Abstract
Pseudomonas aeruginosa is a pathogen in both humans and animals. This bacterium, most often associated with respiratory infections in cystic fibrosis patients, was found to be the causative agent in bovine mastitis outbreaks among 11 Irish dairy herds. Epidemiological findings suggested that the infection was spread to all herds by teat wipes that had been contaminated with this organism. Two molecular-typing strategies were used in an attempt to determine the genomic relationship(s), if any, of the P. aeruginosa strains isolated from the various herds and to verify whether the same strain was responsible for each outbreak. Thirty-six isolates from the mastitis outbreaks were tested and compared to fourteen clinical isolates from Cork University Hospital. With one exception, all outbreak-linked strains produced identical patterns when ribotyped with ClaI and PvuII enzymes. Eight of the clinical isolates gave the same ClaI ribotype pattern as the mastitis-causing strains. However, PvuII proved more discriminatory, with only the outbreak isolates producing identical patterns. Similar results were obtained with RW3A-primed DNA amplification fingerprinting, with all outbreak isolates except one displaying the same fingerprint array. The clinical strains produced several fingerprint patterns, all of which were different from those of the mastitis-causing isolates. Fine-resolution DNA fingerprinting with a fluorescence-labelled RW3A primer also identified a number of low-molecular-weight polymorphisms that would have remained undetected by conventional methods. These data support the view that the same P. aeruginosa strain was responsible for the mastitis outbreaks in all 11 herds.
Pseudomonas aeruginosa is a gram-negative pathogen. This bacterium is frequently responsible for nosocomial infections in humans, particularly in cystic fibrosis (CF) (9) and burn patients. P. aeruginosa has also been identified as an animal pathogen and as the occasional cause of bovine mastitis. In cases of P. aeruginosa mastitis, the bacterium has been detected in contaminated wash hoses in milking parlors, in water and spray nozzles, and in contaminated antibiotic preparations (20). The ability to survive in moist environments contributes greatly to this organism’s ubiquitous presence in nature (5, 11).
Standard laboratory identification of Pseudomonas spp. is usually based on assessment of colony morphology and oxidase tests. Identification at the species level is obtained by using pyocin typing, antibiogram analysis, O-antigen serotyping, and biochemical tests such as the API 20NE (bioMérieux, Marcy l’Etoile, France) identification system (6, 12). Many P. aeruginosa strains, particularly those associated with CF patients, have aberrant phenotypic characteristics, and conventional typing methods often prove ineffective (18). For successful epidemiological evaluation and the development of effective treatment programs, a definitive typing method which facilitates rapid and reliable strain identification is necessary.
It is well recognized that phenotypic typing methods may not correlate with genetic variation (3, 18). However, typing isolates at the molecular level is generally regarded as a sensitive and discriminating approach. Among the molecular typing techniques applied to P. aeruginosa, ribotyping, in which patterns produced after digestion and blotting of ribosomal genes are analyzed, is frequently used (1, 7). Pulsed-field gel electrophoresis is another technique often employed for typing bacterial isolates (7). Other methods based on Southern blotting have used toxA-specific (17) and pilin gene-specific (22) probes. More recently, methods based on PCR have been developed for the comparative typing of P. aeruginosa (1, 7, 8, 10, 13, 14, 16, 21).
Cork Regional Veterinary Laboratory (CRVL) is situated in the most intensive dairy-producing region of Ireland, with over 400,000 dairy cows in its catchment area. Prior to the outbreaks reported in this paper, only one herd outbreak of P. aeruginosa mastitis had been diagnosed by CRVL in 25 years. In that outbreak, contaminated water was implicated as the source of the bacterium (19a). In the autumn of 1995, a pharmaceutical company sales promotion offered free tubs of teat wipes with the purchase of dry cow therapy (DCT) tubes. The tubs of wipes were sealed and contained 120 wipes in 70% ethanol. The purpose of the wipes was to clean and sterilize the teat end before the infusion of DCT antibiotic into the mammary gland via the teat opening. In autumn of 1996, some of these promotional tubs of wipes were still available.
From November 1996 to May 1997, CRVL investigated outbreaks of mastitis in 11 herds. The promotional wipes had been used on all of the herds during the administration of DCT. Tubs of the wipes used for each of 10 of the herds were available for testing at the time of investigation. An unopened tub of wipes with an intact seal was also obtained from the manufacturer. Standard bacteriological examination of both milk samples from infected cows and the wipes was undertaken. P. aeruginosa isolates with consistent API 20NE biotype profiles were identified in all the wipes cultured. The cultured milk samples indicated that the organism was present in all herds, albeit with slight inconsistencies in the API 20NE bioprofiles of some isolates. This organism was easily cultured from acute mastitis cases in both dry and freshly calved cows, but it was more difficult to isolate from chronic mastitis cases in lactating cows, possibly because of low concentrations of the organism and intermittent excretion. All isolates were resistant to the standard antibiotics used in mastitis treatment.
As part of an epidemiological investigation of 11 mastitis outbreaks in which P. aeruginosa had been detected, ribotype analysis and DNA amplification fingerprinting (DAF) were undertaken. This paper presents the findings of these experiments and discusses the utility of molecular surveillance methods.
MATERIALS AND METHODS
Bacterial isolates.
A total of 50 Pseudomonas isolates were used in this study (Table 1), and 36 of the isolates were P. aeruginosa obtained from the bovine mastitis outbreak under investigation. A selection of 14 control Pseudomonas organisms were obtained from the Department of Medical Microbiology, Cork University Hospital (CUH) for comparison. These controls included P. aeruginosa ATCC 27853, Pseudomonas spp. isolated from a variety of clinical cases, and isolates cultured from CF patients. Four of the hospital isolates were identified as P. aeruginosa based on their colony morphology, antibiogram typing, and API 20NE biotyping. The remaining 10, known to be Pseudomonas spp. (gram-negative, oxidase-positive bacilli in often-mucoid colonies with production of green pigment and a characteristic odor) were not further characterized. All isolates were stored on nutrient agar slopes at 4°C and, when required, were grown in tryptic soy broth (Oxoid, Basingstoke, Hampshire, United Kingdom). The isolates were also maintained on cryostat beads (Mast Diagnostics, Merseyside, United Kingdom) at −20°C for long-term storage.
TABLE 1.
Pseudomonas isolates typed with ClaI and PvuII ribotyping and RW3A DNA fingerprinting
| Outbreak case no.a | Strain no.b | Origin of strainc | RW3A DAF | Ribotyped
|
|
|---|---|---|---|---|---|
| ClaI | PvuII | ||||
| 1 | ATCC 27853 | Control | I | I | I |
| CIT-V2 | 2713 | Mastitis outbreak | II | II | II |
| CIT-V3W | 2111 | Mastitis outbreak | II | II | II |
| CIT-V4 | 1278 | Mastitis outbreak | II | II | II |
| CIT-V5 | 3704 | Mastitis outbreak | II | II | II |
| CIT-V6W | 1590 | Sealed tub | II | II | II |
| CIT-V7 | 1670 | Mastitis outbreak | II | II | II |
| CIT-V8W | 1754 | Mastitis outbreak | II | II | ND |
| CIT-V9 | 3507 | Mastitis outbreak | II | II | II |
| CIT-V10W | 1171 | Mastitis outbreak | II | II | II |
| CIT-V11 | 2865 | Mastitis outbreak | II | II | II |
| CIT-V12 | 1945 | Mastitis outbreak | II | II | ND |
| CIT-V13W | 2304 | Mastitis outbreak | II | II | II |
| CIT-V14 | 6023 (677) | Mastitis outbreak | II | II | II |
| CIT-V15 | 3450 (3) | Mastitis outbreak | III | III | III |
| CIT-V16 | 6023 (731) | Mastitis outbreak | II | II | II |
| CIT-V17 | 2375 | Mastitis outbreak | II | II | II |
| CIT-V18 | 2379 | Mastitis outbreak | II | II | II |
| CIT-V19W | 1752 | Mastitis outbreak | II | II | II |
| CIT-V20W | 1663 | Mastitis outbreak | II | II | II |
| CIT-V21 | 1776 | Mastitis outbreak | II | II | II |
| CIT-V22 | 1715 | Mastitis outbreak | II | II | II |
| CIT-V23W | 2166 | Mastitis outbreak | II | II | II |
| CIT-V24 | 1753 (6) | Mastitis outbreak | II | II | ND |
| CIT-V25 | 2851 | Mastitis outbreak | II | II | II |
| CIT-V26 | 2599 | Mastitis outbreak | II | II | II |
| CIT-V27W | 3042 | Mastitis outbreak | II | II | II |
| CIT-V28 | 1753 (2) | Mastitis outbreak | II | II | II |
| CIT-V29 | 6023 (5) | Mastitis outbreak | II | II | II |
| CIT-V30W | 2728 | Mastitis outbreak | II | II | ND |
| CIT-V31 | 6023 (743) | Mastitis outbreak | II | II | II |
| CIT-V32 | 6043 (529) | Mastitis outbreak | II | II | II |
| CIT-V33 | 6023 (712) | Mastitis outbreak | II | II | II |
| CIT-V34 | 6023 (627) | Mastitis outbreak | II | ND | II |
| CIT-V35 | 3040 (601) | Mastitis outbreak | II | II | II |
| CIT-V36 | 3040 (2) | Mastitis outbreak | II | II | II |
| CIT-H1 | 33454 | Ear swab | IV | IV | IV |
| CIT-H2 | 33937 | CF patient | V | II | V |
| CIT-H3 | 33977 | NA | VI | II | VI |
| CIT-H4 | 33977 | NA | VI | II | VI |
| CIT-H5 | 33938 (2) | CF patient | VII | IV | VII |
| CIT-H6 | 33938 | CF patient | VIII | IV | VII |
| CIT-H7 | 33936 | CF patient | IX | II | VIII |
| CIT-H8 | 33967 | Leg ulcer | X | II | VI |
| CIT-H9 | 33424 | NA | VI | II | IX |
| CIT-H10 | 33947 | Leg ulcer | XI | V | X |
| CIT-H11 | 33535 | Ear swab | XII | VI | XI |
| CIT-H12 | 33588 | Leg ulcer | XIII | II | XII |
| CIT-H13 | 33534 | Ear swab | XIV | VI | XIII |
| CIT-H14 | 33617 | NA | VI | II | IX |
V, veterinary isolate; H, hospital isolate; W, wipe isolate.
Numbers in parentheses indicate animal number.
NA, not available; sealed tub, tub unbroached before testing.
ND, not determined.
DNA isolation.
Bacterial cells were grown in 5 ml of tryptic soy broth overnight at 37°C, and DNA was extracted as previously described (23) with minor modifications. Bacterial cells were centrifuged at 14,000 × g for 5 min and washed in 1 ml of 1 M NaCl. Following centrifugation, the cells were washed in 1 ml of a TE buffer (50 mM Tris-HCl [pH 8.0], 50 mM EDTA), centrifuged at high speed, and resuspended in 0.7 ml of the same TE buffer. Two hundred micrograms of lysozyme (Sigma, St. Louis, Mo.) was added, and the mixture was incubated at 37°C for 30 min. Thirty microliters of 20% sodium dodecyl sulfate was added, and the mixture was incubated at 65°C for 10 min. To complete the lysis, 60 μl of proteinase K (10 mg/ml) (Sigma) was added and the mixture was incubated at 37°C for 1 h. Cell lysates were extracted twice with 1 ml of a phenol-chloroform solution. DNA was precipitated from the aqueous phase with 0.33 M NH4-acetate and 2.5 volumes of cold ethanol overnight at −20°C. The precipitated DNA was then dissolved in a TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The integrity of the extracted DNA was assessed by electrophoresis in a 1% agarose gel, and the DNA concentration was measured spectrophotometrically as the A260.
Ribotyping.
The restriction endonucleases ClaI and PvuII (New England Biolabs, Beverly, Mass.) were chosen for digestion of the Pseudomonas genomic DNA. Briefly, 2.0 μg of DNA was cleaved with each enzyme according to the manufacturer’s instructions. The digests were electrophoresed at 25 V overnight in 0.8% SeaKem IDNA agarose gels (FMC, Rockland, Maine) in a GNA 200 apparatus (Pharmacia, Uppsala, Sweden). Digoxigenin (DIG)-labelled lambda phage DNA digested with HindIII (Boehringer Mannheim GmbH, Mannheim, Germany) was used as a molecular weight marker.
After electrophoresis, DNA was transferred to nylon membranes (Sigma-Aldrich, Deisenhofen, Germany) with the VacuGene XL blotting system (Pharmacia), following the protocol recommended by the manufacturer. All membranes were air dried, and DNA was cross-linked by UV irradiation in a Spectrolinker XL 1000 (Spectronics Corporation).
A cDNA probe was prepared from Escherichia coli 16 and 23S rRNA (Boehringer) by using avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.) (2). This probe was DIG labelled by incorporating the DIG-dUTP (Boehringer) nucleotide into the reaction. The probe was then used for hybridization to rRNA sequences at 60°C in a hybridizer (Techne, Cambridge, United Kingdom). Hybridization, washing, and color development of the membrane were carried out as described by Popovic et al. (19).
RW3A-DNA amplification fingerprinting.
All PCRs were performed in final volumes of 50 μl each, containing 300 ng of genomic DNA, 100 pmol of RW3A primer (derived from a repetitive element in the genome of Mycoplasma pneumoniae [25]), 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 9.0], 500 mM KCl, 1% Triton X-100), 8 μl of deoxynucleotide triphosphate mix (consisting of 1.25 mM [each] dATP, dCTP, dGTP, and dTTP), 2.5 mM MgCl2, 0.5% dimethyl sulfoxide (Sigma), and 2.5 U of Taq DNA polymerase (Sigma). The RW3A primer (5′-TCG CTC AAA ACA ACG ACA CC-3′) (4) was synthesized and polyacrylamide gel purified by Eurogentec (Abingdon, United Kingdom). Each reaction was amplified in a MiniCycler (MJ Research) by using the following temperature profile: predenaturation at 94°C for 2 min followed by 35 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min and a final extension step at 72°C for 5 min. Each amplification reaction included a negative control which contained all reagents except the target DNA. Amplified DNA products were resolved by conventional electrophoresis through horizontal 2% agarose gels at 100 V, and the results were visualized and photographed over a UV transilluminator. Analysis of all isolates was performed in duplicate, with some isolates being analyzed in triplicate. All results were reproducible.
GeneScan-mediated DNA fingerprint analysis.
Following analysis by conventional agarose gel electrophoresis of the DNA fingerprints generated with the RW3A primer, a group of these isolates were chosen for GeneScan analysis. Isolates were taken from the outbreak group, which displayed identical patterns, and from the hospital collection, which displayed different patterns, and were used for comparison. For GeneScan analysis, RW3A PCR fingerprinting was performed as outlined above, except that the primer was 5′-end labelled with the fluorescent dye (F) 6-carboxyfluorescine. It was necessary to purify the PCR products with a QIAquick PCR Purification kit (Qiagen, Ltd., West Sussex, United Kingdom) prior to scanning. Samples were prepared for GeneScan analysis by mixing 0.5 μl of the F-labelled PCR-purified product, 0.5 μl of GeneScan 6-carboxy-X-rhodamine-labelled 2500 internal standards (Applied Biosystems, Warrington, United Kingdom), and 12 μl of high-pressure liquid chromatography-purified water. All appropriate cathode and anode buffers for electrophoresis were prepared in an ABI Prism 310 genetic analyzer according to the manufacturer’s recommendations. Each sample was automatically electroinjected into a 50-cm capillary containing a 2.5% polyacrylamide matrix and electrophoresed for 15 min at 11 kV at 30°C. F-labelled PCR products were excited by an argon laser, and the emitted radiation was captured and analyzed by the system software. RW3A-generated DNA amplicons were automatically sized by comparison with the internal standards, and all data were stored in a digital format on a dedicated computer (Macintosh PowerMac; Apple, Cuppertino, Calif.).
RESULTS
Ribotyping.
Genomic DNA from all 50 Pseudomonas isolates was digested with ClaI and PvuII restriction endonucleases. After hybridization with the 16 and 23S rRNA cDNA probe, ribotype patterns were detected in each sample, with bands ranging from 6.5 to 23 kb after digestion with ClaI (Fig. 1) and from 2.3 to 23 kb after digestion with PvuII (data not shown). Some of the bands appeared weak or differed between gels and were not considered further. The latter did not present a problem for interpretation, as the patterns derived from the stronger bands were both stable and reproducible.
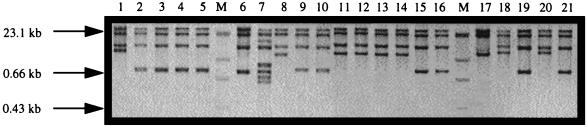
FIG. 1.
Ribotype patterns of a representative selection of Pseudomonas isolates after digestion with ClaI. Molecular weight markers ([DIG]-labelled lambda phage DNA fragments digested with HindIII [Boehringer]) are shown in lanes M. Lane 1, ATCC 27853 (group I); Lane 2, CIT-V2 (II); lane 3, CIT-V3 (II); lane 4, CIT-V4 (II); lane 5, CIT-V5 (II); lane 6, CIT-V6 (II); lane 7, CIT-V15 (III); lane 8, CIT-H1 (IV); lane 9, CIT-H2 (II); lane 10, CIT-H3 (VI); lane 11, CIT-H4 (II); lane 12, CIT-H5 (IV); lane 13, CIT-H6 (IV); lane 14, CIT-H7 (II); lane 15, CIT-H8 (II); lane 16, CIT-H9 (II); lane 17, CIT-H10 (V); lane 18, CIT-H11 (VI); lane 19, CIT-H12 (II); lane 20, CIT-H13 (VI); and lane 21, CIT-H14 (II). Numbers in parentheses denote ribotype groups.
With one exception (CIT-V15; Fig. 1, lane 7), all strains linked to the outbreak produced identical ribotype patterns, suggesting that the 16 and 23S rRNA interspacer regions were conserved among these isolates. ClaI banding patterns for eight of the clinical isolates were also identical to the mastitis-linked strains (Fig. 1, lanes 9, 10, 15, 16, 19, and 21). However, by using PvuII, the outbreak pattern was confined to the mastitis-linked isolates. These eight clinical isolates were further classified into five subgroups, with CIT-H3, -H4, and -H8 comprising the largest group. The isolates CIT-H9 and -H14 comprised the next largest group, while three individual patterns for CIT-H2, -H7, and -H12 were noted (Table 1). Isolate CIT-V15 (Table 1) clearly displayed a ribotype pattern after digestion with ClaI and PvuII enzymes that was different from those of the remaining 35 outbreak isolates and the CUH clinical strains (Fig. 1, lane 7). This finding suggests that on the basis of ribotyping, CIT-V15 cannot be implicated as being involved in the outbreak. This observation was further confirmed by DAF analysis (see below).
RW3A-primed DNA fingerprinting.
Total genomic DNA was extracted from all 50 Pseudomonas isolates by using a modified version of the protocol by Versalovic et al. (23). All strains were successfully amplified by using PCR. Typically, the DNA fragments visualized after ethidium bromide staining ranged from approximately 100 bp to 1.3 kbp (Fig. 2), with an average of seven amplicons per gel lane. Several different groupings existed within the collection, each displaying a unique DNA banding array.
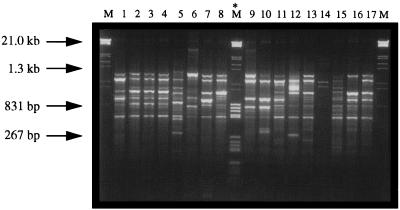
FIG. 2.
DNA fingerprint patterns of representative Pseudomonas isolates typed with the primer RW3A. PCR products (10-μl volumes) were loaded onto a 2% agarose gel in 1× Tris-acetate-EDTA buffer containing 0.1 μg of ethidium bromide per ml. Samples were electrophoresed at 100 V for 90 min. Lanes M contain molecular weight markers (grade III; Boehringer). Lane M* contains a mixture of molecular weight markers (grades III and V). Lane 1, ATCC 27853 (group I); lane 2, CIT-V2 (II); lane 3, CIT-V3 (II); lane 4, CIT-V4 (II); lane 5, CIT-V15 (III); lane 6, CIT-H1 (IV); lane 7, CIT-H2 (V); lane 8, CIT-H3 (VI); lane 9, CIT-H5 (VII); lane 10, CIT-H6 (VIII); lane 11, CIT-H7 (IX); lane 12, CIT-H8 (X); lane 13, CIT-H9 (VI); lane 14, CIT-H10 (XI); lane 15, CIT-H11 (XII); lane 16, CIT-H12 (XIII); and lane 17, CIT-H13 (XIV). Numbers in parentheses denote DAF groups.
The P. aeruginosa ATCC 27853 control strain produced a unique pattern which was designated group I (Fig. 2, lane 1). All of the mastitis outbreak-linked strains had identical patterns and were designated group II (representative isolates are shown in Fig. 2, lanes 2 to 4), with the exception of CIT-V15 (Table 1), which clearly exhibited a unique DNA fingerprint and was designated group III (Fig. 2, lane 5). The fact that these isolates had corresponding DNA patterns is consistent with a clonal origin for these organisms. In comparison, the CUH clinical isolates could be divided into a number of groups, reflecting the genomic diversity of these strains (Fig. 2, lanes 6 through 17).
F-RW3A GeneScan analysis.
As the banding patterns for all mastitis-linked isolates obtained after conventional agarose gel analysis appeared to be identical and because this method is incapable of adequately resolving DNA amplicons of low molecular weight, a fluorescence-based strategy was used to investigate whether the low-molecular-weight range contained polymorphic DNA fragments. The inclusion of an F-labelled RW3A primer in the fingerprinting reaction generated DNA amplicons with fluorescent labels attached after amplification. Use of the GeneScan ABI Prism 310 genetic analyzer offers a finer approach to analyzing DNA fragments in the low-molecular-weight range. Furthermore, all F-labelled amplicons can be detected and automatically sized directly against a suitable internal lane size standard.
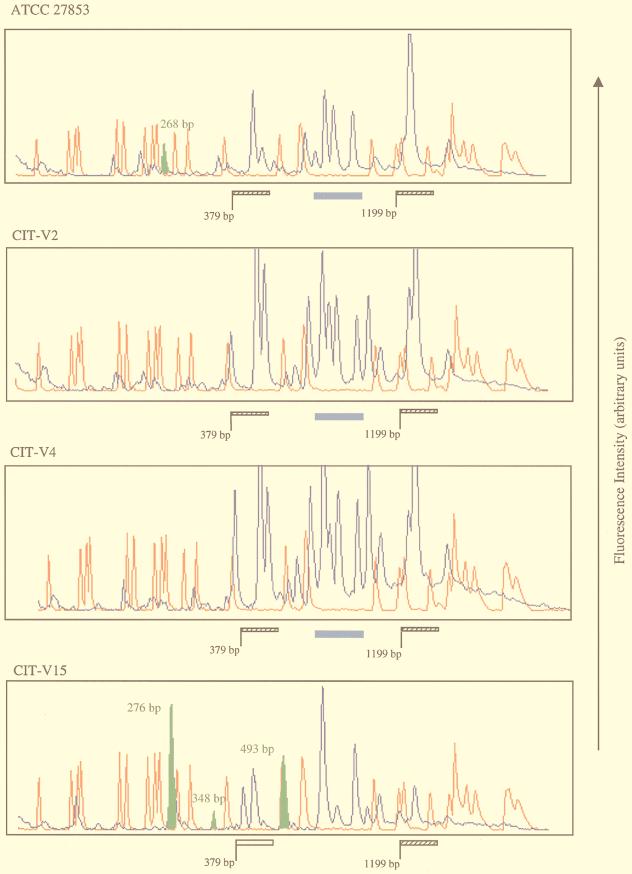
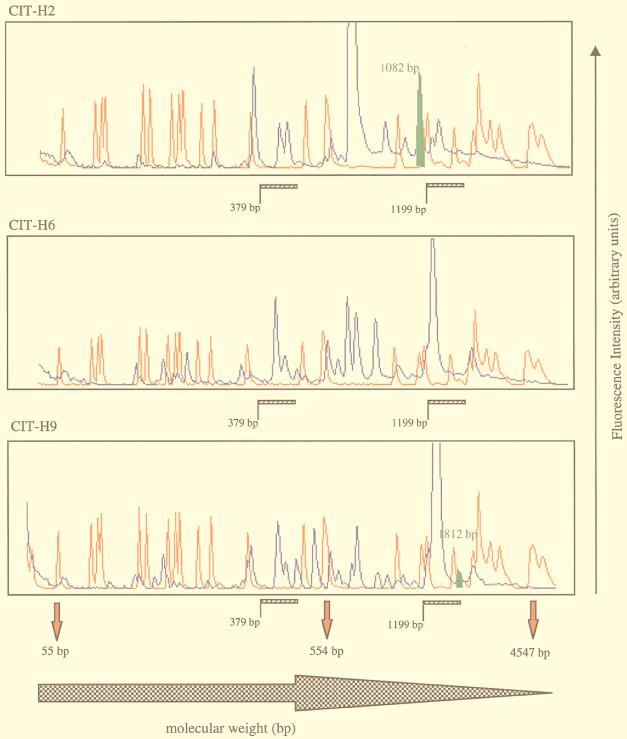
Representative outbreak isolates (groups II and III), clinical isolates (groups V, VIII, and XI), and the control strain (group I) were used for GeneScan analysis. The outbreak isolates were chosen because they displayed identical patterns in standard agarose gels. A selection of the scan patterns is shown in Fig. 3. All of these patterns were scanned from 55 bp to 4.5 kbp. When the mastitis-linked strains were compared, several RW3A-derived DNA fragments (blue peaks) were found to be conserved between all isolates. This observation was also true of other Pseudomonas spp. in the collection. These presumptive monomorphic fragments were conserved at a number of positions within the scan window (indicated by the hatched bars in Fig. 3) and were located in the size ranges from 379 to 488 bp and from 1,199 to 1,740 bp. Furthermore, the peaks obtained with the control strain (ATCC 27853) and with CIT-V15, -H2, -H6, and -H9 showed variable heights. Although all PCR and scan profile conditions were standardized, the latter observation may be consistent with quantitative polymorphisms. Careful examination of the scan traces identified another cluster of bands (indicated by the solid blue bar in Fig. 3), found in only the control strain and the two outbreak strains analyzed. These peaks may represent functionally significant conserved regions within the Pseudomonas genome. Interestingly, the double peak observed between 379 and 488 bp in all traces (Fig. 3, hatched bar) is position shifted in CIT-V15 (Fig. 3, open bar), with lengths of 431 and 446 bp assigned to these DNA fragments, respectively. This finding suggests that when CIT-V15 is compared to the other outbreak strains, a structural genomic difference is revealed within this region, which may be consistent with the results from ribotyping and pulsed-field gel electrophoresis (data not shown) that exclude this isolate from the outbreak-causing groups.
FIG. 3.
GeneScan traces of Pseudomonas isolates. Blue peaks represent 6-carboxyfluorescine-labelled RW3A-generated DNA fragments which were sized by direct comparison with the 6-carboxy-X-rhodamine-labelled internal standards (red peaks). The size range is indicated by the solid red arrowheads below the final trace. Regions of interest within the Pseudomonas genome are indicated by hatched and open bars and a solid blue bar for some isolates. Polymorphic DNA fragments are denoted by the solid green peak with corresponding molecular lengths.
Several polymorphic DNA bands were also identified (Fig. 3), the significance of which is currently under investigation. Finally, comparison of the GeneScan patterns for the CUH clinical isolates demonstrated a greater degree of heterogeneity within the size range scanned. This finding is similarly reflected in the ribotype and original DNA fingerprint data.
DISCUSSION
Eleven outbreaks of bovine mastitis in County Cork in the southern region of Ireland were investigated by standard laboratory methods and refined DNA-based techniques. Laboratory findings and clinical history suggested that P. aeruginosa-contaminated teat wipes were the cause of the mastitis outbreaks. The probable sequence of events was that P. aeruginosa-contaminated wipes were rubbed on the teat, and the bacteria deposited at the teat opening were subsequently introduced into the teat lumen by the nozzle of a DCT antibiotic tube. As no P. aeruginosa outbreaks had been diagnosed following the 1995 lactation period, we suggest that tubs of teat wipes were contaminated with very low concentrations of P. aeruginosa at the manufacturing stage, and multiplication of the bacterium occurred through 1996. Although none of the wipes used for one herd remained at the time of investigation, the putative clonal P. aeruginosa was obtained from this herd directly from milk samples CIT-V2 and CIT-V11 (Table 1).
All strains were first identified as P. aeruginosa by standard laboratory methods. It proved difficult to obtain additional information concerning the relationship between these strains with the commonly used API 20NE biotyping methods. In attempting to establish whether a genetic relationship existed among the isolates cultured, we used two molecular-typing strategies in this investigation. These were ribotyping, which samples the interspacer regions between the 16 and 23S rRNA genes, and DNA fingerprinting, which analyzes the complete genome based on the amplification of conserved motif platforms, such as the M. pneumoniae repeat sequence MP-2 (25).
With the exception of isolate CIT-V15, recovered from one of the infected herds, the molecular-typing methods applied in this study found all P. aeruginosa isolates from this mastitis outbreak to be identical at the genomic level. When all isolates were ribotyped with ClaI, the outbreak isolates displayed conserved patterns. Similarly, eight of the clinical isolates were found to have the same ribotype pattern as the mastitis outbreak isolates. This pattern similarity can be explained by the recognized stability of the inter-ribosomal gene spacer regions of these isolates (1, 15).
Analysis of the DAF patterns by using the MP-2 repeat sequence (25) as a target to amplify interrepeat regions of the Pseudomonas genome was found to be a more sensitive approach, capable of detecting genomic differences among isolates. All mastitis-linked isolates except CIT-V15 displayed identical DNA banding patterns in conventional agarose gels, producing bands ranging from 267 bp to 1.3 kbp. Furthermore, this technique allowed the hospital isolates within the collection to be divided into several groups based on the comparison of ribotype data. Taken together, all of these data supported the clonal nature of these P. aeruginosa-linked mastitis outbreaks.
It is anticipated that sensitive and discriminating typing protocols will become an essential component of an overall infection control policy. A necessary part of any widespread policy based upon molecular-typing methods is the establishment of universally accepted laboratory protocols which allow interlaboratory comparisons. To date, no universal protocol has emerged, and we believe that this will continue to be the case. Recent advances in automated DNA sequencing technologies have facilitated the development of F-labelled DNA fingerprint detection strategies. Essentially, F-labelled PCR products were analyzed in this study by using the capillary-based ABI Prism 310 GeneScan analyzer after in vitro enzyme-mediated amplification. Genome comparisons were made based on the electropherogram output for each isolate, in which all peaks were automatically sized by comparison with known internal standards. Capillary electrophoresis offers several advantages over the cumbersome preparation of polyacrylamide gels required for analogous instruments (e.g., ABI Prism 373/377). Automated F detection clearly provides finer resolution (4, 24) than standard agarose gels. An additional advantage is the possibility of using differentially labelled primers for multiplexing, thereby increasing the sensitivity and discriminating power of this strategy. In a more recent development, it is now possible to directly compare strain genome-derived fingerprints by linking them to an electronic DNA fingerprint, a process which will greatly improve pathogen tracking (5a).
In conclusion, this study examined the genetic relationship of 36 P. aeruginosa isolates which were being investigated by CRVL as the etiological agent in localized mastitis outbreaks in Irish dairy herds. By ribotype analysis and DNA fingerprinting (with conventional agarose and high-resolution electrophoresis) of the genomes of all strains recovered, we found 35 of the isolates to be genetically identical. These data support a clonal nature for this outbreak and highlight the utility of molecular surveillance methods for the investigation of potential infection linkages among organisms. In the future, these strategies will facilitate disease control monitoring and allow a more complete understanding of organism epidemiology and target intervention(s) when appropriate.
ACKNOWLEDGMENTS
We thank Bartley Cryan for supplying the CUH clinical strains, Patrick Wall, Leslie Cotter, Bridget Lucy, and Emma Fanning for valuable comments on the manuscript, and John Murphy for providing technical assistance. The herd owners and private veterinary practitioners are acknowledged for providing information on the outbreaks.
M.D. acknowledges CIT for the award of a postgraduate scholarship. This work was funded in part by the Irish government’s Graduate Training Programme (GTP/97/CR12).
Footnotes
Dedicated to the memory of our colleague Martin O’Dwyer.
REFERENCES
- 1.Bennekov T, Colding H, Ojeniyi B, Bentzon M W, Høiby N. Comparison of ribotyping and genome fingerprinting of Pseudomonas aeruginosa isolates from cystic fibrosis patients. J Clin Microbiol. 1996;34:202–204. doi: 10.1128/jcm.34.1.202-204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg H M, Kiehlbauch J A, Wachsmuth I K. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphisms of rRNA genes. J Clin Microbiol. 1991;29:2368–2374. doi: 10.1128/jcm.29.11.2368-2374.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotter L, Daly M, Greer P, Cryan B, Fanning S. Motif-dependent DNA analysis of a methicillin resistant Staphylococcus aureus (MRSA) collection. Br J Biomed Sci. 1998;52:99–106. [PubMed] [Google Scholar]
- 4.del Vecchio V G, Petroziello J M, Gress M J, McCleskey F K, Melcher G P, Crouch H K, Lupski J R. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J Clin Microbiol. 1995;33:2141–2144. doi: 10.1128/jcm.33.8.2141-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erksine R J, Unflat J G, Eberhart R J, Hutchinson L J, Hicks C R, Spencer S B. Pseudomonas mastitis: difficulties in detection and elimination from contaminated wash-water systems. J Am Vet Med Assoc. 1987;191:811–815. [PubMed] [Google Scholar]
- 5a.Fanning, S. Unpublished data.
- 6.Govan J R W. Pyocin typing of Pseudomonas aeruginosa. Methods Microbiol. 1978;10:61–91. [Google Scholar]
- 7.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández J, Ferrús M A, Hernández M, Owen R J. Arbitrarily primed PCR fingerprinting and serotyping of clinical Pseudomonas aeruginosa strains. FEMS Immunol Med Microbiol. 1997;17:37–47. doi: 10.1111/j.1574-695X.1997.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 9.Høiby N. Pseudomonas aeruginosa infection in cystic fibrosis. Acta Pathol Microbiol Scand Sect B. 1977;28(Suppl.):1–96. [PubMed] [Google Scholar]
- 10.Kersulyte D, Struelens M J, Deplano A, Berg D E. Comparison of arbitrarily primed PCR and macrorestriction (pulsed-field gel electrophoresis) typing of Pseudomonas aeruginosa strains from cystic fibrosis patients. J Clin Microbiol. 1995;33:2216–2219. doi: 10.1128/jcm.33.8.2216-2219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirk J H, Bartlett P C. Nonclinical Pseudomonas aeruginosa mastitis in a dairy herd. J Am Vet Med Assoc. 1984;184:671–673. [PubMed] [Google Scholar]
- 12.Liu P. Changes in somatic antigens of Pseudomonas aeruginosa induced by bacteriophages. J Infect Dis. 1969;119:237–246. doi: 10.1093/infdis/119.3.237. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Davin-Regli A, Bosi C, Charrel R N, Bollet C. Epidemiological investigation of Pseudomonas aeruginosa nosocomial bacteraemia isolates by PCR-based DNA fingerprinting analysis. J Med Microbiol. 1996;45:359–365. doi: 10.1099/00222615-45-5-359. [DOI] [PubMed] [Google Scholar]
- 14.Mahenthiralingam E, Campbell M E, Foster J, Lam J S, Speert D P. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin C, Ichou M A, Massicot P, Goudeau A, Quentin R. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J Clin Microbiol. 1995;33:1461–1466. doi: 10.1128/jcm.33.6.1461-1466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyawaki H, Fujita J, Takigawa K, Negayama K, Yamagishi Y, Yamaji Y, Ouchi K, Nakazawa T, Kawanishi K, Takahara J. Investigation of nosocomial respiratory infection due to Pseudomonas cepacia by arbitrarily primed polymerase chain reaction. Bacteriology. 1995;23:77–83. doi: 10.1016/0732-8893(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 17.Ogle J W, Janda J M, Wood D E, Vasil M L. Characterisation and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987;155:119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- 18.Ojeniyi B, Høiby N. Comparison of different typing methods of Pseudomonas aeruginosa. Antibiot Chemother. 1991;44:13–22. doi: 10.1159/000420292. [DOI] [PubMed] [Google Scholar]
- 19.Popovic T, Bopp C A, Olsvik Ø, Kiehlbauch J A. Ribotyping in molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 573–583. [Google Scholar]
- 19a.Power, E. Unpublished data.
- 20.Radostits O M, Blood D C, Gay C C. Veterinary medicine. 8th ed. London, England: Bailliere Tindall; 1994. p. 598. [Google Scholar]
- 21.Renders N, Römling U, Verbrugh H, van Belkum A. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J Clin Microbiol. 1996;34:3190–3195. doi: 10.1128/jcm.34.12.3190-3195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speert D P, Campbell M E, Farmer S W, Volpel K, Joffe A M, Paranchych W. Use of a pilin gene probe to study molecular epidemiology of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:2589–2593. doi: 10.1128/jcm.27.11.2589-2593.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;24:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster C A, Towner K J, Humphreys H, Ehrenstein B, Hartung D, Grundmann H. Comparison of rapid automated laser fluorescence analysis of DNA fingerprints with four other computer assisted approaches for studying relationships among Acinetobacter isolates. J Med Microbiol. 1996;44:185–194. doi: 10.1099/00222615-44-3-185. [DOI] [PubMed] [Google Scholar]
- 25.Wenzel R, Herrman R. Repetitive DNA in Mycoplasma pneumoniae. Nucleic Acids Res. 1988;16:8337–8350. doi: 10.1093/nar/16.17.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]