Abstract
We aimed to identify the prevalence of thermophilic species of Campylobacter in meats of different species available on the Brazilian commercial market and to determine the genetic diversity, antimicrobial resistance and virulence potential of the isolates. A total of 906 samples, including chicken, beef and pork carcasses and chicken and beef livers, were purchased in retail outlets, and prevalences of 18.7% (46/246), 3.62% (5/138), 10.14% (14/138), 3.62% (5/138) and 4.47% (11/132), respectively, were identified, evidencing the dissemination of genotypes in the main producing macro-regions. Of all isolates, 62.8% were classified as multidrug resistant (MDR), with resistance to amoxicillin-clavulanate (49.4%), tetracycline (51.8%) and ciprofloxacin (50.6%) and co-resistance to macrolides and fluoroquinolones (37.1%). Multivirulent profiles were identified mainly in isolates from chicken carcasses (84.8%), and the emergence of MDR/virulent strains was determined in pork isolates. All isolates except those from chicken carcasses showed a high potential for biofilm formation (71.4% luxS) and consequent persistence in industrial food processing. For chicken carcasses, the general virulence was higher in C. jejuni (54.3%), followed by C. coli (24%) and Campylobacter spp. (21.7%), and in the other meat matrices, Campylobacter spp. showed a higher prevalence of virulence (57.2%). The high rates of resistance and virulence reinforce the existence of strain selection pressure in the country, in addition to the potential risk of strains isolated not only from chicken carcasses, but also from other meat matrices.
Keywords: antimicrobial resistance, chilled meat, frozen meat, RAPD, virulence
1. Introduction
Campylobacter spp. are the most prevalent bacterial etiologic agents in foodborne gastroenteritis in developed countries and were officially responsible for 59.7 cases of campylobacteriosis per 100,000 of the population in 2020 [1,2]. Chicken meat has been the main source of human infection since 2008, but the consumption of meat from other animals also participates in the chain of transmission, since the species that make up the genus are capable of colonizing different farm animals [2,3].
Although campylobacteriosis affects more than 400–500 million people worldwide [4], and Brazil is an important producer and exporter of animal protein worldwide, there is no official record in the country of cases of human Campylobacteriosis in the last 10 years, and there are no regulations for the systematic or regulatory monitoring of the pathogen in meat sold directly to the population. Thus, even with records of the presence of Campylobacter in different products of animal origin in research in other countries [5], these reports are sporadic and cover specific regions and conditions in Brazil, which encourages the search for real knowledge about the presence, number, virulence potential and dissemination of this bacterium in meat from different species, as well as the determining factors of its presence, including the type of sanitary inspection and how the product is marketed, frozen or chilled.
The pathogenesis of Campylobacter infection is complex and not yet fully elucidated, but like other pathogens, in addition to the host–parasite relationship, it possesses the gene apparatus to colonize, invade, produce cytotoxins, and perpetuate itself in the environment [6]. This includes different genes involved in colonization and adhesion (pdlA and cadF), cell invasion (ciaB), motility (flaA), stress adaptation (dnaJ), toxin production (cdtA, cdtB and cdtC) and quorum sensing communication (luxS), which play a significant role in disease development. In addition, being resistant to different classes of antimicrobials demonstrates, along with their prevalence, an increased risk of infecting hosts and causing conditions that are more severe or difficult to treat [7,8].
Considering the prominent position of the Brazilian meat industry in the global market, Brazil’s status as the world’s largest exporter of chicken meat [9], the underreporting in the country regarding contamination and infection by Campylobacter and the scarcity of studies that characterize this pathogen at the epidemiological, molecular and phenotypic levels, we propose a panoramic analysis of the occurrence of Campylobacter in meats marketed in Brazil. We included in the investigation the prevalence of the genus, the main species of public health importance, antimicrobial resistance, virulence potential and genetic diversity.
2. Materials and Methods
2.1. Sampling and Microbiological Analysis
We analyzed 906 meat samples from different species, representative of 53 Brazilian commercial brands, during the period from August 2014 to February 2016, under different types of sanitary inspection (Federal, State and Municipal), qualified for domestic trade (State and Municipal-internal consumer market) and/or export (Federal-external consumer market). Samples were obtained from a total of 31 producers, of which 14 were representative brands of the main chicken carcass markets in the country, collected directly from the commercializing markets, under coordination of the National Health Surveillance Agency (ANVISA). The number of samples analyzed to determine prevalence was established according to Thrusfield [10]:
Table 1 shows how the number of samples, sampling unit and isolation protocol for each type of matrix were defined.
Table 1.
Number, sample types and sampling methods used in the study.
| Meat Matrix | Pexp * | Chilled | Frozen | Total | Isolation Protocol ISO 10272-1:2006 (ISO, 2006) |
Matrix Portion |
|---|---|---|---|---|---|---|
| Chicken carcasses | 80% 1 | 80 | 166 | 246 | A: Rinsing | ⅟2 carcass |
| Pork shank | 10% 2 | 138 | 138 | B: Rinsing | 150 g | |
| Bovine liver | 80% 3 | 132 | 114 | 246 | B: Rinsing | 150 g |
| Chicken liver | 90% 4 | 24 | 114 | 138 | C: Homogenization | 10 g |
| Minced meat | 10% 2 | 138 | 138 | C: Homogenization | 10 g | |
| Total | 512 | 394 | 906 |
Protocol A: for chicken carcasses. The sample aliquot was obtained using the rinsing technique, with half of the carcass placed individually in a sterile plastic bag containing 225 mL of sterile 0.1% peptone water (Difco®). The samples were subjected to a process of agitation and massaging for 60 s, with the most vigorous massaging on the neck, armpit, chest and groin. The product of the lavage was quantitatively and qualitatively analyzed for Campylobacter (Step 1). The other half of the carcass was submitted to freezing in a domestic freezer (−18 to −20 °C) for 30 days for later evaluation of survival and the number of viable Campylobacter cells for samples that were positive in the first stage (Stage 2).
Protocol B: for pork and beef liver samples. The methodology consisted of weighing 150 g of each meat matrix in 100 mL of sterile 0.1% peptone water (Difco®). The samples were submitted to the same rinsing technique as described in Protocol A, and the rinse water was used for counting and analysis of the presence/absence of Campylobacter (Step 1). Step 2 consisted of storing another 150 g of each matrix and evaluating the same parameters after 30 days under freezing in the samples that were positive in the first step.
Protocol C: intended for samples of ground beef and chicken liver. We used 10 g of the matrix diluted in 90 mL of Bolton broth (Oxoid®). The samples were homogenized and analyzed quantitatively and qualitatively for Campylobacter (Step 1). The rest of the matrices were kept at –20 °C for 30 days for analysis of the samples that were positive in the first step (Step 2).
Thirty milliliters of the rinsate was added to 30 mL of doubly concentrated Bolton broth (CM0983, Oxoid, Hampshire, UK) supplemented with an antibiotic mix (SR0183E, Oxoid) and with 5% horse blood (Laborclin, Paraná, Brazil) (Protocols A and B). Protocol C samples were incubated directly (10 g in 90 mL Bolton broth supplemented with 5% defibrinated sheep blood and an antibiotic mixture). Before incubation, 1-mL aliquots of the samples diluted in Bolton broth (protocol C) or in 0.1% peptone water (protocols A and B) were used to perform serial decimal dilutions and quantification of Campylobacter in Campylobacter Blood-Free Selective Agar (Modified CCDA-Preston) (CM0739, Oxoid). Bolton broth tubes were incubated in a microaerobic atmosphere (5–15% O2 and 10% CO2) using a Microaerobac (Probac do Brasil, Sao Paulo, Brazil) at 37 °C for 44 ± 4 h. After Bolton broth incubation, a membrane filtration method was used to plate the samples on Campylobacter Blood-Free Selective Agar (Modified CCDA-Preston) (CM0739, Oxoid) plates supplemented with an antibiotic (SR0155E, Oxoid). Briefly, a 0.65-µm-pore-size cellulose membrane filter (Millipore, MA, USA) was placed on top of the medium, and 300 µL of each enrichment in Bolton broth was added to the plate. After approximately 15 min, the membrane was dry, and it was removed from the agar plate. Modified CCDA-Preston plates were incubated at 37 °C for 44 ± 4 h in a microaerobic atmosphere, as described above.
For quantification, 1-mL aliquots of the samples diluted in Bolton broth or 0.1% peptone water (protocols 1, 2 and 3) were used to perform serial decimal dilutions in 9 mL of 0.1% peptone water (DifcoTM). After this procedure, 100 µL of the respective dilutions were inoculated in CCDA agar plates, supplemented with antibiotics and covered with a 0.65-µm-pore-size cellulose membrane. After removal of the membrane, samples were incubated at 37 °C for 44 ± 4 h in a microaerobic atmosphere, followed by calculations for the expression of quantitative results.
2.2. Molecular Analysis of Specific Genes, Transcript Production and Genetic Similarity
After isolation, two colonies typical of Campylobacter spp. were randomly selected from each modified CDDA-Preston plate for further analysis. The selected colonies were resuspended in Bolton broth (Oxoid) and cultured overnight. Total DNA was extracted using a commercial kit (Wizard Genomic DNA Purification kit, Promega, Madison, WI, USA), according to the manufacturer’s instructions. All PCR reactions performed for species identification, virulence genes, virulence transcripts and similarity analysis are described in Table 2.
Table 2.
Primers, function, amplicon size, PCR conditions, RT-PCR, RAPD-PCR and references.
| Genes | Function | Primers | Sequence 5′ → 3′ | Size (bp) | DNA (ng) |
Primer (pmol) | PCR Condition | Reference |
|---|---|---|---|---|---|---|---|---|
| 16S-rRNA | Gender identification | 16S-rRNA-F 16S-rRNA-R |
ATCTAATGGCTTAACCATTAAAC GGACGGTAACTAGTTTAGTATT |
857 | 30 | 40 | 94 °C—1 min; 25 cycles: 94 °C—1 min, 60 °C—1 min, 72 °C—1 min; 72 °C—7 min | Linton et al. [16] |
| pg | Multiplex PCR: Identification of C. jejuni and C. coli | pg3 pg50 |
GAACTTGAACCGATTTG ATGGGATTTCGTATTAAC |
460 | 20 | 40 | 94 °C—4 min; 25 cycles: 94 °C—1 min, 47 °C—1 min, 72 °C—1 min; 72 °C—7 min | Harmon et al. [17] |
| C | C1 C4 |
CAAATAAAGTTAGAGGTAGAATGT GGATAAGCACTAGCTAGCTGAT |
160 | 20 | ||||
| flaA | Motility | flaA-F flA-R |
ATGGGATTTCGTATTAACAC CTGTAGTAATCTTAAAACATTTTG |
1728 | 20 | 30 | 95 °C—10 min; 35 cycles: 95 °C—1 min, 45 °C—1 min, 72 °C—2 min; 72 °C—10 min | Hänel et al. [18] |
| pdlA | Paracellular invasion | pldA- 361 pldA-726 |
AAGAGTGAGGGAAATTCCA GCAAGATGGCAGGATTATCA |
385 | 20 | 30 | 95 °C—10 min; 35 cycles: 95 °C—1 min, 45 °C—1 min, 72 °C—2 min; 72 °C—10 min | Zheng et al. [19] |
| cadF | Colonization | cadFI-F2B cadFI-R1B |
TTGAAGGTAATTTAGATATG CTAATACCTAAAGTTGAAAC |
400 | 20 | 30 | 95 °C—10 min; 35 cycles: 95 °C—1 min, 45 °C—1 min, 72 °C—2 min; 72 °C—10 min | Zheng et al. [19] |
| ciaB | Intracellular invasion | ciaBI-652 ciaB-1159 |
TGCGAGATTTTTCGAGAATG TGCCCGCCTTAGAACTTACA |
527 | 20 | 30 | 95 °C—10 min; 35 cycles: 95 °C—1 min, 45 °C—1 min, 72 °C—2 min; 72 °C—10 min | Zheng et al. [19] |
| cdtABC | Multiplex PCR: Cytotoxin | cdtA-F cdtA-R cdtB-F cdtB-R cdtC-F cdtC-R |
CTATTACTCCTATTACCCCACC AATTTGAACCGCTGTATTGCTC AGGAACTTTACCAAGAACAGCC GGTGGAGTATAGGTTTGTTGTC ACTCCTACTGGAGATTTGAAAG CACAGCTGAAGTTGTTGTTGGC |
420 531 339 |
80 | 20 | 94 °C—5 min; 30 cycles: 94 °C—1 min, 57 °C—1 min, 72 °C—1 min; 72 °C—5 min | Martinez et al. [20] |
| dnaJ | Thermotolerance | dnaJ F dnaJ R |
AAGGCTTTGGCTCATC CTTTTTGTTCATCGTT |
720 | 20 | 20 | 95 °C—2 min; 30 cycles: 94 °C—1 min, 46 °C—1 min, 72 °C—1 min; 72 °C—5 min | Datta et al. [21] |
| sodB | Oxidative stress | sodB F sodB R |
ATGATACCAATGCTTTTGGTGATTT TAATACGACTCACTATAGGGCATTTGCATA AAAGCTAACTGATCC |
638 | 20 | 20 | 95 °C—2 min; 30 cycles: 94 °C—1 min, 46 °C—1 min, 72 °C—1 min; 72 °C—5 min | Biswas et al. [22] |
| luxS | Quorum-sensing | luxS-1 luxS-2 |
AGGCAAAGCTCCTGGTAAGGCCAA GGATCCGTATAGGTAAGTTCATTTT TGCTCC |
1080 | 50 | 10 | 94 °C—3 min; 30 cycles: 94 °C—30 s, 57 °C—1 min; 72 °C—1 min; 72 °C—10 min | Elvers, Park [23] |
|
HLWL85
1290 |
RAPD-PCR: genetic similarity | HLWL85 1290 |
ACGTATCTGC GTGGATGCGA |
-- | 10 | 30 | 92 °C—2 min; 35 cycles: 92 °C—15 s, 36 °C —1 min; 72 °C—1 min; 1 final cycle at 72 °C—5 min. | Akopyanz et al. [24] |
| ciaB | RT-PCR: Invasion | ATATTTGCTAGCAGCGAAGAG GATGTCCCACTTGTAAAGGTG |
157 | 200 | 4 | 94 °C—3 min; 45 cycles: 94 °C—15 s, 51 °C—20 s, 72 °C—20 s; 72 °C—3 min | Li et al. [15] | |
| dnaJ | RT-PCR: Thermotolerance | AGTGTCGAGCTTAATATCCC GGCGATGATCTTAACATACA |
117 | 200 | 4 | 94 °C—3 min; 45 cycles: 94 °C—15 s, 51 °C—20 s, 72 °C—20 s; 72 °C—3 min | Li et al. [15] | |
| p19 | RT-PCR: Iron uptake under stress |
GATGATGGTCCTCACTATGG CATTTTGGCGTGCCTGTGTA |
206 | 200 | 4 | 94 °C—3 min; 45 cycles: 94 °C—15 s, 51 °C—20 s, 72 °C—20 s; 72 °C—3 min | Birk et al. [14] | |
| sodB | RT-PCR: Oxidative Stress | TATCAAAACTTCAAATGGGG TTTTCTAAAGATCCAAATTCT |
170 | 200 | 4 | 94 °C—3 min; 45 cycles: 94 °C—15 s, 51 °C—20 s, 72 °C—20 s; 72 °C—3 min | Birk et al. [14] |
The ability to produce transcripts for ciaB (invasion), dnaJ (thermotolerance), p19 (iron uptake) and sodB (protection against oxidative stress) was investigated by qualitative reverse transcription PCR (RT-PCR) [14,15]. RNA extraction was performed using the Trizol method according to Li et al. [15]. The RNA concentration used was 200 ng/μL, quantified in a NanoDrop spectrophotometer (Thermo Scientific®, Wilmington, DE, USA).
Reverse transcription was performed with 10 U of RNase inhibitor, 40 U of MMLV-RT (Amersham Biosciences®, Saint Louis, MO, USA), 1X MMLV-RT buffer (Amersham Biosciences®), 200 μM of dNTPs (dGTP, dATP, dTTP and dCTP), 126 pmol of random hexamer oligonucleotides as primers (Invitrogen, Carlsbad, CA, USA), 20 μL of DEPC water (Invitrogen®) and 1 μL of RNA, all maintained at 37 °C for one hour to obtain complementary DNA (cDNA). Subsequently, 3 μL of cDNA was used for amplification in a 25-μL reaction volume, comprising 0.625 U of Taq DNA polymerase, 5 mM MgCl2, 200 μM of dNTPs and 4 pmol of each primer (Table 2) (Invitrogen).
The genetic diversity among the isolates was determined by the RAPD-PCR (Random Amplification of Polymorphic DNA) technique, according to the protocol described in Table 2.
At the end of the reactions, the amplified products were submitted to electrophoresis in 1.5% agarose gel, stained with SYBR safe solution (Invitrogen Brasil Ltd), submitted to a voltage of approximately 8 V/cm and subsequently visualized with UV light in a transilluminator (L.PIX Loccus Biotechnology, Cotia, Brazil).
The RAPD-PCR results were evaluated using the GelCompar II program (Comparative Analysis of Electrophoresis Patterns, version 1.50, Applied Maths Korthrijk, Sint-Martens-Latem, Belgium) with the Dice similarity coefficient, with 1% tolerance for each primer separately. For dendrogram construction, the UPGMA method (unweighted pair group method with arithmetic mean) used the average of all experiments.
2.3. Susceptibility to Antimicrobials
Resistance to antibiotics in Campylobacter was assessed by the disk diffusion method against six antibiotics routinely used to treat infections in humans and in veterinary medicine. Samples were suspended in 0.9% NaCl to obtain 5 × 105 CFU/mL (0.5 McFarland Standard) and seeded on Mueller Hinton agar plates supplemented with 5% defibrinated sheep blood (Laborclin). The following antimicrobials were tested: amoxicillin and clavulanic acid (10 µg), azithromycin (15 µg), ciprofloxacin (5 µg), erythromycin (15 µg), gentamycin (10 µg) and tetracycline (30 µg) (Oxoid®), according to EUCAST [25]. Plates were sealed and incubated at 41 ± 1 °C for 40–48 h in microaerobic conditions as described above. Subsequently, the strains were classified as sensitive (S) or re-susceptible (R) to the antimicrobial tested C. jejuni IAL 2383 and C. jejuni NCTC 11351 were used as positive controls, whereas a blank sample was used as a negative control. For those antimicrobials not classified for Campylobacter, the de-defined standard for Enterobacterales was used.
2.4. Statistical Analysis
To compare rates between groups, the binomial test for two proportions was used at 5% significance (Action Tool (2015) using the R program (R Development Core Team, 2015). The results were used to estimate the public health risk posed by the consumption of the chicken carcasses. Fisher’s exact test was used to compare Campylobacter positivity between the different matrices, forms of preservation (frozen/chilled), inspection systems, commercial brands and years of isolation. The calculations were performed in the GraphPad-Prism 8.0 program.
3. Results
3.1. Prevalence of Campylobacter on Chicken Carcasses and in Meat Matrices
Campylobacter spp. were identified in a total of 18.69% (46/246) of the chicken carcass samples and 5.3% (35/660) of the different types of meat evaluated, including chicken liver and chilled and frozen pork and beef (Table 3).
Table 3.
Prevalence of Campylobacter spp. in different meat matrices and forms of commercialization in Brazil in the period from 2014 to 2016.
| MATRIX | N | Forms of Commercialization (N) |
Campylobacter spp. n/N (%) |
C. coli n/N (%) |
C. jejuni n/N (%) |
TOTAL n/N (%) |
|---|---|---|---|---|---|---|
| Chicken carcasses | 246 | Frozen (166) Chilled (80) |
8/166 (4.82) 2/80 (2.50) a |
9/166 (5.42) 2/80 (2.50) a |
19/166 (11.45) 6/80 (7.50) b |
36/166 (21.69) 10/80 (12.50) I |
| Chicken liver | 138 | Frozen (114) Chilled (24) |
1/114 (0.87) 1/24 (4.16) a |
1/114 (0.87) 1/24 (4.16) a |
1/114 (0.87) --- a |
3/114 (2.63) 2/24 (8.33) II |
| Bovine liver | 246 | Frozen (114) Chilled (132) |
0 7/132 (5.30) a |
0 2/132 (1.51) a |
0 2/132 (1.51) a |
--- 11/132 (8.33) II |
| Minced meat | 138 | Chilled (138) | 5/138 (3.62) a | 0 a | 0 a | 5/138 (3.62) II |
| Pork shank | 138 | Chilled (138) | 6/138 (4.35) a | 5/138 (3.62) a | 3/138 (2.17) a | 14/138 (10.14) II |
| TOTAL | 906 | Chicken carcasses (246) Other meat matrix (660) |
10/246 (4.07) a 20/660 (3.03) a |
11/246 (4.47) a 9/660 (25.71) ab |
25/246 (10.16) b 6/35 (17.14) b |
46/246 (18.69) I 35/660 (5.30) II |
| Frozen (394) Chilled (512) |
9/394 a 21/512 a |
10/394 a 10/512 a |
20/394 a 11/512 a |
39/394 I 42/512 I |
||
| Total (906) | 30/906 (3.31) a | 20/906 a | 31/906 a | 81/906 |
N = total number of samples; n (%) = number of positive samples and percentage. Different numbers in the same column I,II and letters in the same line a,b indicate a statistical difference in each variable analyzed (p < 0.05, Fisher’s exact test).
The prevalence of the different Campylobacter species in chicken carcasses and in the other meat matrices were, respectively, C. jejuni (54.3%, 25/46, and 17.1%, 6/35), C. coli (23.9%, 11/46, and 25.7%, 9/35) and the other Campylobacter spp. (21.7%, 10/46, and 57.1%, 20/35). C. jejuni was the most common species in chicken carcasses (p = 0.001, Fisher test) and Campylobacter spp. in the other breeds (p = 0.002, Fisher test). The prevalence of Campylobacter in chicken liver (3.62%) and beef liver (4.48%) was lower than expected.
All positive samples were confirmed by the quantitative method, with a limit of quantification of 10 CFU/g. In 8/81 (9.9%) samples (6 from chicken carcass and 2 from beef liver), the enumeration was accurate and equivalent to an average of 123 CFU/g (minimum = 10 and maximum = 468 CFU/g). For the other samples, there was confluent growth indicating counts higher than 107 CFU/g [26], which determined statistical equivalence in the levels of contamination by Campylobacter spp. in the different meat matrices (Kruskal–Wallis test, p = 0.352).
Freezing did not represent a form of Campylobacter spp. control, since the prevalence in both forms of commercial preservation was statistically equal (p = 0.412, Fisher test). However, slow (in-home) subsequent freezing of the positive samples did not maintain Campylobacter viability, since in the second stage (samples kept frozen for 30 days after processing, described in the methodology: stage/step 2), we reisolated the bacteria only in 2/46 chicken carcass samples originally marketed frozen (one C. jejuni and one Campylobacter spp.).
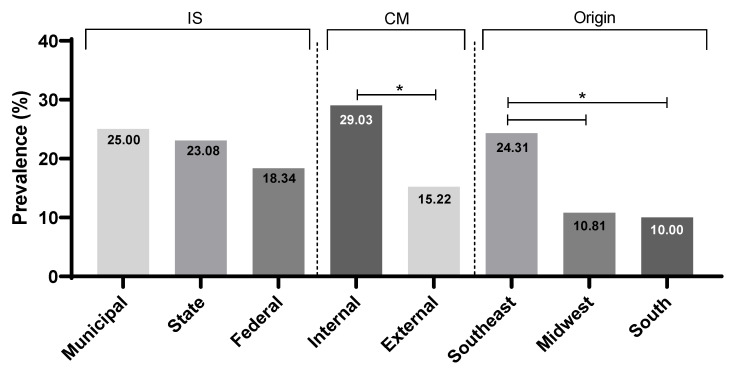
The type of inspection system did not influence the prevalence rates of Campylobacter (p > 0.05), but the positivity rate was lower (p = 0.023) in companies that export their products (28/184; 15.22%) than in those that sell to the domestic market (18/62; 29.03%). We obtained a significantly higher prevalence in the Southeast (35/144; 24.31%) than in the Midwest (10/92; 10.81%; p = 0.011) and South (1/10; 10%; p = 0.045) regions (Figure 1).
Figure 1.
Prevalence of Campylobacter spp. in chicken carcasses according to the inspection system, * p < 0.05, Fisher’s exact test.
3.2. Antimicrobial Resistance Phenotypes
The least effective antimicrobials for controlling the isolated strains were tetracycline (51.85%) and ciprofloxacin (50.62%). We found a significantly higher percentage of resistance to tetracycline among Campylobacter spp. isolates. (66.7%) than among C. coli (35%) (p = 0.043, Fisher test). For ciprofloxacin, high resistance was attributed to strains isolated from chicken carcasses (60.9%, p = 0.045 Fisher test). The highest susceptibility was identified for the antimicrobial’s azithromycin and gentamicin, with 72.84% and 67.9%, respectively (Table 4).
Table 4.
Antimicrobial resistance in Campylobacter strains isolated in the study.
| Antimicrobial | Source N = 81 |
C. jejuni n1 = 25 n2 = 6 |
C. coli n1 = 11 n2 = 9 |
Campylobacter spp. n1 = 10 n2 = 20 |
Total N1 = 46 N2 = 35 |
|---|---|---|---|---|---|
| Chicken carcasses 1 | 14/25 (56%) | 8/11 (72.7%) | 6/10 (60%) | 28/46 (60.9%) a | |
| CIP | Other meat matrices 2 | 2/6 (33.3%) | 3/9 (33.3%) | 8/20 (40%) | 13/35 (37.1%) b |
| Total | 16/31 (51.6%) | 11/20 (55%) | 14/30 (46.7%) | 41/81 (50.61%) | |
| Chicken carcasses 1 | 11/25 (44%) | 5/11 (45.5%) | 4/10 (40%) | 25/46 (54.3%) | |
| AMC | Other meat matrices 2 | 2/6 (33.3%) | 4/9 (44.4%) | 9/20 (45%) | 15/35 (42.9%) |
| Total | 13/31 (41.9%) | 9/20 (45%) | 13/30 (43.3%) | 40/81 (49.4%) | |
| Chicken carcasses 1 | 11/25 (44%) | 2/11 (18.2%) | 4/10 (40%) | 17/46 (37%) | |
| GEN | Other meat matrices 2 | 2/6 (33.3%) | 2/9 (22.2%) | 5/20 (25%) | 9/35 (25.7%) |
| Total | 13/31 (41.9%) | 4/20 (20%) | 9/30 (30%) | 26/81 (32.09%) | |
| Chicken carcasses 1 | 11/25 (44%) | 6/11 (54.5%) | 2/10 (20%) | 19/46 (41.3%) | |
| ERY | Other meat matrices 2 | 1/6 (16,7%) | 2/9 (22.2%) | 6/20 (30%) | 9/35 (25.7%) |
| Total | 12/31 (38.7%) | 8/20 (40%) | 8/30 (26.7%) | 28/81 (34.6%) | |
| Chicken carcasses 1 | 11/25 (44%) | 2/11 (18.2%) | 8/10 (80%) | 21/46 (45,7%) | |
| TET | Other meat matrices2 | 4/6 (66.7%) | 5/9 (55.5%) | 12/20 (60%) | 21/35 (60%) |
| Total | 15/31 (48.4%) | 7/20 (35%) I | 20/30 (66.7%) II | 42/81 (51.9%) | |
| Chicken carcasses 1 | 6/25 (24%) | 4/11 (36.4%) | 3/10 (30%) | 13/46 (28.3%) | |
| AZM | Other meat matrices 2 | 1/6 (16.7%) | 2/9 (22.2%) | 6/20 (30%) | 9/35 (25.7%) |
| Total | 7/31 (22.6%) | 6/20 (30%) | 9/30 (30%) | 22/81 (27.2%) |
N: total number of strains isolated, N1: total number of isolates from chicken carcasses, N2: total number of isolates from other meat matrices, n1: number of isolates from chicken carcasses of each species, n2: number of isolates from other matrices of each species. CIP: Ciprofloxacin, AMC: Amoxillin and clavulanate, GEN: gentamicin, ERY: Erythromycin, TET: Tetracycline, AZM: Azithromycin. p < 0.05 in the column a,b and in the line I,II, Fisher’s exact test.
Analysis of resistance profiles identified 35 types (Table 5), of which those with multidrug resistance (three or more classes of unrelated antimicrobials) were the most common (22/35 profiles, 62.8%, and 44/81 strains, 54.3%), statistically different from the number of susceptible, mono- and co-resistant strains (p < 0.0001). The number of MDR strains did not differ (p < 0.05) according to species: 18/30 (60.0%), 18/31 (58.1%) and 8/20 (40.0%) of the isolates corresponded to Campylobacter spp., C. jejuni and C. coli, respectively. The P35 profile, which includes resistance to all antimicrobials tested, was representative of isolates belonging to the other meat sources, except for chicken carcasses (p = 0.032). Moreover, in the other meat matrices, we detected higher MDR in C. jejuni species versus C. coli (p = 0.031). Co-resistance to macrolides and fluoroquinolones was identified in 13/35 (37.1%) profiles, which included 26/81 (32.1%) strains and did not differ according to the matrix type or species (p < 0.05).
Table 5.
Multidrug resistance profiles of Campylobacter isolated from different meat matrices.
| Profiles | Number of Profiles | Caracter | Origin | Campylobacter spp. | C. jejuni | C. coli | Total 1 | Total 2 (%) | |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 1 | Susceptible | Carcasses | 1 | 1 | 2 | 2 (2.5) A | ||
| Other meat matrix | - | - | - | ||||||
| P2 a P7 | 6 | Monoresistance | Carcasses | - | 4 | 4 | 8 | 16 (19.8) B | |
| Other meat matrix | 4 | - | 4 | 8 | |||||
| P8 a P13 | 6 | Co-Resistance | Carcasses | 5 | 8 | 2 | 15 I | 19 (23.5) B | |
| Other meat matrix | 2 | - | 2 | 4 II | |||||
| P14 a P24 | 11 | MDR (3C) | Carcasses | 2 | 11 | 2 | 15 | 22 (27.2) | 44 (54.3) C |
| Other meat matrix | 5 | 1 | 1 | 7 | |||||
| P25 a P34 | 10 | MDR (4C and 5C) | Carcasses | 2 | 2 | 3 | 7 | 18 (22.2)/ | |
| Other meat matrix | 6 | 4 | 1 | 11 | |||||
| P35 | 1 | MDR (all) | Carcasses | - | - | - | 0 I | 4 (4.9) | |
| Other meat matrix | 3 | - | 1 | 4 II | |||||
| Total | 35 | - | Total | 30 | 31 | 20 | 81 | ||
P: profile, MDR: multidrug resistance, 3/4/5C: number of antimicrobial classes included in the profiles, Total 1: broken down by sample type, Total 2: total number of isolates in the respective profile group, I,II p < 0.05 when comparing each profile group between matrix types, A,B,C p < 0.05 when comparing profile groups, Fisher’s exact test.
3.3. Characterization of Virulence Factors
All strains presented at least one of the virulence genes studied, indicating that they have different degrees of potential virulence. The most identified genes were cadF (62/81), ciaB (50/81), pldA (49/81) and flaA (44/81), and the least identified were sodB and cdtABC (Table 6).
Table 6.
Number and percentage of virulence genes in the 81 strains of Campylobacter spp. isolated from meat products.
| GENE | Chicken Carcasses-n(%) | Total 1 N(%) |
Other Meat Matrix-n(%) | Total 2 N(%) |
TOTAL | ||||
|---|---|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. spp. | C. jejuni | C. coli | C. spp. | N(%) | |||
| ciaB | 23 (92.0) | 7 (63.6) | 7 (70.0) | 37 (80.4) a | 4 (66.6) | 5 (55.5) | 4 (20.0) | 13 (37.1) b | 50 (61.7) |
| pldA | 21 (84.0) | 8 (72.7) | 9 (90.0) | 38 (82.6) a | 3 (50.0) | 2 (22.2) | 6 (30.0) | 11 (31.4) b | 49 (60.5) |
| flaA | 19 (76.0) | 11 (100.0) | 6 (60.0) | 36 (78.3) a | 2 (33.3) | 4 (44.4) | 2 (10.0) | 8 (22.8) b | 44 (54.3) |
| cadF | 24 (96.0) | 11 (100.0) | 9 (90.0) | 44 (95.6) a | 3 (50.0) | 6 (50.0) | 9 (45.0) | 18 (51.4) b | 62 (76.5) |
| cdtA | 14 (56.0) | 3 (27.3) | 4 (40.0) | 21 (45.6) a | 2 (33.3) | 0 | 1 (5.0) | 3 (8.5) b | 24 (29.6) |
| cdtB | 15 (60.0) | 3 (27.3) | 4 (40.0) | 22 (47.8) a | 2 (33.3) | 0 | 1 (5.0) | 3 (8.5) b | 25 (30.9) |
| cdtC | 14 (56.0) | 3 (27.3) | 4 (40.0) | 21 (45.6) a | 1 (16.7) | 0 | 1 (5.0) | 2 (5.7) b | 23 (28.4) |
| luxS | 14 (56.0) | 0 | 1 (10.0) | 15 (32.6) a | 6 (100.0) | 5 (55.5) | 14 (70.0) | 25 (71.4) b | 40 (49.4) |
| dnaJ | 18 (72.0) | 7 (63.6) | 5 (50.0) | 30 (65.3) a | 1 (16.7) | 5 (55.5) | 7 (35.0) | 13 (37.1) b | 43 (53.0) |
| sodB | 2 (8.0) | 0 | 0 | 2 (4.4) a | 0 | 0 | 0 | 0 a | 2 (2.4) |
| TOTAL | 25 (54.3) | 11 (24.0) | 10 (21.7) | 46 (100) | 6 (17.1) | 9 (25.7) | 20 (57.2) | 35 (100) | 81 (100) |
C. spp.: Campylobacter spp.; Total 1 refers to isolates from chicken carcasses. Total 2 refers to isolates from other breeds. Different letters in (a,b) the same row indicate p < 0.05, Fisher’s exact test.
The prevalence of 8/10 virulence genes were evident in samples from broiler carcasses compared to isolates from the other matrices (p < 0.02, Fisher test). The exceptions were the sodB gene, poorly identified in the samples (2/81, 2.5%), and the luxS gene, more prevalent in isolates from other meat sources (25/35, 71.4%; p = 0.001, Fisher test). C. jejuni showed significantly higher virulence percentages for the ciaB (p = 0.042), cdtABC (p = 0.018) and luxS (p = 0.009) genes compared to the other species, and C. coli showed a high prevalence of pldA (p = 0.034), flaA (p = 0.002) and cadF (p = 0.021).
Discrimination into profiles allowed the identification of 50 variations in the virulence characterization of the strains, of which 30 profiles were of isolates exclusively from chicken carcasses, and 19 were from other meats. Only one profile contained one strain from pork shank and one from chicken carcass. We observed that chicken carcasses represented the main source of virulent Campylobacter (p < 0.0001, Fisher test), with 39/46 (84.8%) showing four or more virulence genes, in contrast to the other matrices (9/35; 25.7%). It is worth noting the emergence of virulent strains in the pork shank matrix, which, besides presenting four of the nine strains with four or more virulence genes, showed greater potential for invasion and biofilm formation (p = 0.042), identified by the presence of high percentages for genes ciaB (9/14, 64.3%), pldA (6/14, 42.8%), cadF (9/14, 64.3%) and luxS (12/14, 85.7%). C. jejuni (p = 0.001) and C. coli (p = 0.042) showed higher virulence in relation to Campylobacter spp., and C. jejuni grouped the eight profiles with more virulence genes (V43-50) (Table S1). We also observed that strains from frozen samples were more likely to be more virulent (27/39, 69.2%) than those from chilled samples (14/42, 33.3%) (p = 0.006, OR = 4.0, Fisher test).
The analysis of virulence transcript production was restricted to the 46 strains obtained from chicken carcasses. It is worth noting that 17/46 (36.9%) of the strains did not transcribe any of the genes tested. Transcription of the dnaJ gene (23/46; 50%) was identified in C. jejuni (48%), C. coli (63%) and Campylobacter spp. (40%), with no statistical difference among species (p > 0.05). The production of transcripts of the sodB gene was observed in only 4.3% of isolates, all from C. jejuni. The p19 and ciaB genes were transcribed in 50% and 30.4% of strains, respectively, with no statistical difference among species (p < 0.05). We observed that for C. jejuni, the gene transcription process was more evident, since the joint transcription of all or three of the evaluated genes (except sodB) was exclusive to this species (6/46; 13.0%). The condition of the sample (chilled/frozen) did not interfere in the transcription of the genes studied (p > 0.05).
3.4. Genetic Similarity
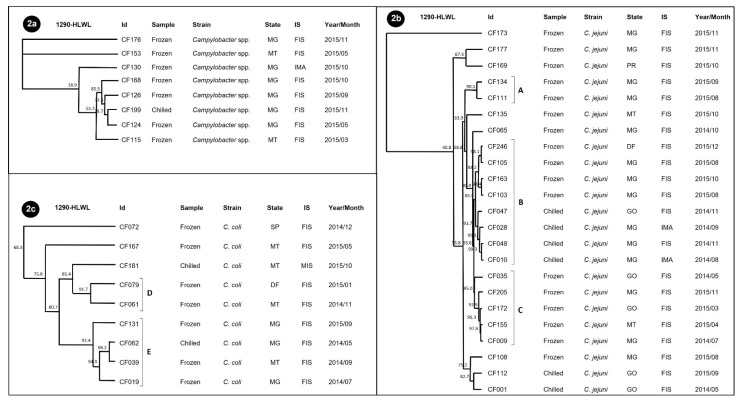
In the evaluation of genetic similarity in chicken carcasses, three dendrograms were discriminated for 40/46 strains, with one for each species: Campylobacter spp. (Figure 2a), C. jejuni (Figure 2b) and C. coli (Figure 2c), and no clones were present. The strains that lacked RAPD profiling included two strains of each species (two C. jejuni, two C. coli and two Campylobacter spp.).
Figure 2.
Dendrogram of the 40 Campylobacter spp. isolates from chilled and frozen chicken carcasses, from data generated by the RAPD-PCR technique with primers 1290 and HLWL, using the average from experiments and the UPGMA method with 85% optimization by the GelCompar program. (2a) Results found for Campylobacter spp. (2b) Results found for C. jejuni: (A) cluster with 90.1% homology; (B) cluster with 85.9% homology; (C) cluster with 85% homology. (2c) Results found for C. coli: (D) cluster with 91.7% homology; (E) cluster with 92.4% homology. SP (São Paulo), MT (Mato Grosso), MG (Minas Gerais).
The strains of Campylobacter spp. were no more than 80% similar, indicating the probable presence of several other Campylobacter species (other than C. jejuni and C. coli). All were unique to the year 2015 in the MG state and with the common presence of the cadF, ciaB and pldA genes and resistance to ciprofloxacin and tetracycline. Three clusters were identified in C. jejuni (profiles of A, B and C). Cluster A grouped two strains with a similarity of 90.1% from frozen samples from Minas Gerais state in August and September 2015, which had in common the ciaB, cadF and pldA genes and resistance to GEN and ERY. Eight strains included cluster B, with 85.9% homology, from different origins (MG, MT and GO), from chilled and frozen samples produced in the years 2014 and 2015, under different inspection systems and distinct virulence and antimicrobial resistance profiles. The five strains that made up group C had in common the fact that they came from frozen chicken carcass samples, along with the presence of the flaA, ciaB, cdt, cadF and pldA genes and resistance to ciprofloxacin.
C. coli strains were grouped into two clusters. For cluster D, three strains from different regions were grouped, which had in common the presence of the flaA, pldA and cadF genes and resistance to ciprofloxacin. Cluster E included four strains with the presence of the cadF gene in common.
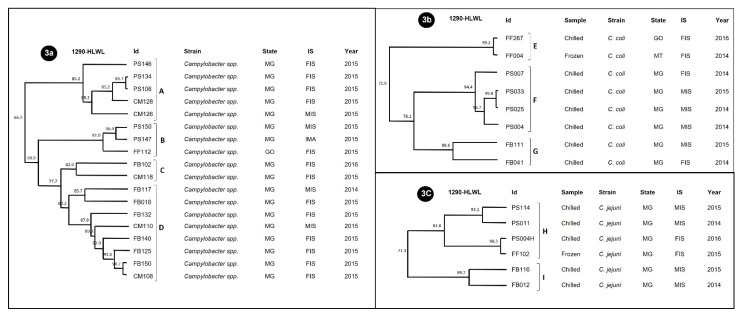
For the other meat matrices, the dendrograms are shown in Figure 3 and included 32/35 strains that were discriminated into four clusters for Campylobacter spp. (A, B, C and D), three for C. coli (E, F and G) and two for C. jejuni (H and I). The strains that did not show a RAPD profile included one from chicken liver (Campylobacter spp.) and two from pork shank (C. coli and Campylobacter spp.).
Figure 3.
Comparative dendrogram of 32 Campylobacter strains for other meat matrices using the Dice similarity coefficient with 1% tolerance and the UPGMA method with 0.80% optimization. (3a) Analysis of Campylobacter spp.; (3b) Analysis of C. coli; (3c) Analysis of C. jejuni. (A–I) clusters formed with homology greater than 80%. CM—ground bovine duckling; FB—bovine liver; FF—chicken liver; PS—pork shank. SP (São Paulo), MT (Mato Grosso) and MG (Minas Gerais).
In Campylobacter spp. (Figure 3a), the distribution of strains from distinct meat matrices within the same RAPD genotype was common. Especially, in cluster B, we detected a 92% similarity between samples originating from pork shank and chicken liver in the year 2015, with the presence of the luxS gene in common.
For C. coli (Figure 3b) and C. jejuni (Figure 3c), we observed that the clusters were discriminated according to the meat matrix, annual production and industrial and sanitary inspection services. In C. coli, the presence of the flaA and cadF genes was common to clusters E and F. In cluster E, two chicken liver strains were grouped together, with 99.2% similarity, from samples stored under refrigeration and freezing, from brands produced in different states (MT and GO), indicating the dissemination of the genotype among different establishments. For C. jejuni, both clusters (H and I) contained the ciaB and luxS genes. Cluster H, composed of three strains from pork shank and one from chicken liver, showed a similarity of 81.8%, demonstrating the presence of common genotypes circulating in different production chains.
4. Discussion
4.1. Campylobacter Positivity
The highest prevalences of the Campylobacter genus were found in chicken carcasses and pork shank and corroborated other recent studies in carcasses and in slaughterhouses [27,28]. However, the prevalence of Campylobacter in liver meat varies widely from country to country and can reach extremely high rates (96%) [29,30]. The lowest values found for beef liver and chicken liver in our study were not expected, since this kind of meat is a maintenance niche for bacteria and a source of foodborne outbreaks [31]. Thus, it is possible that the bacterial injury condition potentiated the acquisition of the viable but nonculturable form, reducing our prevalence values.
C. jejuni was the most frequently isolated pathogen in poultry carcasses, and in meat matrices, Campylobacter spp. were the most prevalent, which are justified by differences in contamination in the pre-slaughter sector, reservoir animals, retail meat origin and regions of origin [32].
Despite the presence of samples with low counts (<1000 CFU/g), the others showed confluent growth indicative of counts higher than 107 CFU/g [26]. According to EU legislation, the maximum count allowed in up to 60% of sampling is 1000 CFU/g. The confluent growth pattern identified in the other samples raises concerns about the level of contamination, providing high susceptibility to infection [33].
The cold chain has been indicated as a way to decrease or control pathogen load in food [34], but freezing of commercial samples was not an effective means of controlling bacteria. Research with C. jejuni in chicken meat demonstrated that the combination of refrigeration and freezing were no substitute for safe handling and proper cooking of poultry [35]. A study by MAPA (2021) demonstrated that the time between product manufacture and laboratory analysis can be decisive in the number of samples showing high counts for Campylobacter spp. [36], but in our study, this factor was not relevant since there was no difference in this period between the negative (mean of 42.69 days) and positive frozen samples (mean of 50.36 days). This allows us to infer that some strains with a greater ability to adapt may be selected under adverse conditions and that freezing does not guarantee a food free of Campylobacter spp.
In parallel, our study found that subsequent slow freezing of the samples for 30 days allowed the recovery of only 2/46 (4.3%), probably due to the long duration of injury and acquisition of the viable but nonculturable form.
Exporting companies presented a lower rate of Campylobacter isolation, probably due to the higher stringency of legislation to meet the foreign market, which ensures greater biosecurity in production activities. The main strategies for pathogen control in the food export industry include the application of specific food safety protocols, such as the Exploratory Program for Research and Estimation of Prevalence of Campylobacter spp., in conjunction with the monitoring and control of Salmonella spp., instituted by the MAPA normative instruction, determined by IN No. 20, of 21 October 2016 [36] and programs for strict hygienic practices (HACCP), which focus on the reduction or absence of the pathogen in the final product [37].
Southeastern Brazil is the country’s second largest chicken-producing region, which justifies the highest percentage of isolation in this location, which includes the states of Minas Gerais and São Paulo [38].
4.2. Resistance of Campylobacter Isolates
Resistance to AMC presented with a relatively high rate and was aggravated by the fact that it is a β-lactam widely used in animal treatment, with several registrations and commercial brands approved by the Ministry of Agriculture (MAPA) [39]. Resistance to AMC has been associated with the production of β-lactamases, and its exacerbated use may contribute to the selection of carbapenem-resistant strains [40]. It is known that Brazil is among the 10 largest consumers of veterinary antimicrobials recognized worldwide and lacks complete reports regarding their use [41], which determines the need for more rigorous control.
The low efficacy of TET is impactful from a clinical point of view, since it is an alternative drug that can be administered in the treatment of campylobacteriosis [42]. In addition, TET has been banned in the country as a growth promoter since the year 2009 [43], but it was used extensively in the past, which may have selected resistant strains that have remained circulating.
For CIP, we also identified low efficacy, which was mainly attributed to chicken carcass strains. The cause of quinolone resistance in the poultry chain is its overuse in production, treatment and disease control, in addition to the spread of resistance in human strains as an aggravating factor [44].
Co-resistance to macrolides and fluoroquinolones in 37.1% of strains raises concerns about treatment for campylobacteriosis, since both represent drugs of choice [45] and are considered critically important antibiotics [46]. The concern is amplified for quinolones, considering their maintenance in South American wastewater due to uncontrolled veterinary practices [47]. For macrolides, the implementation of stringent control measures [48] may have determined the higher susceptibility of our strains to AZI and ERY.
GEN also showed good efficiency, which contributes to the institution of this drug in the therapy in severe cases of campylobacteriosis [49], especially when there is resistance to the drugs of choice (macrolides and fluoroquinolones) [50].
MDR profiles were more prevalent, which intensifies concern about the treatment of campylobacteriosis. The prevalence of MDR strains varies in the literature [51,52]. C. jejuni was the most common MDR species in samples of other meat matrices. Furthermore, resistance to all the tested drugs was identified exclusively in swine and bovine meat matrices. These facts point to the consequences of intensive antibiotic application. which promotes intense selection pressure on MDR Campylobacter at the end of slaughter processing, aggravating the problem at the interface of food consumption and human disease [53].
4.3. Virulence of Campylobacter Isolates
The variability in the virulence potential identified by the diversity of virulence profiles (50) reflects the genetic plasticity of Campylobacter, which represents a genus in an intense process of evolution. The high occurrence of recombination events generates alterations in its genetic material that facilitate gene flow among species of the same genus and drive its high diversity [54].
The higher frequency of cadF, ciaB, pldA and flaA in the strains reflects their virulence potential. The cadF gene plays a primary role in adhesion, which represents a prerequisite for colonization of the host gastrointestinal tract. The ability of Campylobacter to adhere to fibronectin is mediated by adhesins on the surface of the bacterium encoded by this gene [55]. Cell invasion and intestinal colonization are aided by expression of the pldA gene [19]. Furthermore, in animals, the presence of strains positive for this gene assists in the maintenance of commensal relationships [56]. Rivera Amil et al. [57] described that ciaB acts by encoding a protein that functions to destroy microtubules and thus facilitates cell invasion and infection in the host. The flaA gene is highly conserved in Campylobacter and is important for survival under the internal conditions of the gastrointestinal environment [58]. The absence of this gene may reduce colonization ability and alter Campylobacter motility, but it also allows for greater antigenic variation, which represents a strategy for evading the host immune system [59].
The low frequency of sodB indicates that the strains are less able to maintain viability under oxidative stress conditions. This gene encodes the production of SOD (superoxide dismutase) proteins that assist in the survival of Campylobacter under stress conditions. Under heat stress (chilling/freezing), there is an intense production of free radicals by the bacterial cell, which, in the absence of SOD, induces the process of injury and intracellular dehydration [60].
The identification of the dnaJ gene in 53% of strains determines the potential to encode heat-shock-related proteins, which allows for tolerance to sudden temperature variations [61].
The cytolethal distending toxin (CDT) is encoded by three adjacent genes, cdtA, cdtB and cdtC, and is involved in cell cycle arrest in the G2/M phase [62]. This cytotoxin induces a cytoplasmic distension, leading to cell death by apoptosis about three to four days after infection, contributing to the development and pathogenesis of inflammatory diarrhea caused by campylobacteriosis in humans [63]. The low percentage of cdtABC found in our study (<31%) was not expected [64] but is similar to the Angelovski et al. [65] study.
The greatest potential for biofilm formation (luxS gene) was detected in strains from other meat matrices. This gene is the most important in the acquisition of the fundamental sessile form in cell–cell communication (quorum-sensing; QS), in the structuring of biomass and in the recognition and inclusion of bacterial populations [66]. Thus, the higher potential for biofilm formation in strains from the other meat matrices also constitutes one of the main strategies of Campylobacter to resist multiple stress conditions, including low temperatures and competitive exclusion caused by the more prevalent cohabitant microbiota in these matrices. In addition, generalist isolates of C. jejuni possess the ability to survive for long periods under aerobic conditions within the microenvironments present in biofilms, which contributes to their persistence in primary animal protein processing [67].
Strains from chicken carcasses showed a higher virulence potential, which is in agreement with the literature [68], which reinforces that these strains may be under the influence of selection pressure imposed by the conditions maintained in an industrial environment [69]. It is worth mentioning that specifically for pork shank, there is an emergence of virulent strains, which is a warning, since few studies have shown this profile [28].
Strains from frozen samples presented a four-times greater chance of being highly virulent (presence of four or more virulence genes). This fact is alarming, since it indicates that freezing can determine a selection pressure for more virulent strains, since stress conditions impact the virulence and pathogenesis of bacterial strains [70].
The similarity in virulence of C. jejuni and C. coli shows that although many papers indicate C. jejuni as the more virulent species, it is possible that C. coli has the same potential acquired through cohabitation with C. jejuni. Inter-species recombination, especially between C. jejuni and C. coli, plays an important role in the evolution of the genus Campylobacter. In fact, about 18.6% of the allelic variants identified in C. coli exhibit ancestry from C. jejuni, while only 2.3% of C. jejuni alleles were acquired from C. coli, indicating an asymmetric gene flow between the two species and justifying the increased incidence of C. coli-related cases of campylobacteriosis [71].
The discrimination of unique profiles of chicken carcasses and the other meats reflects specific diversities according to origin. Campylobacter is known to exhibit a multi-host lifestyle that reveals its broad genetic diversity. Genetic sequencing analyses have shown the specificity of certain profiles harboring typical origins, termed host specialists, whose presence of an interaction barrier can be identified by the separation of host niches, such as those identified in chicken carcasses in our study. Other strains, called host generalists, may colonize a wide range of animal reservoirs, similar to what we observed for virulence profiles unique to other meat matrices [72].
The injury condition (chilling/freezing) of the isolates represents an important factor in the modulation of the transcription process and can explain the absence of transcripts in 17/46 isolates and the high transcription of stress-related genes (dnaJ and p19).
The lower transcription of ciaB is justified by the modulation of transcription according to the needs of Campylobacter, with ciaB being more expressed when the bacterium finds a favorable invasion condition inside the host [73]. The low transcription of sodB was expected, since only two strains presented the gene and both produced transcripts. The production of transcripts identified exclusively in C. jejuni for at least three of the investigated genes indicates its greater adaptation to adverse conditions (cooling/freezing), which allows better regulation and maintenance of the transcription process.
4.4. Similarity of Campylobacter Isolates
The identification of bands by RAPD analysis was not possible in all strains, due to failure of DNA amplification indicated by the absence of banding or the possible interference of nucleases; both cases can be identified in several strains [74].
For Campylobacter spp. from chicken carcasses, the failure to cluster reflects the presence of distinct species common in some regions, which include simultaneous contamination with C. consius, C. foetus, C. hyoilei, C. lari and C. mucosalis [75].
The formation of clusters containing strains from two or more meat matrices (Figure 3a) in Campylobacter spp. indicates greater proximity between isolates from distinct matrices. Modeling studies on genomic plasticity in Campylobacter demonstrate that this pathogen presents a generalist gradient that allows colonization of new niches and optimizes the ability to survive rapid transitions in different hosts, evidencing that the acquisition of the same strain may originate from more than one source [54].
Persistence of strains in the same state and in different periods has been identified in C. jejuni isolated from chicken carcasses. This maintenance indicates disregard of biosecurity standards and self-control programs coupled with the potential of the strains to establish themselves in the sessile life form and determine recurrent contamination of the final product. Indications of reintroduction and persistence of genetically identical strains have been investigated during the production cycle in broiler and turkeys, and intervention measures such as empty periods and microbial decontamination techniques reduce Campylobacter spp. prevalence in primary processing [76].
The spread of genotypes across different regions of the country was the most identified fact in the strain similarity analysis. This indicates that the perpetuation of similar strains in different regions occurs due to the trade of products and that they can determine the dissemination of these profiles at a global level, considering the importance of the country in the export of animal protein [77].
5. Conclusions
We conclude from this study that multiple factors are involved in Campylobacter epidemiology, allowing a distribution among regions and the persistence of strains in chilled and frozen meats of different origins. The higher prevalences in chicken carcasses indicate the main focus of control in the country, but monitoring in bovine and porcine carcasses should be a central point to extend control. C. jejuni is the most important species in the country, but monitoring should be extended to other Campylobacter species, considering the high levels of resistance and virulence identified and the genetic plasticity of this genus. Selection pressure for emerging freeze-adapted, multivirulent and MDR strains indicates the need for a comprehensive assessment of Campylobacter molecular mechanisms as a measure to support the implementation of widespread surveillance strategies for this pathogen.
Acknowledgments
To ANVISA for the availability of samples to carry out the study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijerph19106087/s1, Table S1: Virulence profiles of 81 Campylobacter strains isolated from meat matrices in Brazil.
Author Contributions
Conceptualization, R.T.d.M. and D.A.R.; methodology, J.R.I. and M.F.T.; software, M.G.T.; validation, M.G.T., J.L.M.P. and G.R.A.F.; formal analysis, M.G.T., C.F.D., M.C.C. and B.d.A.B.; investigation, M.G.T. and R.T.d.M.; resources, D.A.R.; data curation, M.G.T. and R.T.d.M.; writing—original draft preparation, M.G.T., R.T.d.M., C.F.D. and M.C.C.; writing—review and editing, M.G.T., R.T.d.M. and D.A.R.; visualization, R.T.d.M.; supervision, M.G.T.; project administration, R.T.d.M. and D.A.R.; funding acquisition, D.A.R. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by CNPq, grant number 440090/2014-8, and CAPES Financial code 001.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ilktac M., Ongen B., Humphrey T.J., Williams L.K. Molecular and phenotypical investigation of ciprofloxacin resistance among Campylobacter jejuni strains of human origin: High prevalence of resistance in Turkey. APMIS. 2020;128:41–47. doi: 10.1111/apm.13005. [DOI] [PubMed] [Google Scholar]
- 2.EFSA The European Union One Health 2020 Zoonoses Report. EFSA J. 2021;19 doi: 10.2903/j.efsa.2021.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manyi-Loh C., Mamphweli S., Meyer E., Makaka G., Simon M., Okoh A. An Overview of the Control of Bacterial Pathogens in Cattle Manure. Int. J. Environ. Res. Public Health. 2016;13:843. doi: 10.3390/ijerph13090843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heredia N., García S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018;4:250–255. doi: 10.1016/j.aninu.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S., Young S.R., Tong E., Abbott J.W., Womack N., Friedman S.L., McDermott P.F. Antimicrobial Resistance of Campylobacter Isolates from Retail Meat in the United States between 2002 and 2007. Appl. Environ. Microbiol. 2010;76:7949–7956. doi: 10.1128/AEM.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabir S.M.L., Chowdhury N., Asakura M., Shiramaru S., Kikuchi K., Hinenoya A., Neogi S.B., Yamasaki S. Comparison of Established PCR Assays for Accurate Identification of Campylobacter jejuni and Campylobacter coli. Jpn. J. Infect. Dis. 2019;72:81–87. doi: 10.7883/yoken.JJID.2018.340. [DOI] [PubMed] [Google Scholar]
- 7.Wieczorek K., Osek J. Antimicrobial Resistance Mechanisms among Campylobacter. BioMed Res. Int. 2013;2013:1–12. doi: 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farfán M., Lártiga N., Benavides M.B., Alegría-Morán R., Sáenz L., Salcedo C., Lapierre L. Capacity to adhere to and invade human epithelial cells, as related to the presence of virulence genes in, motility of, and biofilm formation of campylobacter jejuni strains isolated from chicken and cattle. Can. J. Microbiol. 2019;65:126–134. doi: 10.1139/cjm-2018-0503. [DOI] [PubMed] [Google Scholar]
- 9.Associação Brasileira de Proteína Animal . ABPA Relatório Anual 2020. Associação Brasileira de Proteína Animal; Rio de Janeiro, Brazil: 2020. [Google Scholar]
- 10.Thrusfield M.V. Epidemiologia Veterinária. Roca; São Paulo, Brazil: 2004. [Google Scholar]
- 11.Food and Agriculture Organization of the United Nations World Health Organization–FAO/WHO Risk assessment of Campylobacter spp. in broiler chickens: Technical Report. Microbiol. Risk Assess. Ser. 2009;12:132. [Google Scholar]
- 12.Noormohamed A., Fakhr M. A Higher Prevalence Rate of Campylobacter in Retail Beef Livers Compared to Other Beef and Pork Meat Cuts. Int. J. Environ. Res. Public Health. 2013;10:2058–2068. doi: 10.3390/ijerph10052058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte R., Hudson J.A., Graham C. Campylobacter in chicken livers and their destruction by pan frying. Lett. Appl. Microbiol. 2006;43:591–595. doi: 10.1111/j.1472-765X.2006.02020.x. [DOI] [PubMed] [Google Scholar]
- 14.Birk T., Wik M.T., Lametsch R., Knøchel S. Acid stress response and protein induction in Campylobacter jejuni isolates with different acid tolerance. BMC Microbiol. 2012;12:174. doi: 10.1186/1471-2180-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y.-P., Ingmer H., Madsen M., Bang D.D. Cytokine responses in primary chicken embryo intestinal cells infected with Campylobacter jejuni strains of human and chicken origin and the expression of bacterial virulence-associated genes. BMC Microbiol. 2008;8:107. doi: 10.1186/1471-2180-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linton D., Lawson A.J., Owen R.J., Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon K.M., Ransom G.M., Wesley I.V. Differentiation ofCampylobacter jejuniandCampylobacter coliby polymerase chain reaction. Mol. Cell. Probes. 1997;11:195–200. doi: 10.1006/mcpr.1997.0104. [DOI] [PubMed] [Google Scholar]
- 18.Hänel I., Müller J., Müller W., Schulze F. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 2004;101:75–82. doi: 10.1016/j.vetmic.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J., Meng J., Zhao S., Singh R., Song W. Adherence to and Invasion of Human Intestinal Epithelial Cells by Campylobacter jejuni and Campylobacter coli Isolates from Retail Meat Products. J. Food Prot. 2006;69:768–774. doi: 10.4315/0362-028X-69.4.768. [DOI] [PubMed] [Google Scholar]
- 20.Martinez I., Mateo E., Churruca E., Girbau C., Alonso R., Fernandezastorga A. Detection of cdtA, cdtB, and cdtC genes in Campylobacter jejuni by multiplex PCR. Int. J. Med. Microbiol. 2006;296:45–48. doi: 10.1016/j.ijmm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Datta S., Niwa H., Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003;52:345–348. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]
- 22.Biswas D., Hannon S.J., Townsend H.G.G., Potter A., Allan B.J. Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry. Int. Microbiol. 2011;14:25–32. doi: 10.2436/20.1501.01.132. [DOI] [PubMed] [Google Scholar]
- 23.Elvers K.T., Park S.F. Quorum sensing in Campylobacter jejuni: Detection of a luxS encoded signalling molecule. Microbiology. 2002;148:1475–1481. doi: 10.1099/00221287-148-5-1475. [DOI] [PubMed] [Google Scholar]
- 24.Akopyanz N., Bukanov N.O., Westblom T.U., Kresovich S., Berg D.E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.EUCAST The European Committee on Antimicrobial Susceptibility Testing (2018) Breakpoint Tables for Interpretation of MICs and Zone Diameters. EUCAST 2018, Version 8. [(accessed on 25 January 2022)]. Available online: https://www.eucast.org/
- 26.Koneman E., Allen S. Diagnostic Microbiology. Lippincott; New York, NY, USA: 2010. [Google Scholar]
- 27.Hungaro H.M., Mendonça R.C.S., Rosa V.O., Badaró A.C.L., Moreira M.A.S., Chaves J.B.P. Low contamination of Campylobacter spp. on chicken carcasses in Minas Gerais state, Brazil: Molecular characterization and antimicrobial resistance. Food Control. 2015;51:15–22. doi: 10.1016/j.foodcont.2014.11.001. [DOI] [Google Scholar]
- 28.Andrzejewska M., Szczepańska B., Śpica D., Klawe J.J. Prevalence, Virulence, and Antimicrobial Resistance of Campylobacter spp. in Raw Milk, Beef, and Pork Meat in Northern Poland. Foods. 2019;8:420. doi: 10.3390/foods8090420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NSW Food Authority Campylobacter in Chicken Liver. [(accessed on 25 January 2022)]; Available online: https://www.foodauthority.nsw.gov.au/about-us/science/market-analysis/chicken-liver.
- 30.Liu S., Kilonzo-Nthenge A., Nahashon S.N., Pokharel B., Mafiz A.I., Nzomo M. Prevalence of Multidrug-Resistant Foodborne Pathogens and Indicator Bacteria from Edible Offal and Muscle Meats in Nashville, Tennessee. Foods. 2020;9:1190. doi: 10.3390/foods9091190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanier W.A., Hale K.R., Geissler A.L., Dewey-Mattia D. Chicken Liver–Associated Outbreaks of Campylobacteriosis and Salmonellosis, United States, 2000–2016: Identifying Opportunities for Prevention. Foodborne Pathog. Dis. 2018;15:726–733. doi: 10.1089/fpd.2018.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narvaez-Bravo C., Taboada E.N., Mutschall S.K., Aslam M. Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int. J. Food Microbiol. 2017;253:43–47. doi: 10.1016/j.ijfoodmicro.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Teunis P.F.M., Bonačić Marinović A., Tribble D.R., Porter C.K., Swart A. Acute illness from Campylobacter jejuni may require high doses while infection occurs at low doses. Epidemics. 2018;24:1–20. doi: 10.1016/j.epidem.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization WHO Campylobacter. [(accessed on 15 January 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/campylobacter#:~:text=Campylobacter species can be killed by heat and,33 million of healthy life years are lost.
- 35.Soro A.B., Whyte P., Bolton D.J., Tiwari B.K. Strategies and novel technologies to control Campylobacter in the poultry chain: A review. Compr. Rev. Food Sci. Food Saf. 2020;19:1353–1377. doi: 10.1111/1541-4337.12544. [DOI] [PubMed] [Google Scholar]
- 36.DIPOA . MAPA Anuário dos Programas de Controle de Alimentos de Origem Animal do DIPOA. Departamento de Inspeção de Produtos de Origem Animal; Brasil, Brazil: 2021. [Google Scholar]
- 37.Wagenaar J.A., Mevius D.J., Havelaar A.H. El agente Campylobacter en la producción animal y las estrategias de control para reducir la incidencia de la campilobacteriosis humana. Rev. Sci. Tech. Oie. 2006;25:581–594. doi: 10.20506/rst.25.2.1680. [DOI] [PubMed] [Google Scholar]
- 38.Associação Brasileira de Proteína Animal . Relatório Anual 2021. ABPA; Brasil, Brazil: 2021. [Google Scholar]
- 39.Brasil MAPA Produtos Veterinários. [(accessed on 2 April 2022)]; Available online: https://mapa-indicadores.agricultura.gov.br/publico/single/?appid=a3e9ce67-d63b-43ff-a295-20123996ead7&sheet=377bdc66-84e3-4782-96b3-81718e4d83aa&lang=pt-BR&opt=ctxmenul&select=clearall.
- 40.Hartmann L., Schieweck O., Greie J.C., Szabados F. Human Campylobacter jejuni and Campylobacter coli Isolates: Demographic Pattern and Antimicrobial Susceptibility to Clinically Important Antimicrobials used in Livestock. J. Med. Microbiol. Diagn. 2018;7:269. doi: 10.4172/2161-0703.1000269. [DOI] [Google Scholar]
- 41.Tiseo K., Huber L., Gilbert M., Robinson T.P., Van Boeckel T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics. 2020;9:918. doi: 10.3390/antibiotics9120918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramires T., de Oliveira M.G., Kleinubing N.R., de Fátima Rauber Würfel S., Mata M.M., Iglesias M.A., Lopes G.V., Dellagostin O.A., da Silva W.P. Genetic diversity, antimicrobial resistance, and virulence genes of thermophilic Campylobacter isolated from broiler production chain. Braz. J. Microbiol. 2020;51:2021–2032. doi: 10.1007/s42770-020-00314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brasil MAPA Instrução Normativa No. 26, de 09 de julho de 2009. Aprova o Regulamento Técnico para a Fabricação, o Controle de Qualidade, a Comercialização e o Emprego de Produtos Antimicrobianos de Uso Veterinário. [(accessed on 2 April 2022)]; Available online: https://sistemasweb.agricultura.gov.br/sislegis/action/detalhaAto.do?method=visualizarAtoPortalMapa&chave=1984822284.
- 44.Ge B., Wang F., Sjölund-Karlsson M., McDermott P.F. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. J. Microbiol. Methods. 2013;95:57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 45.Dai L., Sahin O., Grover M., Zhang Q. New and alternative strategies for the prevention, control, and treatment of antibiotic-resistant Campylobacter. Transl. Res. 2020;223:76–88. doi: 10.1016/j.trsl.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.WHO . WHO List of Critically Important Antimicrobials (CIA) WHO; Geneva, Switzerland: 2018. [Google Scholar]
- 47.Robles-Jimenez L.E., Aranda-Aguirre E., Castelan-Ortega O.A., Shettino-Bermudez B.S., Ortiz-Salinas R., Miranda M., Li X., Angeles-Hernandez J.C., Vargas-Bello-Pérez E., Gonzalez-Ronquillo M. Worldwide Traceability of Antibiotic Residues from Livestock in Wastewater and Soil: A Systematic Review. Animals. 2021;12:60. doi: 10.3390/ani12010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ministério da Agricultura, Pecuária e Abastecimento Instrução Normativa No. 14, de 17 de Maio de 2012. [(accessed on 2 April 2022)]; Available online: https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/insumos-pecuarios/alimentacao-animal/arquivos-alimentacao-animal/legislacao/instrucao-normativa-no-14-de-17-de-maio-de-2012.pdf/view.
- 49.Es-soucratti K., Hammoumi A., Bouchrif B., Asmai R., En-nassiri H., Karraouan B. Occurrence and antimicrobial resistance of Campylobacter jejuni isolates from poultry in Casablanca-Settat, Morocco. Ital. J. Food Saf. 2020;9 doi: 10.4081/ijfs.2020.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitehouse C.A., Zhao S., Tate H. Antimicrobial Resistance in Campylobacter Species: Mechanisms and Genomic Epidemiology. Adv. Appl. Microbiol. 2018;103:1–47. doi: 10.1016/bs.aambs.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Würfel S.D.F.R., da Fontoura Prates D., Kleinubing N.R., Dalla Vecchia J., Vaniel C., Haubert L., Dellagostin O.A., da Silva W.P. Comprehensive characterization reveals antimicrobial-resistant and potentially virulent Campylobacter isolates from poultry meat products in Southern Brazil. LWT. 2021;149:111831. doi: 10.1016/j.lwt.2021.111831. [DOI] [Google Scholar]
- 52.Nisar M., Mushtaq M.H., Shehzad W., Hussain A., Muhammad J., Nagaraja K.V., Goyal S.M. Prevalence and antimicrobial resistance patterns of Campylobacter spp. isolated from retail meat in Lahore, Pakistan. Food Control. 2017;80:327–332. doi: 10.1016/j.foodcont.2017.03.048. [DOI] [Google Scholar]
- 53.Marin C., Lorenzo-Rebenaque L., Moreno-Moliner J., Sevilla-Navarro S., Montero E., Chinillac M.C., Jordá J., Vega S. Multidrug-Resistant Campylobacer jejuni on Swine Processing at a Slaughterhouse in Eastern Spain. Animals. 2021;11:1339. doi: 10.3390/ani11051339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodcock D.J., Krusche P., Strachan N.J.C., Forbes K.J., Cohan F.M., Méric G., Sheppard S.K. Genomic plasticity and rapid host switching can promote the evolution of generalism: A case study in the zoonotic pathogen Campylobacter. Sci. Rep. 2017;7:9650. doi: 10.1038/s41598-017-09483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin S., Joe A., Lynett J., Hani E.K., Sherman P., Chan V.L. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 2004;39:1225–1236. doi: 10.1111/j.1365-2958.2001.02294.x. [DOI] [PubMed] [Google Scholar]
- 56.Ngobese B., Zishiri O.T., El Zowalaty M.E. Molecular detection of virulence genes in Campylobacter species isolated from livestock production systems in South Africa. J. Integr. Agric. 2020;19:1656–1670. doi: 10.1016/S2095-3119(19)62844-3. [DOI] [Google Scholar]
- 57.Rivera-Amill V., Kim B.J., Seshu J., Konkel M.E. Secretion of the Virulence-Associated Campylobacter Invasion Antigens from Campylobacter jejuni Requires a Stimulatory Signal. J. Infect. Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- 58.Wysok B., Wojtacka J. Detection of virulence genes determining the ability to adhere and invade in Campylobacter spp. from cattle and swine in Poland. Microb. Pathog. 2018;115:257–263. doi: 10.1016/j.micpath.2017.12.057. [DOI] [PubMed] [Google Scholar]
- 59.Hendrixson D.R., Akerley B.J., DiRita V.J. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 2001;40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- 60.Stead D., Park S.F. Roles of Fe Superoxide Dismutase and Catalase in Resistance of Campylobacter coli to Freeze-Thaw Stress. Appl. Environ. Microbiol. 2000;66:3110–3112. doi: 10.1128/AEM.66.7.3110-3112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stintzi A., Whitworth L. Investigation of the Campylobacter jejuni Cold-Shock Response by Global Transcript Profiling. Genome Lett. 2003;2:18–27. [Google Scholar]
- 62.Trindade M.M., Perdoncini G., Sierra-Arguello Y.M., Lovato M., Borsoi A., Nascimento V.P. Detecção dos genes codificantes da toxina CDT, e pesquisa de fatores que influenciam na produção de hemolisinas em amostras de Campylobacter jejuni de origem avícola. Pesqui. Veterinária Bras. 2015;35:709–715. doi: 10.1590/S0100-736X2015000800002. [DOI] [Google Scholar]
- 63.Asakura M., Samosornsuk W., Hinenoya A., Misawa N., Nishimura K., Matsuhisa A., Yamasaki S. Development of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the detection and identification of Campylobacter jejuni, Campylobacter coli and Campylobacter fetus. FEMS Immunol. Med. Microbiol. 2008;52:260–266. doi: 10.1111/j.1574-695X.2007.00369.x. [DOI] [PubMed] [Google Scholar]
- 64.Quino W., Caro-Castro J., Hurtado V., Flores-León D., Gonzalez-Escalona N., Gavilan R.G. Genomic Analysis and Antimicrobial Resistance of Campylobacter jejuni and Campylobacter coli in Peru. Front. Microbiol. 2022;12 doi: 10.3389/fmicb.2021.802404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angelovski L., Popova Z., Blagoevska K., Mojsova S., Manovska M.R., Prodanov M., Jankuloski D., Sekulovski P. Isolation Rate of Campylobacter Spp. and Detection of Virulence Genes of Campylobacter jejuni Across the Broiler Chain. Maced. Vet. Rev. 2021;44:149–157. doi: 10.2478/macvetrev-2021-0020. [DOI] [Google Scholar]
- 66.Bandara H.M.H.N., Lam O.L.T., Jin L.J., Samaranayake L. Microbial chemical signaling: A current perspective. Crit. Rev. Microbiol. 2012;38:217–249. doi: 10.3109/1040841X.2011.652065. [DOI] [PubMed] [Google Scholar]
- 67.Mouftah S.F., Cobo-Díaz J.F., Álvarez-Ordóñez A., Mousa A., Calland J.K., Pascoe B., Sheppard S.K., Elhadidy M. Stress resistance associated with multi-host transmission and enhanced biofilm formation at 42 °C among hyper-aerotolerant generalist Campylobacter jejuni. Food Microbiol. 2021;95:103706. doi: 10.1016/j.fm.2020.103706. [DOI] [PubMed] [Google Scholar]
- 68.Borba Martins Peres P.A., Torres de Melo R., Armendaris P.M., Barreto F., Follmann Perin T., Grazziotin A.L., Paz Monteiro G., Pereira Mendonça E. Multi-virulence and phenotypic spread of Campylobacter jejuni carried by 2chicken meat in Brazil. PLoS ONE. 2022. in press . [DOI] [PMC free article] [PubMed]
- 69.Aidley J., Rajopadhye S., Akinyemi N.M., Lango-Scholey L., Bayliss C.D. Nonselective Bottlenecks Control the Divergence and Diversification of Phase-Variable Bacterial Populations. mBio. 2017;8:16. doi: 10.1128/mBio.02311-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes C.N., Passaglia J., Vilela F.P., Pereira da Silva F.M.H.S., Duque S.S., Falcão J.P. High survival rates of Campylobacter coli under different stress conditions suggest that more rigorous food control measures might be needed in Brazil. Food Microbiol. 2018;73:327–333. doi: 10.1016/j.fm.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 71.Backert S. Fighting Campylobacter Infections-Towards a One Health Approach. Volume 6. Springer; Amsterdam, The Netherlands: 2021. [Google Scholar]
- 72.Ben Romdhane R., Merle R. Fighting Campylobacter Infections. Curr. Top. Microbiol. Immunol. 2021;431:818. doi: 10.1007/978-3-030-65481-8_2. [DOI] [PubMed] [Google Scholar]
- 73.Han X., Guan X., Zeng H., Li J., Huang X., Wen Y., Zhao Q., Huang X., Yan Q., Huang Y., et al. Prevalence, antimicrobial resistance profiles and virulence-associated genes of thermophilic Campylobacter spp. isolated from ducks in a Chinese slaughterhouse. Food Control. 2019;104:157–166. doi: 10.1016/j.foodcont.2019.04.038. [DOI] [Google Scholar]
- 74.Blackburn C.D.W., editor. Food Spoilage Microorganisms. Cambridge University Press; Cambridge, UK: 2006. [Google Scholar]
- 75.Lynch Ó.A., Cagney C., McDowell D.A., Duffy G. Occurrence of fastidious Campylobacter spp. in fresh meat and poultry using an adapted cultural protocol. Int. J. Food Microbiol. 2011;150:171–177. doi: 10.1016/j.ijfoodmicro.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 76.Gichure J.N., Kamau Njage P.M., Wambui J.M., Dykes G.A., Buys E.M., Coorey R. Systematic-review and meta-analysis on effect of decontamination interventions on prevalence and concentration of Campylobacter spp. during primary processing of broiler chickens. Food Microbiol. 2022;102:103923. doi: 10.1016/j.fm.2021.103923. [DOI] [PubMed] [Google Scholar]
- 77.Barros A., Novo C.S., Feddern V., Coldebella A., Scheuermann G.N. Determination of Eleven Veterinary Drugs in Chicken Meat and Liver. Appl. Sci. 2021;11:8731. doi: 10.3390/app11188731. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.