to 4 μl of proteins was also separated on a 12.5% homogenous SDS-PAGE Phast-Gel, stained with Coomassie brilliant blue R (Sigma-Aldrich), and destained in methanol-water-acetic acid (40:53:7). N-terminal sequence analysis was carried out at the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge.
RESULTS
Effect of Rcd overexpression on cells in broth culture.
In order to achieve inducible high-level expression of Rcd, the coding sequence under control of the lambda PR promoter (λPR) was inserted into pUC18. A temperature-sensitive cI repressor protein (cIts857) was used to control Rcd expression. The cIts857 protein is inactive at 42°C, so transcription of rcd from the λPR promoter is induced when the growth temperature is shifted from 30 to 42°C. The cIts857 allele was present either on a compatible plasmid, pcIts857 (22) (the two-plasmid system; Fig. 1A), or on the same plasmid as Rcd (the single-shot system; Fig. 1B). As expected, when Rcd was overexpressed with the two-plasmid system in DS941 (a derivative of E. coli K-12 AB1157) and in many other strains used in large-scale fermentations, it prevented colony formation on Iso-Sensitest broth agar, L agar, and minimal agar (data not shown). To our surprise, in L broth culture the same strains responded slowly or not at all to Rcd overproduction. Figure 2A shows that overexpressing Rcd in DS941 leads to a reduction in growth rate, although growth continued for many hours, and the final OD was only slightly less than the control culture containing the plasmid pKS490 which expresses Rcd at extremely low levels.
FIG. 1.
Plasmids used to generate an Rcd-induced quiescent state. (A) The two-plasmid system consists of plasmid B8 with rcd under control of the λPR promoter and pcIts857, which encodes the cIts857 temperature-sensitive repressor. (B) The single-shot vector pCm(ss)-19 contains both the λPR-rcd fusion and the cIts857 temperature-sensitive repressor.
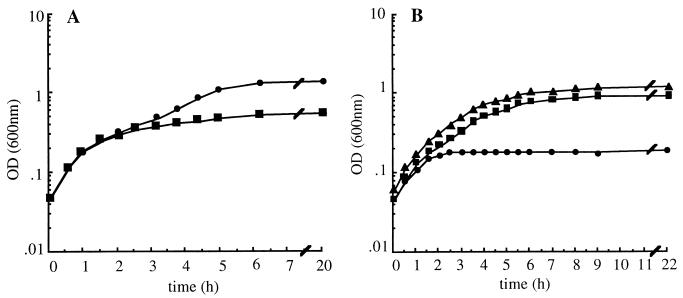
FIG. 2.
Effect of overexpressing Rcd on the growth of E. coli K-12 in broth culture. (A) DS941 transformed with the two-plasmid Rcd expression system (■) and the control plasmid pKS490 (●). (B) DS941hns-205 plasmid-free (▴) or transformed with the two-plasmid Rcd expression system (●) or pUC19 and pcIts857 (■). Cultures were initially grown in L broth at 30°C and then diluted in L broth prewarmed to 42°C at t = 0. Carbenicillin was added each hour to maintain selection for plasmid B8 and pUC19.
A mutation in hns is necessary to generate an Rcd-induced quiescent state in broth culture.
We supposed that differences between the levels of global regulators for cells growing on agar and in liquid culture might explain differences in response to Rcd induction. It has been reported that when H-NS (histone-like nucleoid structuring) protein (2, 33) is overexpressed it causes growth inhibition, filamentation, and nucleoid compaction (24). Since nucleoid condensation is also found in DS941 overexpressing Rcd on agar plates and in broth culture, we speculated that an increased level of H-NS in tandem with Rcd expression might result in growth arrest in broth culture. Much to our surprise, we found the reverse was true: cells in L broth (Fig. 2B) and minimal medium (37) carrying an hns-205::Tn10 mutation (11, 30) entered a stable nongrowing (quiescent) state in response to Rcd induction. Figure 2B shows the effect of overexpressing Rcd with the two plasmid system in DS941hns-205 growing in L broth. Cells growing exponentially at 30°C were subcultured into L broth prewarmed to 42°C to induce Rcd. By 2 to 3 h after the temperature shift there was no further increase in the OD of the culture, whereas control cultures without Rcd showed normal growth kinetics. To confirm that mutation in hns is required for the establishment of the quiescent state, we compared the effect of Rcd overexpression in hns-205 cells with or without pSC1, a multicopy plasmid expressing wild-type H-NS (18). When H-NS was expressed from pSC1, the culture continued to grow well beyond the point at which cells lacking the H-NS-producing plasmid entered a quiescent state (data not shown). We have compared the efficacy of various hns alleles in the establishment of the quiescent state (data not shown). We found that cells carrying the N43 hns::tet (source of strain: I. B. Holland, Institut de Génétique et de Microbiologie, Université Paris-Sud, Orsay, France) and hns::neo (32) alleles entered an Rcd-induced quiescent state, whereas cells carrying an hns-206::amp (6) mutation did not.
Viability of cells in the quiescent state.
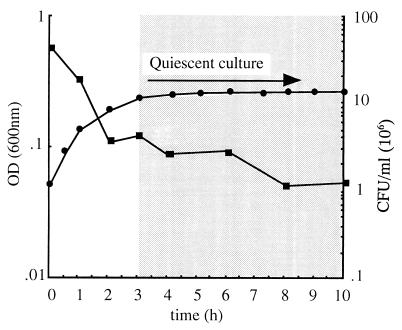
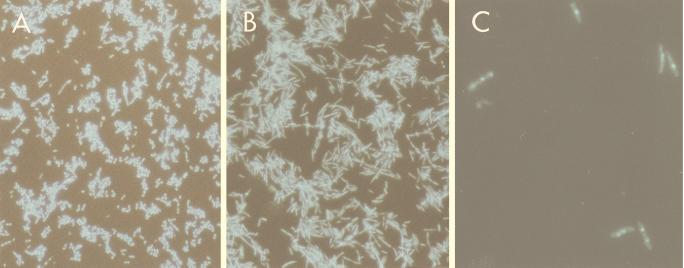
Results consistent with those from the two-plasmid system were also obtained with the single-shot vector, pCm(ss)-19, which carries both the rcd gene under λPR and the cIts857 repressor. Typical quiescent induction kinetics with pCm(ss)-19 are shown in Fig. 3, which also includes data on the viability of quiescent cells assayed by measuring CFU on agar plates at 30°C. We found that CFU initially showed a decline after Rcd induction but then stabilized as the culture remained in quiescence for up to 8 h (Fig. 3). Cell viability was also tested with the Live/Dead Baclight bacterial viability kit (Molecular Probes), which utilizes the fluorescent nucleic acid stains SYTO9 and propidium iodide. After 8 h of culture at 42°C, approximately 25% of the cells per field of view were stained green with SYTO9, suggesting that a significant subpopulation of quiescent cells remained alive (data not shown). Taken together, these results suggest that the viability and the ability to escape from the quiescent state decrease with time, but a relatively stable subpopulation of viable cells remain. Microscopic examination of DAPI-stained quiescent cells showed that the majority of cells were elongated to an average of four times the normal cell length compared to the control strain, where cells were grown under identical conditions but in the absence of Rcd (Fig. 4A and B). A closer examination at a higher magnification revealed that quiescent cells contained very compact nucleoids (Fig. 4C).
FIG. 3.
Overexpression of Rcd from the single-shot vector. DS941hns-205 transformed with the single-shot vector, pCm(ss)-19 (●), was grown initially in L broth at 30°C and then diluted in L broth prewarmed to 42°C at t = 0. Cell viability was tested by the ability to form colonies on Iso-Sensitest broth agar plates with appropriate antibiotic selection for pCm(ss)-19 under repressing conditions (30°C) for Rcd expression (■).
FIG. 4.
Morphology of quiescent cells. DS941hns-205 (control) and DS941hns-205 pCm(ss)-19 cells were cultured at 42°C (inducing conditions when expressing Rcd). Cells were stained with DAPI and visualized by using fluorescent light microscopy. (A) DS941hns-205 (OD600 = 0.316). (B) DS941hns-205 pCm(ss)-19 6 h after transfer to 42°C (OD600 = 0.258). Magnification (A and B), ×400. (C) DS941hns-205 pCm(ss)-19 4 h after transfer to 42°C (OD600 = 0.244). Magnification, ×1,000.
Quiescent cells synthesize proteins preferentially from plasmid-borne genes.
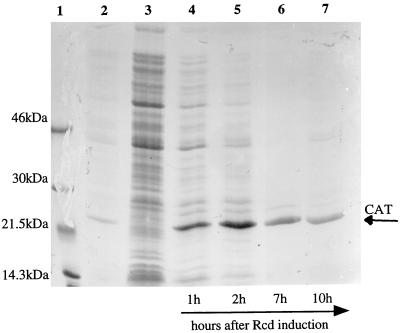
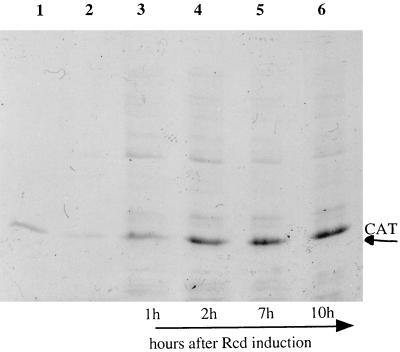
The de novo protein synthetic capacity of quiescent cells was investigated by [35S]methionine pulse-labelling and SDS-PAGE analysis of total protein isolated from DS941hns-205 containing pCm(ss)-19 after induction of Rcd expression. Figure 5 shows an autoradiograph of [35S]methionine-labelled total cell protein separated on an SDS–4 to 15% PAGE gel. It can be clearly seen that quiescent cells continue de novo protein synthesis for up to 10 h (lane 7) after induction of Rcd expression (equivalent to 7 h in the quiescent state). For the first 2 h after Rcd induction a diverse population of proteins continues to be expressed, although the level of expression of a single protein of approximately 23 kDa is disproportionately high (lanes 4 and 5 in Fig. 5). Cultures pulse-labelled 7 or 10 h after Rcd induction show continued, high levels of synthesis of the 23-kDa protein but an extremely low level of production of other proteins. Plasmid pCm(ss)-19 carries the gene for chloramphenicol acetyltransferase (CAT), and N-terminal sequence analysis confirmed that this major protein band was the 25.659-kDa CAT protein. Thus, plasmid gene expression appears to continue at a high level in quiescent cells. Figure 6 shows total proteins from a quiescent broth culture of DS941hns-205 pCm(ss)-19 separated on an SDS–12.5% polyacrylamide gel stained with Coomassie brilliant blue R. The amount of protein loaded on this gel was normalized to a cell culture biomass based on an OD600 of 0.147. Lane 1 contains 0.4 μg of purified CAT protein, which comigrates with the major protein band in quiescent cells. Densitometry of the stained gels shows that CAT protein constitutes up to 40% of protein in these samples.
FIG. 5.
Assessment of the de novo protein synthetic capacity of quiescent cells. Samples (10 ml) from a culture of DS941hns-205 pCm(ss)-19 were labelled for 1 h with [35S]methionine at various times after induction of Rcd expression. Total proteins were separated on an SDS–4 to 15% polyacrylamide gradient gel, and [35S]methionine labelling was revealed by autoradiography. Lanes: 1, Rainbow 14C-methylated protein molecular weight markers; 2, DS941hns-205 pCm(ss)-19 grown at 30°C for 1 h (OD600 = 0.359); 3, DS941hns-205 grown at 42°C for 1 h (OD600 = 0.225); 4, DS941hns-205 pCm(ss)-19 grown at 42°C for 1 h (OD600 = 0.147); 5, DS941hns-205 pCm(ss)-19 grown at 42°C for 2 h (OD600 = 0.211); 6, DS941hns-205 pCm(ss)-19 grown at 42°C for 7 h (OD600 = 0.279); 7, DS941hns-205 pCm(ss)-19 grown at 30°C for 10 h (OD600 = 0.285). An equal volume of total protein preparation was loaded in each lane.
FIG. 6.
Analysis of total protein isolated from quiescent cells. Total protein from DS941hns-205 pCm(ss)-19 was isolated at various times after the induction of Rcd expression. Proteins were separated on an SDS–12.5% polyacrylamide gel and stained with Coomassie brilliant blue R. Lanes: 1, purified CAT protein (0.4 μg); 2, total protein from DS941hns-205 pACYC/cIts857 grown at 42°C for 160 min (OD600 = 0.219); 3, DS941hns-205 pCm(ss)-19 grown at 42°C for 1 h (OD600 = 0.147); 4, for 2 h (OD600 = 0.211); 5, for 7 h (OD600 = 0.279); or 6, for 10 h (OD600 = 0.285).
Chromosomal gene expression is depressed in quiescent cells.
Quiescent cells have condensed nucleoids, which suggests that many chromosomal genes may be switched off. Furthermore, our pulse-labelling experiments indicate a downregulation of chromosomal gene expression in quiescence. We have further investigated this phenomenon by measuring the expression of the chromosomal lacZ gene induced by IPTG in strain DS903hns-205 in the presence and absence of Rcd.
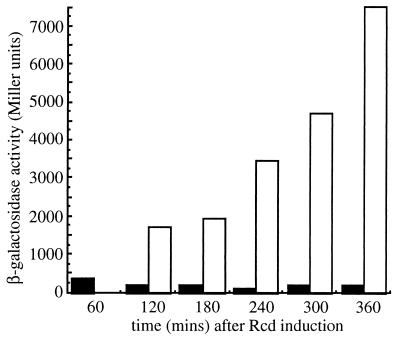
When Rcd was expressed, growth of the culture stopped within 2 h, whereas in the absence of Rcd cells grew normally, eventually entering stationary phase (data not shown). The levels of β-galactosidase in these cultures are shown in Fig. 7, where Miller units are normalized for cell biomass and the data thus represent the protein content per cell. The control culture DS903hns-205, which contained no Rcd, showed a high level of β-galactosidase activity, increasing from 1,700 to 7,500 Miller units. It is common to observe an increase in lacZ expression as a culture enters stationary phase. In contrast, the level of β-galactosidase expression from the chromosomal lacZ gene in the quiescent culture was less than 200 Miller units (Fig. 7). This result is consistent with the downregulation of chromosomal genes in quiescent cells.
FIG. 7.
Chromosomal lacZ gene expression in quiescent cells. DS903hns-205 and DS903hns-205 containing the two-plasmid system for Rcd expression were grown initially in L broth at 30°C and then diluted in L broth prewarmed to 42°C containing 1 mM IPTG to induce expression of the lacZ gene. Carbenicillin (50 μg ml−1) was added each hour to maintain selection for plasmid B8. DS903hns-205 containing the two-plasmid system reached an Rcd-induced quiescent state within 2 h, whereas the control culture continued to grow. β-Galactosidase activity is expressed in Miller units in the presence (black bars) and absence (white bars) of Rcd.
DISCUSSION
We have exploited the ColE1-encoded regulatory transcript, Rcd, to develop an E. coli quiescent cell system for the overexpression of proteins from plasmids. Quiescent cells are generated by the controlled overexpression of Rcd in hns-205 mutant cells in broth culture. The cell density at which growth is arrested can be readily controlled and is achieved under culture conditions where nutrients are not limiting. Quiescent cells remain competent for protein synthesis from plasmid-borne genes, while chromosomal gene expression decreases, possibly as a consequence of nucleoid condensation. The low level of chromosome gene expression is advantageous for the purification of proteins by downstream processes. Furthermore, in the quiescent state the metabolic resources of the cell are channelled toward the expression of plasmid-borne genes. Our data suggest that quiescent cells are extremely productive and that a relatively small biomass can have a high level of protein production.
We have shown that cells are able to synthesize the plasmid-encoded CAT protein for at least 10 h after Rcd induction despite some decline in the rate of synthesis over time. It is known that CAT expression in E. coli increases four- to fivefold when the growth temperature is shifted from 37 to 42°C, possibly as the result of posttranscriptional regulation (15). Despite a temperature shift being involved in Rcd expression, this does not explain the high level of CAT expression observed in quiescent cells. The level of CAT production in the control culture DS941hns-205 pACYC/cIts857 (Fig. 6, lane 2) grown at 42°C for 160 min to a biomass equivalent to that of a quiescent culture produced levels of CAT protein far lower than was observed in quiescent cells. Our data suggest that the CAT promoter is very active in quiescent cells. We are presently developing dedicated expression vectors for use in this system and have identified several additional promoters which also direct high levels of gene expression in cells which have entered quiescence (38).
For rapid establishment of an Rcd-induced quiescent state, the bacterial strain must contain an hns mutation. In this report we used the hns-205::Tn10 allele, which produces a C-terminal-truncated H-NS protein (7, 11). Quiescence was also achieved for strains containing the N43 hns::tet (which also produces C-terminal-truncated H-NS protein) and the hns::neo null mutation (32). However, not all hns mutations appear suited for achieving an Rcd-induced quiescence. For example, overexpressing Rcd in DS941 containing the hns-206::amp null mutation (6) does not stop cell growth. At present, we have no explanation for the allele specificity of hns on entry into quiescence.
Quiescent cell culture uncouples protein synthesis from biomass production and retains cells in a nutrient rich, nonstressful environment favorable for extended protein synthesis of plasmid-borne genes. These cells may provide a useful expression system for “difficult” proteins (e.g., polytopic membrane proteins) which disrupt cell growth and division in conventional culture. There may also be benefits for the expression of benign proteins where a scale-down in the size of fermentations could be achieved without a reduction in product yield. Furthermore, use of quiescent cells means that the segregational instability of cloning vectors is no longer a problem; indeed some plasmids in these cells continue to replicate, amplifying their copy number and the dosage of any cloned gene (30).
We note that the plasmid-encoded cIts857 protein does not show the same high-level accumulation as CAT in quiescent cells. This disparity in protein levels probably reflects the differences between the efficiency of transcription and translation of the CAT and cI genes. It is also possible that the cIts857 protein is unstable and is degraded rapidly at 42°C. In any case it is clear that not all plasmid-borne genes are expressed at the same high level in quiescent cells. We can exploit such differences by using transcription and translation control signals which ensure the high-level expression of the product gene while choosing expression signals of nonproduct genes, such as antibiotic resistance, which lead to low-level expression in quiescence.
The quiescent state is quite distinct from the stationary phase. This difference is demonstrated by our observation that in the former case β-galactosidase levels decline, while in the latter they rise (Fig. 7). Furthermore, an initial attempt to identify promoters that are activated in the quiescent state focused on “gearbox” promoters and used the well-characterized bolAp1 promoter as an example (34). The expression of this promoter is maximal in slow-growing and stationary-phase cells, but we were unable to detect expression from plasmid-borne bolAp1::lacZ fusions in quiescent cells (7a).
The expression system described here is protected by International patent PCT/GB97/00731 (30).
ACKNOWLEDGMENTS
This work was supported by Biotechnology and BioSciences Research Council research grants TO3491 and TO5594 to D.K.S.
We thank Pat Higgins, University of Alabama, for supplying plasmid pSC1 and Philip Oliver, University of Cambridge, for supplying plasmid pcIts857.
REFERENCES
- 1.Amrein K E, Takacs B, Stieger M, Molnos J, Flint N A, Burn P. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc Natl Acad Sci USA. 1995;92:1048–1052. doi: 10.1073/pnas.92.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann B J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colloms S, McCulloch R, Grant K, Neilson L, Sherratt D J. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 6.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 7.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 7a.Eyre, D., D. C. D. Rowe, and D. K. Summers. Unpublished data.
- 8.Flickinger M C, Rouse M P. Sustaining protein synthesis in the absence of rapid cell division: an investigation of plasmid-encoded protein expression in Escherichia coli during very slow growth. Biotechnol Prog. 1993;9:555–572. doi: 10.1021/bp00024a001. [DOI] [PubMed] [Google Scholar]
- 9.Grisshammer R, Tate C G. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- 10.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 11.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 12.Hockney R C. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 1994;12:456–463. doi: 10.1016/0167-7799(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 13.Kaprelyants A S, Gottschal J C, Kell D B. Dormancy in non-sporulating bacteria. FEMS Microbiol Rev. 1993;104:271–286. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy C K. Induction of colicin production by high temperature or inhibition of protein synthesis. J Bacteriol. 1971;108:10–19. doi: 10.1128/jb.108.1.10-19.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S J, Jeon H Y, Kim H B. Chloramphenicol acetyltransferase expression of Escherichia coli is increased at 42°C. Biotechnol Tech. 1997;11:435–438. [Google Scholar]
- 16.Kurland C G, Dong H. Bacterial growth inhibition by overproduction of protein. Mol Microbiol. 1996;21:1–4. doi: 10.1046/j.1365-2958.1996.5901313.x. [DOI] [PubMed] [Google Scholar]
- 17.Matin A. Physiology molecular biology and applications of the bacterial starvation response. J Appl Bacteriol Symp Suppl. 1992;73:49S–57S. doi: 10.1111/j.1365-2672.1992.tb03624.x. [DOI] [PubMed] [Google Scholar]
- 18.McGovern V, Higgins N P, Chiz R S, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1019–1029. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 20.Miroux B, Walker J E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 21.Patient M E, Summers D K. ColE1 multimer formation triggers inhibition of E. coli cell division. Mol Microbiol. 1993;8:1089–1095. doi: 10.1111/j.1365-2958.1993.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 22.Remaut E, Tsao H, Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983;22:103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- 23.Rockabrand D, Blum P. Multicopy plasmid suppression of stationary-phase chaperone toxicity in Escherichia coli by phosphogluconate dehydratase and the N-terminus of DnaK. Mol Gen Genet. 1995;249:498–506. doi: 10.1007/BF00290575. [DOI] [PubMed] [Google Scholar]
- 24.Spurio R, Falconi M, Brandi A, Pon C L, Gualerzi C O. The oligomeric structure of the nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summers D K. The kinetics of plasmid loss. Trends Biotechnol. 1991;9:273–278. doi: 10.1016/0167-7799(91)90089-z. [DOI] [PubMed] [Google Scholar]
- 26.Summers D K. Stability of genetic material in prokaryotes. Biologicals. 1993;21:91–93. doi: 10.1006/biol.1993.1053. [DOI] [PubMed] [Google Scholar]
- 27.Summers D K. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 28.Summers D K, Sherratt D J. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- 29.Summers D K, Beton C W H, Withers H L. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol. 1993;8:1031–1038. doi: 10.1111/j.1365-2958.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- 30.Summers, D. K., and D. C. D. Rowe. 15 March 1997. International patent application, PCT/GB97/00731. Methods and means relating to quiescent cells and uses thereof. Cambridge University Technical Services, Ltd., Cambridge, United Kingdom.
- 31.Turner J R, Robertson C R, Schippa S, Matin A. Use of glucose starvation to limit growth and induce protein production in Escherichia coli. Biotechnol Bioeng. 1992;40:271–279. doi: 10.1002/bit.260400211. [DOI] [PubMed] [Google Scholar]
- 32.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 33.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Seirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 34.Vicente M, Kushner S R, Garrido T, Aldea M. The role of the “gearbox” in the transcription of essential genes. Mol Microbiol. 1991;5:2085–2091. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 35.Vieira J, Messing J. The pUC plasmids an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 36.Visick J E, Clarke S. Repair, refold, recycle: how bacteria can deal with spontaneous and environment damage to proteins. Mol Microbiol. 1995;16:835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 37.Watson, E. A., and D. K. Summers. Unpublished data.