Abstract
Humantenine, an alkaloid isolated from the medicinal herb Gelsemium elegans (Gardner & Chapm.) Benth., has been reported to induce intestinal irritation, but the underlying toxicological mechanisms remain unclear. The object of the present study was to investigate the RNA N6-methyladenosine (m6A) modification and distinct mRNA transcriptome profiles in humantenine-treated HCT116 human colon cancer cells. High-throughput MeRIP-seq and mRNA-seq were performed, and bioinformatic analysis was performed to reveal the role of abnormal RNA m6A modification and mRNA expression in humantenine-induced intestinal cell toxicity. After humantenine treatment of HCT116 cells, 1401 genes were in the overlap of differentially m6A-modified mRNA and differentially expressed mRNA. The Kyoto Encyclopedia of Genes and Genomes and Gene Ontology annotation terms for actin cytoskeleton, tight junctions, and adherens junctions were enriched. A total of 11 kinds of RNA m6A methylation regulators were differentially expressed. The m6A methylation levels of target genes were disordered in the humantenine group. In conclusion, this study suggested that the HCT116 cell injury induced by humantenine was associated with the abnormal mRNA expression of m6A regulators, as well as disordered m6A methylation levels of target genes.
Keywords: humantenine, m6A modification, transcriptome, tight junction, actin cytoskeleton
1. Introduction
Gelsemium elegans (Gardner & Chapm.) Benth. (G. elegans), an evergreen woody vine belonging to the family Loganiaceae, is a traditional medicinal herb with a drug use history of over 1800 years [1]. It is widely distributed in Southeast Asia, especially in Hunan, Guangdong, Fujian, and Yunnan provinces of China. In traditional Chinese medicine, G. elegans is used for the treatment of neuropathic pain, rheumatoid pain, swelling and skin ulcers, and also exerts anti-inflammatory, anti-tumor, anti-anxiety, and immunomodulatory effects [2]. However, G. elegans has been reported to be associated with severe gastrointestinal toxicity, such as vomiting, abdominal pain, diarrhea, or constipation, and even to induce symptoms of the nervous, circulatory, and respiratory systems, such as dizziness, drops in blood pressure, and respiratory paralysis [3]. To date, more than 2000 poisoning deaths have been reported in China [4]. According to the statistics from the Chinese Center for Disease Control and Prevention, the lethality rate of G. elegans was 15.8%, ranking second in the total vegetal food poisoning cases in China from 2004 to 2013 [5]. This lethality rate even reached 47.6% in Guangxi Province in 2015 to 2017, as reported by the foodborne disease incident monitoring and reporting system [6]. The severe side effects of G. elegans limit its clinical application.
There is an urgent need to reveal the toxicity mechanisms of G. elegans in order to allow the safe use of G. elegans, so the isolation and identification of toxic components is important. To date, nearly 200 components have been discovered in G. elegans, including alkaloids, iridoids, lipids, triterpene, steroids, and their glycosides, coumarins, flavones, lignans, phenolic acids, fructoses and their derivatives [7,8]. Among them, the alkaloids include 121 species, and indole alkaloids are the main biologically active components that exert pharmacological and toxicological effects. The most abundant indole alkaloids, from highest to lowest, are koumine, gelsevirine, gelsemine, humantenine, and gelsenicine. A previous study reported that the median lethal dose (LD50) of total alkaloids is 1.5 mg/kg.bw in female mice after intramuscular injection, and 1.2 mg/kg.bw in male rats after intraperitoneal injection [9]. The LD50 of koumine, gelsevirine N-oxide, gelsemine, and gelsenicine was also investigated in rodent experimental models, and the toxicity of humantenine has not been reported.
Previous studies suggested that the biological effects of alkaloids may be related to RNA epigeneticmodification [10,11]. In recent decades, about 160 RNA modifications have been reported, most of which are involved in the transcriptional and post-transcriptional regulation of genes [12]. The modification types include N1-methyladenosine (m1A) [13], 5-methylcytosine (m5C) [14], N6-methyladenosine (m6A), and 7-methylguanosine (m7G) [15], of which m6A is the most abundant and prevalent internal chemical modification in the mRNA of eukaryotes [16]. By means of liquid chromatography with tandem mass spectrometry, it has been reported that m6A has an abundance of 0.1–1.79% modifications per main base in mammals [17]. The m6A methylation modification sites are mostly located in stop codons and 3′ untranslated regions (3′UTRs) of mRNA [18]. In addition, the consensus motif RRACH (R = A/G, H = A/U/C) was found in m6A-modified RNA [19]. Regarding the biological function of RNA m6A modification, it plays an important role in epigenetic regulation and affects gene expression related to RNA splicing, translation, localization, and stability [20]. RNA m6A methylation is involved in a variety of biological processes, such as cell growth, development, metabolism, renewal, differentiation, immunity, and proliferation [21].
RNA m6A methylation is a dynamic and reversible process mainly regulated by m6A methyltransferases (writers), m6A methylation recognition proteins (readers), and m6A demethylases (erasers) together [22]. At the initial stage, the methyl group is transferred to the sixth nitrogen atom of adenine by the methyltransferase complex composed of METTL3, METTL14, and WTAP. The m6A-modified RNA can then be specifically recognized by YTH-domain family protein (YTHDF) and insulin-like growth factor 2 mRNA-binding protein (IGFBP) [23]. The readers play a vital role in the mRNA decay, storage, and stability [24]. At the same time, the m6A modification can be cleared by demethyltransferases, such as ALKBH5 and FTO.
The modification of RNA transcripts has been known since the 1970s. In recent years, the development of methylated RNA immunoprecipitation sequencing (MeRIP-seq) has made it possible to gain a thorough understanding of RNA m6A modification and decipher the corresponding biological functions [18]. In this study, we hypothesized that humantenine may alter RNA m6A modification in HCT116 human colon cancer cells and induce downstream biological adverse effects. High-throughput m6A MeRIP-seq and mRNA-seq were performed to reveal the potential molecular mechanisms of humantenine-induced HCT116 cell toxicity.
2. Materials and Methods
2.1. Chemical Reagent
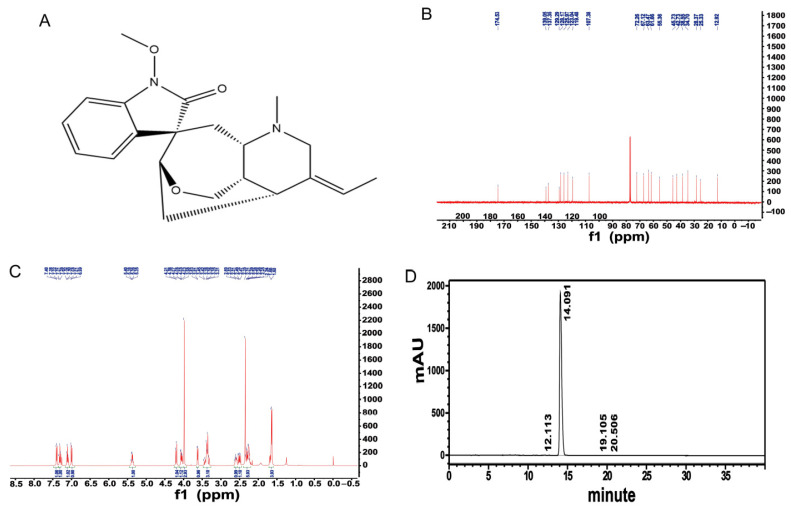
Humantenine (C21H26O3N2) was purchased from Must Bio-Technology Co., Ltd. (Chengdu, China). The chemical structure (Figure 1A) of humantenine was confirmed from the 400 MHz 13C and 1H nuclear magnetic resonance (NMR) spectra (Bruker, Rheinstetten, Germany) (Figure 1B,C). The purity of humantenine was 99.77% (Figure 1D), as determined by high-performance liquid chromatography (HPLC) (Shimadzu, Tokyo, Japan).
Figure 1.
(A) The chemical structure of humantenine. The 400 MHz 13C (B) and 1H (C) NMR spectra of humantenine. (D) The purity of humantenine as determined by HPLC. Chromatographic conditions: Sinachrom ODS-BP (250 × 4.6 mm, 5 μm particle size); mobile phase of acetonitrile: water = 50: 50; column temperature: 35 °C; injection volume: 10 μL. The retention time of humantenine was 14.091 min, and the area percent of humantenine was 99.77%.
2.2. Cell Culture and Treatment
Human colon cancer cell line HCT116 was originally purchased from American Type Culture Collection. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GE Healthcare Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, New York, NY, USA), 100 μg/mL streptomycin sulfate and 100 U/mL penicillin G sodium salt, and then maintained in an incubator (Thermo Fisher, Langenselbold, Germany) with an atmosphere of 5% CO2 at 37 °C. Humantenine was dissolved in dimethyl sulfoxide (DMSO) to a concentration of 400 μM. The final working concentration of DMSO in cell culture experiments was 1%.
2.3. RNA Preparation
The cells were cultured in 9 cm dishes. After humanteinine treatment for 48 h, the cells were washed twice in PBS, and Trizol reagent (Invitrogen, Carlsbad, CA, USA) was added to lyse cells. Total RNA was extracted, and the concentration and integrity were measured via Qubit RNA HS Assay and Agilent 2100 Bioanalyzer (Santa Clara, CA, USA), respectively.
2.4. High-Throughput m6A MeRIP-seq and mRNA-seq
The m6A-modified RNA enrichment and sequencing was performed by E-GENE Tech Co., Ltd. (Shenzhen, China). Firstly, we used the Dynabeads™ mRNA Purification Kit (Invitrogen) to separate mRNA from the 44 μg total RNA. Subsequently, 1× RNA Fragmentation Buffer (Thermo Fisher, Langenselbold, Germany) was used at 70 °C for 5 min to turn purified mRNA into fragments. After purification, 5 μg of m6A antibody (Synaptic Systems, Goettingen, Germany) that had been incubated with 25 μL of protein A/G magnetic beads mixture (Thermo Fisher, Langenselbold, Germany) was taken out and used to enrich RNA. The incubation was then rotated at 4 °C for 4 h. After immunoprecipitation, the beads were washed three times with IP buffer (150 mM NaCl, 10 mM Tris-HCl, 0.1% IGEPAL CA-630 in nuclease-free H2O) for 10 min each time, rotating at 4 °C. Finally, m6A-modified mRNA fragments were extracted with Trizol reagent and purified using an RNA Clean & Concentrator Kit (Zymo Research, Orange, CA, USA). The purified m6A antibody-enriched RNA and some unenriched mRNA fragments were used as input to construct libraries with the VAHTS mRNA-seq V3 Library Prep Kit for Illumina (Vazyme, Nanjing, China) according to the manufacturer’s protocol. The established libraries were analyzed using an Agilent 2100 Bioanalyzer, quantified by real-time PCR, and then sequenced on the NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Three independent samples were used in each group of m6A MeRIP-seq and mRNA-seq.
2.5. Sequencing Data Analysis
The raw data were processed by Trim Galore (Cambridge, UK). The sequencing results were aligned with the human genome reference hg19 using Hisat2 [25], the RNA expression level was analyzed using StringTie (Baltimore, MD, USA) [26], and differential expression was calculated using DEseq [27]. The exomepeak2 (Suzhou, China) [28,29] was applied for m6A peak calling and the detection of differential methylation. In exomepeak2, the Poisson Generalized Linear Model was used to estimate the methylation level and detect differential methylation regions. The exomePeak2′s estimation of sequencing depth size factors was on non-methylation background regions. The consensus of m6A motif sequences was identified by STREME [30]. The STREME algorithm integrated the position weight matrix Markov model to report a useful estimate of the statistical significance of each motif it discovered. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) annotations were done by KOBAS (Beijing, China) [31] and DAVID (Frederick, MD, USA) [32], respectively. The distribution of epitranscriptome profiles was visualized by MetaTX (Suzhou, China) [33]. The m6A conservation and disease association were obtained from ConsRM (Suzhou, China) [34] and RMDisease (Suzhou, China) [35]; the substrates of m6A regulators identified by CLIP techniques were downloaded from starBase v2.0 (Guangzhou, China) [36]. The m6A patterns of gene methylated sites were visualized using Integrative Genomics Viewer (IGV) software. Protein–protein interaction (PPI) analysis was performed using STRING and visualized using Cytoscape.
2.6. Molecular Docking
To explore whether humantenine could interact with differentially expressed regulators of m6A modification, the SYBYL-X 2.0 software (Tripos, St Louis, MO, USA) was applied to perform molecular docking. The 3D structures of proteins were retrieved from the Protein Data Bank (PDB). The protein structures were prepared using SYBYL-X 2.0 to remove water molecules and heteroatoms, add hydrogen atoms, and repair side chains [37]. The 2D structure of humantenine was downloaded from the PubChem compound database and was imported into Chem3D (Waltham, MA, USA) to generate 3D structures according to the energy minimization principle. After being stored in mol2 format, the 3D structure of humantenine was set to the lowest energy conformation to mimic the stable molecular conformation in the natural system [38]. Proteins were docked in Surflex-Dock GeomX mode through the semi-flexible docking method. The total score, which is a comprehensive evaluation of hydrophobic complementarity, polar complementarity, solvation terms, and entropic terms, was deemed a stable interaction when the value was higher than 5 [39].
3. Results
3.1. Transcriptome-Wide Detection of m6A Modification after Humantenine Treatment of HCT116 Cells
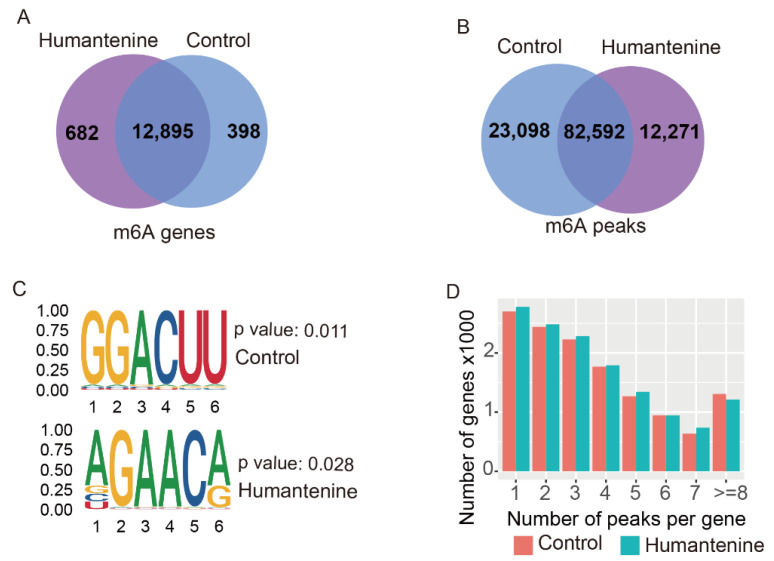
To explore the mechanism of action of humantenine in HCT116 human colon cancer cells, MeRIP-seq analysis and RNA-seq were performed. In the humantenine group, 94,863 m6A peaks containing transcripts of 13,577 genes were identified by the R package exomePeak. Similarly, 105,690 m6A peaks were identified in the control group, representing transcripts of 13,293 genes. In addition, 82,592 peaks corresponding to 12,895 genes were found at the intersection of the humantenine and control groups (Figure 2A,B). We then used STREME to determine whether the m6A consensus sequence of RRACH (where R represents purine, A is m6A, and H is a non-guanine base) was reported in the detection of m6A (Figure 2C). The result showed the existence of some motifs, including the classical consensus sequence in the control and humantenine groups. Notably, the majority of genes had 1–3 m6A methylation peaks, whereas a relatively small number of genes contained four or more m6A methylation peaks in both humantenine and control groups (Figure 2D).
Figure 2.
The m6A modification patterns in humantenine and control groups. (A) Venn diagram showing overlap of the m6A genes from both groups. (B) Venn diagram showing overlap of the m6A peaks from both groups. (C) The motifs for m6A peak regions based on STREME. (D) The distribution of m6A methylation peaks per gene.
3.2. Distribution of m6A Modification in the Transcriptome
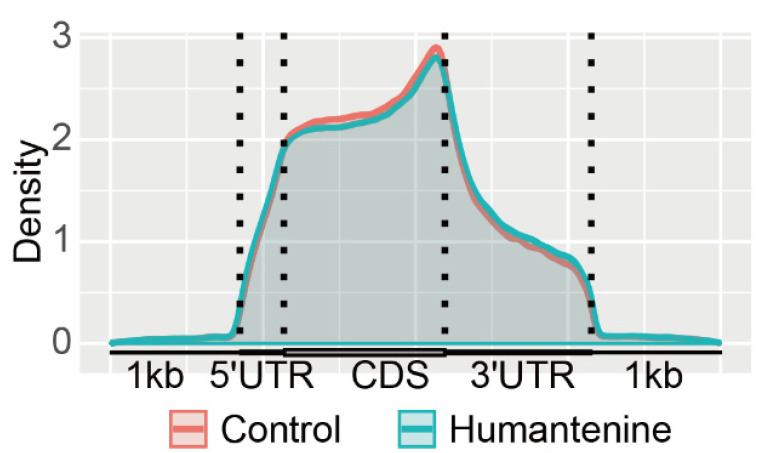
The distributions of m6A methylation in the whole transcriptomes of the humantenine and control groups were analyzed. The result showed that m6A modification tended to enrich in five transcript segments: 5′untranslated region (5′UTR), the start codon segment, coding sequence (CDS), the stop codon segment, and 3′UTR. The m6A peak density increased rapidly between the 5′UTR and the start codon and was relatively gentle in the CDS region. The highest density region was near the stop codon. In the 3′UTR region, the density of the m6A peaks decreased rapidly (Figure 3).
Figure 3.
Density of m6A methylation peaks in mRNA transcripts.
3.3. Differentially Methylated Genes and Differentially Expressed Genes
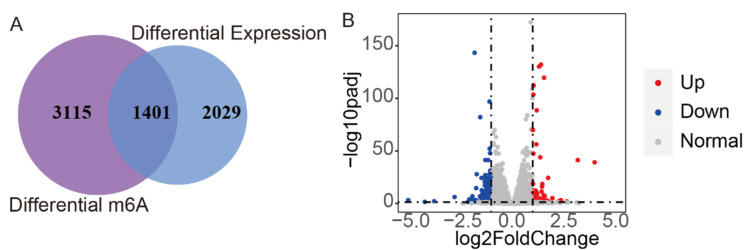
Setting the statistical standard as p ≤ 0.05, a total of 4516 differential m6A-modified mRNAs were found through m6A-seq data, and 3430 mRNAs were determined to be differentially expressed between humantenine and control groups after the analysis of RNA-seq data. In addition, 1401 genes were consistently observed at the overlap of differentially m6A-modified mRNA and differentially expressed mRNA (Figure 4A). When the statistical standard was set as fold change ≥ 1 and p ≤ 0.05, 681 differentially expressed mRNAs were screened out, of which 248 were up-regulated and 433 were down-regulated (Figure 4B).
Figure 4.
(A) Differential m6A modified mRNA and differential mRNA expression in humantenine and control groups. (B) The volcano plot of differentially expressed mRNAs between both groups.
3.4. KEGG and GO Annotation of the Overlap of Differentially m6A-Modified mRNA and Differentially Expressed mRNA
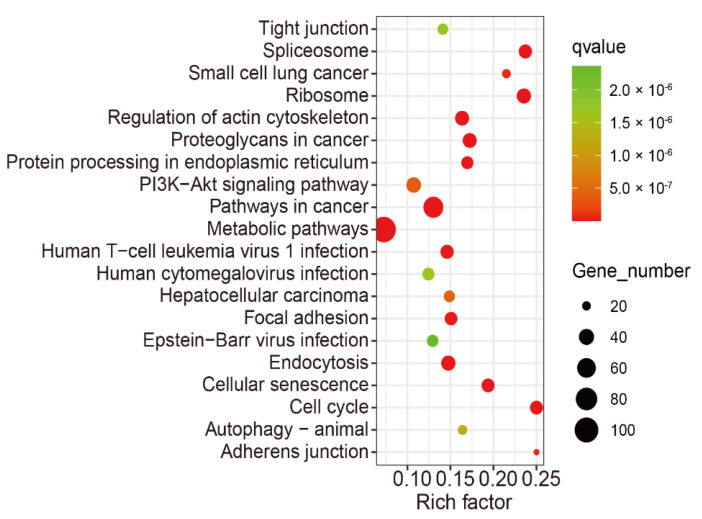
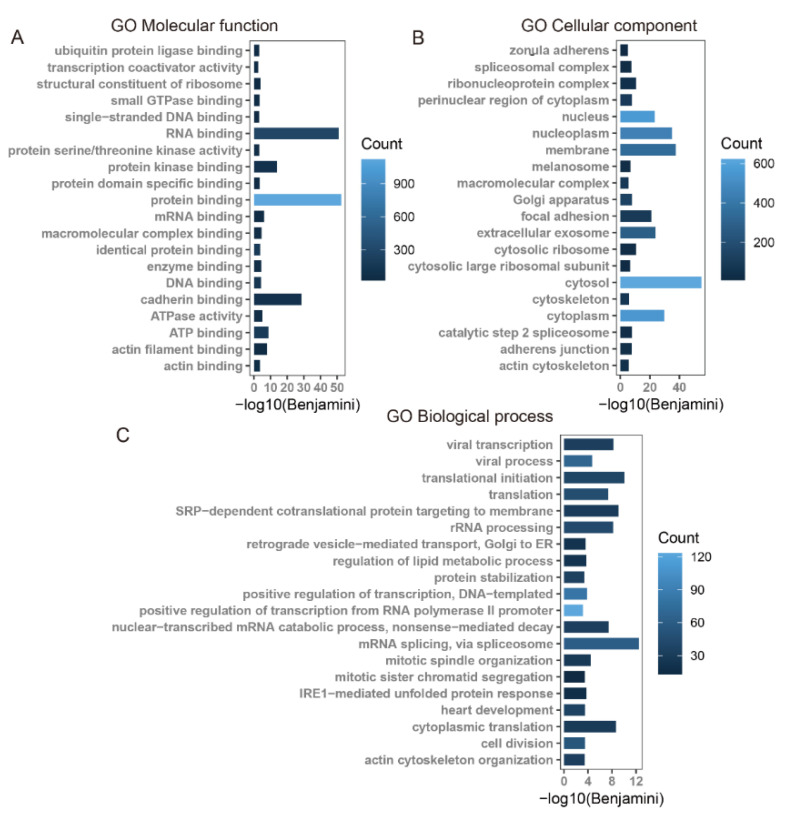
Using KEGG pathway and GO enrichment analysis on the DAVID web server, the 1401 overlapping genes of differentially m6A-modified mRNA and differentially expressed mRNAs were associated with significant pathways and biological functions. The KEGG pathway analysis revealed that these genes were mainly involved in tight junction, regulation of actin cytoskeleton, metabolic pathways, focal adhesion, and adherens junction pathways, and so on (Figure 5). The GO enrichment analysis was classified into three functional types: molecular function (MF), cellular component (CC), and biological process (BP). The MF terms included actin filament binding and actin binding. The CC terms included zonula adherens, cytoskeleton, adherens junction, and actin cytoskeleton. The BP terms included actin cytoskeleton organization (Figure 6).
Figure 5.
The top 20 KEGG analysis enriched pathways of the 1401 overlapped genes of differentially m6A modified mRNA and differentially expressed mRNA.
Figure 6.
GO functional annotation of 1401 overlapped genes of differentially m6A-modified mRNAs and differentially expressed mRNAs. (A) GO molecular function. (B) GO cellular component. (C) GO biological process.
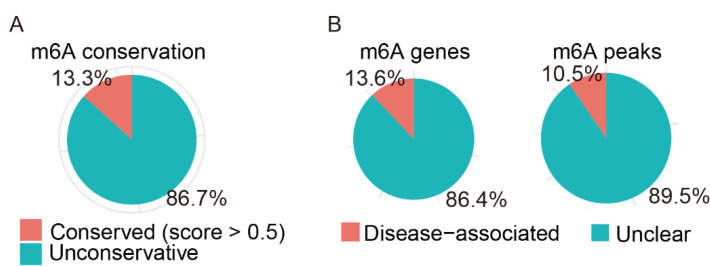
3.5. The m6A Conservation and Disease Association
As shown in Figure 7A, based on the analysis of ConsRM, 86.7% of m6A-modified sites were nonconservative in the 1401 overlapping genes of differentially m6A-modified mRNAs and differentially expressed mRNAs. As shown in Figure 7B, the results of the RMDisease analysis showed that 13.6% of m6A genes and 10.5% of m6A peaks were associated with diseases.
Figure 7.
Analysis of m6A conservation and disease association. (A) The conserved and un-conserved m6A sites in differentially m6A-modified mRNA. (B) The disease-associated m6A genes and peaks in differentially m6A-modified mRNA.
3.6. Gene Expression of Tight Junctions, Adherens Junctions, and Regulation of Actin Cytoskeleton, and Their Potential Regulators
According to the RNA-seq data in HCT116 cells, after treatment with humantenine, the mRNA expression levels of genes associated with tight junctions (CLDN4, CDK4, TJP3, MAGI1), adherens junctions (ACTN1, NECTIN2, WASF1), and regulation of actin cytoskeleton (MYH9, IQGAP3, DIAPH3, WASF1, ARPC5L, DOCK1) were significantly differentially expressed (Table 1). The potential regulators, including parts of writers, readers, and erasers, were identified using CLIP techniques. The results showed that the reader IGF2BP3 was involved in the regulation of all these genes.
Table 1.
The mRNA expression levels of tight junction, adherens junction, regulation of actin cytoskeleton, and their potential regulators.
| Gene | log2FoldChange | padj | Writer | Reader | Eraser | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| METTL3 | HNRNPA2B1 | IGF2BP3 | YTHDC1 | YTHDC2 | YTHDF2 | ALKBH5 | ||||
| Tight junction | CLDN4 | 0.5708 | 8.04 × 10−32 | N | N | Y | N | N | N | N |
| CDK4 | 0.4725 | 4.43 × 10−26 | Y | Y | Y | Y | N | N | N | |
| TJP3 | −0.9036 | 1.84 × 10−67 | N | N | Y | N | N | N | N | |
| MAGI1 | −0.4761 | 6.83 × 10−16 | Y | Y | Y | Y | N | N | N | |
| Adherens junction | ACTN1 | 0.2529 | 3.31 × 10−11 | Y | Y | Y | Y | N | N | N |
| NECTIN2 | 0.2513 | 1.58 × 10−5 | N | Y | Y | Y | N | N | N | |
| WASF1 | −0.4224 | 9.38 × 10−5 | Y | N | Y | Y | N | N | N | |
| Regulation of actin cytoskeleton | MYH9 | 0.3207 | 2.97 × 10−22 | Y | Y | Y | Y | N | N | N |
| IQGAP3 | −0.8892 | 3.82 × 10−16 | N | Y | Y | Y | N | N | N | |
| DIAPH3 | −0.6702 | 2.45 × 10−9 | N | Y | Y | Y | N | N | Y | |
| WASF1 | −0.4224 | 9.38 × 10−5 | Y | N | Y | Y | N | N | N | |
| ARPC5L | −0.3197 | 1.42 × 10−4 | N | Y | Y | Y | N | Y | N | |
| DOCK1 | −0.3172 | 8.64 × 10−6 | Y | N | Y | Y | N | N | N | |
Y means the mRNA was the substrate of the RNA m6A regulators, and N means not.
3.7. Potential RNA m6A Regulators of Differentially Methylated Genes
In order to identify the potential regulators of RNA m6A methylation, we analyzed the mRNA expression levels of 22 RNA m6A methylation writers, readers, and erasers. As shown in Table 2, four writers, six readers, and one eraser were significantly differentially expressed (padj ≤ 0.05). Among them, writers RBM15 and METTL3, reader YTHDF3, and eraser ALKBH5 were up-regulated; writers RBM15B and ZC3H13, and readers HNRNPA2B1, YTHDF2, YTHDC2, IGF2BP3, and YTHDC1 were down-regulated.
Table 2.
The mRNA expression levels of m6A regulators in humantenine-treated HCT116 cells.
| Gene | Regulation | Base Mean | log2FoldChange | padj |
|---|---|---|---|---|
| ALKBH5 | eraser | 5355.58 | 0.5337 | 2.43 × 10−18 |
| HNRNPA2B1 | reader | 31,317.46 | −0.2827 | 3.61 × 10−13 |
| YTHDF2 | reader | 5827.43 | −0.3176 | 1.44 × 10−7 |
| YTHDC2 | reader | 1996.91 | −0.3977 | 9.40 × 10−5 |
| IGF2BP3 | reader | 7360.70 | −0.1831 | 5.89 × 10−4 |
| RBM15B | writer | 4931.66 | −0.2692 | 5.93 × 10−4 |
| YTHDF3 | reader | 4575.69 | 0.2471 | 2.44 × 10−3 |
| YTHDC1 | reader | 4745.43 | −0.1911 | 3.73 × 10−3 |
| RBM15 | writer | 1909.31 | 0.3036 | 6.91 × 10−3 |
| METTL3 | writer | 1801.37 | 0.2508 | 2.14 × 10−2 |
| ZC3H13 | writer | 8932.93 | −0.1169 | 2.65 × 10−2 |
| VIRMA | writer | 6469.09 | 0.1091 | 1.41 × 10−1 |
| FTO | eraser | 2646.90 | 0.1141 | 2.69 × 10−1 |
| IGF2BP1 | reader | 65.30 | 0.5237 | 3.67 × 10−1 |
| YTHDF1 | reader | 5181.16 | 0.0458 | 5.94 × 10−1 |
| HNRNPC | reader | 24,691.72 | −0.0436 | 6.68 × 10−1 |
| METTL14 | writer | 2711.17 | 0.0432 | 7.66 × 10−1 |
| METTL5 | writer | 1640.50 | 0.0410 | 7.80 × 10−1 |
| CBLL1 | writer | 2958.85 | −0.0289 | 8.24 × 10−1 |
| IGF2BP2 | reader | 8415.70 | −0.0156 | 8.55 × 10−1 |
| FMR1 | reader | 2456.26 | −0.0059 | 9.70 × 10−1 |
| WTAP | writer | 2760.03 | 0.0004 | 9.98 × 10−1 |
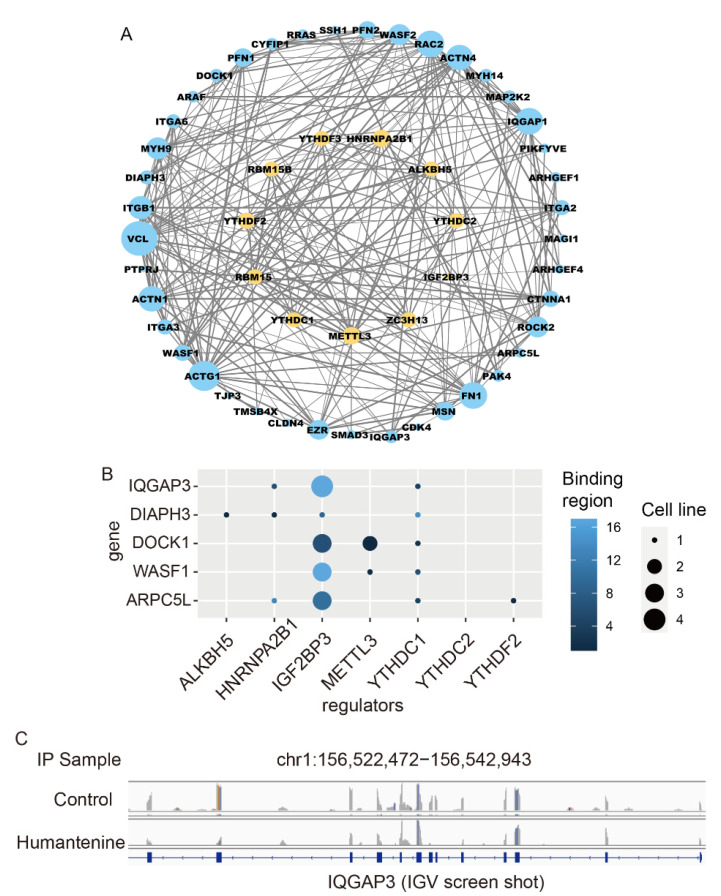
Since the GO and KEGG terms included tight junctions, adherens junctions, and actin cytoskeleton, as shown in Figure 5 and Figure 6, the genes enriched in these terms were collected. We then analyzed the regulatory relationship between these genes and 11 differentially expressed RNA m6A methylation regulators in a PPI network (Figure 8A). The degrees of the 11 regulators ALKBH5, HNRNPA2B1, YTHDF2, YTHDC2, IGF2BP3, RBM15B, YTHDF3, YTHDC1, RBM15, METTL3, and ZC3H13 were 9, 11, 10, 9, 3, 9, 9, 9, 9, 11, and 9, respectively.
Figure 8.
(A) The protein-protein interactions between m6A regulators (yellow circle) and actin cytoskeleton, tight junction, and adherens junction factors (blue circle). (B) The effect of m6A regulators on the differentially expressed genes of actin cytoskeleton. (C) The level of m6A on IQGAP3 mRNA transcripts observed by IGV.
To explore the substrates of RNA m6A methylation regulators, the writer METTL3, readers HNRNPA2B1, IGF2BP3, YTHDC1, YTHDC2, and YTHDF2, and eraser ALKBH5 were selected, as well as the actin cytoskeleton genes IQGAP3, DIAPH3, DOCK1, WASF1, and ARPC5L. The result showed that insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) was involved in the regulation of all five of these actin cytoskeleton genes (Figure 8B). As indicated by the IGV display of IGF2BP3, the m6A methylation level on mRNA transcripts was decreased after humantenine treatment (Figure 8C).
3.8. Molecular Interactions of Humantenine with Differently Expressed Regulators of RNA m6A Modification
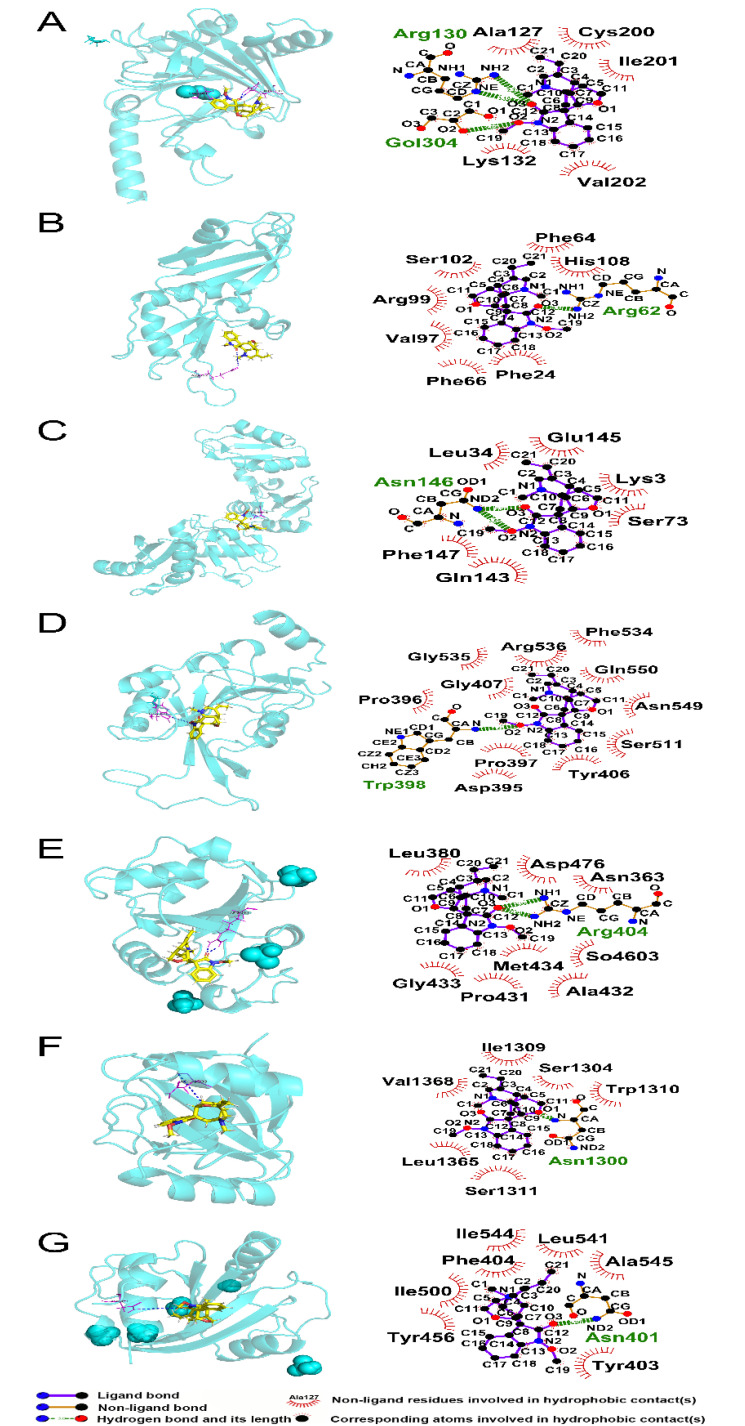
We evaluated the molecular interactions of humantenine with differentially expressed regulators of m6A modification, including ALKBH5, HNRNPA2B1, IGF2BP3, METTL3, YTHDC1, YTHDC2, and YTHDF2. The results are shown in Figure 9 and Table 3. All the total scores were over 5, which suggested stabilized interactions between humantenine and these regulators.
Figure 9.
Molecular interactions between humantenine and differently expressed regulators of m6A modification. Both 3D and 2D interaction models were presented. (A) ALKBH5, (B) HNRNPA2B1, (C) IGF2BP3, (D) METTL3, (E) YTHDC1, (F) YTHDC2 and (G) YTHDF2.
Table 3.
Molecular interactions between humantenine and differently expressed regulators of m6A modification.
| Protein | PDB ID | Total Score | Crach | Polar | H-Bond Number | Residues Involved in H-Bond Formation | Hydrophobic Contacts Number | Residues Involved in Hydrophobic Contacts |
|---|---|---|---|---|---|---|---|---|
| ALKBH5 | 4NJ4 | 6.7567 | −0.8591 | 1.9259 | 3 | Arg130 (2 hydrogen bonds), Gol304 | 5 | Ala127, Lys132, Cys200, Ile201, Val202 |
| HNRNPA2B1 | 5HO4 | 5.1469 | −0.5213 | 0.9835 | 1 | Arg62 | 7 | Phe24, Phe64, Phe66, Val97, Arg99, Ser102, His108 |
| IGF2BP3 | 6FQ1 | 6.5409 | −0.9417 | 1.2075 | 2 | Asn146 (2 hydrogen bonds) | 6 | Lys3, Leu34, Ser73, Gln143, Glu145, Phe147 |
| METTL3 | 5IL2 | 6.09 | −1.0199 | 0.001 | 1 | Trp398 | 11 | Asp395, Pro396, Pro397, Tyr406, Gly407, Ser511, Phe534, Gly535, Arg536, Asn549, Gln550 |
| YTHDC1 | 6ZCN | 5.3546 | −0.3608 | 2.0979 | 2 | Arg404 (2 hydrogen bonds) | 8 | Asn363, Leu380, Pro431, Ala432, Gly433, Met434, Asp476, So4603 |
| YTHDC2 | 6K6U | 6.2361 | −0.9342 | 0 | 1 | Asn1300 | 6 | Ser1304, Ile1309, Trp1310, Ser1311, Leu1365, Val1368 |
| YTHDF2 | 7R5F | 6.2342 | −0.7086 | 0 | 1 | Asn401 | 7 | Tyr403, Phe404, Tyr456, Ile500, Leu541, Ile544, Ala545 |
Theoretically, humantenine bound ALKBH5 via the formation of two hydrogen bonds at Arg 130 and one hydrogen bond at Gol304, and five hydrophobic contacts with Ala127, Lys132, Cys200, Ile201, and Val202 (Figure 9A). Humantenine bound HNRNPA2B1 via the formation of one hydrogen bond at Arg62, and seven hydrophobic contacts with Phe24, Phe64, Phe66, Val97, Arg99, Ser102, and His108 (Figure 9B). Humantenine bound IGF2BP3 via the formation of two hydrogen bonds at Asn146, and six hydrophobic contacts with Lys3, Leu34, Ser73, Gln143, Glu145, and Phe147 (Figure 9C). Humantenine bound METTL3 via the formation of one hydrogen bond at Trp398, and eleven hydrophobic contacts with Asp395, Pro396, Pro397, Tyr406, Gly407, Ser511, Phe534, Gly535, Arg536, Asn549, and Gln550 (Figure 9D). Humantenine bound YTHDC1 via the formation of two hydrogen bonds at Arg404 and eight hydrophobic contacts with Asn363, Leu380, Pro431, Ala432, Gly433, Met434, Asp476, and So4603 (Figure 9E). Humantenine bound YTHDC2 via the formation of one hydrogen bond at Asn1300 and six hydrophobic contacts with Ser1304, Ile1309, Trp1310, Ser1311, Leu1365, and Val1368 (Figure 9F). Humantenine bound YTHDF2 via the formation of one hydrogen bond at Asn401 and seven hydrophobic contacts with Tyr403, Phe404, Tyr456, Ile500, Leu541, Ile544, and Ala545 (Figure 9G).
4. Discussion
The traditional medicine G. elegans exerts excellent pharmacological therapeutic effects in the treatment of a variety of clinical symptoms, especially neuralgia and inflammation. On the other hand, the severe side effects in multiple systems limit its wider application. Therefore, it is in urgent need to reveal the molecular mechanism of G. elegans-induced intestinal irritation, for the clinic drug safety. In the present study, the indole alkaloid humantenine was used in human intestinal cells to analyze the potential epigenetic alteration of RNA m6A modification, and the mRNA transcriptional profile, based on high-throughput m6A MeRIP-seq and mRNA-seq.
Through analysis of m6A MeRIP-seq data, the motif sequence was successfully identified with statistical significance in the region around the m6A peak of the control group and humantenine group. The consensus motif sequence RRACH was observed in the m6A motif region, as proven previously [40,41]. As to the distribution of m6A modification peaks, most genes had 1~3 peaks, and parts of genes contained more peaks, which was consistent with the study by Chen et al. [42]. When the genes were divided into 5′UTR, start codon, CDS, stop codon, and 3′UTR, most m6A modification peaks were in the CDS region, and the highest value was around the stop codon, as frequently reported in the past [40,43]. A higher number of m6A modification sites at the stop codon may be associated with mRNA stability and translation [44].
In the present study, humantenine treatment induced 86.7% RNA m6A modification in un-conserved sites, and 13.6% of genes were associated with diseases, suggesting that this xenobiotic compound might cause intestinal diseases. To explore the biological function change induced by RNA m6A modification, GO and KEGG pathway enrichment analysis was performed. After humantenine treatment in HCT116, a total of 1041 genes was reported in the overlap of the differentially m6A-modified mRNA and differentially expressed mRNA. The top 20 KEGG terms included tight junction, regulation of actin cytoskeleton, and adherens junction. In the GO terms, cadherin binding, actin filament binding, and actin binding were enriched in MF; zonula adherens, focal adhesion, cytoskeleton, adherens junction, and actin cytoskeleton were enriched in CC; actin cytoskeleton organization was enriched in BP. In clinic, diarrhea is the main intestinal side effect of G. elegans, manifested by increased intestinal permeability, which is regulated by intestinal intercellular junction and actin cytoskeleton. The intercellular junction mainly comprises tight junctions and adherens junctions, and acts as the mechanical barrier in defense against exogenous toxic compounds [45]. It also regulates the paracellular transport pathway to maintain intestinal cell integrity [46,47]. Kumar et al. found that Cryptosporidium parvum disrupted intestinal barrier function by downregulating the expression of tight junctions and adherens junctions, resulting in diarrhea [48]. In this study, expression levels of multiple mRNAs involved in tight junctions, adherens junctions, and regulation of actin cytoskeleton were significantly differentially expressed in the humantenine-treated group. These genes encoded proteins of claudin 4, zonula occludens protein 3, actinin α 1, nectin cell adhesion molecule 2, Wiskott–Aldrich syndrome protein family member 1, myosin heavy chain 9, IQ motif containing GTPase activating protein 3 (IQGAP3), diaphanous related formin 3, wasp family member 1, actin related protein 2/3 complex subunit 5, dedicator of cytokinesis 1, and so on. If the junction proteins and actin cytoskeleton were destroyed by humantenine, the paracellular pathway transport would be in disorder, characterized by abnormal osmotic pressure and permeability in intestinal cells [49,50].
RNA m6A modifications were mediated by three specific categories of proteins, namely m6A methyltransferases, m6A demethylases, and m6A-biding proteins. The biological effects of m6A methylation modifications depend on the recognition and binding of m6A-binding proteins [51]. Using the computational prediction method, humantenine was combined with the m6A regulators with total scores higher than 5. Therefore, the present study further tried to identify the m6A regulators of the differentially expressed genes related to intercellular junctions and actin cytoskeleton. A total of 11 m6A regulators were differentially expressed, of which IGF2BP3, an m6A reader exerting the main function of enhancing the stability of mRNA [52], was regarded as the core factor in the regulation of mRNA translation, decay, and storage. For example, IQGAP3 is a Ras GTPase-activating-like protein that belongs to the family of scaffolding proteins and regulates cell adhesion by mediating the actin cytoskeleton [53]. The m6A methylated mRNA of IQGAP3 can be recognized by IGF2BP3, thus maintaining a stable state in nuclear and cytoplasmic conditions. As downstream biological effects, the translation and storage of IQGAP3 mRNA were promoted and the decay of IQGAP3 mRNA was inhibited. In this study, humantenine inhibited the mRNA m6A methylation level of IQGAP3, making it difficult for IGF2BP3 to recognize and bind IQGAP3. As a result, the mRNA stability and expression level of IQGAP3 decreased, which would cause the actin cytoskeleton to collapse and thus enhance intestinal permeability.
5. Conclusions
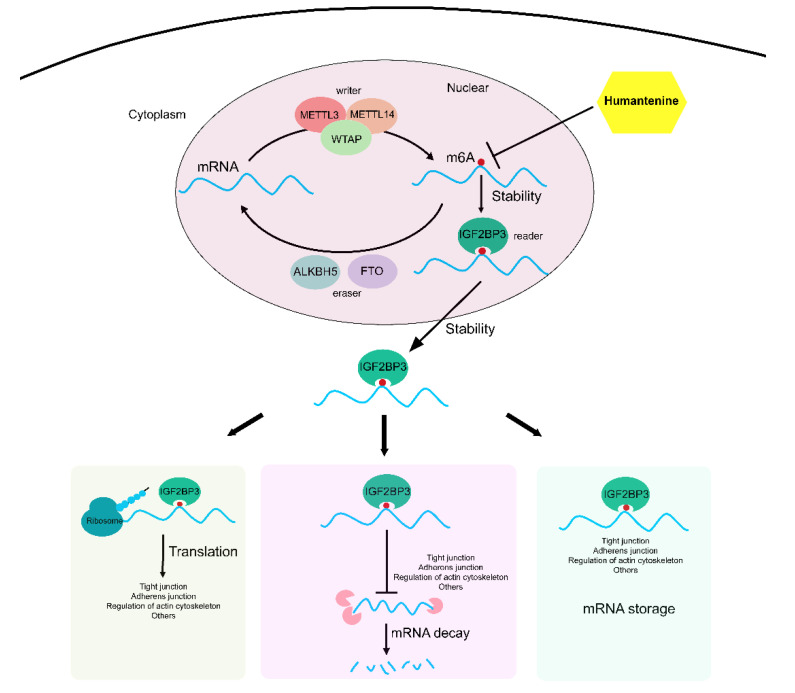
In summary, the present study investigated the mRNA m6A modification and expression profiles of HCT116 human colon cancer cells treated with humantenine. The abnormal mRNA expression of m6A regulators, together with disordered m6A methylation levels of target genes, led to the disruption of mRNA stability, translation, storage, and decay (Figure 10). This study has therefore revealed a possible mechanism for the increased intestinal permeability induced by humantenine.
Figure 10.
Humantenine induced the abnormal m6A methylation of genes involved in tight junctions, adherens junctions, and regulation of actin cytoskeleton. It consequently disrupted mRNA translation, storage, and decay.
Abbreviations
| 3′UTR | 3′ untranslated regions |
| 5′UTR | 5′ untranslated regions |
| CDS | coding sequence (CDS) |
| G. elegans | Gelsemium elegans (Gardner & Chapm.) Benth. |
| GO | Gene Ontology |
| HPLC | high-performance liquid chromatography |
| IGF2BP3 | insulin-like growth factor 2 mRNA-binding protein 3 |
| IGV | Integrative Genomics Viewer |
| IQGAP3 | IQ motif containing GTPase activating protein 3 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| m6A | N6-methyladenosine |
| MeRIP-seq | methylated RNA immunoprecipitation sequencing |
| NMR | nuclear magnetic resonance |
| PPI | protein–protein interaction |
Author Contributions
Conceptualization, A.Z.; methodology, X.C. and X.H.; software, W.B., X.H., C.L. and J.L.; data curation, Y.W., formal analysis, Y.W., W.B., C.L., J.L. and D.Z.; investigation, Y.W., X.C., W.B. and C.L.; visualization, W.B., X.H., C.L. and J.L.; writing—original draft, Y.W. and X.C.; writing—review and editing, A.Z.; supervision, A.Z.; funding acquisition, A.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data is provided in the manuscript. Please contact at zhuan@fjmu.edu.cn for any raw data files and further analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The work is supported by the National Natural Science Foundation of China (82104520), Natural Science Foundation of Fujian Province (2021J05045), Fujian Medical University High-level Talent Research Startup Funding Project (XRCZX2020014), and Open Research Fund of Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education (FMUGIC-202101).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhao Y.T., Wu S.P., Hu C.L., Kang J., Zhao M. Reviews on chemical compositions and pharmacological effect of Gelsemium elegans. Chin. J. Exp. Tradit. Med. Formulae. 2019;25:200–210. [Google Scholar]
- 2.Cao J.J., Yang K., Huang C.Y., Li Y.J., Yu H., Wu Y., Sun Z.L., Liu Z.Y. Pharmacokinetic study of multiple components of Gelsemium elegans in goats by Ultra-performance liquid chromatography coupled to tandem mass spectrometry. J. Anal. Toxicol. 2020;44:378–390. doi: 10.1093/jat/bkz100. [DOI] [PubMed] [Google Scholar]
- 3.Lin H.L., Qiu H.Q., Cheng Y., Liu M.B., Chen M.H., Que Y.X., Que W.C. Gelsemium elegans Benth: Chemical components, pharmacological effects, and toxicity mechanisms. Molecules. 2021;26:7145. doi: 10.3390/molecules26237145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun M.X., Xun Q.Q., Meng W.Q., Cen J.F., Mao G.C., Pei Z.P., Xiao K. Research progress on pharmacology and toxicological mechanism of Gelsemium elegans. J. Toxicol. 2020;34:336–341. [Google Scholar]
- 5.Wang R., Ding F., Gao Y.J., Wang X.Y., Li Q. Epidemiological analysis for vegetal food poisoning in China, 2004–2013. Chin. J. Food Hyg. 2016;28:580–584. [Google Scholar]
- 6.Zhong Y.X., Xie Y.H., Jiang Y.Y., Liu Y.P., Shi M.M., Yang W.M. Analysis of gelsemine poisoning events in the Guangxi Zhuang autonomous region during 2015–2017. Chin. J. Food Hyg. 2019;31:81–83. [Google Scholar]
- 7.Liu Y.C., Sun Z.L., Liu Z.Y. Research progress on non-alkaloids analysis for chemical constituents from Gelsemium. Chin. J. Vet. Drug. 2016;50:55–64. [Google Scholar]
- 8.Mu F.Y., Mu F.W., Mu F.M., Chen Z.H., Ma X.D., Wu D., Guo X.K., Liu C.X. Study on chemical components and pharmacological activity of Gelsemium elegans. Sci. Technol. Innov. 2015;3:75–77. [Google Scholar]
- 9.Tan J.Q., Qiu C.Z., Zheng L.Z. Analgesic effect and no physical dependence of Gelsemium elegans Benth. Pharmacol. Clin. Chin. Mater. Med. 1988;4:24–28. [Google Scholar]
- 10.Li X., Xie X.L., Gu Y.C., Zhang J.M., Song J., Cheng X.F., Gao Y.J., Ai Y.L. Fat mass and obesity-associated protein regulates tumorigenesis of arecoline-promoted human oral carcinoma. Cancer Med. 2021;10:6402–6415. doi: 10.1002/cam4.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z.Y., Zeng J.H., Guo Q., Pu K.M., Yang Y., Chen N.Z., Zhang G., Zhao M.Y., Zheng Q., Tang J.Y., et al. Berberine suppresses stemness and tumorigenicity of colorectal cancer stem-like cells by inhibiting m6A methylation. Front. Oncol. 2021;11:775418. doi: 10.3389/fonc.2021.775418. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A., et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C., et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J.M., Song B.W., Wei Z., Huang D.Y., Zhang Y.X., Su J.L., de Magalhães J.P., Rigden D.J., Meng J., Chen K.Q. m5C-Atlas: A comprehensive database for decoding and annotating the 5-methylcytosine (m5C) epitranscriptome. Nucleic Acids Res. 2022;50:D196–D203. doi: 10.1093/nar/gkab1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song B.W., Tang Y.J., Chen K.Q., Wei Z., Rong R., Lu Z.L., Su J.L., de Magalhães J.P., Rigden D.J., Meng J. m7GHub: Deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics. 2020;36:3528–3536. doi: 10.1093/bioinformatics/btaa178. [DOI] [PubMed] [Google Scholar]
- 16.Kadumuri R.V., Janga S.C. Epitranscriptomic code and its alterations in human disease. Trends Mol. Med. 2018;24:886–903. doi: 10.1016/j.molmed.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones J.D., Monroe J., Koutmou K.S. A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. Wiley Interdiscip. Rev. RNA. 2020;11:e1586. doi: 10.1002/wrna.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z.J., Luo K.X., Zou Z.Y., Qiu M.G.Y., Tian J.K., Sieh L., Shi H.L., Zou Y.X., Wang G., Morrison J., et al. Genetic analyses support the contribution of mRNA N6-methyladenosine (m6A) modification to human disease heritability. Nat. Genet. 2020;52:939–949. doi: 10.1038/s41588-020-0644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao G., Li H.B., Yin Z., Flavell R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erson-Bensan A.E., Begik O. m6A modification and implications for microRNAs. MicroRNA. 2017;6:97–101. doi: 10.2174/2211536606666170511102219. [DOI] [PubMed] [Google Scholar]
- 22.Paramasivam A., Priyadharsini J.V. Epigenetic modifications of RNA and their implications in antiviral immunity. Epigenomics. 2020;12:1673–1675. doi: 10.2217/epi-2020-0307. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W., Qi X.Q., Liu L.N., Ma S.Q., Liu J.W., Wu J. Epigenetic regulation of m6a modifications in human cancer. Mol. Ther-Nucl. Acids. 2020;19:405–412. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 25.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng J., Cui X.D., Rao M.K., Chen Y.D., Huang Y.F. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29:1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y.J., Chen K.Q., Song B.W., Ma J.M., Wu X.Y., Xu Q.R., Wei Z., Su J.L., Liu G., Rong R., et al. m6A-Atlas: A comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 2021;49:D134–D143. doi: 10.1093/nar/gkaa692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics. 2021;37:2834–2840. doi: 10.1093/bioinformatics/btab203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu D.C., Luo H.T., Huo P.P., Wang Z.H., Zhang S., He Z.H., Wu Y., Zhao L.H., Liu J.J., Guo J.C., et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao X.L., Sherman B.T., da Huang W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Chen K.Q., Wei Z., Coenen F., Su J.L., Meng J. MetaTX: Deciphering the distribution of mRNA-related features in the presence of isoform ambiguity, with applications in epitranscriptome analysis. Bioinformatics. 2021;37:1285–1291. doi: 10.1093/bioinformatics/btaa938. [DOI] [PubMed] [Google Scholar]
- 34.Song B.W., Chen K.Q., Tang Y.J., Wei Z., Su J.L., de Magalhães J.P., Rigden D.J., Meng J. ConsRM: Collection and large-scale prediction of the evolutionarily conserved RNA methylation sites, with implications for the functional epitranscriptome. Brief. Bioinform. 2021;22:bbab088. doi: 10.1093/bib/bbab088. [DOI] [PubMed] [Google Scholar]
- 35.Chen K.Q., Song B.W., Tang Y.J., Wei Z., Xu Q.R., Su J.L., de Magalhães J.P., Rigden D.J., Meng J. RMDisease: A database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021;49:D1396–D1404. doi: 10.1093/nar/gkaa790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H., Zhang L., Zhu A., Liu X.Y., Wang T.X., Wan M.Q., Yang X.W., Zhang Y.T., Zhang Y.B. Metabolic profiling of nuciferine in vivo and in vitro. J. Agric. Food Chem. 2020;68:14135–14147. doi: 10.1021/acs.jafc.0c04468. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y.Q., Zhang X., Shen X.F., Wang S., Wang Q., Yang X.W. Computational and experimental characterization of isomers of escin-induced renal cytotoxicity by inhibiting heat shock proteins. Eur. J. Pharmacol. 2021;908:174372. doi: 10.1016/j.ejphar.2021.174372. [DOI] [PubMed] [Google Scholar]
- 39.Zhu A., Sun Y.Q., Zhong Q.W., Yang J.L., Zhang T., Zhao J.W., Wang Q. Effect of euphorbia factor L1 on oxidative stress, apoptosis, and autophagy in human gastric epithelial cells. Phytomedicine. 2019;64:152929. doi: 10.1016/j.phymed.2019.152929. [DOI] [PubMed] [Google Scholar]
- 40.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 41.Harper J.E., Miceli S.M., Roberts R.J., Manley J.L. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y.M., Zhou C.X., Sun Y.Y., He X.Z., Xue D. m6A RNA modification modulates gene expression and cancer-related pathways in clear cell renal cell carcinoma. Epigenomics. 2020;12:87–99. doi: 10.2217/epi-2019-0182. [DOI] [PubMed] [Google Scholar]
- 43.Luo G.Z., MacQueen A., Zheng G.Q., Duan H.C., Dore L.C., Lu Z.K., Liu J., Chen K., Jia G.F., Bergelson J., et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014;5:5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Lu Z.K., Gomez A., Hon G.C., Yue Y.N., Han D.L., Fu Y., Parisien M., Dai Q., Jia G.F., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee A., White N., van der Walle C.F. The intestinal zonula occludens toxin (ZOT) receptor recognises non-native ZOT conformers and localises to the intercellular contacts. FEBS Lett. 2003;555:638–642. doi: 10.1016/S0014-5793(03)01348-6. [DOI] [PubMed] [Google Scholar]
- 46.Farhadi A., Banan A., Fields J., Keshavarzian A. Intestinal barrier: An interface between health and disease. J. Gastroenterol. Hepatol. 2003;18:479–497. doi: 10.1046/j.1440-1746.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- 47.Tervonen A., Ihalainen T.O., Nymark S., Hyttinen J. Structural dynamics of tight junctions modulate the properties of the epithelial barrier. PLoS ONE. 2019;14:e0214876. doi: 10.1371/journal.pone.0214876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar A., Chatterjee I., Anbazhagan A.N., Jayawardena D., Priyamvada S., Alrefai W.A., Sun J., Borthakur A., Dudeja P.K. Cryptosporidium parvum disrupts intestinal epithelial barrier function via altering expression of key tight junction and adherens junction proteins. Cell. Microbiol. 2018;20:e12830. doi: 10.1111/cmi.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guttman J.A., Finlay B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta. 2009;1788:832–841. doi: 10.1016/j.bbamem.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 50.Pérez-Reytor D., Jaña V., Pavez L., Navarrete P., García K. Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front Microbiol. 2018;9:2248. doi: 10.3389/fmicb.2018.02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y.C., Shi Y.F., Shen H.F., Xie W.Z. m6A-binding proteins: The emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020;13:35. doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell. Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T., Wang S., Kaibuchi K. IQGAPs as key regulators of actin-cytoskeleton dynamics. Cell Struct. Funct. 2015;40:69–77. doi: 10.1247/csf.15003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data is provided in the manuscript. Please contact at zhuan@fjmu.edu.cn for any raw data files and further analysis.