Abstract
A robust generalisation of the Gumbel distribution is proposed in this article. This family of distributions is based on the T-X paradigm. From a list of special distributions that have evolved as a result of this family, three separate models are also mentioned in this article. A linear combination of generalised exponential distributions can be used to characterise the density of a new family, which is critical in assessing some of the family’s properties. The statistical features of this family are determined, including exact formulations for the quantile function, ordinary and incomplete moments, generating function, and order statistics. The model parameters are estimated using the maximum likelihood method. Further, one of the unique models has been systematically studied. Along with conventional skewness measures, MacGillivray skewness is also used to quantify the skewness measure. The new probability distribution also enables us to determine certain critical risk indicators, both numerically and graphically. We use a simulated assessment of the suggested distribution, as well as apply three real-world data sets in modelling the proposed model, in order to ensure its authenticity and superiority.
1 Introduction
The employment of traditional probability models to anticipate real-life occurrences is causing increasing dissatisfaction among applied practitioners. Tail characteristics and goodness of fit metrics may have a constraining tendency, which could be one of the reasons. As a response, in recent years, there has been a substantial rise in the generalisation of well-known probability distributions. The challenge is to find such versatile families that can fit both skew and symmetric data. It’s important to realize that the majority of generalised distributions described in the literature are constructed using the generalised classes approach (G-class) and the compounding principle. In [1], the authors provided a concise review of literature regarding generalization of distributions and transformation through versatile parameter induction techniques. We offer to the readers a few, but not exhaustive, lists in chronological sequence: [2–14].
According to [15], Emil J. Gumbel originated the use of the Gumbel distribution (GuD) on data bearing extreme values. By “extreme data” we mean the behaviour of a random variable that occurs at the sample threshold level and is seen using intense data and insights. In the reference [16], the authors defined the GuD, alternatively known as extreme value distribution-type I (γ = 0), as the predominant model for quantifying extreme occurrences such as flood frequency analysis, droughts, earthquakes, sea currents and wind speed in order to understand the trajectory, magnitude, and pattern of complex phenomena. Environmental sciences, geology, accelerated life testing, meteorology, risk assessment and epidemiology are just a few of the a set of diverse fields where it might well be utilized. The authors in [17] showed that the score statistics of global sequence alignment follows a Gumbel distribution. In the reference [18], a comprehensive list of real life scenarios to which GuD can be applicable is provided by the authors. To learn more, see references [19–24].
Earliest generalizations of GuD was reported by [25] by introducing a shape parameter to Gumbel distribution. [26] provided a generalization of GuD based on the asymptotic distribution of the mth extreme, tracing back to [27]. [28] emphasized on a trivial choice of distribution since the GuD with only location and scale parameter yields narrower confidence intervals and has also the danger of underestimating the level of return. In the reference [29], a unique modification of GuD was proposed. It is based on the logarithmic transformation of an odd Weibull variable and is defined as
| (1) |
where Δ = e(x−μ)/σ, −∞ ≤ x ≤ + ∞, −∞ ≤ μ ≤ + ∞ and 0 ≤ βσ ≤ + ∞.
Since then, researchers adopted a more formalistic approach to generalize GuD. Some notable generalizations include [30] to define Beta-Gumbel (BGu), [31] to propose Kumaraswamy-gumbel (KumGu), [32] to define exponentiated-Gumbel (EGu), [33] to define exponentiated-generalized Gumbel (EGGu), just to mention some.
[14] proposed a simplified approach to generalize any continuous distribution viz. a viz. the transformed-transformer (T-X) family, which has become an indispensable part of modern distribution theory. Let z(t) be the probability density function (pdf) and Z(t) be the cumulative distribution function (cdf) of a random variable (av) T such that tϵ(a1, a2) with support −∞ < a1 < a2 < ∞. Let W[Z(x)] act as generator function of the cdf Z(x) of any baseline av such that K[Z(x)] is differentiable and increasing, lies in the defined range, i.e. a1 ≤ K[Z(x)] ≤ a2 such that when x → −∞ as K[Z(x)] → a1 and x → + ∞ as K[Z(x)] → a2.
| (2) |
The pdf corresponding to Eq (2) is
| (3) |
To generalize any continuous distribution, the methodology defined by the cdf in Eq (2) has become indispensable part of modern distribution theory. In the same vein, Al-Aqtash et al. [34] introduced the Gumbel-X family of distributions. Let z(t) be the density function (pdf) and Z(t) be the distribution function (cdf) of an arbitrary variable (av) T such that tϵ(a1, a2) with support −∞ < a1 < a2 < ∞. Let K[Z(x; φ)] = log [(Z(x; φ))/1−(Z(x; φ))] act as the generator function of the cdf of any baseline av such that K[Z(x; φ)] fulfills the defined criterion. For μ = 0, the cdf of Gumbel-X family is given as
| (4) |
where Δ = Z(x)/(1 − Z(x)).
This study introduces a new class of distributions following the T-X methodology, viz. a viz. the exponentiated Gumbel-G (EGuG) family of distributions. This is achieved by replacing the link function K[Z(x; φ)] = log[−log(1 − Z(x; φ))] in T-X family. It is worthy to remember that the link function log[−log(1 − Z(x; φ))] = log[−log(Z(x; φ))] and either of the mentioned form can be employ. EGuG family has thus far not been reported in the literature. We choose EGu distribution to define new family due to its superiority over the ordinary GuD because of presence of shape parameter θ that entails the improvements in tail of the distribution. Moreover, to the best of our knowledge, majority of the extreme value theory literature is supported by data from meteorology, geology, seismology, and hydrology (see references [15, 18–22, 24, 30–34]). The health implications of climate-related shifts in extreme event exposure, on the other hand, have not been explored. This study’s theoretical investigation will presumably fill this void in existing literature. Additionally, log[−log(1 − Z(x; φ))] function involves double log transformation and cannot be employed on GuD, which somehow makes the link function redundant. Following the success of the proposed generator to generalize Logistic and Normal distributions, we use this generator to define EGuG distribution. We study some of its mathematical properties and provide general properties and application of one of its specific model.
This article is outlined as follows: In Section 2, we define the EGuG family and present some of its special models. In order to optimise the structure of the generalisation being proposed, we provide the linear representation of EGuG density along with some of the mathematical properties of the family such as shapes of density and hazard rate function, moments and generating function, order statistics and estimation of model parameters. In Section 3, we choose Nadarajah-Haghighi (NH) distribution as baseline model to form EGuNH distribution whose mathematical properties as well as some risk measures are established. A simulation study is also conducted for some parametric combinations. Section 4 comprises of the numerical illustrations based on three life data sets. In Section 5, the article’s concluding thoughts are summed up.
2 The EGuG family
Let T follows the EGu av with μ = 0 and shape parameters θ ≥ 0 and σ ≥ 0, say EGu (θ, σ), then its cdf is given by
| (5) |
The corresponding pdf to Eq (5) is given as
| (6) |
For any baseline distribution with cdf Z(x; Φ) and pdf z(x; Φ) = dZ(x; Φ)/dx, the cdf of EGuG family is given as
| (7) |
where θ, σ are shape parameters and φ is the vector of baseline parameter.
The pdf corresponding to Eq (7) is given by
| (8) |
where Z(x; φ) is the baseline cdf and z(x; φ) is the baseline pdf. Furthermore, the dependence on the vector φ of the parameters might be omitted at times and simply write Z(x) = Z(x; φ) and z(x) = z(x; φ). Henceforth, X ∼ EGu−G(θ, σ; φ) denotes an av having density Eq (8).
The survival function (sf), hazard rate function (hrf) and cumulative hazard rate function (chrf) of this new family are, respectively, given by
and
Simulating the EGuG family is simply done by inverting Eq (7) as follows: If U has a uniform U(0, 1) distribution, then
| (9) |
has the density function Eq (8), where QZ(.) = Z−1(.) is the baseline quantile function (qf).
2.1 Special models
Eq (7) can be useful in modelling real life survival data with different shapes of hrf. Table (1) lists −log[Z(x; φ)] and the associated parameters for some special distributions.
Table 1. Distributions and corresponding −log[Z(x; φ)] functions.
| Distribution | −log[Z(x; φ)] | φ |
|---|---|---|
| Burr XII (x > 0) | −log[1 − (1 + xa)−b] | (a, b) |
| Weibull (x > 0) | (a, b) | |
| Normal (−∞ < x < ∞) | (μ, σ) | |
| Nadarajah Haghighi (x > 0) | (α, λ) | |
| Rayleigh (x > 0) | (a) | |
| Exponential (x > 0) | (α) | |
| Power function (0 < x < a) | (a, b) | |
| Fréchet (x > 0) | (λ, σ) | |
| Inverted Rayleigh (x > 0) | (λ, σ) | |
| Burr III (x > 0) | z log[1 + x−c] | (c, z) |
| Pareto (δ < x < ∞) | −log[1 − (δ/x)λ] | (δ, λ) |
Here three special models of EGuG family of distribution are defined.
2.1.1 EGu-Weibull(EGuW)
The EGu-W model is defined from Eq (7) by taking Z(x; φ) = 1 − exp{−axb}, z(x; φ) = abxb−1 exp{−axb}, as cdf and pdf of the baseline Weibull distribution with a, b > 0, respectively.
The cdf and pdf of EGu-W distribution are, respectively, given by
and
where θ, σ and b are shape parameters while a is scale parameter.
2.1.2 EGu-BurrXII(EGuBXII)
Let us consider the parent distribution as BXII with power parameters a, b > 0 by taking Z(x; φ) = 1−(1 + xa)−b, z(x; φ) = abxa−1(1 + xa)−b−1 be the cdf and pdf of the distribution.
The cdf and pdf of EGu-BXII distribution are, respectively, given by
and
| (10) |
where θ, σ, a and b are shape parameters.
2.1.3 EGu-Nadarajah Haghighi(EGuNH)
Consider to take Nadaraah Haghighi (NH) as baseline distribution with cdf as Z(x; φ) = [1 − exp{1−(1 + λx)β}] and pdf as z(x; φ) = λβ(1 + λx)β−1exp{1 − (1 + λx)β}. Then, the cdf and pdf of EGuNH reduces to
| (11) |
and
| (12) |
where θ, σ and α are shape parameters while λ is scale parameter.
2.2 Useful expansion for the EGuG cdf
We provide a useful expansion for Eq (7) in terms of linear combinations of exp-G distribution. For a random baseline cdf Z(x), an av is said to have the exp-G distribution having parameter ℓ such that ℓ > 0, say Y ∼ exp-G (ℓ), if its pdf and cdf are given as
respectively. Thus, several properties of the proposed model can be derived from those properties of the exp-G distribution studied by the authors in [3–8], to mention few.
By expanding Eq (7) using binomial and power series expansion, the resultant expression is given
| (13) |
Using Mathematica software, it can be verified that we can start the limit of integers (i, j) from 1 instead of 0 in above equation. Further, we can write [−log Z(x; φ)]−i/σ as since .
Now consider, for any real parameter c and zϵ(0, 1), the following formula holds:
| (14) |
where P0(c) = 1/2; P1(c) = c(3c + 5)/24; P2(c) = c(c2 + 5c + 6)/48 etc is the stirling’s polynomial. Then, the cdf F(x) in Eq (13) can be expressed (using Eq (14) and generalized binomial expansion) as
| (15) |
where .
By differentiating Eq (15), we obtain
| (16) |
where hm(x) = mZm−1(x; Φ) z(x; Φ) is the exp-G density function with power parameter m.
2.3 Shapes of density and hazard function
Analytical descriptions of density and hrf forms are conceivable. The roots of the equation represent the EGuG density’s critical points:
| (17) |
The equation is used to find the EGuG hrf’s crucial points.
| (18) |
Any numerical software can be used to examine Eqs (17) and (18) to determine the local maximum and minimum and inflexion points.
2.4 Moments
The first formula for the sth moment of X follows from Eq (16) as
| (19) |
where Setting s = 1 in Eq (refrthmoment1pdfmix1) can provide explicit expression for the mean of several parent distributions.
A second alternative formula for is obtained from Eq (19) in terms of the baseline qf as
| (20) |
where
The central moments (μt) and cumulants (κt) of X can follow from Eq (19) as and , respectively, where . The skewness and kurtosis can be calculated from the third and fourth standardized cumulants.
The sth incomplete moment of X can be determined from Eq (16) as
| (21) |
The last integral can be computed for most G distributions.
A crucial applicability of the first incomplete moment m1(⋅) has to do with the Bonferroni and Lorenz curves, which are extremely beneficial in a variety of fields. For a given probability π, they are given by and , respectively, where m1(y) comes from Eq (refincompletepdfmix1) with s = 1 and q = Q(π) follows from Eq (9). The Lorenz and Bonferroni curve for EGuNH are displayed graphically (Figs 13 and 14, subsequently).
The totality of excursions from the mean and median is used to estimate the degree of scatter in a population and is defined by and , respectively, where is the mean and M = Q(0.5) is the median. These measures can be expressed as and , where is given by Eq (refcdfEGuG).
The moment generating function (mgf) of X can be expressed as
where Mm(t) is the mgf of Ym. Hence, M(t) can be determined from the exp-G generating functions.
2.5 Order statistics
Order statistics are used in a wide range of statistical theory and practise. Let X1, …, Xn is a random sample from the EGu-G distribution and Xi: n denote the ith order statistic. Then, pdf of Xi:n can be written as
Inserting Eq (refcdfEGuG) and Eq (refpdfEGuG) in the last equation, and expanding it as in section (3.1), we get
| (22) |
where
and
2.6 Estimation
The three alternate approaches for inference are point estimation, interval estimation, and hypothesis tests. Several approaches for parameter point estimation have been published in the literature, the most extensively utilised of which is the maximum likelihood method. MLEs (maximum likelihood estimates) have properties that can be used to construct confidence ranges for model parameters. Large sample theory provides simple approximations that work well in repeated sampling for these estimations. The normal approximation for MLEs can be tackled analytically or computationally in distribution theory.
We use the optimum likelihood method to estimate the unknown parameters of the new distribution. Let x1, ⋯, xn be n observations from the EGu-G family given by Eq (8) with parameter vector Θ = (θ, β; Φ)⊤. The log-likelihood ℑ = ℑ(Θ) for Θ is given by
| (23) |
Eq (refmleegug) can be maximized either directly by using the R (optim function), SAS (NLMixed procedure) or Ox (MaxBFGS function), or then by solving the nonlinear likelihood equations by differentiating it. The components of the score vector U(Θ) are
Setting these equations to zero and solving them simultaneously yields the MLEs of the family parameters.
The observed information matrix for the parameter vector Θ = (θ, σ, Φk)⊤ is given by
whose elements can be determined by using any mathematical software. Under normal conditions of regularity, the multivariate normal distribution, where is the observed information analysed at , can be used to estimate confidence ranges for model parameters. Furthermore, we may use likelihood ratio (LR) statistics to assess the EGuG model to any of its specific models.
3 Properties of EGuNH
In comparison to Gamma, Weibull, and exponentiated exponential distributions, NH distribution (also known as extended exponential distribution) is the preferred option for zero inflated data. The cdf and pdf of NH distribution has already been defined in Section (3.1.3). For λ = 1, we define the cdf and pdf of EGuNH distribution as
| (24) |
and
| (25) |
where C = 1 − (1 + x)α and θ, σ and α are shape parameters.
Henceforth, we denote by X a av having density (25). The sf and hrf of X has the form
and
3.1 Shapes of density and hazard rate function of EGuNH distribution
The crucial points of the pdf of X are obtained from the equation:
Similarly, the critical points of the hrf of X are obtained from the equation:
Some plots of the density of EGuNH for selected parameter values are presented in Figs 1–4 while plots of the hrf of EGuNH for random parameter values are presented (Figs 5–8). It is apparent that the density of EGuNH can be reversed-J, unimodal, and symmetrical. Similarly, EGuNH hazard rate shapes may tend to be increasing, decreasing, bathtub, or upside-down bathtub. The new model is much superior at fitting data sets in a variety of risk evaluation scenarios.
Fig 1. Plots of EGuNH density for a variety of parameter combinations.
Fig 4. Plots of EGuNH density for a variety of parameter combinations.
Fig 5. Plots of EGuNH hazard rate for a variety of parameter combinations.
Fig 8. Plots of EGuNH hazard rate for a variety of parameter combinations.
Fig 2. Plots of EGuNH density for a variety of parameter combinations.
Fig 3. Plots of EGuNH density for a variety of parameter combinations.
Fig 6. Plots of EGuNH hazard rate for a variety of parameter combinations.
Fig 7. Plots of EGuNH hazard rate for a variety of parameter combinations.
3.2 Central properties of EGuNH distribution
In this section, some useful expressions for the linear expansion, moments and incomplete moments of EGuNH distribution have been deduced using the Eq (16).
Proposition 1.
| (26) |
where
. Recalling the result defined in Eq (15) as
A straightforward differentiation of the above result yields density by
| (27) |
The result in (27) is the linear expansion of NH densities. Hence, we shall derive several core properties of EGuNH using the major result of Eq 27.
Proposition 2.
Let W be a av with density ψ(x;m, α). Then, several properties of W can follow from those of X. The sth ordinary moment of X can be written as
| (28) |
where and
Utilizing the results derived in Eq (19), the sth moments are defined in (28).
Proposition 3.
The sth incomplete moment expression can be written as
| (29) |
where
Following the results defined in Eq (21), the sth incomplete moments are defined in (29). The skewness and kurtosis of X can be calculated from the third and fourth standardized cumulants. The classical skewness (Fig 9) and kurtosis plots (Fig 10) of the EGuNH distribution are displayed. Additionally, we provide the graphical illustration of MacGvillary skewness (MGs), which is based on quantile approach, in Figs 11 and 12. These plots reveal that the parameters θ and σ play a decisive role in modeling the skewness and kurtosis behaviors of X.
Fig 9. Bowley skewness of EGuNH.
Fig 10. Moors kurtosis of EGuNH.
Fig 11. MacGillivray skewness of EGuNH for a variety of parameter combinations.
Fig 12. MacGillivray skewness of EGuNH for a variety of parameter combinations.
MacGillivray [35] proposed another method to evaluate the skewness measure based on the qf and is defined as
where uϵ(0, 1), Q(.) is the qf defined in (32).
The MG skewness plots are very sensitive for extremely small values of parameter θ and σ which certainly signifies longer tails of EGuNH. Likewise, plots of the Lorenz (Fig 13) and Bonferroni (Fig 14) curves of EGuNH distribution for some random values are displayed. These plots reveal how the distribution parameters affect inequality measures which can be used to establish some orderings, an essential feature for applied statisticians. Some descriptive statistics related to EGuNH are presented in Tables 2 and 3, respectively.
Fig 13. Plots of the Lorenz curves of EGuNH distribution.
Fig 14. Plots of the Bonferroni curves of EGuNH distribution.
Table 2. Descriptive measures of EGuNH for some parameter values.
| Parameter values | Descriptives | ||||
|---|---|---|---|---|---|
| (θ, σ, α) | Q1 | Q2 | Q3 | B | M |
| (2.1, 0.1, 1.5) | 0.275 | 0.294 | 0.311 | −0.060 | 1.248 |
| (2.1, 0.29, 1.5) | 0.255 | 0.310 | 0.361 | −0.041 | 1.231 |
| (85.5, 0.979, 0.85) | 1.990 | 2.182 | 2.395 | 0.053 | 1.241 |
| (6.5, 0.33, 1.35) | 0.39 | 0.436 | 0.482 | −0.003 | 1.234 |
| (1.5, 1.9, 2.15) | 0.012 | 0.189 | 0.451 | 0.192 | 0.866 |
| (0.91, 2.1, 1.5) | 0 | 0.048 | 0.497 | 0.805 | 1.314 |
| (1.7, 9.1, 1.5) | 0 | 0.604 | 2.536 | 0.524 | 0.757 |
| (0.7, 5.1, 1.3) | 0 | 1.394 | 1.996 | 0.667 | 1.757 |
| (1.7, 0.1, 0.85) | 0 | 0.314 | 2.267 | 0.609 | 0.888 |
Table 3. Moments and moment ratios of EGuNH for some parameter combinations.
| Parameter values | Moments and moments ratio | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (θ, σ, α) | E(x) | E(x2) | E(x3) | E(x4) | V(x) | σ(x) | CV | CS | CK |
| (0.83, 2.1, 1.5) | 0.225 | 0.233 | 0.241 | 0.299 | 0.141 | 0.401 | 1.322 | 1.589 | 4.253 |
| (0.91, 2.1, 1.5) | 0.295 | 0.263 | 0.291 | 0.365 | 0.176 | 0.420 | 1.422 | 1.680 | 4.944 |
| (1.7, 9.1, 1.5) | 1.343 | 4.258 | 15.684 | 63.478 | 2.456 | 1.567 | 1.167 | 1.680 | 4.944 |
| (2.1, 0.1, 1.5) | 0.292 | 0.086 | 0.026 | 0.008 | 0.001 | 0.027 | 0.094 | 2.505 | 6.661 |
| (2.1, 0.29, 1.5) | 0.306 | 0.100 | 0.034 | 0.012 | 0.006 | 0.078 | 0.254 | -0.285 | 2.957 |
| (1.5, 1.9, 2.15) | 0.597 | 0.767 | 1.219 | 2.205 | 0.410 | 0.640 | 1.072 | 88.455 | 47.815 |
| (6.5, 0.33, 1.35) | 0.436 | 0.194 | 0.089 | 0.041 | 0.005 | 0.068 | 0.157 | -0.044 | 3.206 |
| (85.5, 0.979, 0.85) | 2.163 | 4.764 | 10.965 | 24.462 | 0.088 | 0.297 | 0.137 | 0.437 | 3.289 |
3.3 Acturial measures EGuNH: Value at risk
The theory of finance is based upon risk evaluation. Investors are particularly interested to invest in entities in which there is minimum risk (specified with high degree of confidence) of losing money. In finance, value at risk (VaR) is the most extensively used metric for assessing liability. It is also known as quantile risk measure or quantile premium principle of the distribution of aggregate losses. It is characterised by a level of assurance “q” (usually at 95% and 99%). To a layman, VaR answers a simple question that “What is the worst case scenario that can happen in a particular investment?”
If X has pdf (25), then VaR is the qth quantile of its cdf (24), defined as
| (30) |
3.4 Acturial measures EGuNH: Expected shortfall
Despite of the popularity of VaR measures, there are many shortcomings (see [36]). To counter inherent problems in VaR, Artzner et al. [37, 38] proposed the use of expected shortfall (ES). Expected shortfall quantifies the average loss in states beyond the VaR level. ES has a number of aliases such as “conditional VaR”, “mean excess loss” or “tail VaR”. We define the ES as follows
| (31) |
For a combination of various parameter values, plots of VaRs (Fig 15) and ESs (Fig 16) are displayed respectively.
Fig 15. Plots of the VaR of EGuNH distribution for some random parameter values.
Fig 16. Plots of the ES of EGuNH distribution for some random parameter values.
3.5 Parameter estimation of EGuNH
The log-likelihood function ℑ for the vector of parameters Θ = (θ, σ, α)⊤ for the model defined in (25) is given by
The components of the score vector U(Θ) are
The MLE of Θ can also be obtained by solving the nonlinear equations Uθ = 0, Uσ = 0 and Uα = 0. Because these equations cannot be solved analytically, the estimates can be calculated numerically using statistical software.
3.6 Simulation study of EGuNH distribution
The qf of the EGuNH distribution has an explicit form as follows
| (32) |
Here, we use Monte Carlo simulations to demonstrate the performance and correctness of maximum likelihood estimations of the EGuNH parameters by inverting Eq (24) to generate a sample data from the model. The simulation study is perform for sample sizes n = 50, 100, 200, 500, and parameter combinations: I: θ = 0.2, σ = 0.75 and α = 0.5, II: θ = 2.2, σ = 0.45 and α = 0.5, III: θ = 3.4, σ = 0.75 and α = 1.5 and IV: θ = 3.4, σ = 1.35 and α = 1.5. This study is carried out for N = 2000 times, each with given n and computed the average estimates (AEs) as well as their average biases (Bias), mean squared errors (MSEs) and coverage probabilities (CPs) of the MLEs.
The AEs, Bias, MSEs and CPs for the parameters θ, σ and α are given in Tables (4)–(7). The empirical findings suggests that the bias and MSEs decreases as sample size increases. Further, the empirical CPs are quite close to the nominal level of 95%. As a result, MLEs and their approximate findings can be used to evaluate and build approximated confidence intervals of the EGuNH distribution parameters θ, σ and α.
Table 4. AEs, Biases, MSEs and CPs for combination-I.
| n = 50 | n = 100 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 0.279 | 0.081 | 0.447 | 0.99 | 0.274 | 0.074 | 0.436 | 0.98 |
| σ | 0.927 | -1.661 | 0.691 | 1.00 | 0.895 | -1.651 | 0.683 | 0.97 |
| α | 0.925 | -0.241 | 0.494 | 0.98 | 0.817 | -0.203 | 0.441 | 0.92 |
| n = 200 | n = 500 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 0.272 | 0.072 | 0.429 | 0.96 | 0.272 | 0.051 | 0.297 | 0.94 |
| σ | 0.784 | -1.350 | 0.669 | 0.95 | 0.757 | -1.115 | 0.662 | 0.95 |
| α | 0.629 | -0.220 | 0.359 | 0.95 | 0.548 | -0.204 | 0.320 | 0.96 |
Table 7. AEs, Biases, MSEs and CPs for combination-IV.
| n = 50 | n = 100 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 3.877 | -0.281 | 0.097 | 0.97 | 3.595 | -0.223 | 0.019 | 0.92 |
| σ | 0.889 | 0.021 | 0.025 | 1.00 | 0.827 | 0.010 | 0.020 | 0.94 |
| α | 2.541 | 0.058 | 0.033 | 0.97 | 2.230 | 0.030 | 0.028 | 0.99 |
| n = 200 | n = 500 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 3.495 | -0.019 | 0.010 | 0.96 | 3.410 | -0.013 | 0.009 | 0.95 |
| σ | 0.765 | 0.005 | 0.016 | 0.97 | 0.751 | 0.003 | 0.008 | 0.96 |
| α | 1.915 | 0.015 | 0.014 | 0.95 | 1.507 | 0.011 | 0.010 | 0.95 |
Table 5. AEs, Biases, MSEs and CPs for combination-II.
| n = 50 | n = 100 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 2.307 | -0.031 | 0.027 | 1.00 | 2.197 | -0.023 | 0.012 | 0.92 |
| σ | 0.499 | -0.021 | 0.011 | 0.91 | 0.495 | -0.005 | 0.002 | 0.98 |
| α | 0.511 | 0.018 | 0.018 | 0.99 | 0.510 | 0.010 | 0.005 | 0.93 |
| n = 200 | n = 500 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 2.196 | -0.014 | 0.002 | 0.97 | 2.198 | -0.011 | 0.001 | 0.95 |
| σ | 0.501 | 0.001 | 0.001 | 0.95 | 0.498 | 0.001 | 0.001 | 0.94 |
| α | 0.508 | 0.008 | 0.003 | 0.94 | 0.501 | 0.000 | 0.002 | 0.95 |
Table 6. AEs, Biases, MSEs and CPs for combination-IV.
| n = 50 | n = 100 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 3.507 | -0.201 | 0.027 | 0.98 | 3.405 | -0.123 | 0.019 | 0.93 |
| σ | 0.769 | 0.021 | 0.015 | 0.97 | 0.761 | 0.010 | 0.013 | 0.99 |
| α | 2.541 | 0.058 | 0.023 | 0.90 | 2.130 | 0.030 | 0.018 | 0.97 |
| n = 200 | n = 500 | |||||||
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| AEs | Bias | MSEs | CPs | AEs | Bias | MSEs | CPs | |
| θ | 3.405 | -0.014 | 0.010 | 0.96 | 3.401 | -0.011 | 0.007 | 0.96 |
| σ | 0.755 | 0.005 | 0.006 | 0.94 | 0.753 | 0.003 | 0.004 | 0.95 |
| α | 1.615 | 0.015 | 0.004 | 0.96 | 1.511 | 0.011 | 0.002 | 0.94 |
4 Applications of the EGuNH distribution
Statistical methods that fail to account for all of the uncertainties in the model are prone to produce an overly optimistic assessment of future extremes, are frequently contradicted by observed extreme events in a variety of scientific fields. The current literature regarding extreme value theory is full of such models in which data sets are meteorology data such as earthquakes, floods, rains, droughts, hurricanes etc. (see [15–32]). On the contrary, health hazards is an area of extreme value theory which should be explored. Death, damage, or disease; exacerbation of underlying medical disorders; and negative effects on mental health are some of the health hazards of climate-related increases in exposure to extreme occurrences.
In this section, we provide some applications of the EGuNH model on three real life phenomenons, two of which related to health hazards in extreme value theory. We estimate the unknown parameters of the distributions by the principal of maximum likelihood. We compute the log-likelihood function evaluated at the MLEs () using the method of a limited-memory quasi-Newton code for bound-constrained maximization (L-BFGS-B). In order to select the best probability model, a variety of criteria for evaluating information (ICs) can be considered. We considered the following well-known ICs: the maximized log-likelihood (), Akaike Information criterion (AIC), Anderson-Darling (A⋆), Cramér-von Mises (W⋆) and Kolmogorov-Smirnov measures (D⋆; P-value (p⋆)), where lower values of all these statistics except higher p⋆ values of K-S, indicate good fits. The required computations are carried out using the R script AdequacyModel which is freely accessible from http://cran.r-project.org/web/packages/AdequacyModel/AdequacyModel.pdf.
The fits of the EGuNH distribution is compared with other competitive models which are given in Table 8. The parameters are all positive real numbers of these densities.
Table 8. The comparative fitted models.
4.1 Meteorology data
Meteorological phenomena are weather events that most individuals are affected by, due to changes in extreme weather and climatic events, such as earthquakes, heat waves, floods, hurricanes, droughts etc. The present data is taken from [44], denoted by D1, gives the time in days between successive serious earthquakes world-wide. An earthquake is included if its magnitude was at least 7.5 on the Richter scale, or if over 1000 people were killed. There were 63 earthquakes recorded altogether, and so 62 recorded waiting times. The data are: 840, 157, 145, 44, 33, 121, 150, 280, 434, 736, 584, 887, 263, 1901, 695, 294, 562, 721,40, 1336, 335, 1354, 454,139, 780, 203, 436, 30, 246, 1617, 638, 937, 735,76, 710, 36, 667,384, 129, 46, 402, 194, 40, 556, 99, 9, 209, 599, 38, 365, 92, 82, 220, 759, 304, 83, 319, 375, 832, 460, 567, 328.
4.2 Cancer data
According to [45], extreme events have the potential to disrupt the delivery of cancer care. For example, some deadly carcinogens may be released into communities as a result of hurricanes and wild fires; industry shutdowns may result in a shortage of life-saving medical equipment in hospitals, causing shortages in cancer facilities across the country; and infrastructure collapse may limit access to patients undergoing cancer therapies. The following two data, denoted by cancer 1 (D2) and cancer 2 (D3) are related to cancer patients.
Cancer data 1. The survival times, in weeks, of 33 patients who succumbed to Acute Myelogenous Leukemia are the subject of D2. This data was recently studied by the authors in [46] The data are: 65, 156, 100, 134, 16, 108, 121, 4,39, 143, 56, 26, 22, 1, 1, 5, 65, 56, 65, 17, 7, 16, 22, 3, 4, 2, 8, 4, 3, 30, 4,43.
Cancer data 2. D3 signifies the number of patients suffering from blood cancer. The Saudi Cancer Registry (SCR) provides such information, covering the time period from 1994 to the present day. The data is extracted from a report [47] which concerns an overview of cancer incidence statistics for Saudi Arabia in 2012. The data are: 1277, 1290, 1357, 1369, 1408, 1455, 1478, 1549, 115, 181, 255, 418, 441, 461, 516, 739, 743, 789, 807, 865, 924, 983, 1024, 1062, 1063, 1165, 1191, 1222, 1222, 1251, 1578, 1578, 1599, 1603, 1605, 1696, 1735, 1799, 1815, 1852. The descriptive statistics for each of the three data sets are given in Table 9.
Table 9. The descriptive statistics related to D1, D2 and D3.
| Data | Sample Size | Arithmetic Mean | Standard Deviation | Lowest | Highest | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|
| 1 | 62 | 437.21 | 399.93 | 9 | 1901 | 1.50 | 2.52 |
| 2 | 32 | 42.07 | 46.95 | 1 | 156 | 1.12 | 0.03 |
| 3 | 40 | 1137 | 481.60 | 115 | 1852 | -0.49 | -0.73 |
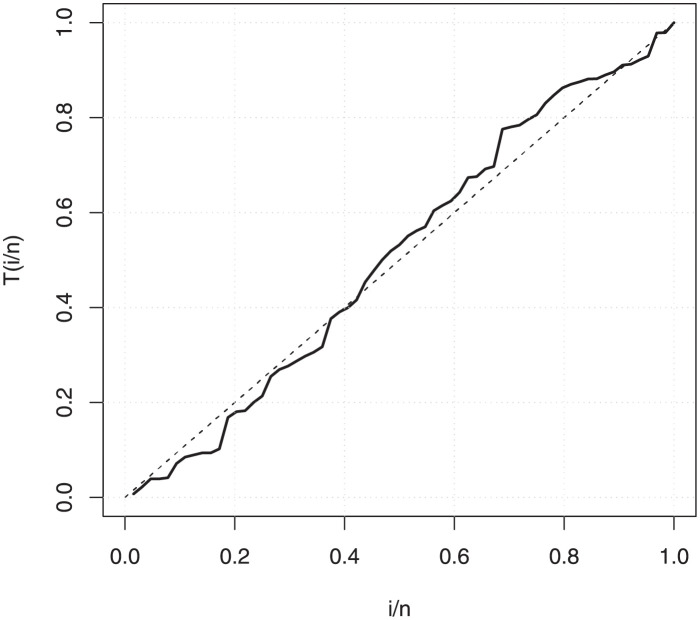
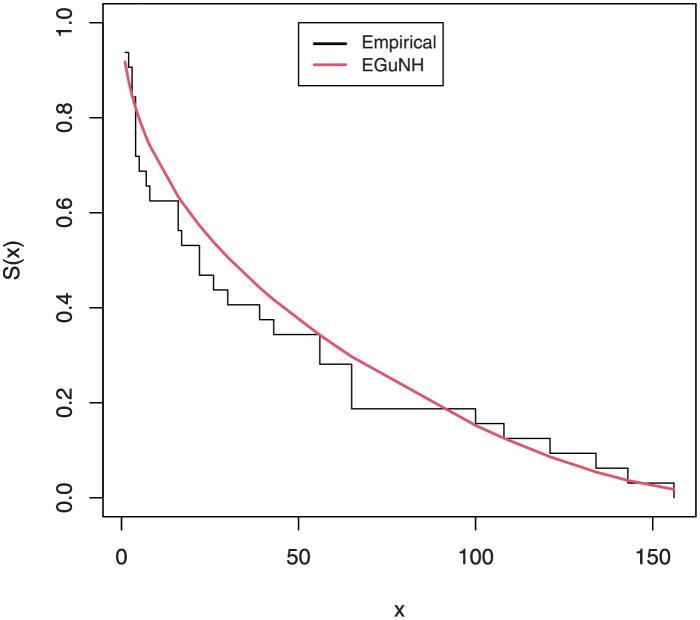
The empirical findings of all the three data are suggestive of the heavy tailed data. The TTT plots Figs (17)–(19) for the data sets are given. In particular, the TTT plots show bathtub, increasing and decreasing hrf, allowing us to fit EGuNH model on these data sets. The approximated hrf Figs (20)–(22) for each data point correlates to the TTT graphs. Table 10 summarizes the results of the MLEs and their related standard errors (in parentheses) of the model parameters for the proposed model while the ICs are listed in Table 11 for the D1, D2 and D3, respectively. It is customary to supplement the analytical result defined in Tables 9 and 10, by displaying it graphically. Hence, the estimated pdfs Figs (23)–(25), PP–plots Figs (26)–(28), estimated cdfs Figs (29)–(31) and estimated sfs Figs (32)–(34) for the three data sets are given. On the given data sets, the numerical values authenticates that the EGuNH model provides the best fit as compared to the other models.
Fig 17. TTT plot for D1.
Fig 19. TTT plot for D3.
Fig 20. Estimated hrf for D1.
Fig 22. Estimated hrf plots for D3.
Table 10. MLEs with their respective SEs (in parenthesis) for D1, D2 and D3.
| Data 1 | Data 2 | Data 3 | |
|---|---|---|---|
| Distribution | MLEs | MLEs | MLEs |
| EGuNH | 3.655,11.787,0.275 | 0.071,0.031,0.076 | 22.753,20.069,0.469 |
| (θ,σ,α) | (2.217),(2.371),(0.073) | (0.015),(0.003),(0.001) | (5.093),(2.193),(0.011) |
| ENH | 1.285,0.764,0.004 | 1.256,1.716,0.011 | 4.515,0.971,0.002 |
| (θ,α,λ) | (0.229),(0.117),(0.001) | (0.178),(0.556),(0.005) | (0.987),(0.066),(0.002) |
| GaNH | 0.295,7.164,3.874 | 1.6,1.256,0.013 | 1.146,3.802,0.002 |
| (θ,aα,λ) | (1.018),(6.192),(2.987) | (0.542),(0.174),(0.007) | (0.094),(0.615),(0.518) |
| LxNH | 8.918,0.080,18.577 | 1.470,3.660,0.006 | 13.499,0.111,0.456 |
| (θ,α,λ) | (3.211),(0.027),(58.410) | (0.126),(0.930),(0.002) | (3.672),(0.025),(0.643) |
| TLNH | 1.499,0.600,0.003 | 1.374,1.5490.007 | 11.215,0.371,0.009 |
| (θ,α,λ) | (0.262),(0.084),(0.987) | (0.173),(0.405),(0.003) | (6.699),(0.072),(0.017) |
| MONH | 5.162,0.886,0.003 | 3.067,0.962,0.038 | 42.021,0.827,0.005 |
| (θ,α,λ) | (0.559),(0.131),(0.001) | (0.159),(0.560),(0.055) | (2.127),(0.081),(0.311) |
| NH | 1.054,0.212 | 2.616,0.006 | 0.067,158.079 |
| (α,λ) | (0.104),(0.298) | (0.728),(0.012) | (0.095),(89.711) |
Table 11. The statistics , AIC, BIC, A⋆, W⋆, D⋆ and p⋆ for D1, D2 and D3.
| Distribution | AIC | BIC | A ⋆ | W ⋆ | D ⋆ | p ⋆ | |
|---|---|---|---|---|---|---|---|
| Data set 1 | |||||||
| EGuNH | 441.19 | 889.90 | 896.33 | 0.41 | 0.05 | 0.08 | 0.79 |
| GaNH | 444.78 | 895.56 | 901.94 | 1.05 | 0.17 | 0.11 | 0.54 |
| LxNH | 448.86 | 903.71 | 910.14 | 1.58 | 0.25 | 0.18 | 0.45 |
| TLNH | 442.73 | 891.24 | 897.67 | 0.61 | 0.09 | 0.10 | 0.68 |
| ENH | 442.67 | 890.98 | 897.74 | 0.46 | 0.07 | 0.09 | 0.77 |
| MONH | 441.90 | 890.14 | 897.01 | 0.34 | 0.05 | 0.09 | 0.78 |
| NH | 442.27 | 891.75 | 897.15 | 0.31 | 0.06 | 0.08 | 0.78 |
| Data set 2 | |||||||
| EGuNH | 149.027 | 304.054 | 308.451 | 0.567 | 0.08 | 0.111 | 0.817 |
| GaNH | 150.203 | 306.406 | 310.803 | 0.588 | 0.09 | 0.122 | 0.729 |
| LxNH | 152.672 | 311.345 | 315.741 | 0.860 | 0.14 | 0.126 | 0.687 |
| TLNH | 150.104 | 306.205 | 310.602 | 0.565 | 0.08 | 0.127 | 0.681 |
| ENH | 150.101 | 306.202 | 310.599 | 0.581 | 0.08 | 0.126 | 0.692 |
| MONH | 150.036 | 306.071 | 310.468 | 0.593 | 0.09 | 0.143 | 0.534 |
| NH | 152.875 | 310.751 | 312.005 | 0.636 | 0.10 | 0.159 | 0.487 |
| Data set 3 | |||||||
| EGuNH | 307.586 | 621.173 | 626.240 | 1.401 | 0.226 | 0.151 | 0.317 |
| ENH | 310.775 | 627.566 | 632.616 | 1.912 | 0.319 | 0.199 | 0.084 |
| GaNH | 308.249 | 622.499 | 627.566 | 1.494 | 0.243 | 0.155 | 0.284 |
| LxNH | 313.300 | 632.601 | 637.668 | 2.176 | 0.369 | 0.172 | 0.183 |
| TLNH | 316.845 | 639.691 | 644.757 | 2.868 | 0.500 | 0.184 | 0.131 |
| MONH | 307.698 | 622.019 | 627.107 | 1.626 | 0.235 | 0.161 | 0.289 |
| NH | 400.885 | 805.770 | 809.184 | 2.518 | 0.433 | 0.607 | 0.000 |
Fig 23. Estimated density for D1.
Fig 25. Estimated density plot for D3.
Fig 26. PP plot for D1.
Fig 28. PP plot for D3.
Fig 29. Estimated cdf plot for D1.
Fig 31. Estimated cdf plot for D3.
Fig 32. Estimated sf plot for D1.
Fig 34. Estimated sf plot for D3.
Fig 18. TTT plot for D2.
Fig 21. TTT plot for D2.
Fig 24. Estimated plots of density for D2.
Fig 27. PP plot for D2.
Fig 30. Estimated cdf plot for D2.
Fig 33. Estimated sf plot of density for D2.
The variance-covariance matrices of the MLEs of the EGuNH distribution for D1 is
The variance-covariance matrices of the MLEs of the EGuNH distribution for D2 is
The variance-covariance matrices of the MLEs of the EGuNH distribution for D3 is
4.2.1 Numerical calculations of VaRs and ESs
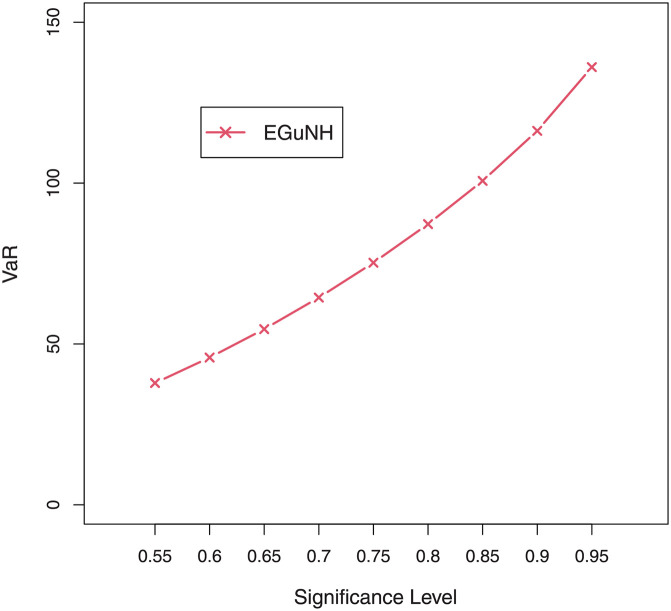
We were able to further investigate EGuNH’s application to these risk measures thanks to the results reported in Section 4. To quantify the volatility associated with these measures, we take the values of MLEs of D1, D2 and D3., respectively, from Table 11. Higher risk measures indicate heavier tails, while lower risk measures indicate a model with a much lighter tail tendency. It’s pertinent to mention that the EGuNH model yielded significantly more impressive results than others, implying that the model has a longer tail. The numerical findings of VaRs and ESs for data 1, data 2, and data 3 of the proposed model at respective level of significance (LoS) are shown in Table 12. The summarized output of these risk measures (VaRs in Figs 35–37 and ESs in Figs 38–40), graphically, for the reader’s expedience.
Table 12. Numerical measures of VaRs and ESs of EGuNH for D1, D2 and D3.
| LoS | Data 1 | Data 2 | Data 3 | |||
|---|---|---|---|---|---|---|
| VaRs | ESs | VaRs | ESs | VaRs | ESs | |
| 0.55 | 327.4119 | 146.4568 | 37.86083 | 12.29339 | 1125.354 | 727.9202 |
| 0.60 | 373.4939 | 163.4189 | 45.77802 | 14.74769 | 1201.044 | 764.1592 |
| 0.65 | 425.7544 | 181.5435 | 54.61479 | 17.46849 | 1282.559 | 800.8571 |
| 0.70 | 486.2076 | 201.0896 | 64.41790 | 20.46624 | 1372.164 | 838.4086 |
| 0.75 | 557.9849 | 222.4155 | 75.25685 | 23.75171 | 1473.216 | 877.2859 |
| 0.80 | 646.3563 | 246.0443 | 87.25890 | 27.33904 | 1591.174 | 918.1102 |
| 0.85 | 761.2607 | 272.8042 | 100.69662 | 31.25081 | 1736.442 | 961.7964 |
| 0.90 | 925.2607 | 304.1764 | 116.24233 | 35.52842 | 1927.397 | 1009.9029 |
| 0.95 | 1210.5273 | 343.4825 | 136.04737 | 40.26890 | 2210.153 | 1065.6140 |
Fig 35. Estimated VaRs for D1.
Fig 37. Estimated VaRs for D3.
Fig 38. Estimated ES for D1.
Fig 40. Estimated ES for D3.
Fig 36. Estimated VaRs for D2.
Fig 39. Estimated ES for D2.
5 Concluding remarks
We propose and study the EGuG model and obtain a wide range of mathematical and statistical modelling methods to characterise the model’s structural and dynamic aspects including properties such as quantile function, ordinary and incomplete moments, mean deviations, bonferroni and lorenz curves, generating function and order statistics. The parameters of the family are estimated by the method of maximum likelihood. An extended exponential distribution is taken as baseline model to propose EGuNH distribution. Some simulations are performed to check the asymptotic properties of the estimates. Three applications to real data set are presented to illustrate the potentiality of the proposed models. For future research, the proposed model can further be extended using compounding. We expect that the modification may facilitate in estimating analytically tractable Bayesian estimates of the reliability function under different priors.
Acknowledgments
The authors would seek this opportunity to thank the respected comments made by the reviewers which greatly help in the overall presentation of the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1. Tahir MH, Nadarajah S, Parameter induction in continuous univariate distributions: Well-established G families. An. Acad. Bras. Ciênc. (2015) 87: 539–568. doi: 10.1590/0001-3765201520140299 [DOI] [PubMed] [Google Scholar]
- 2. Azzalini A, A class of distributions which includes the normal ones. Scandanavian Journal of Statistics (1985)12: 171–178. [Google Scholar]
- 3. Mudholker GS, Srivastava DK, Exponentiated Weibull family for analyzing bathtub failure data. IEEE Transactions of Reliability (1993)42: 299–302 doi: 10.1109/24.229504 [DOI] [Google Scholar]
- 4. Marshall AW, Olkin I, A new method for adding parameters to a family of distributions with application to the exponential and Weibull families Biometrika 84 (1997), pp. 641–652. [Google Scholar]
- 5. Gupta RC, Gupta PL, Gupta RD, Modeling failure time data by Lehman alternatives Commun. Stat. Theory Methods 27 (1998), pp. 887–904. doi: 10.1080/03610929808832134 [DOI] [Google Scholar]
- 6. Gupta RD, Kundu D, Exponentiated exponential family: An alternative to gamma and Weibull distributions Biometrical Journal 42 (2001), pp. 117–130 doi: [DOI] [Google Scholar]
- 7. Eugene N, Lee C, Famoye F, Beta-normal distribution and its applications Commun. Stat. Theory Methods 31 (2002), pp. 497–512. doi: 10.1081/STA-120003130 [DOI] [Google Scholar]
- 8. Nadarajah S, Kotz S, The exponentiated type distributions Acta Applicandae Mathematica, 92 (2006a), pp.97–111. doi: 10.1007/s10440-006-9055-0 [DOI] [Google Scholar]
- 9. Gleaton JU, Lynch JD, Properties of generalized log-logistic families of lifetime distributions J. Probab. Stat. Sci. 4 (2006), pp. 51–64. [Google Scholar]
- 10. Gupta RD, Kundu D, Generalized exponential distribution Aust. N. Z. J. Stat. 41 (1999), 173–188. [Google Scholar]
- 11. Zografos K, Balakrishnan N, On families of beta- and generalized gamma generated distributions and associated inference Stat. Methodol. 6 (2009), pp. 344–362. doi: 10.1016/j.stamet.2008.12.003 [DOI] [Google Scholar]
- 12. Cordeiro GM, DeCastro M, A new family of generalized distributions J. Stat. Comput. Simul. 81 (2011), 883–898. doi: 10.1080/00949650903530745 [DOI] [Google Scholar]
- 13. Cordeiro GM, Ortega EMM, Cunha DCC, The exponentiated generalized class of distributions J. Data. Sci. 11 (2013), 1–27. doi: 10.6339/JDS.2013.11(1).1086 [DOI] [Google Scholar]
- 14. Alzaatreh A, Famoye F, Lee C, A new method for generating families of continuous distributions Metron 71 (2013), pp. 63–79. doi: 10.1007/s40300-013-0007-y [DOI] [Google Scholar]
- 15.Coles S, An Introduction to Statistical Modeling of Extreme Values Springer, NewYork, 2001.
- 16. Fisher RA, Tippet LHC, Limiting form of the frequency distribution of the largest or smallest number of the sample Proceedings of the Cambridge Philosophical society, (1928) 24:190–190. doi: 10.1017/S0305004100015681 [DOI] [Google Scholar]
- 17. Sardiu M, Alves G, Yu YK, Score statistics of global sequence alignment from the energy distribution of a modified directed polymer and directed percolation problem Physical Review E (2005), 72(6). doi: 10.1103/PhysRevE.72.061917 [DOI] [PubMed] [Google Scholar]
- 18. Kotz S, Nadarajah S, Extreme Value Distribution: Theory and Applications Imperial college Press; (2000), London. [Google Scholar]
- 19. Nadarajah S, Choi D, Maximum daily rainfall in South Korea J. Earth Syst. Sci., 116(4), August 2007, 311–320. doi: 10.1007/s12040-007-0028-0 [DOI] [Google Scholar]
- 20. De M, A new unbiased plotting position formula for the gumbel distribution Stochastic environmental research and risk assessment (2000), 14:1–7. [Google Scholar]
- 21. Solomon O, Prince O, Flood frequency analysis of Osse river using Gumbel’s distribution Civil and environmental research (2013), 3(10). [Google Scholar]
- 22. Clarke RT, Fitting and testing the significance of linear trends in Gumbel distributed data Hydrology and Earth system sciences (2002), 6(1):17–24. doi: 10.5194/hess-6-17-2002 [DOI] [Google Scholar]
- 23. Salinas V, Perez P, Gonzalez E, Vaquera H, Goodness of fit tests for the Gumbel distribution with type-II right censor data Revista Colombiana de Estadistica (2012), 35(3): 409–424. [Google Scholar]
- 24. Miladinovic B, Tsokos CP, Senstivity of the Bayesian reliability estimates for the modified Gumbel failure model International journal of Reliability, quality and safety engineering (2009), 16(4):331–341. doi: 10.1142/S0218539309003423 [DOI] [Google Scholar]
- 25.Ojo MO, On a generalization of the Gumbel distribution Statistics on the Internet (2000).
- 26. Adeymei S, Ojo MO, A generalization of the Gumbel distribution Kragujevac Journal of Mathematics (2003), 25: 19–29. [Google Scholar]
- 27. Gumbel EJ, Les valeurs extremes des distributions statistiques Ann. Inst. H Poincare, 5, (1934), 115. [Google Scholar]
- 28. Persson K, Ryden J, Exponentiated Gumbel distribution for the estimation of return levels of significant wave height Journal of Environmental Statistics (2010), 1(3): 1–12. [Google Scholar]
- 29. Cooray K, Generalized Gumbel distribution Journal of Applied Statistics (2010), 37(1): 171–179. doi: 10.1080/02664760802698995 [DOI] [Google Scholar]
- 30. Nadarajah S, Kotz S, The Beta-Gumbel distribution Mathematical Problems in Engineering (2004),4:323–332. doi: 10.1155/S1024123X04403068 [DOI] [Google Scholar]
- 31. Cordiero GM, Nadarajah S, Ortega EMM, The Kumaraswamy Gumbel distribution Statistical Methods Application (2012), 21: 139–168. doi: 10.1007/s10260-011-0183-y [DOI] [Google Scholar]
- 32. Nadarajah S, The exponentiated Gumbel distribution with climate application Environmetrics (2006), 17: 13–23. doi: 10.1002/env.739 [DOI] [Google Scholar]
- 33. Andrade T, Rodriguez H, Bourguignon M, Cordeiro GM, The exponentiated generalized Gumbel distribution Revista Colombiana de Estadistica (2015), 38(1): 123–143. doi: 10.15446/rce.v38n1.48806 [DOI] [Google Scholar]
- 34. Al-Aqtash R, Lee C, Famoye F, Gumbel-Weibull distribution: Properties and applications J. Mod. Appl. Stat. Methods (2014), 13:201–225. doi: 10.22237/jmasm/1414815000 [DOI] [Google Scholar]
- 35.Macgillivray HL, Skewness and Asymmetry: Measures and Orderings The Annals of Statistics, September 1986. 10.1214/aos/1176350046 [DOI]
- 36. McNeil AJ, Frey R, Embrechts P, Quantitative risk management: concepts, techniques and tools Princeton University Press; (2005). [Google Scholar]
- 37. Artzner P, Delbaen F, Eber JM, Heath D, Thinking Coherently Risk (1997) November, 10(11):68–71. [Google Scholar]
- 38. Artzner P, Application of coherent risk measures to capital requirements in insurance North American Actuarial Journal (1999), 3(2):11–25. doi: 10.1080/10920277.1999.10595795 [DOI] [Google Scholar]
- 39. Ramirez FAP, Guerraa RR, Canterleb DR, Cordeiro GM, The Logistic Nadarajah Haghighi distribution and its associated regression model for reliability applications Reliability Engineering and system safety (2020). doi: 10.1080/03610926.2014.980516 [DOI] [Google Scholar]
- 40. Sangsanit Y, Bodhisuwan W, The Topp-Leone generator of distributions: Properties and inferences Songklanakarin Journal of Science and Technology (2016), 38(5): 537–548. doi: 10.14456/sjst-psu.2016.69 [DOI] [Google Scholar]
- 41. Lemonte AJ, A new exponential-type distribution with constant, decreasing, increasing, upside-down bathtub and bathtub-shaped failure rate function Computational Statistical and Data Analysis (2013), 62:149–170. doi: 10.1016/j.csda.2013.01.011 [DOI] [Google Scholar]
- 42. Lemonte AJ, Cordeiro GM, Arenas GM, A new useful three-parameter extension of the exponential distribution Statistics (2016) 50:312–337. [Google Scholar]
- 43. Nadarajah S, Haghighi F, An extension of the exponential distribution Statistics: A Journal of Theoretical and Applied Statistics 45(6):543–558. doi: 10.1080/02331881003678678 [DOI] [Google Scholar]
- 44. Castillo E, Hadi AS, Balakrishnan N, Sarabia M, ExtremeValue and related models withApplications in engineering and science Wiley Science: A John Wiley Sons Publication [Google Scholar]
- 45. Nogueira LM, Sahar LE, Jason A, Jemal A, Yabroff KR, Association Between Declared Hurricane Disasters and Survival of Patients With Lung Cancer Undergoing Radiation Treatment JAMA (2019), 322(3):269–271. doi: 10.1001/jama.2019.7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jamal F, Abuzaid AH, Tahir MH, Nasir MA, Khan S, Mashwani WK, New Modified Burr III Distribution, Properties and Applications Math. Comput. Appl. 2021. 26(82). 10.3390/mca26040082 [DOI] [Google Scholar]
- 47. Bazarbashi S, Eid HA, Minguet J, Cancer Incidence in Saudi Arabia: 2012 Data from the Saudi Cancer Registry Asian Pacific Journal of Cancer Prevention (2017). doi: 10.22034/APJCP.2017.18.9.2437 [DOI] [PMC free article] [PubMed] [Google Scholar]