Abstract
Streptomyces ambofaciens is prone to genetic instability involving genomic rearrangements at the extremities of the chromosomal DNA. An amplified DNA sequence (ADS205), including an open reading frame (orfPS), is responsible for the reversible loss of spiramycin production in the mutant strain NSA205 (ADS205+ Spi−). The product of orfPS is homologous to polyketide synthase systems (PKSs) involved in the biosynthesis of erythromycin and rapamycin and is overexpressed in strain NSA205 compared with the parental strain RP181110. As PKSs and fatty acid synthase systems have the same precursors, we tested the possibility that overexpression of orfPS also affects lipid metabolism in strain NSA205. This report focuses on comparative analysis of lipids in strain RP181110, the mutant strain NSA205, and a derivative, NSA228 (ADS205− Spi+). NSA205 showed a dramatically depressed lipid content consisting predominantly of phospholipids and triacylglycerols. This lipid content was globally restored in strain NSA228, which had lost ADS205. Furthermore, strains RP181110 and NSA205 presented similar phospholipid and triacylglycerol compositions. No abnormal fatty acids were detected in NSA205.
Streptomycetes are filamentous bacteria which exhibit a complex cycle of morphological differentiation and produce a wide range of secondary metabolites with clinical, veterinary, and agricultural uses. In the streptomycetes, many characteristics associated with biochemical differentiation are genetically unstable. In Streptomyces ambofaciens, a spiramycin producer (24), numerous dispensable characteristics of secondary metabolism, such as pigmentation, aerial mycelium formation, and antibiotic production, are genetically unstable (14, 31). Most described instabilities consist of chromosomal deletions, often associated with large-scale DNA amplifications (2, 16, 17). These rearrangements are found at the extremities of the chromosome. In a previous work, non-spiramycin-producing derivatives of strain RP181110 carrying amplified DNA sequences (ADS) of various sizes were isolated. All of these ADS were shown to be located within a unique region (named AUD205 [for amplifiable unit of DNA]) of the wild-type chromosome. Studies of the progeny of one of these strains, NSA205, which harbors the largest ADS (ADS205 [89 kb]) and is defective in spiramycin production (Spi−), revealed that this amplification was correlated with the Spi− phenotype, and both were unstable. Indeed, in all cases studied, the loss of ADS205 was accompanied by restored spiramycin production (4). ADS205 corresponded to the amplification of a particular sequence (AUD205), which does not contain the cluster of spiramycin biosynthetic genes (5, 15, 27). Sequencing of part of AUD205 permitted identification of an open reading frame whose expression was higher in NSA205 than in the parental strain RP181110. The predicted product of orfPS presents homologies with polyketide synthases (PKS) responsible for erythromycin and rapamycin biosynthesis (1, 7, 29). In their initial stages, all polyketides are synthesized by a mechanism that is very similar to that of fatty acid biosynthesis (11, 22). Simple acyl precursor units such as acetyl coenzyme A (CoA), propionyl-CoA, and butyryl-CoA are activated in malonyl-CoA, methylmalonyl-CoA, or ethylmalonyl-CoA by two kinds of key enzymes (acylkinases coupled with phosphotransferases and carboxylases) in Streptomyces. Polyketide synthase catalyzes successive condensation of activated precursors to a long carbon chain. As fatty acid and polyketide biosyntheses are mechanistically related and often use the same precursor molecules, we tested the hypothesis that amplification of AUD205, which was shown to greatly reduce spiramycin production, also interacts with lipid metabolism of strain NSA205.
Herein we report a comparative analysis of the lipid compositions of strains RP181110, NSA205, and NSA228, a derivative of strain NSA205 which has lost the amplification of ADS205 and has restored spiramycin production (4). The global lipid content of S. ambofaciens NSA205 was strongly depressed: the phospholipid content was dramatically depleted and there was no observed accumulation of neutral lipids, such as triacylglycerols, compared with the parental strain RP181110. Lipid content was globally restored in strain NSA228, and it presented a sixfold-higher level of spiramycin production.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
S. ambofaciens RP181110 was obtained from Rhône-Poulenc and derived from strain ATCC 15154 (5). S. ambofaciens NSA205, a non-spiramycin-producing mutant derived from RP181110, has a deletion associated with an amplification of 89 kb (ADS205) (see Fig. 1). Strain NSA205 is genetically unstable and can undergo additional deletion events leading (in seven independent cases) to restoration of spiramycin production, loss of ADS205, and partial deletion of AUD205. NSA228 is one of seven derivatives (4). It has restored spiramycin production, has lost the amplification of ADS205, and contains a partial AUD205 deletion (see Fig. 1) (4, 5). Bacillus subtilis ATCC 6633 was used as an indicator strain for determination of antibiotic production. Spores of S. ambofaciens were maintained in 20% glycerol at −20°C, and 100 ml of preculture medium (the same as fermentation medium) was inoculated with 0.1 ml of S. ambofaciens (about 108 spores/ml) and incubated in 500-ml Erlenmeyer flasks on a rotary shaker (250 rpm) at 28°C for 48 h. About 0.1 g of biomass per liter from the resulting preculture was used to inoculate the fermentation medium. The fermentation medium (18) contained dextrins (30 g/liter), KH2PO4 (14 mM), CoCl2 · 6H2O (0.3 mg/liter), NaCl (20 g/liter), NH4Cl (37 mM), MgSO4 · 7H2O (1 g/liter), and ZnSO4 · 7H2O (15 mg/liter). The pH in the flasks was maintained by CaCO3 (5 g/liter), which was replaced by CaCl2 · 2H2O (1 g/liter) in the 3-1 jar fermentor (CMF 100; α-Alpha-Laval Chemap AG, Volketswil, Switzerland). During the 6-day fermentation, the temperature was controlled at 28°C and the pH was held at 7.0 ± 0.1 by KOH (2 M) and HCl (2 M). The fermentor’s agitation rate was maintained at 300 rpm, and dissolved oxygen saturation was 30% ± 5%. The antibiotic detection medium contained glucose (8 g/liter), yeast extract (3 g/liter), meat extract (3 g/liter), KH2PO4 (100 mM), peptone trypsic (10 g/liter), and Bacto Agar (8 g/liter) and was adjusted to pH 7.3. Extractions of genomic DNA were performed with mycelia grown in fermentors for 12 h at 30°C and resuspended in Hickey-Tresner liquid medium (25).
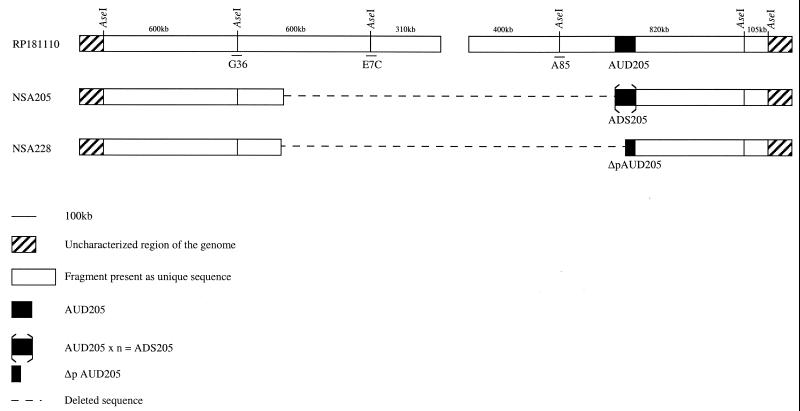
FIG. 1.
AseI restriction map of the ends of the S. ambofaciens RP181110 chromosomal DNA. AUD205 is unique in strain RP181110 and is partially deleted in strain NSA228 (Δp AUD205). Tandem reiteration of AUD205 in strain NSA205 leads to ADS205. The AseI-linking cosmids G36, E7C, and A85 are also indicated. The left extremity of the deletion was not precisely determined in the mutants.
Determinations of dextrin and ammonium concentrations.
Dextrin concentration was determined according to the method described by Hanson and Phillips (10). Culture medium ammonium concentration was determined with an ammonia electrode (Orion Research, Boston, Mass.), with ammonium chloride as the standard.
DNA extraction, endonuclease restriction, and Southern analyses.
Total DNA was extracted as previously described by Leblond et al. (15). Restriction enzymes were purchased from Boehringer (Mannheim, Germany) and used as recommended by the supplier. DNA fragments were separated on agarose gels according to the method of Sambrook et al. (28). Southern blotting, prehybridization, and hybridization were carried out as previously described (6). The cosmids used as probes correspond to recombinant cosmids resulting from a genomic library of S. ambofaciens DSM40697 (15) (see Fig. 1). Cosmids 20A12, 21B10, 24D1, 21E10, and 23E11 permitted detection of almost the entire AUD205 region. Cosmids A85, E7C, and G36 are AseI-linking cosmids (15). Labeling of probes and detection of specific hybrids were performed with the DIG DNA labeling and detection kit under conditions recommended by the supplier (Boehringer).
Measurement of growth.
The cell dry weight (CDW) was measured by monitoring the absorbance at 660 nm (A660) with a spectrophotometer (DU 7500; Beckman), according to the method described by Lebrihi et al. (19). For each strain under these conditions, 1 g CDW per liter corresponded to an A660 of 3 in the culture medium.
Antibiotic production.
Spiramycin titers of culture broths were estimated by the agar plate diffusion assay, with B. subtilis as the indicator. The three antibiotic forms of spiramycin (I, II, and III) were identified by high-pressure liquid chromatography (HPLC) (model 590; Millipore Waters) with a μ-Bondapak C18 reversed-phase column (3.9 by 300 mm, 5-μm particle diameter) (Millipore Waters). Spiramycin was supplied by Rhône-Poulenc-Rorer (Vitry-sur-Seine, France). Analysis was conducted under isocratic conditions with acetonitrile-sulfuric acid (1%) (76:24 [vol/vol]) as the mobile phase. The sample (20 μl) was subjected to a 0.8-ml min−1 flow rate. Spiramycin forms were detected at 238 nm with an LC Spectrophotometer (model 481; Millipore Waters) and were quantified with an integrator (model 745; Millipore Waters).
Extraction of lipids.
Total lipid extracts were obtained from powdered, freeze-dried cells by extraction with chloroform-methanol (2:1 [vol/vol]) as described by Folch et al. (8). All lipid samples were dried, weighed, and stored at −70°C under nitrogen gas prior to analysis.
Separation of lipids.
Total lipid extracts dissolved in chloroform were fractionated by column chromatography (Silica Gel 60; Merck, Darmstadt, Germany). Neutral lipids were eluted with chloroform, glycolipids were eluted with acetone-chloroform at 95:5 (vol/vol), and phospholipids were eluted with methanol-chloroform at 90:10 (vol/vol). Neutral lipids or total lipid extracts were separated and identified (Rf values compared with those of reference substances) by thin-layer chromatography (TLC) on precoated Silica Gel 60 TLC plates (Merck) with hexane-diethyl ether-acetic acid (90:10:1 [vol/vol/vol]) as the solvent. Spots were homogeneously sprayed with 40% (vol/vol) perchloric acid in water and carbonized at 180°C. After development, the plates were scanned with a Shimadzu-9000 densitometer (600 nm) in order to quantify neutral lipids against standards (Sigma-Aldrich Chemie, Steinheim, Germany).
Phospholipids were separated by two-dimensional TLC. The solvent for the first dimension was chloroform-methanol-ammonia (65:25:5 [vol/vol/vol]). The solvent for the second dimension was chloroform-methanol-acetic acid-water (80:12:15:4 [vol/vol/vol/vol]). Phospholipids were identified by the following staining procedures: phosphorus-containing lipids with ammonium molybdate stain (Sigma-Aldrich Chemie), amino-nitrogen-containing lipids with ninhydrin stain, mannose-containing lipids with anisaldhehyde detection, and acidic phospholipids with rhodamine 6G stain. Quantitative determinations of phospholipid families were carried out by scanning plates (545 nm) sprayed with the ammonium molybdate stain, using standards (Fluka Biochemika).
Fatty acid analysis.
Aliquots of phospholipids and triacylglycerols with a standard were converted into fatty acid methyl esters (FAMEs) by transmethylation (20). Lipids in chloroform were dried under nitrogen gas, and 1 ml of BF3/methanol reagent (14%) (Sigma) was added. The resulting mixture was heated at 90°C for 45 min. After cooling, 1 ml of water was added and FAMEs were extracted into hexane (three extractions of 2 ml each). FAMEs were concentrated in hexane and separated by gas chromatography (GC) with a Chrompack model CP-9001 GC equipped with a Carbowax 20M (Ohio Valley) OV-1 capillary column (30 m by 0.25 mm by 0.5 μm) and a flame ionization detector. The sample injector was operated in the split mode at a ratio of 1 to 20. Carrier flow of nitrogen was maintained at 100 kPa of pressure. Injector and detector temperatures were set at 250 and 260°C, respectively. The oven temperature during a chromatographic run (40 min) was ramped from 165 to 200°C at 1°C/min, and the oven was subsequently cooled. A 1-μl sample was injected. Chromatograms were integrated with the Chromatography Data System (Maestro; Chrompack). FAME composition was determined by measuring peak areas after identification with standards and by comparing relative retention times.
Short-chain fatty acid measurements.
Short-chain fatty acids in the medium were quantified by GC (model EL; Intersmat IGC 121) with a Propak Q80 column, 100 mesh (2 mm by 2 m), and a flame ionization detector. The column temperature was held constant at 180°C. Known amounts of standard short-chain fatty acids were used as an external standard for calibration procedures. The external standard (8 μl) or sample supernatant was mixed with 2 μl of internal standard (methanol-HCl (6 M)-distilled water (4:6.2:89.8 [vol/vol/vol]). A 2-μl subsample of the resulting mixture was injected for analysis.
Ethyl acetate extracts of culture supernatants.
Cultures of S. ambofaciens RP181110, NSA205, and NSA228, grown in chemically defined medium for 5 days at 28°C, were centrifuged for 15 min at 6,000 rpm. The supernatants (200 ml) were adjusted to pH 9.0 with concentrated NH4OH and were extracted twice with 1 volume of ethyl acetate. The organic phases were combined and concentrated by rotary evaporation. Extracts were analyzed on silica gel TLC plates (Merck) which were developed with chloroform-ethanol-ammonium acetate (15%) (85:15:1 [vol/vol/vol]), pH 7.0. Anisaldehyde-sulfuric acid-ethanol (1:1:9 [vol/vol/vol] spray heated for 1 min at 110°C was used to vizualize the compounds. The indicator strain, B. subtilis, was seeded into antibiotic medium and run on unstained TLC plates to test inhibition of compounds. Inhibition zones were developed by overnight incubation of the plates at 37°C. Spiramycin was used as the standard to identify spiramycin forms produced by the three S. ambofaciens strains.
RESULTS
Molecular characterization of strains NSA205 and NSA228.
As mentioned above, strain NSA205 carries an ADS which was spontaneously lost to generate NSA228. Physical mapping studies revealed that strains NSA205 and NSA228 share an extensive deletion (>400 kb) with different right endpoints, since AUD205 is partially deleted in strain NSA228. Finally, Southern experiments using the spiramycin resistance genes and some of the spiramycin biosynthetic genes as probes revealed that neither mutant is deleted for these genes, which are located outside of the unstable region (4, 5, 15).
BamHI restriction patterns of strains RP181110, NSA205, and NSA228 were hybridized with different probes derived from the unstable region to test whether some additional deletion event occurred during the conservation of strains NSA205 and NSA228. Both mutant strains were found to be deleted for the regions revealed by probes A85 and E7C (Fig. 1). On the other hand, the region revealed by probe G36 was present in both mutants, as confirmed by the presence of a hybridization pattern identical to that of strain RP181110. Finally, hybridization patterns obtained by the probes belonging to AUD205 confirmed the amplification of this region in strain NSA205 and its partial deletion in strain NSA228. Thus, no additional deletion event was detected.
All of these results, combined with those already reported (5), allowed us to conclude that strains NSA205 and NSA228 are probably deleted at both chromosomal extremities and that strain NSA228 harbors a partial deletion of AUD205 (Fig. 1).
Growth of organisms.
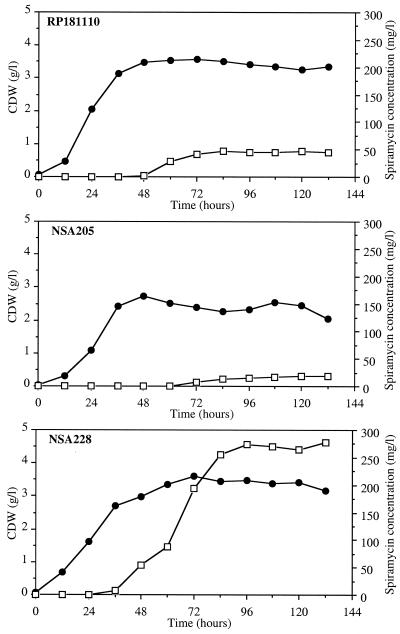
Growth was based on submerged culture conditions with a chemically defined medium which permits reproducible growth and spiramycin production (18). Precultures in the exponential phase of growth were used to inoculate fermentors. All fermentations were carried out in triplicate for each strain. The growth curves presented in Fig. 2 correspond to mean values deduced from the three independent experiments. No initial lag phase was observed in the fermentors. The three strains showed similar growth in each experiment, although the maximum amount of biomass obtained in each culture was higher for strains RP181110 and NSA228 (3.5 g/liter) than for strain NSA205 (2.25 g/liter). Carbon and nitrogen sources (dextrins and ammonium) were consumed at the same rate by each strain during the exponential growth phase (qdextrins = 0.17 g h−1 g−1 CDW; qNH4+ = 0.6 mmol h−1 g−1 CDW). In each culture, the stationary phase started when the nitrogen source, which was the limiting growth factor, was totally depleted (data not shown). During the stationary phase of growth, dextrins were slowly consumed (0.03 g h−1 g−1 CDW) by each strain. No significant difference in carbon source utilization was observed between the parental strain and NSA205. In contrast, ammonium was released sooner for strain NSA205 (72 h) than for the others (108 h). Specific yield of ammonium production was higher in NSA205 (4.5 mmol g−1 CDW) than in strains RP181110 and NSA228 (1 mmol g−1 CDW). This indicated that cellular lysis associated with proteolysis was higher in strain NSA205 than in strains RP181110 and NSA228.
FIG. 2.
Growth (●) and spiramycin production (□) of S. ambofaciens RP181110, NSA205, and NSA228 on a chemically defined medium in a 3-liter batch fermentor. Results are means of three independent experiments with standard deviations of 20 to 30%.
Spiramycin production.
During fermentation, spiramycin production was quantified by the disc diffusion method. HPLC analysis of culture broth indicated that in all cases, the three forms of spiramycin (I, II, and III) were detected in the same proportions (data not shown). In strains RP181110 and NSA228, spiramycin production occurred after 48 and 36 h of growth, respectively. This period corresponded to the end of the transition phase of growth (Fig. 2). Specific spiramycin production rates were 0.4 and 1.7 mg h−1 g−1 CDW, respectively (Fig. 2). Strain NSA228 had a sixfold-higher specific production yield (74 mg g−1 CDW) than strain RP181110 (13 mg g−1 CDW). Strains RP181110 and NSA228 produced spiramycin for 36 and 60 h, respectively. Dary et al. (4) did not detect spiramycin production after 7 days of growth of NSA205 in a complex medium. Under our conditions, late (72 h of growth) spiramycin production occurred in NSA205 for a period of 12 h (Fig. 2). However, specific production yield and specific production rate were significantly lower in strain NSA205 (5 mg g−1 CDW and 0.3 mg h−1 g−1 CDW, respectively) than in strains RP181110 and NSA228 (Fig. 2). This weak production could be due to the medium used for fermentation or to the loss of amplification of ADS205, which was shown to be unstable and lost at high frequency (4). Hence, mycelium of NSA205 was harvested at different times (between 24 and 144 h) and genomic DNA was extracted and digested with BamHI to visualize the amplification, which was detected at each time tested (data not shown). However, the possibility of heterogeneity of genomes in NSA205 fermentation could not formally be excluded.
Analysis of short-chain fatty acid excretion in the medium.
To test whether the non-spiramycin-producing phenotype of strain NSA205 resulted from high excretion of short fatty acid precursors, GC analysis was conducted on culture medium. Excretion of acetate only was detected (data not shown). For each strain, it reached a maximum level of about 75 mg/liter after 144 h of growth. No significant level of propionate, butyrate, or isobutyrate was detected. These results suggest that inhibition of spiramycin production in strain NSA205 does not result from higher excretion of short-chain fatty acids required for antibiotic production.
Formation of intracellular lipids during growth.
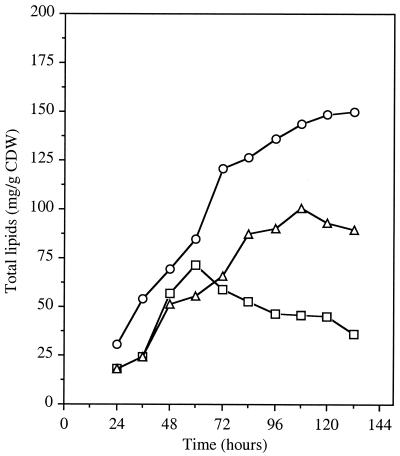
Variations in intracellular total lipid content were investigated during a 6-day fermentation of strains RP181110, NSA205, and NSA228. The lipid content continually increased in strains RP181110 and NSA228 (Fig. 3). However, in each independent experiment, the maximum lipid content observed in strain NSA228 (90 mg g−1 CDW) was significantly lower than in strain RP181110 (150 mg g−1 CDW). The lipid content of strain NSA205 was found to be highly depressed. Indeed, in each experiment, lipid accumulation stopped after 60 h of incubation (Fig. 3) and continually decreased until the end of fermentation. The maximum level of lipids obtained in this strain was about 72 mg g−1 CDW, after 60 h of incubation.
FIG. 3.
Formation of total lipids during growth of S. ambofaciens RP181110 (○), NSA205 (□), and NSA228 (▵) cultivated in a synthetic medium. Results are means of three independent experiments with standard deviations of 15 to 20%.
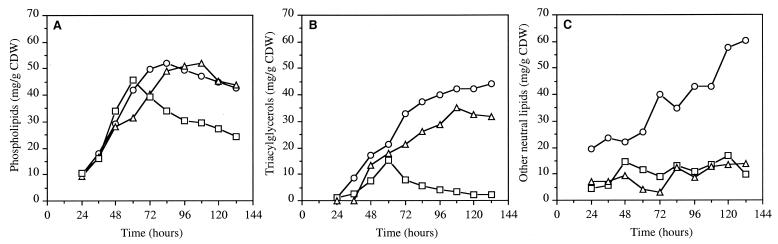
Specific analysis of lipid families revealed three fractions which constituted the majority of total lipids: phospholipids, neutral fractions, and glycolipids. The glycolipid fraction was in the minority in each strain (data not shown), and no more analysis was conducted on this family. Until 60 h of growth, accumulation of phospholipids was observed in each strain (Fig. 4A); it was about 42, 32, and 46 mg g−1 CDW in strains RP181110, NSA228, and NSA205, respectively. While the phospholipid content of strains RP181110 and NSA228 continued to increase between 60 and 80 h and remained steady throughout the stationary phase, the level of phospholipids decreased dramatically (about 40%) in strain NSA205, as was observed for total lipids (Fig. 4A).
FIG. 4.
Formation of major lipid families (phospholipids [A], triacylglycerols [B], and other neutral lipids [C] during growth of S. ambofaciens RP181110 (○), NSA205 (□), and NSA228 (▵). Results are means of two independent experiments with standard deviations of 10% for phospholipids, 20% for triacylglycerols, and 30% for other neutral lipids.
Analysis of neutral fractions separated by TLC revealed two families. The first corresponded to triacylglycerols, which were detected and identified with carbonization of standards. The second was visible on TLC without treatment. These lipids were yellow-orange colored and probably contained pigments. Triacylglycerols were detected in small amounts during the exponential and transitional phases of growth (Fig. 4B). At 84 h, increasing lipid content in RP181110 and NSA228 corresponded to the accumulation of neutral lipids. In contrast, in strain NSA205, triacylglycerols increased to 15 mg g−1 CDW until 60 h and thereafter showed a significant decrease. The fraction containing pigments was not quantified directly, but a rough estimation was performed by using the difference between the neutral fraction and the triacylglycerols, which were quantified with standards. The fraction containing pigments increased significantly between 60 and 132 h for RP181110 and was significantly lower and stable throughout fermentation in strains NSA228 and NSA205 (Fig. 4C). Small amounts of free fatty acids, monoacylglycerols, and diacylglycerols were detected at every stage of growth in the neutral fraction, as determined by TLC.
In conclusion, NSA205 had a greatly depressed lipid content from 60 h of fermentation. Further, except for the fraction containing pigments, lipid content was globally restored in strain NSA228.
Evolution of the different phospholipid families during growth.
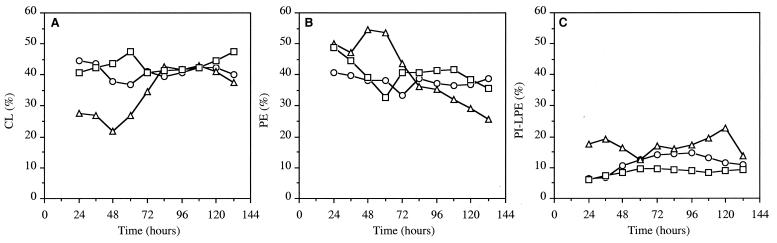
The phospholipid fraction was analyzed by two-dimensional TLC during fermentation of strains RP181110, NSA205, and NSA228. Four major families were observed: cardiolipin (CL) (Fig. 5A), phosphatidylethanolamine (PE) (Fig. 5B), and phosphatidylinositol and lysophosphatidylethanolamine (PI-LPE) (Fig. 5C). The last two were not separated on silica gel plates and were quantified together. Two other minor families were detected: phosphatidic acid and phosphatidylinositolmannoside. RP181110 and NSA205 had the same relative CL proportion during fermentation (between 38 and 45%) (Fig. 5A), as well as similar PE (Fig. 5B) and PI-LPE (Fig. 5C) profiles. Thus, strain NSA205 seemed to be quantitatively affected in all phospholipid content. The most striking differences were observed with strain NSA228; these differences concerned the four families of phospholipids. In each experiment, the relative proportions of CL in strain NSA228 varied during fermentation (Fig. 5A). Indeed, at 24 h CL represented only 27% of the phospholipids. Its relative proportion increased until 84 h, at which time it constituted 40% of the phospholipids, as in strains RP181110 and NSA205. The PE proportions were steady between 24 and 60 h, after which they continually decreased to represent 25% of the phospholipids at the end of fermentation (Fig. 5B). Differences were also observed for PI-LPE proportions (Fig. 5C), which were higher in strain NSA228. Although the phospholipid content was globally restored in strain NSA228, the differences described above suggest that the additional deletion affecting at least AUD205 may have resulted in modifications of phospholipid composition.
FIG. 5.
Relative proportions of the major phospholipid families of S. ambofaciens RP181110 (○), NSA205 (□), and NSA228 (▵). (A) CL. (B) PE. (C) PI-LPE. Results are means of two independent experiments with standard deviations of 5 to 10%.
Fatty acid composition.
The fatty acid composition of triacylglycerols and phospholipids was investigated by GC, with suitable standards, in fermentations of strains RP181110, NSA205, and NSA228. The profiles in each family showed a relative representation of fatty acids within the C12 to C20 range of carbon chain length for each strain. The branched-chain fatty acids corresponded to the iso (i) and anteiso (ai) series derivatives of C15:0, C16:0, and C17:0 acids. The mean proportions of phospholipid fatty acid constituents observed at three times during growth (48, 72, and 120 h) are shown in Table 1. Spiramycin production began in strains RP181110 and NSA228 at 48 h. The 72-h point corresponded to the phase of production, and spiramycin production was stopped at 120 h (Fig. 2).
TABLE 1.
Fatty acid composition of phospholipids of S. ambofaciens NSA205, and NSA228 grown in fermentation medium
| Incubation time (h) | Strain | % Total fatty acida
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| i-C15:0 | i-C16:0 | C16:0 | C18:0 | C20:0 | ai-C15:0 | C16:1 | ai-C17:0 | C18:1 | C18:2 | ||
| 48 | RP181110 | 0.4 | 10.1 | 28.7 | 28.3 | 1.4 | 4.8 | 2.5 | 1.7 | 15.2 | 6.7 |
| NSA205 | 2.6 | 6.1 | 28.6 | 25.6 | 1.9 | 5.2 | 4.3 | 1.6 | 15.2 | 8.6 | |
| NSA228 | 0.4 | 7.4 | 26.2 | 26.2 | 3.3 | 5.4 | 3.9 | 3.9 | 14.8 | 8.3 | |
| 72 | RP181110 | 5.6 | 20.5 | 16.5 | 13.2 | 5.1 | 14.5 | 6.3 | 6.8 | 7.3 | 4.1 |
| NSA205 | 3.7 | 17.5 | 19.0 | 15.4 | 3.2 | 11.1 | 3.9 | 7.2 | 11.7 | 7.0 | |
| NSA228 | 4.4 | 12.2 | 21.2 | 19.1 | 6.1 | 8.1 | 6.5 | 4.2 | 11.5 | 6.4 | |
| 120 | RP181110 | 0 | 30.8 | 15.1 | 6.0 | 1.1 | 27.4 | 0.0 | 10.8 | 5.0 | 3.7 |
| NSA205 | 4.4 | 14.8 | 20.9 | 18.1 | 3.6 | 12.0 | 4.2 | 6.7 | 9.4 | 5.7 | |
| NSA228 | 0.9 | 6.8 | 28.5 | 26.2 | 2.6 | 4.8 | 2.6 | 2.2 | 15.8 | 9.3 | |
i, isoacid; ai, anteisoacid. The number to the left of the colon is the number of carbons atoms; the number to the right of the colon is the number of double bonds. The ratio of (iso + saturated) to (anteiso + unsaturated) fatty acids is presented. Values are means of two independent fermentations for each strain. Sums of percentages may not be exactly 100% due to rounding of percentages of individual fatty acids.
The majority of fatty acids underwent variations during growth. Between 48 and 72 h, the variations observed in each strain had similar tendencies. Conversely, fatty acid proportions differed between 72 and 120 h. For example, the i-C16:0, ai-C15:0, and ai-C17:0 fatty acids increased in strain RP181110 while their relative proportions decreased in the mutant strain NSA228. The fatty acid profiles of triacylglycerols revealed no significant differences. Indeed, during fermentation, variations in fatty acid proportions for one strain and between strains were less important and concerned only a few fatty acids (data not shown). The most striking differences were observed for C16:1 and ai-C17:0, which were less concentrated in RP181110 than in NSA228 and NSA205 at each time.
Finally, no abnormal fatty acids were detected in strains NSA205 and NSA228. Therefore, the depressed lipid content observed in strain NSA205 does not correlate with synthesis of an abnormal, potentially harmful, fatty acid.
Ethyl acetate extracts of fermentation broths.
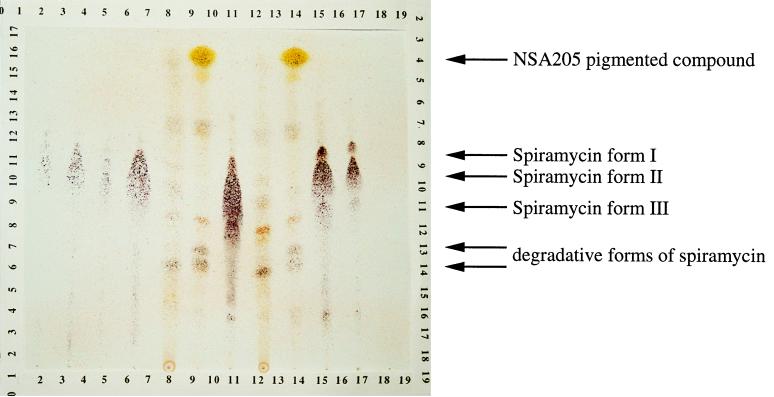
As no abnormal fatty acid was detected in strain NSA205, the depressed lipid content could have resulted from competition for precursors between orfPS and fatty acid synthase. In order to detect the putative orfPS product, ethyl acetate extracts of fermentation broths from each strain were analyzed by TLC (Fig. 6) and bioassayed with B. subtilis as the indicator. Before staining, a yellow-orange compound with an Rf of 0.9 was detected in the supernatant of strain NSA205. This compound was not detected in the two other strain supernatants. With anisaldehyde sulfuric spray, spiramycin forms (Rf between 0.4 and 0.7) were detected as violet-brown and were significantly higher in strain NSA228 (Fig. 6). The yellow-orange compound produced by strain NSA205 showed a strong yellow color with anisaldehyde sulfuric spray (Fig. 6). A small amount of this compound was observed in strain RP181110 but not in strain NSA228. Spiramycin forms and degradation products (Rf [inferior], 0.4) were detected with bioassays. The compound produced only by NSA205 was not active against B. subtilis.
FIG. 6.
TLC of ethyl acetate extracts from 200-ml culture supernatants of S. ambofaciens strains. Lanes 1 to 4, spiramycin standards (Sigma) (1, 0.125 mg; 2, 0.25 mg; 3, 0.065 mg; 4, 0.5 mg); lanes 5 and 8, RP181110 extracts; lanes 6 and 9, NSA205 extracts; lanes 7, 10, and 11, NSA228 extracts.
DISCUSSION
The aim of this investigation was to determine whether the lipid metabolism of strain NSA205 could be affected by amplification of AUD205, which contains a PKS gene (orfPS). The lipid content of three strains (RP181110, NSA205, and NSA228) was determined. RP181110 is the parental strain. NSA205 contains a deletion associated with a large amplification of 89 kb containing orfPS. Strain NSA228 was isolated from the progeny of strain NSA205. This strain was isolated for its capacity to produce spiramycin. All strains selected from the progeny of strain NSA205 for the ability to produce spiramycin had lost ADS205 and presented a partial deletion of AUD205 (4).
Spiramycin production of each strain was analyzed in a chemically defined medium. The spiramycin production level of the parental strain RP181110 was considered the reference. Although on a complex medium NSA205 was found to be a non-spiramycin-producing strain (4), under culture conditions favorable for spiramycin production, very low production was detected later, compared with that of the other strains. Strain NSA228 produced spiramycin to a sixfold-higher level than the reference strain. The high spiramycin production of NSA228 may result from an additional deletion event affecting at least AUD205. Dary et al. (4) showed that the Spi− phenotype was unstable in strain NSA205 and was lost at a high frequency in its lineage. A loss of ADS205 in only a few genomes could explain the detection of spiramycin during the cultivation of NSA205, since some of them (like the NSA228 genomes) are responsible for production 15-fold higher than that of NSA205.
Lipids of streptomycetes are widely reported in the literature (3, 9, 12, 21, 23, 26). Lipid composition is strongly influenced by environmental conditions and the physiological state of the bacteria (30). Therefore, cultures in batch fermentors were controlled for different parameters (pH, dissolved oxygen, temperature) and allowed adequate growth for lipid analysis and comparison. These studies showed that phospholipids and neutral lipids were in the majority. The global lipid content of strains RP181110 and NSA228 increased with culture age. Conversely, lipids were synthesized for only 60 h in strain NSA205 and then were greatly depleted. Analysis of the major constituents of total lipid content indicated that phospholipids and triacylglycerols, both dependent on long-chain fatty acid biosynthesis, decreased and were responsible for the total lipid depletion in strain NSA205. A qualitative interaction between the synthases which could produce abnormal, potentially harmful, fatty acids should be responsible for the depressed lipid content in NSA205. Yu and Hopwood (32) described an early growth inhibition of strains in which expression of sets of whiE PKS genes were probably responsible for interference with fatty acid biosynthesis by competition or by production of abnormal fatty acids. Hence, the phospholipid families and their fatty acid composition, like triacylglycerol composition, were investigated for each strain. No abnormal fatty acids were detected, and the phospholipid composition was similar to that of the parent.
Reduction of overall lipid content in strain NSA205 could be due to amplification of orfPS. Indeed, strain NSA228 showed restored lipid content except for the pigment fraction. As the reduction of this fraction affected both mutants, it was probably associated with the large deletion present in both strains (Fig. 1). As Aigle et al. (1) observed, orfPS was overtranscribed in NSA205 compared with RP181110 during both the stationary and rapid phases of growth, and because no abnormal fatty acid was detected, we postulated that orfPS overexpression could result in a metabolic drain of the acyl precursors (acetyl-CoA and malonyl-CoA) shared by the PKS and fatty acid synthase systems. Therefore, supernatants of the three strains were analyzed. Ethyl acetate extracts revealed the presence of a great amount of a pigmented compound in the supernatant of strain NSA205. Only a small amount of this pigmented compound was detected in strain RP181110, and it was not detected in strain NSA228. Although its structure remains to be elucidated, this compound could belong to the large class of polyketides. These results support the hypothesis of a metabolic drain of precursors from strain NSA205. As the new compound has not been purified and quantified, it is difficult to speculate on whether it accounts for the decrease in the amount of lipids. Acetyl-CoA carboxylase is considered to be one of the key enzymes leading to the formation of malonyl-CoA in S. ambofaciens, but other pathways have been studied (13). Future experiments will attempt to compare the specific activities of these enzymes in cellular extracts of both strain RP181110 and strain NSA205. Moreover, monitoring the incorporation of [14C]acetate into lipids and compounds extracted with ethyl acetate from strain NSA205 could confirm the drain of acyl precursors from lipids to a polyketide during the stationary phase of growth.
ACKNOWLEDGMENT
This work was supported by a grant from the French Ministry of Education and Research.
REFERENCES
- 1.Aigle B, Schneider D, Morilhat C, Vandewiele D, Dary A, Holl A-C, Simonet J-M, Decaris B. An amplifiable and deletable locus of Streptomyces ambofaciens RP181110 contains a very large gene homologous to polyketide synthase genes. Microbiology. 1996;142:2815–2824. doi: 10.1099/13500872-142-10-2815. [DOI] [PubMed] [Google Scholar]
- 2.Birch A, Häusler A, Hütter R. Genome rearrangement and genetic instability in Streptomyces spp. J Bacteriol. 1990;172:4138–4142. doi: 10.1128/jb.172.8.4138-4142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan P J. Mycobacterium and other actinomycetes. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, England: Academic Press; 1988. pp. 203–285. [Google Scholar]
- 4.Dary A, Bourget N, Girard N, Simonet J-M, Decaris B. The amplication of a particular DNA sequence reversibly prevents spiramycin production in Streptomyces ambofaciens RP181110. Res Microbiol. 1992;143:99–112. doi: 10.1016/0923-2508(92)90039-q. [DOI] [PubMed] [Google Scholar]
- 5.Dary A, Kaiser P, Bourget N, Thompson C J, Simonet J-M, Decaris B. Large genomic rearrangements of the unstable region in Streptomyces ambofaciens are associated with major changes in global gene expression. Mol Microbiol. 1993;10:759–769. doi: 10.1111/j.1365-2958.1993.tb00946.x. [DOI] [PubMed] [Google Scholar]
- 6.Demuyter P, Leblond P, Decaris B, Simonet J-M. Characterization of two spontaneously amplifiable units of DNA in Streptomyces ambofaciens. J Gen Microbiol. 1988;134:2001–2007. doi: 10.1099/00221287-134-7-2001. [DOI] [PubMed] [Google Scholar]
- 7.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 8.Folch J, Lees M, Stanley G H. A simple method for the isolation and purification of total lipid from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 9.Gräfe U, Rheinhardt R, Krebs D, Roth M, Noack D. Altered lipid composition in a non-differentiating derivative of Streptomyces hygroscopicus. J Gen Microbiol. 1982;129:2693–2698. doi: 10.1099/00221287-128-11-2693. [DOI] [PubMed] [Google Scholar]
- 10.Hanson R S, Phillips J A. Chemical composition. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 328–364. [Google Scholar]
- 11.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosyntheiss. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 12.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laakel M, Lebrihi A, Khaoua S, Schneider F, Lefebvre G, Germain P. A link between primary and secondary metabolism: malonyl-CoA formation in Streptomyces ambofaciens growing on ammonium ions or valine. Microbiology. 1994;140:1451–1456. [Google Scholar]
- 14.Leblond P, Demuyter P, Moutier L, Laakel M, Decaris B, Simonet J-M. Hypervariability, a new phenomenon of genetic instability, related to DNA amplification in Streptomyces ambofaciens. J Bacteriol. 1989;171:419–421. doi: 10.1128/jb.171.1.419-423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblond P, Fischer G, Francou F-X, Berger F, Guérineau M, Decaris B. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol Microbiol. 1996;19:261–271. doi: 10.1046/j.1365-2958.1996.366894.x. [DOI] [PubMed] [Google Scholar]
- 16.Leblond P, Demuyter P, Simonet J-M, Decaris B. Genetic instability and hypervariability in Streptomyces ambofaciens: towards an understanding of mechanism of genome plasticity. Mol Microbiol. 1990;4:689–696. doi: 10.1111/j.1365-2958.1990.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 17.Leblond P, Demuyter P, Simonet J-M, Decaris B. Genetic instability and associated genomic plasticity in Streptomyces ambofaciens: PFGE evidence for large DNA alterations in a limited genomic region. J Bacteriol. 1991;173:4229–4233. doi: 10.1128/jb.173.13.4229-4233.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebrihi A, Lamsaif D, Lefebvre G, Germain P. Effect of ammonium ions on spiramycin biosynthesis in Streptomyces ambofaciens. Appl Microbiol Biotechnol. 1992;37:382–387. doi: 10.1007/BF00210997. [DOI] [PubMed] [Google Scholar]
- 19.Lebrihi A, Lefebvre G, Germain P. Phosphate repression of cephamycin and clavulanic acid production by Streptomyces clavuligerus. Appl Microbiol Biotechnol. 1987;26:130–135. [Google Scholar]
- 20.Morrison W R, Smith L M. Preparation of fatty acid methyl esters and dimethy-acetals from lipids with boron trifluoride-methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 21.Olukoshi E R, Packter N M. Importance of stored triacylglycerols in Streptomyces: possible carbon source for antibiotics. Microbiology. 1994;140:931–943. doi: 10.1099/00221287-140-4-931. [DOI] [PubMed] [Google Scholar]
- 22.Omura S, Takeshima H, Nakagawa A, Miyazawa J, Pirou T, Lukacs G. Studies on the biosynthesis of 16 membered macrolide antibiotics using carbon-13 magnetic resonance spectroscopy. Biochemistry. 1977;16:2860–2866. doi: 10.1021/bi00632a009. [DOI] [PubMed] [Google Scholar]
- 23.Packter N M, Olukoshi E R. Ultrastructural studies of neutral lipid localisation in Streptomyces. Arch Microbiol. 1995;164:420–427. doi: 10.1007/BF02529740. [DOI] [PubMed] [Google Scholar]
- 24.Pinnert-Sindico S, Ninet L, Preud’Homme J, Cosar C. A new antibiotic: spiramycin. Antibiot Annu. 1955;1954/1955:724–757. [Google Scholar]
- 25.Pridham T G, Anderson P, Foley C, Lindenfelser L A L, Hesseltine C W, Benedict R G. A selection of media for maintenance and taxonomic study of Streptomyces. Antibiot Annu. 1957;1956/1957:947–953. [PubMed] [Google Scholar]
- 26.Pospisil S, Rezanka T. Changes in fatty acid branching and unsaturation of Streptomyces cinnamonensis as a response to NaCl concentration. Folia Microbiol. 1994;39:187–190. [Google Scholar]
- 27.Richardson M A, Khustoss S, Huber M L B, Ford L, Godfrey O, Rao R N. Cloning of spiramycin biosynthesis genes and their use in constructing Streptomyces mutants defective in spiramycin biosynthesis. J Bacteriol. 1990;172:3790–3798. doi: 10.1128/jb.172.7.3790-3798.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortés J, Lester J B, Böhm G A, Staunton J, Leadlay P E. The biosynthetic gene cluster for the polyketide immunosupressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verma J N, Khuller G K. Lipids of actinomycetes. Adv Lipid Res. 1983;20:257–307. [Google Scholar]
- 31.Volff J-H, Vandewiele D, Simonet J-M, Decaris B. Ultraviolet light, mitomycin C and nitrous acid induce genetic instability in Streptomyces ambofaciens ATCC 23877. Mutat Res. 1993;287:141–156. doi: 10.1016/0027-5107(93)90008-4. [DOI] [PubMed] [Google Scholar]
- 32.Yu T-W, Hopwood D A. Ectopic expression of the Streptomyces coelicolor whiE genes for polyketide spore pigment synthesis and their interaction with the act genes for actinorhodin biosynthesis. Microbiology. 1995;141:2779–2791. doi: 10.1099/13500872-141-11-2779. [DOI] [PubMed] [Google Scholar]