Abstract
RNA interference (RNAi) has been exploited by scientists worldwide to make a significant contribution in the arena of sustainable agriculture and integrated pest management. These strategies are of an imperative need to guarantee food security for the teeming millions globally. The already established deleterious effects of chemical pesticides on human and livestock health have led researchers to exploit RNAi as a potential agri-biotechnology tool to solve the burning issue of agricultural wastage caused by pests and pathogens. On the other hand, CRISPR/Cas9, the latest genome-editing tool, also has a notable potential in this domain of biotic stress resistance, and a constant endeavor by various laboratories is in progress for making pathogen-resistant plants using this technique. Considerable outcry regarding the ill effects of genetically modified (GM) crops on the environment paved the way for the research of RNAi-induced double-stranded RNAs (dsRNA) and their application to biotic stresses. Here, we mainly focus on the application of RNAi technology to improve disease resistance in plants and its relevance in today’s CRISPR-dominated world in terms of exogenous application of dsRNAs. We also focused on the ongoing research, public awareness, and subsequent commercialization of dsRNA-based biocontrol products.
Keywords: biopesticides, biotic stress, CRISPR/Cas9, dsRNAs, gene edited, GMO, RNAi
Introduction
Agricultural sector is one of the major contributors to the economy of lower-middle-income countries such as India. Yet each year, biotic stresses such as insects, fungi, bacteria, and nematodes cause substantial losses in crop productivity and overall agricultural production. Conventional breeding methods proved to be time-consuming and inefficient to come up with disease-resistant smart crops for a sustainable agricultural system, calling for the need of plant biotechnological interventions. RNA interference (RNAi), discovered more than a decade ago, is one of the central players of gene regulation and mounting defense against viral infections in plants. Through RNAi pathway, long double-stranded RNAs (dsRNA) are processed into small interfering RNAs (siRNA), which specifically binds and cleaves the targeted viral messenger RNAs (mRNA) in the cytosol, leading to an effective protection of plants (Mezzetti et al., 2020). This potential multipurpose tool is being exploited on a large scale to interfere with the pest–pathogen genes by processes such as host-induced gene silencing (HIGS) (Nowara et al., 2010) and spray-induced gene silencing (SIGS) (Koch et al., 2016) and can make a remarkable contribution in the domain of integrated pest management (Koch et al., 2019). Since the conventional RNAi method involves the production of genetically modified (GM) crops where exogenous dsRNAs are expressed to induce RNAi pathway within, anti-GMO groups has raised concerns regarding the safe consumption of these plant products carrying heterologous genetic material mostly from bacteria, fungi, etc. (Zotti et al., 2018). Although gradually, the stigma against RNAi-based pest–pathogen-resistant plants is reducing worldwide as quite a lot of virus-resistant plants (Squash, Papaya, Plum) have gained approval to be cultivated outside the European Union (for example, in North and South American countries, cis/transgenic crops which do not carry antibiotic resistance genes are not considered as GMOs; Waltz, 2015; Khalid et al., 2017; Limera et al., 2017). On the other hand, clustered regularly interspaced short palindromic repeats (CRISPR/Cas), the most efficient genome editing tool, is in the headlines now as required insertion and deletion (in/del) of nucleotides can be done in the genomic loci very precisely and it can be applied to generate stable knockout plant lines with improved disease resistance against pests and pathogens (Charpentier and Dounda, 2013; Mat Jalaluddin et al., 2019). In many countries, CRISPR/Cas-food crops are not categorized under GMO status because even if the whole procedure involves a transient transgenic state, eventual sexual propagation leads to transgene-free crops by the law of segregation (Schenke and Cai, 2020). Although on a separate note, in case of woody plants, transgene can remain unmodified for over 20 years (Walter et al., 2005) and sexual propagation is not an option for obtaining homozygous single-copy clone. Vegetative propagation on a large scale for field plantation followed by copy number determination using inverse polymerase chain reaction (PCR) can be a suitable solution for ensuring genetic stability of transgenes in woody plants (Stefano et al., 2016). Plant biotechnology field has upheld many instances showing that RNAi is unique in many respect catering to a broader domain of requirements of breeders for improved crop variety as compared to CRISPR/Cas (Mezzetti et al., 2020). For instance, the most recent and talked about innovation in the field of pest management is the exogenous (topical) application of dsRNAs as the most effective strategy for gene silencing (San Miguel and Scott, 2016; Worrall et al., 2019). It is being considered worldwide as a more practical and realistic method as biopesticides since a lot of noteworthy researches have been done to improve several aspects of exogenously applied dsRNAs such as stability, efficacy, and persistence. The modes of application are foliar sprays, seed treatments, root drenching, and many more; all are commercially lucrative options because of the comparatively low production cost, high specificity and the most important aspect is improved biosafety than other categories of biopesticides and chemical pesticides (Zotti et al., 2018; Cagliari et al., 2019; Bramlett et al., 2020; Rodrigues and Petrick, 2020).
In this review, we will delve deep into the details of both RNAi and CRISPR/Cas technology, how these technologies are being used for improving disease resistance in plants against pests such as insects, fungi, and nematodes and how RNAi technology is still relevant in terms of exogenous application of dsRNAs in this new era of CRISPR/Cas.
Two Ground-Breaking Gene-Editing Tools
RNA Interference: The Revolutionary Gene-Silencing Technique
Target-specific gene silencing has gained immense popularity among the scientific community over the years. Out of many gene-silencing techniques, RNAi has been proven to be very effective in targeting and degrading a particular mRNA of a gene. When an organism is exposed to a foreign genetic material such as DNA or RNA, it immediately started mounting an immune response against it. At the very core of such type of sequence directed immune response is RNAi. In the early 1995, Guo and Kemphues tried to use the antisense-strand targeting Caenorhabditis elegans par-1 gene to silence its expression, which is also known as antisense-mediated gene silencing (Guo and Kemphues, 1995). They found not only the exogenous antisense strand but also the sense RNA strand was able to induce silencing. The observation by Fire et al. (1998) first led to the idea that dsRNA is a potent trigger for RNAi-mediated gene silencing, and it is much more effective as compared to a single-stranded RNA. They coined the term “RNA-interference” because they showed for the first time that exogenous introduction of RNA (both sense and antisense) can interfere with the function of an endogenous gene. For such remarkable discovery, the authors were awarded the noble prize in physiology or medicine in 2006. Later, it had been found out that this silencing effect could be transmitted in the germ lines and thereby could be passed on to several generations of C. elegans (Fire et al., 1998; Grishok et al., 2000). This silencing phenomenon is also known as post-transcriptional gene silencing in eukaryotes (De Carvalho et al., 1992), co-suppression in plants (Napoli et al., 1990), and quelling in fungi (Romano and Macino, 1992).
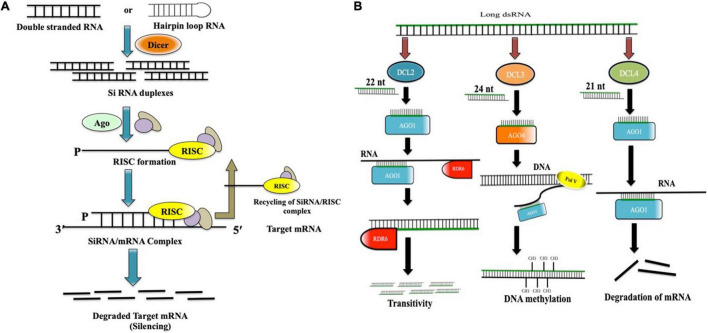
There are mainly three types of non-coding RNAs, which are involved in the downregulation of gene expression: microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs) (Hamilton and Baulcombe, 1999; Pasquinelli et al., 2000; Bartel, 2009; Aalto and Pasquinelli, 2012). Although all these three types have differences in their biogenesis, they all share a few similarities in their mode of action. There are mainly three steps in the RNAi-mediated gene-silencing process (Figure 1A). First, the long dsRNAs or the hairpin loop RNA (hpRNA) gets processed into 21–23 base-pair long nucleotide strand by Dicer (a ribonuclease enzyme type-III) (Ketting et al., 2001). Arabidopsis thaliana has four types of Dicer-like (DCL) paralogs. DCL1 is responsible for the synthesis of miRNAs, whereas DCL2, DCL3, and DCL4 synthesize 22, 24, and 21 nucleotide-long siRNA, respectively (Xie et al., 2004; Henderson et al., 2006). In the second step, these short duplex siRNAs unwind to form one guide strand, which is supposed to guide in targeting the complementary endogenous mRNA, and the other passenger strand, which eventually gets degraded. In final step, the guide strand gets associated with the argonaute proteins (AGOs) to form the RNA-induced silencing complex (RISC) and further guides the RISC complex to locate, bind, and degrade the complementary mRNAs in the cell (Ketting, 2011). Although it is noteworthy to mention that in Arabidopsis, processing of dsRNA varies greatly upon the interaction with each of the DCL paralogs. Generally, DCL4 generates 21 nucleotide-long siRNAs, which gets loaded on argonaute 1 (AGO1) and further guides in targeting complementary mRNAs in cytoplasm (PTGS) (Hamilton and Baulcombe, 1999; Mi et al., 2008). DCL2 generates 22 nucleotide-long siRNAs, which when loaded on AGO1, changes its conformation, and recruits RNA-dependent RNA polymerase 6(RDR6) to the 3′ end of the target, finally transcribing the target mRNA into another duplex siRNA (secondary siRNA). This process is known as transitive silencing (Dalmay et al., 2000). DCL3, on the other hand, creates 24 nucleotide-long siRNA, which when gets loaded with AGO4, recruits DNA methyl transferase, and triggers DNA methylation (Wassenegger et al., 1994; Chan et al., 2004; Figure 1B).
FIGURE 1.
(A) In RNAi-mediated gene silencing, long dsRNA/hpRNA gets chopped off into short (21–23 nucleotides) interfering RNA duplexes (siRNAs). siRNA duplexes unwind and one of the strands (Guide strand) gets incorporated into the RISC complex, which finally degrades the targeted complementary mRNA. RISC complex gets recycled again; (B) Dicer-mediated processing of long dsRNA in Arabidopsis thaliana. DCL2 generates 22 nucleotide-long siRNAs, which interacts with AGO1 and recruits RDR6 to induce transitive silencing. DCL3 generates 24 nucleotide-long siRNAs, which interacts with AGO4 and induces DNA methylation by recruiting DNA PolV, whereas DCL4 generates 21 nucleotide-long siRNAs, which interacts with AGO1 and directs the cleavage of complementary mRNA.
The basic mechanism and the key proteins involved in RNAi pathway are more or less evolutionarily conserved among prokaryotic and eukaryotic organisms (Shabalina and Koonin, 2008). It has been successfully applied to crops to confer resistance against a wide variety of biotic stresses such as insects, fungi, nematodes, and viruses. However, there are many drawbacks in using RNAi as a gene-silencing tool. For example, RNAi machinery is mostly active in the cytoplasm, and it is really difficult to target the long non-coding RNAs in the nucleus (Fatica and Bozzoni, 2014). A major concern over using RNAi is its potential risk of off-target silencing. It has been proved that siRNAs are capable of targeting and silencing non-target mRNAs which have a very few sequence similarities with the target mRNA (Carthew and Sontheimer, 2009). Another disadvantage is that sometimes, these off-target silencing results in a phenotype that dominates the required phenotype for which RNAi construct was designed on the first place (Franceschini et al., 2014).
RNAi for Improving Disease Resistance in Plants
Since its discovery, RNAi has been the primary choice for gene silencing across a wide array of biological fields, but inarguably, RNAi has been extensively used as a plant-protection platform over the years. Plants being a sessile organism have to withstand a wide variety of biotic and abiotic stresses that severely affects its growth and yield. Depending on the suitable environmental condition, various biotic stresses such as insects, fungi, and nematodes attack their host plants and cause severe loss in agricultural yield. Just like plants, insects have RNAi machinery primarily to mount defense against viruses (Matranga and Zamore, 2007). RNA interference in insects can be triggered by exogenous application of dsRNAs in plants by means of dsRNA injection or by feeding insects with dsRNA containing artificial diets (Baum et al., 2007) or by developing transgenic plants expressing dsRNAs targeting vital insect genes (Vélez and Fishilevich, 2018; Zotti et al., 2018; Di Lelio et al., 2022). Once this fact was established, researchers started concentrating their focus on improving the integrated pest management system by synthesizing dsRNAs targeting insects that cause severe damage to important crops. Here, we have discussed different approaches (including GMO/HIGS/SIGS) that involve “RNAi-as the central mechanism” to improve disease resistance in plants.
RNAi Against Insects
Brown planthopper Nilaparvata lugens is the one of the deadliest insect pests for rice. Li et al. (2011) were able to induce RNAi by targeting vacuolar ATP synthase subunit E of Nilaparvata lugens by means of dsRNA ingestion (Li et al., 2011). Cotton plant, expressing CYP6AE14 dsRNA, was generated to acquire enhanced resistance to cotton bollworm Helicoverpa armigera (Mao et al., 2011). In vitro transcribed dsRNA was successfully applied in citrus and grapevine trees by injecting the trunks or drenching the roots, targeting the arginine kinase of two psyllids (Hunter et al., 2012). In a study Bolognesi et al. (2012) first characterized the mechanism of action of dsRNA targeted against the western corn rootworm larvae. The cotton aphid Aphis gossypii is a lethal pest to many agriculturally important crops that helps in virus transmission. They are also capable of developing resistance against a wide range of insecticides. This resistance against organophosphorus insecticides was reduced significantly by means of orally delivered dsRNA targeting carboxylesterase gene (Gong et al., 2014). Transplastomic potato plants (Solanum tuberosum), expressing dsRNAs, were developed that targeted the β-actin gene of the Colorado potato beetles (Zhang J. et al., 2015) and thereby showed significant resistance against this detrimental pest (Table 1).
TABLE 1.
RNAi-targeted editing in plants against insects.
| Insects | Target gene | Plant | Phenotype | References |
| Helicoverpa armigera (Cotton bollworm) | Ecdysone receptor | Tobacco | Reduction of growth followed by death | Zhu et al., 2012 |
| Diabrotica virgifera (Western corn rootworm) | V-ATPaseA | Maize | Stunted growth | Baum et al., 2007 |
| Spodoptera exigua | Ecdysone receptor | Tobacco | Death rate increased | Zhu et al., 2012 |
| Sitobion avenae (Grain aphid) | Salivary proteins DSR32/DSR33 | Wheat | Death rate increased | Wang et al., 2015 |
| Glossina morsitans morsitans | Transferrin | Pea, clover, alfalfa | Mortality rate significantly low | Walshe et al., 2009 |
| Acyrthosiphon pisum | Aquaporin | Pea, clover, alfalfa | Osmotic pressure increased | Shakesby et al., 2009 |
| Acyrthosiphon pisum | SHP | Pea, clover, alfalfa | Reduced fertility | Will and Vilcinskas, 2015 |
| Acyrthosiphon pisum | V-ATPase E | Pea, clover, alfalfa | dsRNA degradation in saliva | Christiaens and Smagghe, 2014 |
| Nilaparvata lugens (brown plant hopper) | Trehalose PO4 synthase | Rice | Lethality | Chen et al., 2010 |
| Aphis gossypii (Cotton aphid) | AgOBP2 | Cotton | Failure of recognizing host | Rebijith et al., 2016 |
| Sitobion avenae (Grain aphid) | Catalase gene CAT | Wheat | Survival rate reduced | Deng and Zhao, 2014 |
| Sitobion avenae Grain aphid | Cytochrome c oxidase | Wheat | Increased mortality | Zhang et al., 2013 |
| Greenbug Schizaphis graminum | Salivary protein C002 | Wheat | Lethal | Zhang Y. et al., 2015 |
| Peregrinus maidis | V-ATPase B&D | Corn | Reduced fertility | Yao et al., 2013 |
| Lygus lineolaris | Apoptosis inhibitor | Cotton, alfalfa, beans | Digestion of dsRNA | Allen and Walker, 2012 |
| Phyllotreta striolata | Arginine kinase | Cruciferae crops | Retarded development/increased mortality | Zhao et al., 2008 |
RNAi Against Fungi
In eukaryotic evolutionary hierarchy, the fungi are a major kingdom, which happens to have the RNAi machinery alongside the plants and animals. Although there are some exceptions, several lineages such as Saccharomyces cerevisiae, some close relative yeasts, and filamentous fungi such as Cryptococcus gattii and Ustilago maydis have lost their RNAi pathway during evolution (Drinnenberg et al., 2011; Nicolás et al., 2013). The reason for loosing RNAi was that S. cerevisiae and other filamentous fungi were infected with the “killer virus,” which kills neighboring cells that possess an active RNAi, while providing immunity to the host cell itself. Hence, the benefit of having RNAi was proved to be selective disadvantage, thus creating RNAi-deficient fungi (Drinnenberg et al., 2011). The RNAi mechanism in other fungi such as Neurospora crassa was first described as quelling by Romano and Macino (1992). Creating transgenic plants expressing dsRNA targeted against fungal genes soon proved to be a promising approach for plant protection (Koch and Kogel, 2014). In a study with barley, Fusarium graminearum infection was controlled by spraying 791 nucleotide-long dsRNA (CYP3-dsRNA) (Koch et al., 2016). Mycotoxins from fungus are considered to be very harmful for both plants and animals. The mycotoxin content of F. graminearum was significantly reduced by transforming it with inverted repeat transgenes (IRTs) containing mycotoxin-specific regulatory genes (McDonald et al., 2005; Table 2).
TABLE 2.
RNAi-targeted editing in plants against fungi.
| Fungi | Target gene | Plant | Phenotype | References |
| Puccinia triticina | PtMAPK1 | Wheat | Significant reduction of fungal growth and disease suppression | Panwar et al., 2013 |
| Blumeria graminis f.sp. tritici | MLO | Wheat | Inhibition of fungal growth | Riechen, 2007 |
| F. graminearum | IRT containing mycotoxin regulatory sequences | Grain and legume crops | Significant reduction in mycotoxin production | McDonald et al., 2005 |
| Blumeria graminis | Avra10 | Barley and wheat | Inhibition of fungal growth | Nowara et al., 2010 |
| Phytophthora parasitica | PnPMA1 | Arabidopsis | Elucidation of a compatible interaction between Arabidopsis and P. parasitica | Wang et al., 2011 |
| Fusarium verticillioides | GUS | Tobacco | Reduced expression of GUS | Tinoco et al., 2010 |
| Phytophthora parasitica var. nicotianae | Glutathione S-transferase | Tobacco | Increase resistance of Nicotiana to infection | Hernández et al., 2009 |
| Puccinia striiformis f. sp. tritici | PSTha12J12 | Barley and wheat | Significant improvement in rust resistance | Yin et al., 2011 |
| Botrytis cinerea | MAP Kinase Bmp3 | Lettuce | Delay in conidial germination, reduction of necrotic lesions | Spada et al., 2021 |
| Botrytis cinerea | DCL1, DCL2 | Tomato, Strawberry, Grape, Lettuce, Onion, Rose | Significant inhibition in gray mold disease | Wang M. et al., 2016 |
| Botrytis cinerea | DND1 | Tomato and Potato | Reduced susceptibility to Botrytis | Sun et al., 2017 |
| Botrytis cinerea | BcTOR | Arabidopsis, Potato, Tomato | Enhanced resistance against gray mold | Xiong et al., 2019 |
RNAi Against Nematodes
Since the discovery of RNAi in model nematode C. elegans Fire et al. (1998), an overwhelming amount of work has been done to further widen our knowledge regarding the nematode genomics. Out of several plant-parasitic nematodes (PPNs), root-knot nematode such as Meloidogyne incognita causes significant crop loss across the globe (Sasser et al., 1984). Among the several essential proteins required by root-knot nematodes, 16D10 was found out to be a very effective target gene for controlling the infection. This particular gene was identified and subsequently silenced using RNAi and thus suppressed nematode development (reproduction) by 74–81% (Huang et al., 2006). Mi-Rpn7gene is a potential target for controlling root-knot nematode M. incognita. Knocking down of Mi-Rpn7 using RNAi resulted in 55.2–66.5% reduction in infectivity (Niu et al., 2012). Dutta et al. (2015) succeeded in making a transgenic tomato line targeting M. incognita-specific protease gene cathepsin L-cysteine proteinase (Mi-cpl-1). RNAi-mediated suppression of Mi-cpl-1 resulted in significant reduction in infectivity by the nematode (Dutta et al., 2015; Table 3).
TABLE 3.
RNAi-targeted editing in plants against nematodes.
| Nematodes | Target gene | Plant | Phenotype | References |
| Meloidogyne incognita | Splicing factor and integrase | Tobacco | >90% reduction in established nematodes | Yadav et al., 2006 |
| Heterodera glycines | Prp-17, Cpn-1, mRNA splicing factor | Soybeans | Significant reduction in number of nematode eggs | Li et al., 2010 |
| Radopholus similis | Rs-cb-1 | Tobacco | Inhibition of development, reduced pathogenicity | Li et al., 2015 |
| Ditylenchus destructor | Unc-15 | Sweet potato | Reduced rate of infection area | Fan et al., 2015 |
| Meloidogyne enterolobii | MeTCTP | Tomato | Attenuation in parasitism | Zhuo et al., 2017 |
| Pratylenchus vulnus | Pv010 | Walnut | Significant reduction in number of nematodes per root | Walawage et al., 2013 |
| Heterodera glycines | Spliceosomal SR protein, ribosomal protein | Soybean | Significant reduction in number of female cysts | Klink et al., 2009 |
| Meloidogyne chitwoodi | Mc16D10L | Potato | Significant reduction (∼68%) in number of egg masses | Dinh et al., 2014a |
| Heterodera schachtii | 3B05, 4G06, 8H07, and 10A06 | Arabidopsis | 23–64% reduction in number of mature nematode females | Sindhu et al., 2009 |
| Heterodera glycines | Major sperm protein (MSP) | Soybeans | 68% reduction in eggs per gram of root tissue | Steeves et al., 2006 |
| Meloidogyne javanica | Putative transcription factor, MjTis11 | Tobacco | Consistent silencing of MjTis11 | Fairbairn et al., 2007 |
| Meloidogyne chitwoodi | Mc16D10L | Arabidopsis | Significant reduction (∼60%) in number of egg masses | Dinh et al., 2014b |
CRISPR/Cas: The New Era of Genome Editing
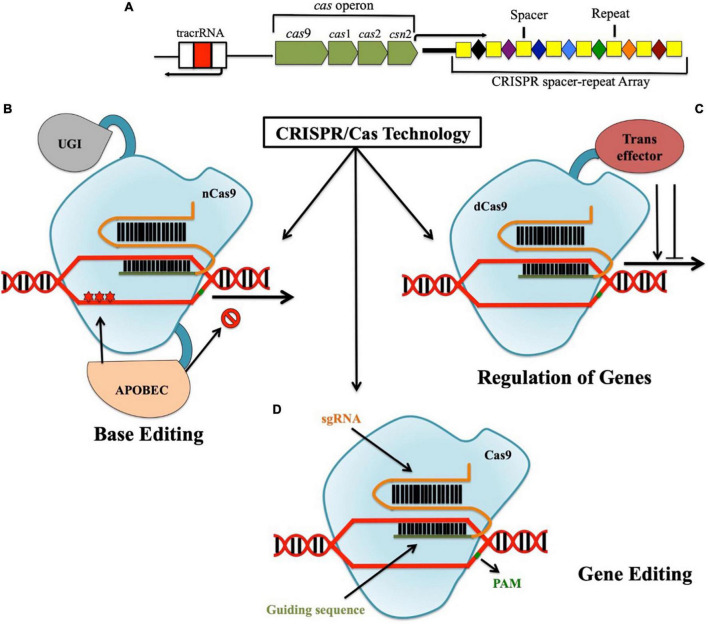
Clustered regularly interspaced short palindromic repeats was first discovered as some sort of “unusual structure” by Japanese researchers in 1987 while working with the iap gene in Escherichia coli (Ishino et al., 1987). Later in 1993, it was first characterized by Francisco Mojica from the University of Alicante, Spain (Mojica and Rodriguez-Valera, 2016). It was detected in various bacteria and archaea and are thought to be involved in the regulation of their genes (Mojica et al., 2002). More than a decade later, it was described as a bacterial adaptive defense system that contains a CRISPR loci comprising of CRISPR, DNA-targeting spacers, and Cas operon (Bolotin et al., 2005; Barrangou et al., 2007; Figure 2A). There are three distinct stages through which CRSIPR/Cas mediated adaptive immunity works, namely, adaptation, maturation, and interference. In the “adaptation stage,” new spacers from the invading organisms such as virus get directly incorporated into the CRISPR array (Barrangou et al., 2007), which finally transcribes the precursor CRISPR RNA (pre-crRNA). With the aid of trans-activating CRISPR RNA (tracrRNA) coupled with CRISPR-associated csn1 protein and RNaseIII, these pre-crRNA becomes mature crRNA (maturation stage) carrying the individual spacer flanked by repetitive sequence. Finally in the “Interference stage,” these mature crRNAs along with Cas proteins form ribonucleoprotein (RNP) complexes that target and cleave foreign nucleic acid sequences, which are complimentary to the crRNA-encoded sequence (Deltcheva et al., 2011; Jinek et al., 2012; Zhang et al., 2012; Doudna and Charpentier, 2014). Since the discovery that CRISPR/Cas coupled with designed guide RNA can be used to specifically target and cleave any stretch of DNA sequence, it revolutionized the modern genetic engineering field (Jinek et al., 2012; Burmistrz et al., 2020). In fact, CRISPR-edited Sicilian Rouge tomatoes with high concentration of γ-aminobutyric acid (GABA) were the first genome-edited crop (approval in December, 2020) that entered the market of Japan successfully by Sanatech, a start-up from University of Tsukuba in September, 2021 (Waltz, 2022). Apart from conventional gene editing, using impaired Cas9 enzymes such as nuclease-dead Cas9 (dCas9; non-functional endonuclease activity) (Moradpour and Abdulah, 2020) and Cas9 nickase (nCas9; mutated nuclease domain of Cas9 for facilitating single-strand cut) (Adli, 2018), this powerful tool is capable of achieving gene regulation and targeted base editing, respectively (Chaudhuri et al., 2022; Figures 2B–D).
FIGURE 2.
CRISPR locus in genome comprises of CRISPR spacer-repeat array, Cas operon, and tracrRNA. Apart from being able to specifically target nucleotides in the genome (gene editing), using impaired Cas9 enzymes such as dCas9 and nickase Cas9, gene regulation and targeted base editing without double-strand breaks can also be achieved, respectively. (A) CRISPR locus in genome; (B) CRISPR/Cas9-mediated gene editing; (C) CRISPR/dCas9-mediated gene regulation; (D) CRISPR/nCas9-mediated base editing.
CRISPR/Cas System in the Generation of Pest-Resistant Crops
Invasion of plants by bacteria, fungi, nematodes, and all other sort of pests poses immense menace to agriculture. Application of agrochemicals and pesticides is the conventional source of resistance used by the farmers. But these agrochemicals gradually pass through the food chain and finally end up harming human health and cattle at large. Pathogens on the other hand are evolving at a steady rate to acquire immunity against these agrochemicals, which makes the production and use of agrochemicals a big challenge to society. One means to bypass this hurdle is the generation of pathogen-resistant plants through recombinant DNA technology where resistance genes from wild-type species can be introduced through standardized plant-breeding technologies (Wang F. et al., 2016). But sometimes alongside the desirable trait, unwanted traits also enter in the host genome. CRISPR has an excellent track record of genetic modification and has been successful in overcoming potential biotic stress challenges from fungi, bacteria, viruses, etc. (Pillay, 2020). A promising work by CRISPR/Cas9 on rice was the knockout of the gene-encoding mitogen-activated protein kinase-5 (OsMPK5), which enhanced plant disease resistance (Hillary and Ceasar, 2019). Researchers have found that conferring resistance to plants from RNA viruses is hard, because the spCas9 (nuclease from Streptococcus pyogenes) recognizes dsDNA solely. But with due perseverance, two enzymes were discovered, namely, FnCas9 (nuclease from Francisella novicida) and LwaCas13a (nuclease from Leptotrichia wadei), which can or could recognize RNA. As opposed to RNAi which confers GMO status, CRISPR/Cas9 has proven to be capable of implementing the modifications required to confer resistance against biotic stresses (in/del) at the nucleotide level. Following the laws of segregation, the CRISPR cassette gets discarded, finally generating plants without the GMO tag (Waltz, 2022).
CRISPR/Cas Against Viruses
Geminiviruses, the damaging family of DNA viruses, have always been an immense threat to plants. Recently, CRISPR/Cas technology has been successfully applied to plant system for achieving resistance against geminiviruses by three eminent scientific groups. Rep, coat CP proteins, and the conserved non-coding intergenic region (IR) are the essential components regulating the virulence of geminivirus. ORFs encoding these three components were targeted by Zahir Ali and his group using engineered sgRNAs, resulting in reduced viral load in the case of tomato yellow leaf curl virus (TYLCV) (Ali et al., 2015). A number of two other research groups also targeted various sequences of the geminivirus genome, which collectively reduced the virulence of the plant pathogen (Baltes et al., 2015; Ji et al., 2015). Zahir Ali also worked on three different viruses: TYLCV, beet curly top virus (BCTV), and Merremia mosaic virus (MeMV) simultaneously with engineered sgRNA using the same protocol mentioned above. The results suggest that all sgRNAs exhibit interference activity resulting in delayed or reduced viral DNA accumulation in the Nicotiana plant. Sanger sequencing also confirmed that TYLCV-specific IR-sgRNA containing the TAATATTAC sequence could target both TYLCV and MeMV genome, proving that a single sgRNA can simultaneously target multiple viruses (Ali et al., 2015).
CRISPR/Cas Against Fungi and Bacteria
Another potential destroyer of crop yield is fungal pathogens. Deadly plant fungal diseases such as smut, rust, rot, and mildew cause severe loss to agriculture every year. Over and above, the fatal mycotoxins from mycotoxigenic fungi, which accumulate as secondary metabolites in the tissues of the host plant, pose fatal health threats to human and animals from contaminated food and fodder. CRISPR/Cas9 approach is being applied nowadays to target the editing of potential candidate genes, which enhance plant resistance against fungal pathogens (Borrelli et al., 2018). Detailed survey reveals multiple bacterial plant pathogens that cause severe crop diseases which eventually result in heavy agricultural damage (Schloss and Handelsman, 2004). Since bacterial diseases are generally asymptomatic, so their control poses to be a humongous challenge for the agricultural scientists. The most common procedures adopted against bacterial diseases are biopesticides, genetic methods, etc. (Kerr, 2016). There are classifications among phytopathogenic bacteria. Some are crop-specific, e.g., Clavibacter michiganensis causing tomato ring rot disease, polyphagous-specific, e.g., Ralstonia solanacearum causing disease in dicots and monocots both together, and the last one being “kingdom crosser,” e.g., Dickeya dadantii, falling in the genre entomo-phytopathogen, attacking both plants and animals (Borrelli et al., 2018). Table 4 gives a detailed idea of the CRISPR/Cas9 tool used to combat DNA/RNA viruses, fungal and bacterial diseases for plant protection.
TABLE 4.
CRISPR/Cas-targeted editing in plants against viruses, fungi, and bacteria.
| Type | Pathogen | Target plant | Target gene and its function | References |
| Virus | Rice Tungro Spherical Virus (RSTV) |
Oryza sativa L. japonica |
Eukaryotic translation initiation factor 4G (elF4G). This is the host factor for the translation of RNA virus. | Macovei et al., 2018 |
| Virus | Turnip Mosaic Virus (TuMV) |
Arabidopsis thaliana |
Eukaryotic translation initiation factor 4E (elF(iso)4E. This is the host factor for the translation of RNA virus. |
Pyott et al., 2016 |
| Virus | Cucumber Vein Yellowing Virus (CVYV), Zucchini Yellow Mosaic Virus (ZYMV), Papaya Ring Spot Mosaic Virus-W (PRSV-W). |
Cucumis sativus |
Eukaryotic translation initiation factor 4E (elF4E). This is the host factor for the translation of RNA virus. |
Chandrasekaran et al., 2016 |
| Virus | Bean Yellow Dwarf Virus (BeYDV) |
Arabidopsis thaliana and Nicotiana benthamiana |
Coat protein (CP), Replication Association protein (Rep), Intergeneric Region (IR). Carry our Rolling Cycle Amplification (RCA) mechanism. |
Ji et al., 2015 |
| Virus | Beet Severe Curly Top Virus (BSCTV) | Nicotiana benthamiana | Long Intergenic Region (LIR), Replication Association protein (Rep)/RepA. Carry our Rolling Cycle Amplification (RCA) mechanism. | Baltes et al., 2015 |
| Virus | Tomato Yellow Leaf Curl Virus (TYLCV), Beet Curly Top Virus (BCTV), Merremia Mosaic Virus (MeMV) | Nicotiana benthamiana | Coat protein (CP), Replication Association protein (Rep), Intergeneric Region (IR). Carry our Rolling Cycle Amplification (RCA) mechanism. | Ali et al., 2015 |
| Virus | Tomato Yellow Leaf Curl Virus (TYLCV), Merremia Mosaic Virus (MeMV), Cotton Leaf Curl Kokhran Virus (CLCuKoV) | Nicotiana benthamiana | Coat protein (CP), Replication Association protein (Rep), Intergeneric Region (IR). Carry our Rolling Cycle Amplification (RCA) mechanism. | Ali et al., 2015 |
| Virus | Turnip Mosaic Virus (TuMV) | Nicotiana benthamiana | Coat protein (CP), Green Fluorescent protein 1, Green Fluorescent Protein 2, Helper Component proteinase silencing suppressor (HC-Pro) | Aman et al., 2018 |
| Virus | Cucumber Mosaic Virus (CMV), Tobacco Mosaic Virus (TMV). | Arabidopsis thaliana and Nicotiana benthamiana | Open Reading Frame 1,2,3 (ORF 1,2,3), Coat protein (CP) and 3′ Untranslated Terminal Repeat (3′-UTR) | Zhang et al., 2018 |
| Fungi | Rice Blast Disease (Magnaporthe oryzae) | Oryza sativa L. japonica | Ethylene Responsive Factor (ERF922). Several stress responses implicate transcription factor | Wang F. et al., 2016 |
| Fungi | Rice Blast Disease (Magnaporthe oryzae) | Oryza sativa L. japonica | Exocyst Component Complex (SEC3A). This acts as the subunit of the exocyst complex in rice. | Ma et al., 2018 |
| Fungi | Powdery mildew (Blumeria graminis f.sp. tritici) | Triticum aestivum | Mildew-Resistant Locus-A1(MLO-A1). This susceptible gene is associated with disease formation. | Wang et al., 2014 |
| Fungi | Black Pod Disease (Phytophthora tropicalis) | Theobroma cacao | Non-expressor of pathogenesis-related3 (NPR3). Regulates the immune system. | Fister et al., 2018 |
| Fungi | Powdery Mildew (Oidium neolycopersici) | Solanum lycopersicon | Mildew-Resistant Locus1 (MLO1). Responsible for the vulnerability toward this disease. | Nekrasov et al., 2017 |
| Fungi | Powdery Mildew (Erysiphe necator) | Vitis vinifera | Mildew-Resistant Locus-7 (MLO-7). This susceptible gene is associated with disease formation. | Malnoy et al., 2016 |
| Fungi | Gray mold (Botrytis cinerea) | Vitis vinifera | WRKY52. This transcription factor is associated with biotic stress response. | Wang et al., 2018 |
| Bacteria | Bacterial Blight (Xanthomonas oryzae pv. oryzae) | Oryza sativa | SWEET13. This is the sucrose transporter gene. | Li et al., 2012; Zhou et al., 2015 |
| Bacteria | Fire blight (Erwinia amylovora) | Malus domestica | DspE-interacting proteins of Malus (DIPM-1, DIPM-2, DIPM-4) involved in fire blight disease as a susceptibility factor | Malnoy et al., 2016 |
| Bacteria | Citrus canker (Xanthomonas citri) | Citrus paradisi | Lateral Organ Boundaries (LOB1), gene promotes pathogen growth, pustule formation. | Jia et al., 2017 |
| Bacteria | Citrus canker (Xanthomonas citri) |
Citrus sinensis Osbeck
|
Lateral Organ Boundaries (LOB1), gene promotes pathogen growth, pustule formation | Peng et al., 2017 |
CRISPR/Cas9 technology for crop protection needs to be thoroughly explored by the researchers globally. It becomes an extremely handy tool for crop protection if it is already known what kind of modifications required in specific genes to confer disease resistance. Resistance to a certain disease can be achieved by CRISPR-mediated knockout of the target DNA and activation of the host cell’s DNA repair process [using non-homologous end joining (NHEJ)] to bring out the desired inactivating mutations. Thus, CRISPR can be proven useful in inactivating a single target gene or plethora of gene families using single gRNA that can target several sites or by multiplex method where multiple gRNAs are introduced simultaneously. There is another contrary case as well, where generation of resistance is caused by the involvement of particular allelic variants. Here, homologous directed repair (HDR)-based cell repair comes into action along with CRISPR-DNA cleavage. It has been inferred that the application of HDR has a promising potential alongside the NHEJ in insertion of mutations using CRISPR/Cas9 tool. Yet, multiple aspects need to be considered and worked upon if CRISPR/Cas9 technology is to be applied on a large scale in plant-pathogen defense scenario.
Relevance of RNAi in Today’s World: In Terms of Exogenous Application of RNAi-Inducing dsRNAs
A tried and tested crop protection platform, the RNAi strategy, has been ruling the domain of plant biotic stress resistance for more than two decades. Initially, this technology was used to generate GM or transgenic plants (Table 5; Mat Jalaluddin et al., 2019) that expressed dsRNAs, overall which bestowed the plants with a natural defense mechanism having the capacity to resist invasions of viruses and other hostile organisms (Taning et al., 2020). It has been quite a while that the production of GMOs/transgenics has led to a lot of hue and cry all over the world. In addition, as goes the age old saying that “Necessity is the mother of invention,” the huge uproar of the public and the anti-GMO scientific community led to the discovery of alternative approaches that skip transgenics and directly apply RNA molecules (dsRNA-containing end-use products, dsRNA-EPs) exogenously to trigger the required RNAi response. Various methods of exogenous application such as spraying, root/seed soaking, mechanical inoculation, trunk injection, and petiole absorption are adopted by the scientists as alternative to the controversial GM strategy of knocking down target genes to control plant damage by biotic stress (Dalakouras et al., 2020). Exogenously applied dsRNA on plants has the capacity to initiate RNAi-mediated gene silencing of the invading pest or the pathogen genes. The researches by various scientific groups (Koch et al., 2016; Konakalla et al., 2016; Kaldis et al., 2018) have demonstrated that the dsRNAs are actively absorbed by the plant cells and are processed into siRNAs which consequently extend the time of prevention of some plant diseases. The efficacy of the RNAi-based strategies for pest control is dependent on multiple factors such as the mechanisms of host–pathogen interactions, structural characteristics, RNAi machinery, and so on. Some shortcomings of exogenous RNAi application have been noticed in case of some pathogenic fungi, where dsRNA is not absorbed by the specific pathogen and secondary siRNA amplification is absent (Song et al., 2018; Kettles et al., 2019). The effectiveness of the exogenously applied RNAi also depends on the mode of delivery of the dsRNA biomolecules, the most common modes being spray method of mechanical inoculation. This mode of delivery varies as per the type of plant selected and the targeted pest (Dalakouras et al., 2018). Exogenous application of naked dsRNAs, liposome-protected dsRNA, and artificial extracellular vesicles (EV) that might be complexed with protein carrier or nanoparticles has brought out significant downregulation of transgenes and various plant endogenes according to many reports (Jiang et al., 2014; Numata et al., 2014; Lau et al., 2015; Dalakouras et al., 2016; Huang et al., 2019).
TABLE 5.
List of RNAi crops commercialized successfully (till 2021).
| Crop | Year of approval | Targeted Gene | Trait |
| Tobacco [Vector Tobacco (USA) Ltd Vector 21–41] | 2006 | QPTase enzyme | Resistance against plum pox virus, Reduced nicotine |
| Plum (USDA-ARS- Appalachian Fruit Research Station C5) | 2009 | CP gene of PPV | Resistance against Plum pox virus |
| Papaya (University of Florida X17- 2) | 2011 | CP gene of PRSV | Resistance against papaya ringspot virus |
| Soybean (Monsanto MON 87705) | 2014 | FATB1-A, FAD2-1A | High oleic soybean oil, Low saturated fatty acid |
| Alfalfa (Monsanto/Forage Genetics KK179) | 2014 | CCOMT | Reduction in lignin content |
| Apple (Okanagan specialty fruits NF872) | 2014 | Apo5, Ppo2, Gpo3, pSR7 | Non-browning |
| Potato (J. R. Simplot E12, E24, F10, F37, J3, J55, J78, G11, H37, H50) | 2014 | Asn1 and Ppo5 | Reduction in black spot, low acrylamide potential |
| Potato (J. R. Simplot W8) | 2014 | Rpi-vnt1, R1, PhL, Vinv | Late blight resistance, low acrylamide potential, reduction in black spot, lowered reducing sugar |
| Maize (MON 87411, Monsanto) | 2015 | DvSnf7 | Western corn rootworm resistance |
| Potato (J. R. Simplot V11) | 2015 | PhL, R1, Ppo5, Asn1 | Lowered reducing sugar, reduction in black spot, potential blight resistance, late low acrylamide |
| Apple (Okanagan specialty fruits GD743 and GS784) | 2016 | Apo5, Ppo2, Gpo3, pSR7 | Non-browning |
| Potato (J. R. Simplot X17 and Y9) | 2016 | PhL, R, Asn 1, Ppo5, Rpi-vn1, Vinv | Lowered reducing sugars, low acrylamide potential, late blight resistance, reduced black spot |
In the present-day scenario, the use of nanocarriers becomes highly prevalent commercially for crop protection against pathogens. They are predominantly used as sprayable RNA-based biocontrol pesticides (Kunte et al., 2020). So, RNAi technology and nanotechnology have been merged effectively to produce maximum benefits in pest and pathogen control industries. A mention worthy progress from laboratory to field is the generation of Bioclay or “layered double hydroxide clay nanoparticles” used as a potential vehicle to deliver or apply dsRNA (Mitter et al., 2017; Fletcher et al., 2020). Recently, it has been reported that DNA nanostructures can act as efficient RNA carriers using Watson and Crick base pairing and can serve as an excellent tool to deliver siRNA in plant cells (Zhang et al., 2019). A lot of research is being carried out regarding RNA-based biocontrol compounds, and most likely, they will be applied in the field of agriculture especially in greenhouse purposes using the appropriate delivery methods.
RNAi-Based Biopesticides: Commercialization, Regulatory Guidelines, and Acceptance by Public
Research and analysis of the RNAi-based biocontrol sector have unfolded its immense potential in the development of the agricultural sector, off course with a patenting trend. With the formulation, discovery, patenting, and commercialization of sprayable RNA-based biopesticides, the recent investments in the research and development (R&D) of RNA based co-formulants have increased drastically (Table 6; Mat Jalaluddin et al., 2019; Bramlett et al., 2020; Christiaens et al., 2020; Giudice et al., 2021). The flag bearer here is the global giant Monsanto whose brand “Biodirect” is already in the process of developing RNA-based biopesticides to control pests, followed by other multinationals such as Bayer, Syngenta, and so on (Taning et al., 2020). The interest shown by these multinational companies in the domain of RNA-based biopesticides has motivated a plethora of agritech start-up companies to develop and function by utilizing the existing technology to curve their own niche in this agribusiness sector. Start-ups such as Greenlight Biosciences, AgroRNA, and RNAgri have started a humongous production of dsRNA at an affordable price. Greenlight Biosciences have developed an innovative and fruitful cell-free bioprocessing technique, which generates RNA sequences in a completely scalable method that will guarantee the availability of dsRNA at affordable rates for field application without bearing the GMO tag. Another budding start-up company AgroSpheres has emphasized on the protection of dsRNA against degradation along with enhanced target delivery to achieve maximum crop protection. This has been achieved by developing bioparticles made up of miniscule spherical cells that do not possess any chromosome and can encapsulate dsRNA. Recent reports state that GreenLight and AgroSpheres have collaborated to further work on the above-mentioned bioparticles to create a protective shield around the dsRNA to prevent degradation from RNases and ultraviolet radiations. Inspired by these recent developments, another biotech start-up Nanosur has specialized in producing modified RNA (MdsRNA) products that can be translocated smoothly and fast through cell membranes, thus improving efficacy and minimizing degradation. Following the footsteps of Nanosur, another start-up TrilliumAg has started its own agriculture platform called Agrisome, which excels in production of self-assembled protein-based nanoparticles called modified RNA molecules (MV-RNA), possessing improved stability and efficient target delivery. These RNAi-based biopesticides have become a great hit among public since they spare the agricultural crops from the GMO tag (Shew et al., 2017). Utmost endeavor has been made by the researchers to make these biocontrol methods a perfect fit into the current integrated pest management system which have been strategized to protect crops from biotic stress and be least depended on conventional methods with adverse effects (Taning et al., 2020).
TABLE 6.
List of patent applications related to RNAi-inducing exogenous application of dsRNAs.
| Application number | Invention details | Assignee | Category | Field of application | Legal status |
| CA 2790211(2011) CN201180012795 (2011) EP20110753916 (2011) US 13/042,856 (2011) US 14/015,715 (2013) US 14/015,785 (2013) US 13/583,302 (2011) EP20170152830 (2011) PCT/US2011/027528 (2011) |
Regarding topical application of mixture containing RNAs, organosilicone surfactant and cationic lipid applied onto surface of plants | Monsanto Technology LLC | Industry | Herbicide resistance | Granted in US (2015), CN (2015) and EP (2017) |
| CA 2848680 (2012) CN 201280054179 (2012) EP20120832415 (2012) US 13/612,941 (2012) PCT/US2012/054862 (2012) |
Regarding a process and composition for modulating Acetyl- CoA carboxylase in weed species | Monsanto Technology LLC | Industry | Weed control | Granted in US (2016) |
| CA 2848685 (2012) CN 201280053985 (2012) EP20120831945 (2012) US 13/612,948 (2012) PCT/US2012/054883 (2012) |
Regarding a process and composition for modulating glutamine synthetase in weed species | Monsanto Technology LLC | Industry | Weed control | Granted in EP (2017), US (2016) |
| PCT/US2016/014344 (2016) | Regarding insecticidal composition containing organosilicone surfactant/cationic lipid, dsRNA complementary to leptinotarsa genes | Monsanto Technology LLC | Industry | Pest control | Entry into the national phase |
| US 14/037,750 (2013) PCT/US2013/062293 (2013) | Regarding transfection of A. tumefaciens crude containing hpRNA targeting BCTV and related curtovirus genes in sugar beets and tomato | Secretary of agriculture, United States | Government | Virus resistance | Granted in US (2017) |
| PCT/EP2007/000287 (2007) CA 2627795 (2007) EP20070700223 (2007) EP20110157036 (2007) EP20110157045 (2007) US 12/087,537 (2007) |
Regarding dsRNA spray carrying adjuvant and surfactant for insect control | Devgen NV | Industry | Insect control | Granted in EP (2012, 2017) |
| PCT/US2012/050687 (2012) CA 2842709 (2012) CN 201280039977 (2012) EP20120823842 (2012) US 13/585,947 (2012) |
Regarding formulation of dsRNA, water, plant hormone Brassinosteroid applied to the leaf surface of plants | Syngenta Participations AG | Industry | Pest/pathogen control | Granted in US (2016), CN (2016) and EP (2016) |
| PCT/EP2004/013049 (2004) EP20040818796 (2004) US 10/579,503 (2004) |
Regarding addition of naked, unpackaged dsRNA or siRNA to the feed of plant sap-sucking insect | CSIRO Bayer BioScience NV (Transfer of rights to Bayer in 2012) | Government, Industry | Pest control | Granted in EP (2011) |
| EP20140870784 (2014) PCT/AU2014/000255 (2014) CA 2934289 (2014) US 15/106,548 (2014) |
Regarding a naked dsRNA loaded upon LDH clay nanosheets | The University of Queensland | University/Research Institute | Virus resistance | Application submitted |
| MY-154890-A (2007) | Regarding a process for protecting orchids from CYMM virus using bacterial crude containing dsRNA | University of Malaya | University/Research Institute | Virus resistance | National (MY) patent |
| US 11/107,370 (2005) | Regarding preparation of a solution comprising recombinant A. tumefaciens containing a nucleic acid sequence complementary to the sequence of a target virus | The Samuel Roberts Noble Foundation | University/Research Institute | Virus resistance | Granted in US (2009) |
| PCT/US2014/026301 (2014) US 14/776,583 (2014) | Regarding topical introduction of dsRNAs having a sequence identical to an EIN2 gene to plant surface | Monsanto Technology LLC | Industry | Delayed senescence in flowers | Application submitted |
| CA 2873828 (2013) CN 201380039346 (2013) EP20130794339 (2013) US 13/901,326 (2013) US 14/403,491 (2013) PCT/IL2013/050447 (2013) |
Regarding introduction of exogenous RNA into seeds using a kit containing naked dsRNA and a priming solution | A.B. Seeds Ltd | Industry | RNA carrier | Application submitted |
| US 15/123,139 (2015) PCT/JP2015/057221 (2015) EP20150758536 (2015) |
Regarding transformation of a plant using carrier peptide containing a complex of nucleic acid molecules and a penetrating polycationic sequence. | Riken | University/Research Institute | RNA carrier | Application submitted |
| PCT/IL2016/050877 (2016) | Regarding a method of delivery into a plant cell using a composition of polycationic polymers, anionic surfactants, cuticle penetrating agent and polynucleotides | Forrest Innovations Ltd | Industry | RNA carrier | Application submitted |
After the discovery of a new product and before its approval for the commercial use, there lay a huge procedure to be followed and that is its potential risk assessment by various biosafety committees. Since dsRNA-based pest control method follows a novel mode of action, biosafety studies have been specially strategized to acquire accurate data regarding the target selectivity and all potential risks involving these novel dsRNAs (Arpaia et al., 2017). Risk assessment in this domain is mainly concerned with the adverse effects that might occur to the non-target organisms (NTOs) exposed, which might lead to the overall negative repercussions on the environment. The mode of action of these biopesticides should be thoroughly analyzed and their adverse effects on the NTOs should be inferred, so that those adverse chains of events can be minimized. When RNAi-inducing dsRNAs accumulate in optimum concentration in the system of these NTOs, it triggers the endogenous RNAi machineries within NTOs, resulting in unwanted gene knockdowns leading to health hazards. All possibilities of such events have to be ruled out through a series of experimental field trials to claim the RNA-based biocontrol compound to be safe for use. Some sequence-independent events might also occur inside the NTOs upon exposure to non-specific dsRNA, causing potential harm to them (Lundgren and Duan, 2013). Another major concern regarding biopesticides (dsRNA) is the formulations used to improve its retention power and delivery inside the target organism. NTOs overexposed with these newly formulated dsRNAs fail to degrade them with their intrinsic nucleases as the cumulative dose amount of formulated dsRNAs is far more magnified and various risk assessment committees have been formed to address several concerns regarding these RNA-based biopesticides. The Federal Insecticide, Fungicide, and Rodenticide Act (FIFRA) Science Advisory Panel took an active initiative to address the potential ecological risks and human health risks and the ways and methods to minimize both. The European Food and Safety Authority (EFSA) also has a commendable role in assessing the risk factors of RNAi-based GM plants. The crux of this discussion is if proper regulatory guidelines are demarcated, then RNA-based biopesticides can prove to be both cost-effective and time-saving to prepare as compared to synthetic pesticides.
In many countries in the world including a third world country such as India, major contribution to economy is made by the agricultural sector. A considerable loss in crop yield happens annually due to various environmental stresses and pest–pathogen attack. So, applications of these RNAi-based biopesticides are of utmost importance. The agriculture industry, as a whole, is a pretty complex structure with multiple layers of middlemen in every stratum. Starting from the R&D scientists who discover these novel biopesticides, till it reaches the hands of local farmers, the product has to pass through field trials, approval of proper biosafety committees for commercialization followed by manufacturing units of agritech start-up farms, distribution agencies, and through the rest of stakeholders. The utmost need of the hour is proper communication between every stratum of this complex sector, so that mass awareness can be created regarding the usage of these RNAi-based biopesticides. Earlier, the worldwide GM controversy has taught us all a valuable lesson that educating and enlightening the common man is generally least bothered about. So, the industry and the R&D sector are the most required bridge between both the farmers and the common consumers. They should come forward and clear out the air regarding all pros and cons of the newly launched biopesticides in the market. The ultimate aim should be to emphasize on the need of proper research and evidence-based product approval by authorized biosafety committees to introduce these novel tools to be implemented for a sustainable agricultural system.
Conclusion
RNA interference, a simple natural mechanism of eukaryotes, has been vastly exploited and extrapolated in the field of agriculture. The products generated (by endogenous/exogenous application) and marketed vouch its immense potential in the domain of crop protection. This technique has already curved its own niche in integrated pest management and sustainable agriculture. On the other hand, relatively new breakthrough genome-editing technology CRISPR/Cas is on its way to prove its efficacy in making disease-resistant smart crops without a “GMO” tag. Novel strategies for the above two technologies are needed on an urgent basis globally, to guarantee food security for the present and future generations. RNAi crops generated through HIGS are gradually paving the way to alternative approaches such as exogenously applied dsRNAs. But before commercialization, all the products need to pass through the acid test of assessment and regulation by biosafety committees. Exogenously applied dsRNA or “Biopesticides” possess immense potential to address the societal concerns regarding chemical pesticides and reluctance toward GMOs. Its successful application in pest management makes RNAi still relevant today. The success of these biopesticides finally lies on mass awareness of public and effective communication to expedite their commercialization.
Author Contributions
KH conceptualized and designed the manuscript. KH and AC wrote the manuscript. MZA, MM, and AD critically read and edited the manuscript. KH, AC, MZA, MM, and AD approved its submission. All the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
- AGO
argonaute
- CRISPR
clustered regularly interspaced short palindromic repeats
- DBD
DNA-binding domains
- DCL
Dicer like
- DSB
double-stranded breaks
- dsRNA
double-stranded RNA
- GenEd
gene edited
- GM
genetically modified
- HIGS
host-induced gene silencing
- hpRNA
hairpin loop RNA
- HR
homologous recombination
- miRNA
microRNA
- NHEJ
non-homologous end joining
- PAM
proto-spacer adjacent motif
- piRNA
PIWI-interacting RNA
- PTGS
post-transcriptional gene silencing
- RDR
RNA-dependent RNA polymerase
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RVD
repeat variable di-residue
- SIGS
spray-induced gene silencing
- siRNA
small interfering RNA
- SSN
sequence-specific nucleases
- tracrRNA
trans-activating CRISPR RNA.
Funding
This work was supported by both National Institute of Plant Genome Research, New Delhi, India and Department of Biotechnology, Ministry of Science & Technology, Government of India (No- BT/PR34369/AGIII/103/1176/2019). KH received INSPIRE fellowship from Department of Science and Technology, Ministry of Science and Technology, Govt. of India.
References
- Aalto A. P., Pasquinelli A. E. (2012). Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 24 333–340. 10.1016/j.ceb.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9:1911. 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z., Abulfaraj A., Idris A., Ali S., Tashkandi M., Mahfouz M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16 1–11. 10.1186/s13059-015-0799-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. L., Walker W. B. (2012). Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 58 391–396. 10.1016/j.jinsphys.2011.12.014 [DOI] [PubMed] [Google Scholar]
- Aman R., Ali Z., Butt H., Mahas A., Aljedaani F., Khan M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19 1–9. 10.1186/s13059-017-1381-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia S., Birch A. N. E., Kiss J., van Loon J. J. A., Messéan A., Nuti M., et al. (2017). Assessing environmental impacts of genetically modified plants on non-target organisms: the relevance of in planta studies. Sci. Total Environ. 583 123–132. 10.1016/j.scitotenv.2017.01.039 [DOI] [PubMed] [Google Scholar]
- Baltes N. J., Hummel A. W., Konecna E., Cegan R., Bruns A. N., Bisaro D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1 4–7. 10.1038/NPLANTS.2015.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J. A., Bogaert T., Clinton W., Heck G. R., Feldmann P., Ilagan O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25 1322–1326. 10.1038/nbt1359 [DOI] [PubMed] [Google Scholar]
- Bolognesi R., Ramaseshadri P., Anderson J., Bachman P., Clinton W., Flannagan R., et al. (2012). Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7:e47534. 10.1371/journal.pone.0047534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 1066 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Borrelli V. M. G., Brambilla V., Rogowsky P., Marocco A., Lanubile A. (2018). The enhancement of plant disease resistance using crispr/cas9 technology. Front. Plant Sci. 9:1245. 10.3389/fpls.2018.01245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlett M., Plaetinck G., Maienfisch P. (2020). RNA-based biocontrols—a new paradigm in crop protection. Engineering 6 522–527. 10.1016/j.eng.2019.09.008 [DOI] [Google Scholar]
- Burmistrz M., Krakowski K., Krawczyk-balska A. (2020). RNA-targeting CRISPR – cas systems and their applications. Int. J. Mol. Sci. 21:1122. 10.3390/ijms21031122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliari D., Dias N. P., Galdeano D. M., dos Santos E. Á, Smagghe G., Zotti M. J. (2019). Management of Pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10:1319. 10.3389/fpls.2019.01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W. L., Zilberman D., Xie Z., Johansen L. K., Carrington J. C., Jacobsen S. E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1336. 10.1126/science.1095989 [DOI] [PubMed] [Google Scholar]
- Chandrasekaran J., Brumin M., Wolf D., Leibman D., Klap C., Pearlsman M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17 1140–1153. 10.1111/mpp.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E., Dounda J. A. (2013). Rewriting a genome. Tha Real Og. Nat. Biotechnol. 495 50–51. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A., Halder K., Datta A. (2022). Classification of CRISPR/Cas system and its application in tomato breeding. Theor. Appl. Genet. 135 367–387. 10.1007/s00122-021-03984-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang D., Yao Q., Zhang J., Dong X., Tian H., et al. (2010). Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 19 777–786. 10.1111/j.1365-2583.2010.01038.x [DOI] [PubMed] [Google Scholar]
- Christiaens O., Smagghe G. (2014). The challenge of RNAi-mediated control of hemipterans. Curr. Opin. Insect Sci. 6 15–21. 10.1016/j.cois.2014.09.012 [DOI] [PubMed] [Google Scholar]
- Christiaens O., Whyard S., Vélez A. M., Smagghe G. (2020). Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11:451. 10.3389/fpls.2020.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras A., Jarausch W., Buchholz G., Bassler A., Braun M., Manthey T., et al. (2018). Delivery of hairpin rnas and small rnas into woody and herbaceous plants by trunk injection and petiole absorption. Front. Plant Sci. 9:1253. 10.3389/fpls.2018.01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras A., Wassenegger M., Dadami E., Ganopoulos I., Pappas M. L., Papadopoulou K. (2020). Genetically modified organism-free RNA interference: exogenous application of RNA molecules in plants1[open]. Plant Physiol. 182 38–50. 10.1104/pp.19.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras A., Wassenegger M., McMillan J. N., Cardoza V., Maegele I., Dadami E., et al. (2016). Induction of silencing in plants by high-pressure spraying of In vitro-synthesized small RNAs. Front. Plant Sci. 7:1327. 10.3389/fpls.2016.01327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Mueller E., Baulcombe D. C. (2000). Potato virus X Amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12 369–379. 10.1105/tpc.12.3.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho F., Gheysen G., Kushnir S., Van Montagu M., Inze D., Castresana C. (1992). Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J. 11 2595–2602. 10.1002/j.1460-2075.1992.tb05324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Zhao Z. (2014). Influence of catalase gene silencing on the survivability of sitobion avenae. Arch. Insect Biochem. Physiol. 86 46–57. 10.1002/arch.21161 [DOI] [PubMed] [Google Scholar]
- Di Lelio I., Barra E., Coppola M., Corrado G., Rao R., Caccia S. (2022). Transgenic plants expressing immunosuppressive dsRNA improve entomopathogen efficacy against Spodoptera littoralis larvae. J. Pest Sci. 10.1007/s10340-021-01467-z [Epub ahead of print]. [DOI] [Google Scholar]
- Dinh P. T. Y., Brown C. R., Elling A. A. (2014a). RNA interference of effector gene Mc16D10L confers resistance against meloidogyne chitwoodi in Arabidopsis and potato. Phytopathology 104 1098–1106. 10.1094/PHYTO-03-14-0063-R [DOI] [PubMed] [Google Scholar]
- Dinh P. T. Y., Zhang L., Brown C. R., Elling A. A. (2014b). Plant-mediated RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in diverse genetic backgrounds of potato and reduces pathogenicity of nematode offspring. Nematology 16 669–682. 10.1163/15685411-00002796 [DOI] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Drinnenberg I. A., Fink G. R., Bartel D. P. (2011). Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333:1592. 10.1126/science.1209575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta T. K., Papolu P. K., Banakar P., Choudhary D., Sirohi A., Rao U. (2015). Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 6:260. 10.3389/fmicb.2015.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn D. J., Cavallaro A. S., Bernard M., Mahalinga-Iyer J., Graham M. W., Botella J. R. (2007). Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226 1525–1533. 10.1007/s00425-007-0588-x [DOI] [PubMed] [Google Scholar]
- Fan W., Wei Z., Zhang M., Ma P., Liu G., Zheng J., et al. (2015). Resistance to Ditylenchus destructor infection in sweet potato by the expression of small interfering RNAs targeting unc-15, a movement-related gene. Phytopathology 105 1458–1465. 10.1094/PHYTO-04-15-0087-R [DOI] [PubMed] [Google Scholar]
- Fatica A., Bozzoni I. (2014). Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 15 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fister A. S., Landherr L., Maximova S. N., Guiltinan M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in theobroma cacao. Front. Plant Sci. 9:268. 10.3389/fpls.2018.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S. J., Reeves P. T., Hoang B. T., Mitter N. (2020). A perspective on RNAi-based biopesticides. Front. Plant Sci. 11:51. 10.3389/fpls.2020.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A., Meier R., Casanova A., Kreibich S., Daga N., Andritschke D., et al. (2014). Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc. Natl. Acad. Sci. U.S.A. 111 4548–4553. 10.1073/pnas.1402353111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G., Moffa L., Varotto S., Cardone M. F., Bergamini C., De Lorenzis G., et al. (2021). Novel and emerging biotechnological crop protection approaches. Plant Biotechnol. J. 19 1495–1510. 10.1111/pbi.13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. H., Yu X. R., Shang Q. L., Shi X. Y., Gao X. W. (2014). Oral delivery mediated rna interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, aphis gossypii glover. PLoS One 9:e102823. 10.1371/journal.pone.0102823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Tabara H., Mello C. C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287 2494–2497. 10.1126/science.287.5462.2494 [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81 611–620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Zhang X., Lu C., Johnson L., Meyers B. C., Green P. J., et al. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. 10.1038/ng1804 [DOI] [PubMed] [Google Scholar]
- Hernández I., Chacón O., Rodriguez R., Portieles R., López Y., Pujol M., et al. (2009). Black shank resistant tobacco by silencing of glutathione S-transferase. Biochem. Biophys. Res. Commun. 387 300–304. 10.1016/j.bbrc.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Hillary V. E., Ceasar S. A. (2019). Application of CRISPR/Cas9 genome editing system in cereal crops. Open Biotechnol. J. 13 173–179. 10.2174/1874070701913010173 [DOI] [Google Scholar]
- Huang C. Y., Wang H., Hu P., Hamby R., Jin H. (2019). Small RNAs – Big Players in Plant-Microbe Interactions. Cell Host Microbe 26 173–182. 10.1016/j.chom.2019.07.021 [DOI] [PubMed] [Google Scholar]
- Huang G., Allen R., Davis E. L., Baum T. J., Hussey R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103 14302–14306. 10.1073/pnas.0604698103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. B., Glick E., Paldi N., Bextine B. R. (2012). Advances in RNA interference: DsRNA treatment in trees and grapevines for insect pest suppression. Southwest. Entomol. 37 85–87. 10.3958/059.037.0110 [DOI] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakatura A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhang H., Zhang Y., Wang Y., Gao C. (2015). Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1 1–4. 10.1038/nplants.2015.144 [DOI] [PubMed] [Google Scholar]
- Jia H., Zhang Y., Orbović V., Xu J., White F. F., Jones J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15 817–823. 10.1111/pbi.12677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Ding L., He B., Shen J., Xu Z., Yin M., et al. (2014). Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 6 9965–9969. 10.1039/c4nr03481c [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science 337 816–822. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis A., Berbati M., Melita O., Reppa C., Holeva M., Otten P., et al. (2018). Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol. Plant Pathol. 19 883–895. 10.1111/mpp.12572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr A. (2016). Biological control of crown gall. Australas. Plant Pathol. 45 15–18. 10.1007/s13313-015-0389-9 [DOI] [Google Scholar]
- Ketting R. F. (2011). The many faces of RNAi. Dev. Cell 20 148–161. 10.1016/j.devcel.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E. J., Bernstein E., Sijen T., Hannon G. J., Plasterk R. H. A. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15 2654–2659. 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles G. J., Hofinger B. J., Hu P., Bayon C., Rudd J. J., Balmer D., et al. (2019). SRNA profiling combined with gene function analysis reveals a lack of evidence for cross-kingdom RNAi in the wheat – Zymoseptoria tritici pathosystem. Front. Plant Sci. 10:892. 10.3389/fpls.2019.00892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid A., Zhang Q., Yasir M., Li F. (2017). Small RNA based genetic engineering for plant viral resistance: application in crop protection. Front. Microbiol. 8:43. 10.3389/fmicb.2017.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink V. P., Kim K. H., Martins V., MacDonald M. H., Beard H. S., Alkharouf N. W., et al. (2009). A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta 230 53–71. 10.1007/s00425-009-0926-2 [DOI] [PubMed] [Google Scholar]
- Koch A., Kogel K. H. (2014). New wind in the sails: Improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12 821–831. 10.1111/pbi.12226 [DOI] [PubMed] [Google Scholar]
- Koch A., Biedenkopf D., Furch A., Weber L., Rossbach O., Abdellatef E., et al. (2016). An RNAi-based control of Fusarium graminearum Infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12:e1005901. 10.1371/journal.ppat.1005901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A., Höfle L., Werner B. T., Imani J., Schmidt A., Jelonek L., et al. (2019). SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non-host plants. Mol. Plant Pathol. 20 1636–1644. 10.1111/mpp.12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konakalla N. C., Kaldis A., Berbati M., Masarapu H., Voloudakis A. E. (2016). Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 244 961–969. 10.1007/s00425-016-2567-6 [DOI] [PubMed] [Google Scholar]
- Kunte N., McGraw E., Bell S., Held D., Avila L. A. (2020). Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 76 26–41. 10.1002/ps.5588 [DOI] [PubMed] [Google Scholar]
- Lau S. E., Schwarzacher T., Othman R. Y., Harikrishna J. A. (2015). dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 15:194. 10.1186/s12870-015-0577-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen Q., Lin Y., Jiang T., Wu G., Hua H. (2011). RNA interference in Nilaparvata lugens (Homoptera: Delphacidae) based on dsRNA ingestion. Pest. Manag. Sci. 67 852–859. 10.1002/ps.2124 [DOI] [PubMed] [Google Scholar]
- Li J., Todd T. C., Oakley T. R., Lee J., Trick H. N. (2010). Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta 232 775–785. 10.1007/s00425-010-1209-7 [DOI] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M. H., Weeks D. P., Yang B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30 390–392. 10.1038/nbt.2199 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang K., Xie H., Wang D. W., Xu C. L., Huang X., et al. (2015). Cathepsin B cysteine proteinase is essential for the development and pathogenesis of the plant parasitic nematode Radopholus similis. Int. J. Biol. Sci. 11 1073–1087. 10.7150/ijbs.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limera C., Sabbadini S., Sweet J. B., Mezzetti B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8:1418. 10.3389/fpls.2017.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren J. G., Duan J. J. (2013). RNAi-based insecticidal crops: potential effects on nontarget species. Bioscience 63 657–665. 10.1525/bio.2013.63.8.8 [DOI] [Google Scholar]
- Ma J., Chen J., Wang M., Ren Y., Wang S., Lei C., et al. (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 69 1051–1064. 10.1093/jxb/erx458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei A., Sevilla N. R., Cantos C., Jonson G. B., Slamet-Loedin I., Čermák T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16 1918–1927. 10.1111/pbi.12927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M., Viola R., Jung M. H., Koo O. J., Kim S., Kim J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. 10.3389/fpls.2016.01904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. B., Tao X. Y., Xue X. Y., Wang L. J., Chen X. Y. (2011). Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 20 665–673. 10.1007/s11248-010-9450-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mat Jalaluddin N. S., Othman R. Y., Harikrishna J. A. (2019). Global trends in research and commercialization of exogenous and endogenous RNAi technologies for crops. Crit. Rev. Biotechnol. 39 67–78. 10.1080/07388551.2018.1496064 [DOI] [PubMed] [Google Scholar]
- Matranga C., Zamore P. D. (2007). Small silencing RNAs. Curr. Biol. 17 789–793. 10.1016/j.cub.2007.07.014 [DOI] [PubMed] [Google Scholar]
- McDonald T., Brown D., Keller N. P., Hammond T. M. (2005). RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant Microbe Interact. 18 539–545. 10.1094/MPMI-18-0539 [DOI] [PubMed] [Google Scholar]
- Mezzetti B., Smagghe G., Arpaia S., Christiaens O., Dietz-Pfeilstetter A., Jones H., et al. (2020). RNAi: what is its position in agriculture? J. Pest Sci. 93 1125–1130. 10.1007/s10340-020-01238-2 [DOI] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter N., Worrall E. A., Robinson K. E., Li P., Jain R. G., Taochy C., et al. (2017). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 3:16207. 10.1038/nplants.2016.207 [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M., Rodriguez-Valera F. (2016). The discovery of CRISPR in archaea and bacteria. FEBS J. 283 3162–3169. 10.1111/febs.13766 [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M., Díez-Villaseñor C., Soria E., Juez G., Francisco J., Mojica M., et al. (2002). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36 244–246. 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- Moradpour M., Abdulah S. N. A. (2020). CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol. J. 18 32–44. 10.1111/pbi.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289. 10.2307/3869076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Wang C., Win J., Lanz C., Weigel D., Kamoun S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482. 10.1038/s41598-017-00578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás F. E., Torres-Martínez S., Ruiz-Vázquez R. M. (2013). Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 9:e1003089. 10.1371/journal.ppat.1003089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Jian H., Xu J., Chen C., Guo Q., Liu Q., et al. (2012). RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success. Eur. J. Plant Pathol. 134 131–144. 10.1007/s10658-012-9971-y [DOI] [Google Scholar]
- Nowara D., Schweizer P., Gay A., Lacomme C., Shaw J., Ridout C., et al. (2010). HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22 3130–3141. 10.1105/tpc.110.077040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata K., Ohtani M., Yoshizumi T., Demura T., Kodama Y. (2014). Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 12 1027–1034. 10.1111/pbi.12208 [DOI] [PubMed] [Google Scholar]
- Panwar V., McCallum B., Bakkeren G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81 595–608. 10.1007/s11103-013-0022-7 [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Peng A., Chen S., Lei T., Xu L., He Y., Wu L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15 1509–1519. 10.1111/pbi.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay M. (2020). Genome Editing Technologies for Crop Improvement, 2nd Edn, ed. Kang M. S. (Wallingford: CABI; ). [Google Scholar]
- Pyott D. E., Sheehan E., Molnar A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17 1276–1288. 10.1111/mpp.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebijith K. B., Asokan R., Hande H. R., Kumar N. K. K., Krishna V., Vinutha J., et al. (2016). RNA Interference of odorant-binding protein 2 (OBP2) of the cotton aphid, Aphis gossypii (Glover), resulted in altered electrophysiological responses. Appl. Biochem. Biotechnol. 178 251–266. 10.1007/s12010-015-1869-7 [DOI] [PubMed] [Google Scholar]
- Riechen J. (2007). Establishment of broad-spectrum resistance against Blumeria graminis f.sp. tritici in Triticum aestivum by RNAi-mediated knock-down of MLO. J. Verbraucherschutz Leb. 2 120–120. 10.1007/s00003-007-0282-8 [DOI] [Google Scholar]
- Rodrigues T. B., Petrick J. S. (2020). Safety considerations for humans and other vertebrates regarding agricultural uses of externally applied RNA Molecules. Front. Plant Sci. 11:407. 10.3389/fpls.2020.00407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N., Macino G. (1992). Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6 3343–3353. 10.1111/j.1365-2958.1992.tb02202.x [DOI] [PubMed] [Google Scholar]
- San Miguel K., Scott J. G. (2016). The next generation of insecticides: DsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 72 801–809. 10.1002/ps.4056 [DOI] [PubMed] [Google Scholar]
- Sasser J. N., Carter C. C., Hartman K. M. (1984). Standardization Of Host Suitability Studies And Reporting Of Resistance To To Root-Knot Nematodes. Raleigh, NC: North Carolina State University, 7. [Google Scholar]
- Schenke D., Cai D. (2020). Applications of CRISPR/Cas to improve crop disease resistance: beyond inactivation of susceptibility factors. iScience 23:101478. 10.1016/j.isci.2020.101478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Handelsman J. (2004). Status of the microbial census. Microbiol. Mol. Biol. Rev. 68 686–691. 10.1128/mmbr.68.4.686-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S. A., Koonin E. V. (2008). Origins and evolution of eukaryotic RNA interference The miRNA and siRNA machinery. Trends Ecol Evol 23 578–587. 10.1016/j.tree.2008.06.005.Origins [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakesby A. J., Wallace I. S., Isaacs H. V., Pritchard J., Roberts D. M., Douglas A. E. (2009). A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem. Mol. Biol. 39 1–10. 10.1016/j.ibmb.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Shew A. M., Danforth D. M., Nalley L. L., Nayga R. M., Tsiboe F., Dixon B. L. (2017). New innovations in agricultural biotech: consumer acceptance of topical RNAi in rice production. Food Control 81 189–195. 10.1016/j.foodcont.2017.05.047 [DOI] [Google Scholar]
- Sindhu A. S., Maier T. R., Mitchum M. G., Hussey R. S., Davis E. L., Baum T. J. (2009). Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 60 315–324. 10.1093/jxb/ern289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. S., Gu K. X., Duan X. X., Xiao X. M., Hou Y. P., Duan Y. B., et al. (2018). Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 19 2543–2560. 10.1111/mpp.12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada M., Pugliesi C., Fambrini M., Pecchia S. (2021). Silencing of the Slt2-Type MAP Kinase Bmp3 in botrytiscinerea by application of exogenous dsRNA affects fungal growth and virulence on Lactuca sativa. Int. J. Mol. Sci. 22 5362. 10.3390/ijms22105362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves R. M., Todd T. C., Essig J. S., Trick H. N. (2006). Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct. Plant Biol. 33 991–999. 10.1071/FP06130 [DOI] [PubMed] [Google Scholar]
- Stefano B., Patrizia B., Cerboneschi M., Massimo G. (2016). Inverse PCR and quantitative PCR as Alternative methods to southern blotting analysis to assess transgene copy number and characterize the integration site in transgenic woody plants. Biochem. Genet. 54 291–305. 10.1007/s10528-016-9719-z [DOI] [PubMed] [Google Scholar]
- Sun K., van Tuinen A., van Kan J. A. L., Wolters A. M. A., Jacobsen E., Visser R. G. F., et al. (2017). Silencing of DND1 in potato and tomato impedes conidial germination, attachment and hyphal growth of Botrytis cinerea. BMC Plant Biol. 17:235. 10.1186/s12870-017-1184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taning C. N. T., Arpaia S., Christiaens O., Dietz-Pfeilstetter A., Jones H., Mezzetti B., et al. (2020). RNA-based biocontrol compounds: current status and perspectives to reach the market. Pest Manag. Sci. 76 841–845. 10.1002/ps.5686 [DOI] [PubMed] [Google Scholar]
- Tinoco M. L. P., Dias B. B. A., Dall’Astta R. C., Pamphile J. A., Aragão F. J. L. (2010). In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 8:27. 10.1186/1741-7007-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez A. M., Fishilevich E. (2018). The mysteries of insect RNAi: A focus on dsRNA uptake and transport. Pest. Biochem. Physiol. 151 25–31. 10.1016/j.pestbp.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Walawage S. L., Britton M. T., Leslie C. A., Uratsu S. L., Li Y. Y., Dandekar A. M. (2013). Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14:668. 10.1186/1471-2164-14-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshe D. P., Lehane S. M., Lehane M. J., Haines L. R. (2009). Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 18 11–19. 10.1111/j.1365-2583.2008.00839.x [DOI] [PubMed] [Google Scholar]
- Walter C., Find J. I., Grace L. J. (2005). “Somatic embryogenesis and genetic transformation in Pinus radiata,” in Protocols for Somatic Embryogenesis in Woody Plants, eds Jain S. M., Gupta P. K. (Dordrecht: Springer; ), 11–24. 10.1007/1-4020-2985-3_2 [DOI] [Google Scholar]
- Waltz E. (2015). Nonbrowning GM apple cleared for market. Nat. Biotechnol. 33 326–327. 10.1038/nbt0415-326c [DOI] [PubMed] [Google Scholar]
- Waltz E. (2022). GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 40 9–11. 10.1038/d41587-021-00025-3 [DOI] [PubMed] [Google Scholar]
- Wang D., Liu Q., Li X., Sun Y., Wang H., Xia L. (2015). Double-stranded RNA in the biological control of grain aphid (Sitobion avenae F.). Funct. Integr. Genomics 15 211–223. 10.1007/s10142-014-0424-x [DOI] [PubMed] [Google Scholar]
- Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-Targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. 10.1371/journal.pone.0154027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Lin F. M., Thomma B. P. H. J., da Huang H., Jin H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. 10.1038/nplants.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Pan Q., He F., Akhunova A., Chao S., Trick H., et al. (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Cris. J. 1 65–74. 10.1089/crispr.2017.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- Wang Y., Meng Y., Zhang M., Tong X., Wang Q., Sun Y., et al. (2011). Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 12 187–201. 10.1111/j.1364-3703.2010.00659.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Heimes S., Riedel L., Sänger H. L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76 567–576. 10.1016/0092-8674(94)90119-8 [DOI] [PubMed] [Google Scholar]
- Will T., Vilcinskas A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57 34–40. 10.1016/j.ibmb.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Worrall E. A., Bravo-Cazar A., Nilon A. T., Fletcher S. J., Robinson K. E., Carr J. P., et al. (2019). Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front. Plant Sci. 10:265. 10.3389/fpls.2019.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L. K., Gustafson A. M., Kasschau K. D., Lellis A. D., Zilberman D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. 10.1371/journal.pbio.0020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Liu M., Zhuo F., Yin H., Deng K., Feng S., et al. (2019). Host-induced gene silencing of BcTOR in Botrytis cinerea enhances plant resistance to grey mould. Mol. Plant Pathol. 20 1722–1739. 10.1111/mpp.12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B. C., Veluthambi K., Subramaniam K. (2006). Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148 219–222. 10.1016/j.molbiopara.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Yao J., Rotenberg D., Afsharifar A., Barandoc-Alviar K., Whitfield A. E. (2013). Development of RNAi methods for peregrinus maidis, the corn planthopper. PLoS One 8:e70243. 10.1371/journal.pone.0070243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Jurgenson J. E., Hulbert S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant-Microbe Interact. 24 554–561. 10.1094/MPMI-10-10-0229 [DOI] [PubMed] [Google Scholar]
- Zhang H., Demirer G. S., Zhang H., Ye T., Goh N. S., Aditham A. J., et al. (2019). DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl. Acad. Sci. U.S.A. 116 7543–7548. 10.1073/pnas.1818290116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Khan S. A., Hasse C., Ruf S., Heckel D. G., Bock R. (2015). Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347 991–994. 10.1126/science.1261680 [DOI] [PubMed] [Google Scholar]
- Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., Graham S., et al. (2012). Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 45 303–313. 10.1016/j.molcel.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhou Y., Wang H., Jones H. D., Gao Q., Wang D., et al. (2013). Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants. BMC Genomics 14:560. 10.1186/1471-2164-14-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Z., Shi M., Holmes E. C. (2018). Using metagenomics to characterize an expanding virosphere. Cell 172 1168–1172. 10.1016/j.cell.2018.02.043 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan J., Sun J. R., Chen J. L. (2015). Cloning and RNA interference analysis of the salivary protein C002 gene in Schizaphis graminum. J. Integr. Agric. 14 698–705. 10.1016/S2095-3119(14)60822-4 [DOI] [Google Scholar]
- Zhao Y., Yang G., Wang-Pruski G., You M. (2008). Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur. J. Entomol. 105 815–822. 10.14411/eje.2008.108 [DOI] [Google Scholar]
- Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82 632–643. 10.1111/tpj.12838 [DOI] [PubMed] [Google Scholar]
- Zhu J. Q., Liu S., Ma Y., Zhang J. Q., Qi H. S., Wei Z. J., et al. (2012). Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One 7:e38572. 10.1371/journal.pone.0038572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo K., Chen J., Lin B., Wang J., Sun F., Hu L., et al. (2017). A novel Meloidogyne enterolobii effector MeTCTP promotes parasitism by suppressing programmed cell death in host plants. Mol. Plant Pathol. 18 45–54. 10.1111/mpp.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti M., dos Santos E. A., Cagliari D., Christiaens O., Taning C. N. T., Smagghe G. (2018). RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest. Manag. Sci. 74 1239–1250. 10.1002/ps.4813 [DOI] [PubMed] [Google Scholar]