Abstract
To assess Radiomics and Machine Learning Analysis in Liver Colon and Rectal Cancer Metastases (CRLM) Growth Pattern, we evaluated, retrospectively, a training set of 51 patients with 121 liver metastases and an external validation set of 30 patients with a single lesion. All patients were subjected to MRI studies in pre-surgical setting. For each segmented volume of interest (VOI), 851 radiomics features were extracted using PyRadiomics package. Nonparametric test, univariate, linear regression analysis and patter recognition approaches were performed. The best results to discriminate expansive versus infiltrative front of tumor growth with the highest accuracy and AUC at univariate analysis were obtained by the wavelet_LHH_glrlm_ShortRunLowGray Level Emphasis from portal phase of contrast study. With regard to linear regression model, this increased the performance obtained respect to the univariate analysis for each sequence except that for EOB-phase sequence. The best results were obtained by a linear regression model of 15 significant features extracted by the T2-W SPACE sequence. Furthermore, using pattern recognition approaches, the diagnostic performance to discriminate the expansive versus infiltrative front of tumor growth increased again and the best classifier was a weighted KNN trained with the 9 significant metrics extracted from the portal phase of contrast study, with an accuracy of 92% on training set and of 91% on validation set. In the present study, we have demonstrated as Radiomics and Machine Learning Analysis, based on EOB-MRI study, allow to identify several biomarkers that permit to recognise the different Growth Patterns in CRLM.
Keywords: EOB-MRI study, colorectal liver metastases, radiomics
1. Introduction
Colorectal cancer (CRC) is one of the main common cancer worldwide, representing about the 10% of new detected cancers in 2020 [1]. Moreover, it is estimated that in 2040 its prevalence will rise quickly to >3 million cases per year [2,3]. Although the management of patients within a multidisciplinary team have improved the clinical outcome thanks to a closer patient follow-up to obtain an earlier detection of metastatic disease and an improvement in the efficacy of systemic therapies, and of surgical indications and –quality, based on a better patient selection, however metastatic spread is the main cause of death [4,5,6,7,8]. In addition, at the diagnosis, about 20% of patients have liver metastases (CRLM), while approximately 40–50% of patients will develop CRLM during surveillance, either after primary tumor resection/multimodal therapy or after the resection of CRLM [8,9,10,11,12,13,14,15,16]. Another key is related to the fact that about 60% of patients can expect to have liver recurrence even after a R0 resection of the primary CRLM. Several researches have demonstrated as patients with (a) T3/T4 CRC status, (b) local positive node, and more than 3 CRLM have higher possibility of recurrence [17,18,19]. In addition, several researches have suggested that the distinct growth pattern of CRLM could be linked to liver recurrence and overall survival (OS) after conversion therapy and resection [20,21,22]. With the increasing frequency of CRLM resections, the histology of the front morphology of CRLM is accessible and different growth patterns have been reported: in the “desmoplastic” or “encapsulated” model, tumour cells are separated from the liver parenchyma by a fibrotic stroma border that is distinguished by a ‘pushing’ or ‘expansive’ pattern, in which the hepatic plaques adjacent to the metastases are flattened with no intermediate fibrotic tissue. The model called “invasive” or “replacement” in which cancer cells infiltrate normal surrounding liver parenchyma, replace the hepatocytes and the sinusoidal stromal [20,21,22]. A recent review [22] assessed 17 clinical studies, including either chemonaïve and chemotherapy-treated patients, showed that in the 82.4% a statistically significant favourable outcome was reported for patients with desmoplastic CRLM while in the 66.7% a significantly unfavourable outcome for patients with a predominantly replacement-type CRLM was demonstrated [22].
In this context, it is evident that the possibility to identify biomarkers that allow, during imaging studies, a proper growth pattern characterization consents a better patient treatment selection [23,24,25,26,27,28,29]. Since, radiomics allows to evaluate tissue at microscopic level, in order to obtain quantitative data to employ as biomarkers [30,31,32,33,34,35,36,37,38,39], this analysis allow to increase diagnostic, prognostic and predictive accuracy in oncological setting [40,41,42,43,44,45,46,47,48]. In fact, Radiomic is created to be employed in decision support of precision medicine, using standard of care images [49,50,51,52,53,54,55,56,57]. Although, several researches have assessed the MRI features of CRLM [58,59,60], at the best of our knowledge no study have evaluated the ability of Radiomics analysis based on MRI studies to characterize CRLM growth pattern. The purpose of this study is to assess the Radiomics and Machine Learning Analysis Based on MRI in the evaluation of CRLM growth pattern.
2. Materials and Methods
Local Ethical Committee board accepted this retrospective study, renouncing to the patient informed consent. Patients were selected by radiological database considering the temporal period from January 2018 to June 2021 and the following inclusion criteria: (1) liver pathological proven metastases; (2) MRI study of high quality in pre-surgical setting and (3) a follow-up Computed Tomography (CT) scan of at least six months after surgical liver resection.
The patient cohort included a training set of 51 patients with 121 liver metastases, and an external validation set of 30 patients with a single lesion. The validation cohort was provided by “Careggi Hospital”, Florence, Italy. The patient characteristics were reported in Table 1.
Table 1.
Characteristics of the study population (81 patients).
| Patient Description | Numbers (%)/Range |
|---|---|
| Gender | Men 53 (65.4%) |
| Women 28 (34.6%) | |
| Age | 61 y; range: 35–82 y |
| Primary cancer site | |
| Colon | 52 (64.2%) |
| Rectum | 29 (35.8%) |
| Prior Chemotherapy | 81 (100%) |
| Hepatic metastases description | |
| Patients with single nodule | 52 (64.2%) |
| Patients with multiple nodules | 29 (35.8%)/range: 2–13 metastases |
| Nodule size (mm) | mean size 36.4 mm; range 7–58 mm |
| Mucinous carcinoma | 25 (30.9%) |
| RAS mutation | 42 (51.9%) |
| Liver Recurrence | 19 (23.5%) |
2.1. MR Imaging Protocol
MR studies were performed with two 1.5T MR scanners: Magnetom Symphony (Siemens, Erlangen, Germany) and Magnetom Aera (Siemens).
The MRI study protocol included conventional sequences, T1 weighted (W), without contrast medium administration, and T2-W, Diffusion Weighted Imaging (DWI) with seven b values in order to obtain functional parameters with mono-exponential model and T1-W sequences after the administration of contrast medium. In Table 2 we reported MR study protocol.
Table 2.
MR acquisition protocol.
| Sequence | Orientation | TR/TE/FA (ms/ms/deg.) |
AT (min) |
Acquisition Matrix |
ST/Gap (mm) |
FS |
|---|---|---|---|---|---|---|
| Trufisp T2-W | Coronal | 4.30/2.15/80 | 0.46 | 512 × 512 | 4/0 | without |
| HASTE T2-W | Axial | 1500/90/170 | 0.36 | 320 × 320 | 5/0 | Without and with (SPAIR) |
| HASTE T2w | Coronal | 1500/92/170 | 0.38 | 320 × 320 | 5/0 | without |
| SPACE T2W FS | Axial | 4471/259/120 | 4.20 | 384 × 450 | 3/0 | With (Spair) |
| In-Out phase T1-W | Axial | 160/2.35/70 | 0.33 | 256 × 192 | 5/0 | without |
| DWI | Axial | 7500/91/90 | 7 | 192 × 192 | 3/0 | without |
| Vibe T1-W |
Axial | 4.80/1.76/30 | 0.18 | 320 × 260 | 3/0 | with (SPAIR) |
Note: TR = Repetition time, TE = Echo time, FA = Flip angle, AT = Acquisition time, ST = Slice thickness, FS = Fat suppression, SPAIR = Spectral adiabatic inversion recovery.
According to the different phase of patient management, our study protocol includes the possibility to administrate a liver-specific contrast agent (in pre surgical setting) and a non-liver-specific contrast agent (in characterization and staging phase). In this study we assessed images obtained employing a liver-specific contrast agent (0.1 mL/kg of Gd-EOB-BPTA—Primovist, Bayer Schering Pharma, Berlin, Germany). A power injector (Spectris Solaris® EP MR, MEDRAD Inc., Indianola, IA, USA) was used to administrate the contrast agent using an infusion rate of 2 mL/s.
After contrast medium administration, VIBE T1-weighted FS (SPAIR) sequences were acquired in different phases of contrast study: arterial (35 s delay), portal/venous (90 s), transitional (120 s), and hepatospecific (EOB) phase (20 min).
2.2. MRI Post-Processing
For each volume of interest, 851 radiomic features were extracted as median values on the segmented volume by two expert radiologists in abdominal imaging and MRI, using the PyRadiomics tool [61] and as previously reported in [62,63] and as reported in [https://readthedocs.org/projects/pyradiomics/downloads/ (accessed on 21 December 2021)].
2.3. Reference Standard
Histopathologic data, from routine report were used as the reference standard for determining metastases growth pattern. Lesions with desmoplastic growth pattern were defined as expansive; lesions with predominantly replacement-type were defined infiltrative.
2.4. Statistical Analysis
Intraclass correlation coefficient was used to evaluate the variability by two radiologists. The non-parametric Kruskal-Wallis test was performed to recognize differences statistically significant of radiomics metrics median values in the identification of tumor growth front (expansive versus infiltrative).
Chi square test was used to evaluate if the tumor growth front (expansive versus infiltrative) was correlated with the local recurrence occurrence.
Receiver operating characteristic (ROC) and Youden index was used to calculate the optimal cut-off for each metric and area under curve (AUC), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and accuracy.
A feature selection was made to delete the redundant and non informative metrics considering the following findings: metrics significant by Kruskal-Wallis test and with an accuracy when considered alone major of 75%. A linear regression analysis was made to calculate the linear regression model of all significant metrics.
Moreover, approaches of artificial intelligence were considered to define the best classifier of all significant metrics. The classifiers considered included support vector machine (SVM), k-nearest neighbors (KNN), artificial neural network (NNET), and decision tree (DT)) [63]. The best model was selected considering the highest area under ROC curve and the highest accuracy. Each classifier was trained with a 10-k fold cross validation and was validated using the external validation set.
McNemar test was used to evaluate that the results of the dichotomy tables were statistically significant.
A p-value < 0.05 was considered as significant. The analysis was made considering the Statistics and Machine Toolbox of MATLAB R2021b (MathWorks, Natick, MA, USA).
3. Results
No correlation statistically significant was relived between the tumor growth front and the local recurrence occurrence (p-value = 0.82 at Chi square test).
On univariate analysis (Table 3), a variable number of metrics were statistically significant which were different when extracted from different MR sequences: 15 significant predictors extracted from T2W SPACE; 8 significant predictors extracted from the arterial phase; 9 significant predictors extracted from the portal phase; 8 significant predictors extracted from the EOB phase.
Table 3.
Univariate analysis results to predict mucinous type.
| T2W SPACE | Arterial Phase | Portal Phase | EOB-Phase | |
|---|---|---|---|---|
| wavelet_HLL_firstorder_Median | wavelet_LHH_glrlm_ShortRunLowGrayLevelEmphasis | wavelet_LHH_glrlm_ShortRunLowGrayLevelEmphasis | wavelet_HLL_glcm_InverseVariance | |
| AUC | 0.71 | 0.69 | 0.80 | 0.78 |
| Sensitivity | 0.79 | 0.95 | 0.84 | 0.84 |
| Specificity | 0.73 | 0.51 | 0.77 | 0.82 |
| PPV | 0.83 | 0.77 | 0.85 | 0.89 |
| NPV | 0.67 | 0.85 | 0.74 | 0.76 |
| Accuracy | 0.77 | 0.79 | 0.82 | 0.83 |
| Cut-off | −0.39 | 0.12 | 0.12 | 0.46 |
The best results (Table 3) with the highest accuracy and AUC at univariate analysis to discriminate expansive versus infiltrative front of tumor growth were obtained by the wavelet_LHH_glrlm_ShortRunLowGrayLevelEmphasis from portal phase sequence with accuracy of 82%, sensitivity of 84%, specificity of 77%, PPV and NPV of 85% and 74%, respectively, and a cut-off value of 0.12. The results were statistically different from those obtained with the T2W and arterial phase MR sequences while were to those obtained from EOB phase sequence with the wavelet_HLL_glcm_InverseVariance that obtained and AUC of 78% and an accuracy of 83%.
Linear regression model increased the performance obtained respect to the univariate analysis (see Table 4) for each sequence except that for EOB-phase sequence. The best results were obtained by a linear regression model of 15 significant features extracted by the T2W SPACE sequence with accuracy of 90%, a sensitivity of 95%, a specificity of 95%, a PPV and a NPV of 80% and 89% respectively. These results were statistically different from the univariate analysis results and compared with the results of metrics extracted from other MR sequences (p-value < 0.01 on McNemar’s test). Table 5 reported the coefficients of metrics and intercept of the best linear regression model.
Table 4.
Linear regression with significant features.
| Linear Regression of Significant Features Extracted by | AUC | Sensitivity | Specificity | PPV | NPV | Accuracy | Cut-Off |
|---|---|---|---|---|---|---|---|
| T2W SPACE | 0.90 | 0.95 | 0.80 | 0.89 | 0.90 | 0.89 | 1.51 |
| arterial phase | 0.74 | 0.89 | 0.89 | 0.93 | 0.83 | 0.89 | 1.45 |
| portal phase | 0.88 | 0.80 | 0.89 | 0.92 | 0.73 | 0.83 | 1.58 |
| EOB-phase | 0.55 | 0.88 | 0.56 | 0.77 | 0.74 | 0.76 | 8.81 |
Table 5.
Linear regression model coefficients.
| Features | Coefficients | p-Value |
|---|---|---|
| Intercept | −10.99 | 0.01 |
| original_shape_SurfaceVolumeRatio | −1.13 | 0.24 |
| wavelet_HLL_glcm_InverseVariance | 13.96 | 0.01 |
| wavelet_HLL_firstorder_Median | 0.14 | 0.06 |
| wavelet_HLL_glrlm_ShortRunEmphasis | 38.72 | 0.00 |
| wavelet_HLL_glrlm_RunPercentage | −38.39 | 0.00 |
| wavelet_LHL_gldm_DependenceNonUniformityNormalized | −7.33 | 0.61 |
| wavelet_LHL_glcm_InverseVariance | −3.19 | 0.51 |
| wavelet_LHL_firstorder_Kurtosis | 0.01 | 0.04 |
| wavelet_LHL_glrlm_ShortRunEmphasis | −24.29 | 0.21 |
| wavelet_LHL_glrlm_RunPercentage | 46.40 | 0.00 |
| wavelet_LHL_glrlm_RunLengthNonUniformityNormalized | −14.58 | 0.15 |
| wavelet_LLH_glcm_Imc1 | −0.31 | 0.87 |
| wavelet_LLL_firstorder_Uniformity | 6.76 | 0.17 |
| wavelet_LLL_firstorder_Minimum | 0.01 | 0.00 |
| wavelet_LLL_glrlm_GrayLevelNonUniformityNormalized | −5.61 | 0.28 |
Furthermore, using pattern recognition approaches, the diagnostic performance to discriminate the expansive versus infiltrative front of tumor growth increased again and the best classifier was a weighted KNN trained with the 9 significant metrics extracted from the portal phase sequence obtaining 92% accuracy on training set and 91% at validation set with an AUC of 0.97 and 0.99, respectively analysis (see Table 6). These results were statistically different from the univariate analysis results and linear regression analysis (p-value < 0.01 on McNemar’s test).
Table 6.
Pattern recognition analysis with significant features.
| The Best Classifier (KNN) Results with Significant Features Extracted by | Dataset | AUC | Accuracy | Sensitivity | Specificity | Training Time [s] | Model Type and Parameters |
|---|---|---|---|---|---|---|---|
| T2W SPACE | Training set | 0.90 | 0.89 | 0.84 | 0.92 | 11.1 | Decision Fine Tree; Maximum number of splits: 100; split criterion: Gini’s diversity index; optimizer options: Hyperparameter options disabled |
| Validation set | 0.88 | 0.86 | 0.86 | 0.86 | |||
| arterial phase | Training set | 0.97 | 0.91 | 0.91 | 0.91 | 2.34 | Weighted KNN; number of neighbors: 10; distance metric: Euclidean; distance weight: squared inverse |
| Validation set | 0.96 | 0.89 | 0.85 | 0.91 | |||
| portal phase | Training set | 0.97 | 0.92 | 0.84 | 0.97 | 9.74 | |
| Validation set | 0.99 | 0.91 | 0.81 | 0.96 | |||
| EOB-phase | Training set | 0.96 | 0.90 | 0.91 | 0.89 | 13.4 | |
| Validation set | 0.95 | 0.80 | 0.67 | 1.00 |
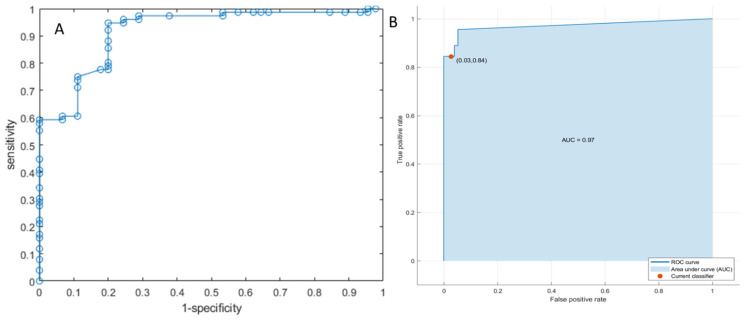
Figure 1 showed the ROC curve of linear regression model of 15 significant features by T2w sequence and the ROC curve of a KNN trained with the 9 significant predictors extracted from the portal phase.
Figure 1.
ROC curve of linear regression model of 15 significant features by T2w sequence in (A) and ROC curve of a KNN trained with the 9 significant predictors extracted from the portal phase in (B).
All results of the dichotomy tables were statistically significant (p-value < 0.01 at McNemar test).
4. Discussion
Since liver is the most common site of distant metastases in patients with CRC and, surgical resection is the only curative treatment, it is clear that the possibility to identify several prognostic features of CRLM to guide the proper patient management, remains a critical key open question. In addition, according to current principles for oncologic liver surgery, liver resection aims to remove the lesion with adequate margins and with sufficient liver remnant volume. This needs a multi-parametric patient assessment for a correct evaluation of the lesion and functional liver status [56,57,58]. Several features could guide the surgical procedure, comprising liver functional status, patient general status, tumour size, localization, and vascular infiltration.
Also, today, several researches have suggested that the distinct CRLM growth pattern is associated with differences in tumor local recurrence and OS. The rationale behind the need to know CRLM growth patterns is at least twofold: on one hand, the patterns may be useful for prognostication and may aid treatment decisions; on the other hand, it is likely that understanding these distinct patterns of metastatic progression will provide important insights into the biological mechanisms that support tumour growth in the liver [22]. Fernández Moro et al. [22] reported that microvessel co-option, besides providing vascular supply, renders metastases of the replacement type resistant to anti-angiogenic therapy. Furthermore, the authors suggested an alternative approach for treating replacement-type metastases. It is worth noting that several studies published before FDA approval of anti-angiogenic therapy found a favourable prognosis for patients with desmoplastic-type CRLM, suggesting that there are important mechanistic insights beyond differences in angiogenesis to be gained from the different growth patterns [22].
In this scenario, it is clear that having the possibility, during an imaging study, to identify biomarkers that could correlate with the growth of the lesion, allows for better treatment selection [58,59,60]. At the best of our knowledge no one study has evaluated the Radiomics and Machine Learning Analysis Based on MRI study, in the assessment of CRLM growth pattern. With regard to univariate analysis, a variable number of metrics were statistically significant: 15 extracted from T2W SPACE; 8 from the arterial phase; 9 extracted from the portal phase; 8 extracted from the EOB phase. The best results with the highest accuracy and AUC at univariate analysis to discriminate expansive versus infiltrative front of tumor growth were obtained by the wavelet_LHH_glrlm_ShortRunLowGrayLevelEmphasis from portal phase.
With regard to linear regression model, this increased the performance obtained respect to the univariate analysis for each sequence except that for EOB-phase sequence. The best results were obtained by a linear regression model of 15 significant features extracted by the T2W SPACE. These results were statistically different from the univariate analysis results. Furthermore, using pattern recognition approaches, the diagnostic performance to discriminate the expansive versus infiltrative front of tumor growth increased again and the best classifier was a weighted KNN trained with the 9 significant metrics extracted from the portal phase. Our results were confirmed by validation external dataset.
However, when we evaluated the tumor growth pattern and the local recurrence, we found no statistically significant correlation. This result could be explained by the number of patients under examination and the short follow-up period.
To date, several researches have assessed Radiomics analysis and CRLM, with regard to mutational status, prognosis and recurrence [32,33,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81]. Andersen et al. showed as homogeneity features were correlated to worse OS [65]. Lubner et al. assessed KRAS status, showing an inversely correlation with the skewness degree, while they showed an association between entropy and OS [67]. In our previous studies we showed that radiomics features obtained by EOB-MRI phase, arterial and portal phase so as by T2-W sequences, allow to predict clinical outcomes following liver resection in CRLM [80,81].
This study has several weaknesses: (1) the small sample size, although the analysis was done on a homogeneous group and on all single lesion; (2) the retrospective nature, (3) a manual segmentation. Furthermore, we not evaluated: (4) the impact of chemotherapy on our data, while we assessed all single study protocol phase, in order to identify the more accurate sequence in the assessment of growth pattern and our results were confirmed by a validation external dataset.
5. Conclusions
In the present study, we have demonstrated as Radiomics and Machine Learning Analysis, based on EOB-MRI study, allow to identify several biomarkers that permited to recognise the different Growth Patterns in CRLM.
The best results with the highest accuracy and AUC at univariate analysis were obtained by the wavelet_LHH_glrlm_ShortRunLowGrayLevelEmphasis from portal phase. Linear regression model increased the performance obtained respect to the univariate analysis for each sequence, except that for EOB-phase sequence. Furthermore, using pattern recognition approaches, the best classifier was a weighted KNN trained with the 9 significant metrics extracted from the portal phase.
Acknowledgments
The authors are grateful to Alessandra Trocino, librarian at the National Cancer Institute of Naples, Italy. Moreover, for the collaboration, authors are grateful for the re-search support to Paolo Pariate, Martina Totaro and Andrea Esposito of Radiology Division, Istituto Nazionale Tumori IRCCS Fondazione Pascale—IRCCS di Napoli, 80131 Napoli, Italy.
Author Contributions
Conceptualization, V.G.; Data curation, V.G.; Formal analysis, R.F.; Investigation, V.G., F.D.M., C.C., M.M.R., M.G., A.A., A.O., F.T., M.C.B., V.M., F.I. and A.P.; Methodology, V.G., R.F., F.D.M., C.C., M.M.R., M.G., A.A., A.O., F.T. and A.P; Writing–original draft, V.G. and R.F.; Writing – review & editing, V.G. and R.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study aligned with National appropriate guidelines and procedures. The Local Ethical Committee board approved this retrospective study.
Informed Consent Statement
This study aligned with National appropriate guidelines and procedures. Renouncing the need for informed patient consent given the study nature.
Data Availability Statement
Some data are available at link https://zenodo.org/record/6496965#.Ympr5tpBy3A.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International Agency for Research on Cancer (IARC), GLOBOCAN 2020: Colorectal Cancer, Number of New Cases in 2020, Both Sexes, All Ages, Vol. 2020. [(accessed on 20 December 2021)]. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact-sheet.pdf.
- 2.Gunter M.J., Alhomoud S., Arnold M., Brenner H., Burn J., Casey G., Chan A.T., Cross A.J., Giovannucci E., Hoover R., et al. Meeting report from the joint IARC–NCI interna-tional cancer seminar series: A focus on colorectal cancer. Ann. Oncol. 2019;30:510–519. doi: 10.1093/annonc/mdz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Cancer Information System (ECIS), Incidence and Mortality Estimates. 2020. [(accessed on 20 December 2021)]. Available online: https://ecis.jrc.ec.europa.eu/explorer.php?$0-0$1-AEE$2-All$4-1,2$3-All$6-0,85$5-2008,2008$7-7$CEstByCancer$X0_8-3$CEstRelativeCanc$X1_8-3$X1_9-AE27$CEstBySexByCancer$X2_8-3$X2_-1-1.
- 4.Fusco R., Granata V., Sansone M., Rega D., Delrio P., Tatangelo F., Romano C., Avallone A., Pupo D., Giordano M., et al. Validation of the standardized index of shape tool to analyze DCE-MRI data in the assessment of neo-adjuvant therapy in locally advanced rectal cancer. Radiol. Med. 2021;126:1044–1054. doi: 10.1007/s11547-021-01369-1. [DOI] [PubMed] [Google Scholar]
- 5.Granata V., Fusco R., De Muzio F., Cutolo C., Setola S.V., Grassi R., Grassi F., Ottaiano A., Nasti G., Tatangelo F., et al. Radiomics textural features by MR imaging to assess clinical outcomes following liver resection in colorectal liver metastases. Radiol. Med. 2022;26 doi: 10.1007/s11547-022-01477-6. [DOI] [PubMed] [Google Scholar]
- 6.Rega D., Granata V., Romano C., D’Angelo V., Pace U., Fusco R., Cervone C., Ravo V., Tatangelo F., Avallone A., et al. Watch and wait approach for rectal cancer following neoadjuvant treatment: The experience of a high volume cancer center. Diagnostics. 2021;11:1507. doi: 10.3390/diagnostics11081507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schicchi N., Fogante M., Palumbo P., Agliata G., Pirani P.E., Di Cesare E., Giovagnoni A. The sub-millisievert era in CTCA: The technical basis of the new radiation dose approach. Radiol. Med. 2020;125:1024–1039. doi: 10.1007/s11547-020-01280-1. [DOI] [PubMed] [Google Scholar]
- 8.Granata V., Grassi R., Fusco R., Izzo F., Brunese L., Delrio P., Avallone A., Pecori B., Petrillo A. Current status on response to treatment in locally advanced rectal cancer: What the radiologist should know. Eur. Rev. Med. Pharmacol. Sci. 2020;24:12050–12062. doi: 10.26355/eurrev_202012_23994. [DOI] [PubMed] [Google Scholar]
- 9.Park S.H., Kim Y.S., Choi J. Dosimetric analysis of the effects of a temporary tissue expander on the radiotherapy technique. Radiol. Med. 2020;126:437–444. doi: 10.1007/s11547-020-01297-6. [DOI] [PubMed] [Google Scholar]
- 10.Rosa C., Caravatta L., Di Tommaso M., Fasciolo D., Gasparini L., Di Guglielmo F.C., Augurio A., Vinciguerra A., Vecchi C., Genovesi D. Cone-beam computed tomography for organ motion evaluation in locally advanced rectal cancer patients. Radiol. Med. 2021;126:147–154. doi: 10.1007/s11547-020-01193-z. [DOI] [PubMed] [Google Scholar]
- 11.Bertocchi E., Barugola G., Nicosia L., Mazzola R., Ricchetti F., Dell’Abate P., Alongi F., Ruffo G. A comparative analysis between radiation dose intensification and conventional fractionation in neoadjuvant locally advanced rectal cancer: A monocentric prospective observational study. Radiol. Med. 2020;125:990–998. doi: 10.1007/s11547-020-01189-9. [DOI] [PubMed] [Google Scholar]
- 12.Fornell-Perez R., Vivas-Escalona V., Aranda-Sanchez J., Gonzalez-Dominguez M.C., Rubio-Garcia J., Aleman-Flores P., Rodríguez L., Porcel-De-Peralta G., Loro J. Primary and post-chemoradiotherapy MRI detection of extramural venous invasion in rectal cancer: The role of diffusion-weighted imaging. Radiol. Med. 2020;125:522–530. doi: 10.1007/s11547-020-01137-7. [DOI] [PubMed] [Google Scholar]
- 13.Cusumano D., Meijer G., Lenkowicz J., Chiloiro G., Boldrini L., Masciocchi C., Dinapoli N., Gatta R., Casà C., Damiani A., et al. A field strength independent MR radiomics model to predict pathological complete response in locally advanced rectal cancer. Radiol. Med. 2020;126:421–429. doi: 10.1007/s11547-020-01266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fusco R., Sansone M., Granata V., Grimm R., Pace U., Delrio P., Tatangelo F., Botti G., Avallone A., Pecori B., et al. Diffusion and perfusion MR parameters to assess preoperative short-course radiotherapy response in locally advanced rectal cancer: A comparative explorative study among Standardized Index of Shape by DCE-MRI, intravoxel incoherent motion- and diffusion kurtosis imaging-derived parameters. Abdom. Radiol. 2018;44:3683–3700. doi: 10.1007/s00261-018-1801-z. [DOI] [PubMed] [Google Scholar]
- 15.Granata V., Caruso D., Grassi R., Cappabianca S., Reginelli A., Rizzati R., Masselli G., Golfieri R., Rengo M., Regge D., et al. Structured Reporting of Rectal Cancer Staging and Restaging: A Consensus Proposal. Cancers. 2021;13:2135. doi: 10.3390/cancers13092135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granata V., Fusco R., Reginelli A., DelRio P., Selvaggi F., Grassi R., Izzo F., Petrillo A. Diffusion kurtosis imaging in patients with locally advanced rectal cancer: Current status and future perspectives. J. Int. Med. Res. 2019;47:2351–2360. doi: 10.1177/0300060519827168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrillo A., Fusco R., Granata V., Filice S., Sansone M., Rega D., Delrio P., Bianco F., Romano G.M., Tatangelo F., et al. Assessing response to neo-adjuvant therapy in locally advanced rectal cancer using Intra-voxel Incoherent Motion modelling by DWI data and Standardized Index of Shape from DCE-MRI. Ther. Adv. Med. Oncol. 2018;10:1758835918809875. doi: 10.1177/1758835918809875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fusco R., Granata V., Rega D., Russo C., Pace U., Pecori B., Tatangelo F., Botti G., Izzo F., Cascella M., et al. Morphological and functional features prognostic factor of magnetic resonance imaging in locally advanced rectal cancer. Acta Radiol. 2018;60:815–825. doi: 10.1177/0284185118803783. [DOI] [PubMed] [Google Scholar]
- 19.Fusco R., Petrillo M., Granata V., Filice S., Sansone M., Catalano O., Petrillo A. Magnetic resonance imaging evaluation in neoadjuvant therapy of locally advanced rectal cancer: A systematic review. Radiol. Oncol. 2017;51:252–262. doi: 10.1515/raon-2017-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van den Eynden G.G., Bird N.C., Majeed A.W., Van Laere S., Dirix L.Y., Vermeulen P.B. The histological growth pattern of colorectal cancer liver metastases has prognostic value. Clin. Exp. Metastasis. 2012;29:541–549. doi: 10.1007/s10585-012-9469-1. [DOI] [PubMed] [Google Scholar]
- 21.Eefsen R.L., Vermeulen P.B., Christensen I.J., Laerum O.D., Mogensen M.B., Rolff H.C., Van Den Eynden G.G., Høyer-Hansen G., Osterlind K., Vainer B., et al. Growth pattern of colorectal liver metastasis as a marker of recurrence risk. Clin. Exp. Metastasis. 2015;32:369–381. doi: 10.1007/s10585-015-9715-4. [DOI] [PubMed] [Google Scholar]
- 22.Moro C.F., Bozóky B., Gerling M. Growth patterns of colorectal cancer liver metastases and their impact on prognosis: A systematic review. BMJ Open Gastroenterol. 2018;5:e000217. doi: 10.1136/bmjgast-2018-000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petralia G., Zugni F., Summers P.E., Colombo A., Pricolo P., Grazioli L., Colagrande S., Giovagnoni A., Padhani A.R. Italian Working Group on Magnetic Resonance Whole-body magnetic resonance imaging (WB-MRI) for cancer screening: Recommendations for use. Radiol. Med. 2021;126:1434–1450. doi: 10.1007/s11547-021-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiloiro G., Cusumano D., de Franco P., Lenkowicz J., Boldrini L., Carano D., Barbaro B., Corvari B., Dinapoli N., Giraffa M., et al. Does restaging MRI radiomics analysis improve pathological complete response prediction in rectal cancer patients? A prognostic model development. Radiol. Med. 2022;127:11–20. doi: 10.1007/s11547-021-01421-0. [DOI] [PubMed] [Google Scholar]
- 25.Granata V., Grassi R., Fusco R., Setola S.V., Belli A., Ottaiano A., Nasti G., La Porta M., Danti G., Cappabianca S., et al. Intrahepatic cholangiocarcinoma and its differential diagnosis at MRI: How radiologist should assess MR features. Radiol. Med. 2021;126:1584–1600. doi: 10.1007/s11547-021-01428-7. [DOI] [PubMed] [Google Scholar]
- 26.Granata V., Bicchierai G., Fusco R., Cozzi D., Grazzini G., Danti G., De Muzio F., Maggialetti N., Smorchkova O., D’Elia M., et al. Diagnostic protocols in on-cology: Workup and treatment planning. Part 2: Abbreviated MR protocol. Eur. Rev. Med. Pharmacol. Sci. 2021;25:6499–6528. doi: 10.26355/eurrev_202111_27094. [DOI] [PubMed] [Google Scholar]
- 27.Karmazanovsky G., Gruzdev I., Tikhonova V., Kondratyev E., Revishvili A. Computed tomography-based radiomics approach in pancreatic tumors characterization. Radiol Med. 2021;12 doi: 10.1007/s11547-021-01405-0. [DOI] [PubMed] [Google Scholar]
- 28.Granata V., Fusco R., Costa M., Picone C., Cozzi D., Moroni C., La Casella G., Montanino A., Monti R., Mazzoni F., et al. Preliminary Report on Computed Tomography Radiomics Features as Biomarkers to Immunotherapy Selection in Lung Adenocarcinoma Patients. Cancers. 2021;13:3992. doi: 10.3390/cancers13163992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granata V., Fusco R., Barretta M.L., Picone C., Avallone A., Belli A., Patrone R., Ferrante M., Cozzi D., Grassi R., et al. Radiomics in hepatic metastasis by colorectal cancer. Infect. Agents Cancer. 2021;16:39. doi: 10.1186/s13027-021-00379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco R., Piccirillo A., Sansone M., Granata V., Rubulotta M., Petrosino T., Barretta M., Vallone P., Di Giacomo R., Esposito E., et al. Radiomics and artificial intelligence analysis with textural metrics extracted by contrast-enhanced mammography in the breast lesions classification. Diagnostics. 2021;11:815. doi: 10.3390/diagnostics11050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fusco R., Granata V., Mazzei M.A., Di Meglio N., Del Roscio D., Moroni C., Monti R., Cappabianca C., Picone C., Neri E., et al. Quantitative imaging decision support (QIDSTM) tool consistency evaluation and radiomic analysis by means of 594 metrics in lung carcinoma on chest CT scan. Cancer Control. 2021;28:1073274820985786. doi: 10.1177/1073274820985786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granata V., Fusco R., Avallone A., De Stefano A., Ottaiano A., Sbordone C., Brunese L., Izzo F., Petrillo A. Radiomics-Derived Data by Contrast Enhanced Magnetic Resonance in RAS Mutations Detection in Colorectal Liver Metastases. Cancers. 2021;13:453. doi: 10.3390/cancers13030453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granata V., Fusco R., Risi C., Ottaiano A., Avallone A., De Stefano A., Grimm R., Grassi R., Brunese L., Izzo F., et al. Diffusion-Weighted MRI and Diffusion Kurtosis Imaging to Detect RAS Mutation in Colorectal Liver Metastasis. Cancers. 2020;12:2420. doi: 10.3390/cancers12092420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petralia G., Summers P.E., Agostini A., Ambrosini R., Cianci R., Cristel G., Calistri L., Colagrande S. Dynamic contrast-enhanced MRI in oncology: How we do it. Radiol. Med. 2020;125:1288–1300. doi: 10.1007/s11547-020-01220-z. [DOI] [PubMed] [Google Scholar]
- 35.Ria F., Samei E. Is regulatory compliance enough to ensure excellence in medicine? Radiol. Med. 2020;125:904–905. doi: 10.1007/s11547-020-01171-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhang A., Song J., Ma Z., Chen T. Combined dynamic contrast-enhanced magnetic resonance imaging and diffusion-weighted imaging to predict neoadjuvant chemotherapy effect in FIGO stage IB2–IIA2 cervical cancers. Radiol. Med. 2020;125:1233–1242. doi: 10.1007/s11547-020-01214-x. [DOI] [PubMed] [Google Scholar]
- 37.Crimì F., Capelli G., Spolverato G., Bao Q.R., Florio A., Rossi S.M., Cecchin D., Albertoni L., Campi C., Pucciarelli S., et al. MRI T2-weighted sequences-based texture analysis (TA) as a predictor of response to neoadjuvant chemo-radiotherapy (nCRT) in patients with locally advanced rectal cancer (LARC) Radiol. Med. 2020;125:1216–1224. doi: 10.1007/s11547-020-01215-w. [DOI] [PubMed] [Google Scholar]
- 38.Kirienko M., Ninatti G., Cozzi L., Voulaz E., Gennaro N., Barajon I., Ricci F., Carlo-Stella C., Zucali P., Sollini M., et al. Computed tomography (CT)-derived radiomic features differentiate prevascular mediastinum masses as thymic neoplasms versus lymphomas. Radiol. Med. 2020;125:951–960. doi: 10.1007/s11547-020-01188-w. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L., Kang L., Li G., Zhang X., Ren J., Shi Z., Li J., Yu S. Computed tomography-based radiomics model for discriminating the risk stratification of gastrointestinal stromal tumors. Radiol. Med. 2020;125:465–473. doi: 10.1007/s11547-020-01138-6. [DOI] [PubMed] [Google Scholar]
- 40.Gurgitano M., Angileri S.A., Rodà G.M., Liguori A., Pandolfi M., Ierardi A.M., Wood B.J., Carrafiello G. Interventional Radiology ex-machina: Impact of Artificial Intelligence on practice. Radiol. Med. 2021;126:998–1006. doi: 10.1007/s11547-021-01351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scapicchio C., Gabelloni M., Barucci A., Cioni D., Saba L., Neri E. A deep look into radiomics. Radiol. Med. 2021;126:1296–1311. doi: 10.1007/s11547-021-01389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J., Jiang H., Gu D., Niu M., Fu F., Han Y., Song B., Tian J. Radiomics in liver diseases: Current progress and future opportunities. Liver Int. 2020;40:2050–2063. doi: 10.1111/liv.14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saini A., Breen I., Pershad Y., Naidu S., Knuttinen M.G., Alzubaidi S., Sheth R., Albadawi H., Kuo M., Oklu R. Radiogenomics and Radiomics in Liver Cancers. Diagnostics. 2018;9:4. doi: 10.3390/diagnostics9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathew R.P., Sam M., Raubenheimer M., Patel V., Low G. Hepatic hemangiomas: The various imaging avatars and its mimickers. Radiol. Med. 2020;125:801–815. doi: 10.1007/s11547-020-01185-z. [DOI] [PubMed] [Google Scholar]
- 45.Nardone V., Reginelli A., Grassi R., Boldrini L., Vacca G., D’Ippolito E., Annunziata S., Farchione A., Belfiore M.P., Desideri I., et al. Delta radiomics: A systematic review. Radiol. Med. 2021;126:1571–1583. doi: 10.1007/s11547-021-01436-7. [DOI] [PubMed] [Google Scholar]
- 46.Brunese L., Brunese M.C., Carbone M., Ciccone V., Mercaldo F., Santone A. Automatic PI-RADS assignment by means of formal methods. Radiol. Med. 2021;127:83–89. doi: 10.1007/s11547-021-01431-y. [DOI] [PubMed] [Google Scholar]
- 47.van der Lubbe M.F.J.A., Vaidyanathan A., de Wit M., Burg E.L.V.D., Postma A.A., Bruintjes T.D., Bilderbeek-Beckers M.A.L., Dammeijer P.F.M., Bossche S.V., Van Rompaey V., et al. A non-invasive, automated diagnosis of Menière’s disease using radiomics and machine learning on conventional magnetic resonance imaging: A multicentric, case-controlled feasibility study. Radiol. Med. 2021;127:72–82. doi: 10.1007/s11547-021-01425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Granata V., Fusco R., Setola S.V., De Muzio F., Dell’ Aversana F., Cutolo C., Faggioni L., Miele V., Izzo F., Petrillo A. CT-Based Radiomics Analysis to Predict Histopathological Outcomes Following Liver Resection in Colorectal Liver Metastases. Cancers. 2022;14:1648. doi: 10.3390/cancers14071648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santone A., Brunese M.C., Donnarumma F., Guerriero P., Mercaldo F., Reginelli A., Miele V., Giovagnoni A., Brunese L. Radiomic features for prostate cancer grade detection through formal verification. Radiol. Med. 2021;126:688–697. doi: 10.1007/s11547-020-01314-8. [DOI] [PubMed] [Google Scholar]
- 50.Agazzi G.M., Ravanelli M., Roca E., Medicina D., Balzarini P., Pessina C., Vermi W., Berruti A., Maroldi R., Farina D. CT texture analysis for prediction of EGFR mutational status and ALK rearrangement in patients with non-small cell lung cancer. Radiol. Med. 2021;126:786–794. doi: 10.1007/s11547-020-01323-7. [DOI] [PubMed] [Google Scholar]
- 51.Chianca V., Albano D., Messina C., Vincenzo G., Rizzo S., Del Grande F., Sconfienza L.M. An update in musculoskeletal tumors: From quantitative imaging to radiomics. Radiol. Med. 2021;126:1095–1105. doi: 10.1007/s11547-021-01368-2. [DOI] [PubMed] [Google Scholar]
- 52.Qin H., Que Q., Lin P., Li X., Wang X.R., He Y., Chen J.Q., Yang H. Magnetic resonance imaging (MRI) radiomics of papillary thyroid cancer (PTC): A comparison of predictive performance of multiple classifiers modeling to identify cervical lymph node metastases before surgery. Radiol. Med. 2021;126:1312–1327. doi: 10.1007/s11547-021-01393-1. [DOI] [PubMed] [Google Scholar]
- 53.Benedetti G., Mori M., Panzeri M.M., Barbera M., Palumbo D., Sini C., Muffatti F., Andreasi V., Steidler S., Doglioni C., et al. CT-derived radiomic features to discriminate histologic characteristics of pancreatic neuroendocrine tumors. Radiol. Med. 2021;126:745–760. doi: 10.1007/s11547-021-01333-z. [DOI] [PubMed] [Google Scholar]
- 54.Granata V., Fusco R., Setola S.V., Raso M.M., Avallone A., De Stefano A., Nasti G., Palaia R., Delrio P., Petrillo A., et al. Liver radiologic findings of chemotherapy-induced toxicity in liver colorectal metastases patients. Eur. Rev. Med Pharmacol. Sci. 2019;23:9697–9706. doi: 10.26355/eurrev_201911_19531. [DOI] [PubMed] [Google Scholar]
- 55.Granata V., Fusco R., Maio F., Avallone A., Nasti G., Palaia R., Albino V., Grassi R., Izzo F., Petrillo A. Qualitative assessment of EOB-GD-DTPA and Gd-BT-DO3A MR contrast studies in HCC patients and colorectal liver metastases. Infect. Agents Cancer. 2019;14:40. doi: 10.1186/s13027-019-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granata V., Fusco R., Castelguidone E.D.L.D., Avallone A., Palaia R., Delrio P., Tatangelo F., Botti G., Grassi R., Izzo F., et al. Diagnostic performance of gadoxetic acid–enhanced liver MRI versus multidetector CT in the assessment of colorectal liver metastases compared to hepatic resection. BMC Gastroenterol. 2019;19:129. doi: 10.1186/s12876-019-1036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds I.S., Furney S.J., Kay E.W., McNamara D.A., Prehn J.H., Burke J.P. Meta- analysis of the molecular associations of mu-cinous colorectal cancer. Br. J. Surg. 2019;106:682e691. doi: 10.1002/bjs.11142. [DOI] [PubMed] [Google Scholar]
- 58.Reynolds I.S., O’Connell E., Fichtner M., McNamara D.A., Kay E.W., Prehn J.H.M., Furney S.J., Burke J.P. Mucinous adenocarcinoma is a pharmacogenomically distinct subtype of colorectal cancer. Pharmacogenom. J. 2019;20:524–532. doi: 10.1038/s41397-019-0137-6. [DOI] [PubMed] [Google Scholar]
- 59.Mccawley N., Clancy C., O’Neill B.D.P., Deasy J., McNamara D.A., Burke J.P. Mucinous Rectal Adenocarcinoma Is Associated with a Poor Response to Neoadjuvant Chemoradiotherapy: A Systematic Review and Meta-analysis. Dis. Colon Rectum. 2016;59:1200–1208. doi: 10.1097/DCR.0000000000000635. [DOI] [PubMed] [Google Scholar]
- 60.Granata V., Fusco R., Avallone A., Catalano O., Piccirillo M., Palaia R., Nasti G., Petrillo A., Izzo F. A radiologist’s point of view in the presurgical and intraoperative setting of colorectal liver metastases. Futur. Oncol. 2018;14:2189–2206. doi: 10.2217/fon-2018-0080. [DOI] [PubMed] [Google Scholar]
- 61.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.-C., Pieper S., Aerts H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granata V., Fusco R., De Muzio F., Cutolo C., Setola S.V., Dell’Aversana F., Ottaiano A., Nasti G., Grassi R., Pilone V., et al. EOB-MR Based Radiomics Analysis to Assess Clinical Outcomes following Liver Resection in Colorectal Liver Metastases. Cancers. 2022;14:1239. doi: 10.3390/cancers14051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J.W.L., Andrearczyk V., Apte A., Ashrafinia S., Bakas S., Beukinga R.J., Boellaard R., et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fusco R., Sansone M., Filice S., Carone G., Amato D.M., Sansone C., Petrillo A. Pattern Recognition Approaches for Breast Cancer DCE-MRI Classification: A Systematic Review. J. Med. Biol. Eng. 2016;36:449–459. doi: 10.1007/s40846-016-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beckers R.C.J., Trebeschi S., Maas M., Schnerr R.S., Sijmons J.M.L., Beets G.L., Houwers J.B., Beets-Tan R.G.H., Lambregts D.M.J. CT texture analysis in colorectal liver metastases and the surrounding liver parenchyma and its potential as an im-aging biomarker of disease aggressiveness, response and survival. Eur. J. Radiol. 2018;102:15–21. doi: 10.1016/j.ejrad.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 66.Andersen I.R., Thorup K., Andersen M.B., Olesen R., Mortensen F.V., Nielsen D.T., Rasmussen F. Texture in the monitoring of regorafenib therapy in patients with colorectal liver metastases. Acta Radiol. 2019;60:1084–1093. doi: 10.1177/0284185118817940. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H., Li W., Hu F., Sun Y., Hu T., Tong T. MR texture analysis: Potential imaging biomarker for predicting the chemotherapeutic response of patients with colorectal liver metastases. Abdom. Radiol. 2018;44:65–71. doi: 10.1007/s00261-018-1682-1. [DOI] [PubMed] [Google Scholar]
- 68.Lubner M.G., Stabo N., Lubner S.J., Del Rio A.M., Song C., Halberg R.B., Pickhardt P.J. CT textural analysis of hepatic metastatic colorectal cancer: Pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Gastrointest. Radiol. 2015;40:2331–2337. doi: 10.1007/s00261-015-0438-4. [DOI] [PubMed] [Google Scholar]
- 69.Simpson A.L., Doussot A., Creasy J.M., Adams L.B., Allen P.J., DeMatteo R.P., Gönen M., Kemeny N.E., Kingham T.P., Shia J., et al. Computed Tomography Image Texture: A Noninvasive Prognostic Marker of Hepatic Recurrence After Hepa-tectomy for Metastatic Colorectal Cancer. Ann. Surg. Oncol. 2017;24:2482–2490. doi: 10.1245/s10434-017-5896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ganeshan B., Miles K.A., Young R.C., Chatwin C.R. Hepatic Enhancement in Colorectal Cancer: Texture Analysis Correlates with Hepatic Hemodynamics and Patient Survival. Acad. Radiol. 2007;14:1520–1530. doi: 10.1016/j.acra.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 71.Rahmim A., Bak-Fredslund K.P., Ashrafinia S., Lu L., Schmidtlein C.R., Subramaniam R.M., Morsing A., Keiding S., Horsager J., Munk O.L. Prognostic modeling for patients with colorectal liver metastases incorporating FDG PET radiomic fea-tures. Eur. J. Radiol. 2019;113:101–109. doi: 10.1016/j.ejrad.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dercle L., Lu L., Schwartz L.H., Qian M., Tejpar S., Eggleton P., Zhao B., Piessevaux H. Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway. JNCI J. Natl. Cancer Inst. 2020;112:902–912. doi: 10.1093/jnci/djaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ravanelli M., Agazzi G.M., Tononcelli E., Roca E., Cabassa P., Baiocchi G.L., Berruti A., Maroldi R., Farina D. Texture features of colorectal liver metastases on pretreatment contrast-enhanced CT may predict response and prognosis in patients treated with bevacizumab-containing chemotherapy: A pilot study including comparison with standard chemotherapy. Radiol. Med. 2019;124:877–886. doi: 10.1007/s11547-019-01046-4. [DOI] [PubMed] [Google Scholar]
- 74.Taghavi M., Staal F.C., Simões R., Hong E.K., Lambregts D.M., van der Heide U.A., Beets-Tan R.G., Maas M. CT radiomics models are unable to predict new liver metastasis after successful thermal ablation of colorectal liver metastases. Acta Radiol. 2021:02841851211060437. doi: 10.1177/02841851211060437. [DOI] [PubMed] [Google Scholar]
- 75.Granata V., Fusco R., Catalano O., Filice S., Amato D.M., Nasti G., Avallone A., Izzo F., Petrillo A. Early Assessment of Colorectal Cancer Patients with Liver Metastases Treated with Antiangiogenic Drugs: The Role of Intravoxel Incoherent Motion in Diffusion-Weighted Imaging. PLoS ONE. 2015;10:e0142876. doi: 10.1371/journal.pone.0142876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Granata V., Fusco R., Petrillo A. Additional Considerations on Use of Abbreviated Liver MRI in Patients with Colorectal Liver Metastases. Am. J. Roentgenol. 2021;217:W1. doi: 10.2214/AJR.21.25652. [DOI] [PubMed] [Google Scholar]
- 77.Esposito A., Buscarino V., Raciti D., Casiraghi E., Manini M., Biondetti P., Forzenigo L. Characterization of liver nodules in patients with chronic liver disease by MRI: Performance of the Liver Imaging Reporting and Data System (LI-RADS v.2018) scale and its comparison with the Likert scale. Radiol. Med. 2019;125:15–23. doi: 10.1007/s11547-019-01092-y. [DOI] [PubMed] [Google Scholar]
- 78.Bozkurt M., Eldem G., Bozbulut U.B., Bozkurt M.F., Kılıçkap S., Peynircioğlu B., Çil B., Ergün E.L., Volkan-Salanci B. Factors affecting the response to Y-90 microsphere therapy in the cholangiocarcinoma patients. Radiol. Med. 2020;126:323–333. doi: 10.1007/s11547-020-01240-9. [DOI] [PubMed] [Google Scholar]
- 79.Shin N., Choi J.A., Choi J.M., Cho E.-S., Kim J.H., Chung J.-J., Yu J.-S. Sclerotic changes of cavernous hemangioma in the cirrhotic liver: Long-term follow-up using dynamic contrast-enhanced computed tomography. Radiol. Med. 2020;125:1225–1232. doi: 10.1007/s11547-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 80.Granata V., Fusco R., Avallone A., Cassata A., Palaia R., Delrio P., Grassi R., Tatangelo F., Grazzini G., Izzo F., et al. Abbreviated MRI protocol for colorectal liver metastases: How the radiologist could work in pre surgical setting. PLoS ONE. 2020;15:e0241431. doi: 10.1371/journal.pone.0241431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Granata V., Fusco R., De Muzio F., Cutolo C., Setola S.V., Aversana F.D., Ottaiano A., Avallone A., Nasti G., Grassi F., et al. Contrast MR-Based Radiomics and Machine Learning Analysis to Assess Clinical Outcomes following Liver Resection in Colorectal Liver Metastases: A Preliminary Study. Cancers. 2022;14:1110. doi: 10.3390/cancers14051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some data are available at link https://zenodo.org/record/6496965#.Ympr5tpBy3A.