Abstract
Heterogeneity in cell signaling pathways is increasingly appreciated as a fundamental feature of cell biology and a driver of clinically relevant disease phenotypes. Understanding the causes of heterogeneity, the cellular mechanisms used to control heterogeneity, and the downstream effects of heterogeneity in single cells are all key obstacles for manipulating cellular populations and treating disease. Recent advances in genetic engineering, including multiplexed fluorescent reporters, have provided unprecedented measurements of signaling heterogeneity, but these vast data sets are often difficult to interpret, necessitating the use of computational techniques to extract meaning from the data. Here, we review recent advances in computational methods for extracting meaning from these novel data streams. In particular, we evaluate how machine learning methods related to dimensionality reduction and classification can identify structure in complex, dynamic datasets, simplifying interpretation. We also discuss how mechanistic models can be merged with heterogeneous data to understand the underlying differences between cells in a population. These methods are still being developed, but the work reviewed here offers useful applications of specific analysis techniques that could enable the translation of single-cell signaling data to actionable biological understanding.
Introduction
Genetically identical cells, exposed to identical stimuli, exhibit markedly different responses. One key manifestation of this non-genetic heterogeneity is in signaling pathway activation, where individual cells display a range of behaviors in both basal activity and responsiveness to a stimulus. The drivers that control signaling heterogeneity and the behavioral outcomes of signaling heterogeneity are not well characterized, and fundamental questions remain. Heterogeneity in signaling activity is thought to drive cellular behaviors, some of which are binary, such as division or cell death [1–3]. Thus, one area of research focuses on what control points and mechanisms translate continuous variation in some components into binary decisions downstream (Figure 1A). Another focus of research is determining to what extent heterogeneity represents randomness, caused by a cell’s intrinsic inability to faithfully interpret a stimulus. Alternatively, perhaps different responses are caused by pre-existing differences among cells with some poised to strongly activate a signaling pathway (Figure 1B–E). This is an important biological question with deep implications relating to the limitations of signaling pathways and communication in multicellular organisms. Non-genetic heterogeneity also has urgent clinical implications. Heterogeneity drives some of the most disastrous features of cancer, including metastasis [4], invasion [5], and the emergence of drug-resistant cells [6,7]. Therefore, understanding the causes of heterogeneous cell signaling and identifying ways to control it are crucial steps toward effective cancer therapy [8]. To start to answer these questions, researchers have used an array of live-cell microscopy techniques to measure behaviors in single cells. However, interpretation of these data has proven difficult, and an array of modeling techniques have emerged to make meaning out of heterogeneous, single-cell data.
Figure 1:
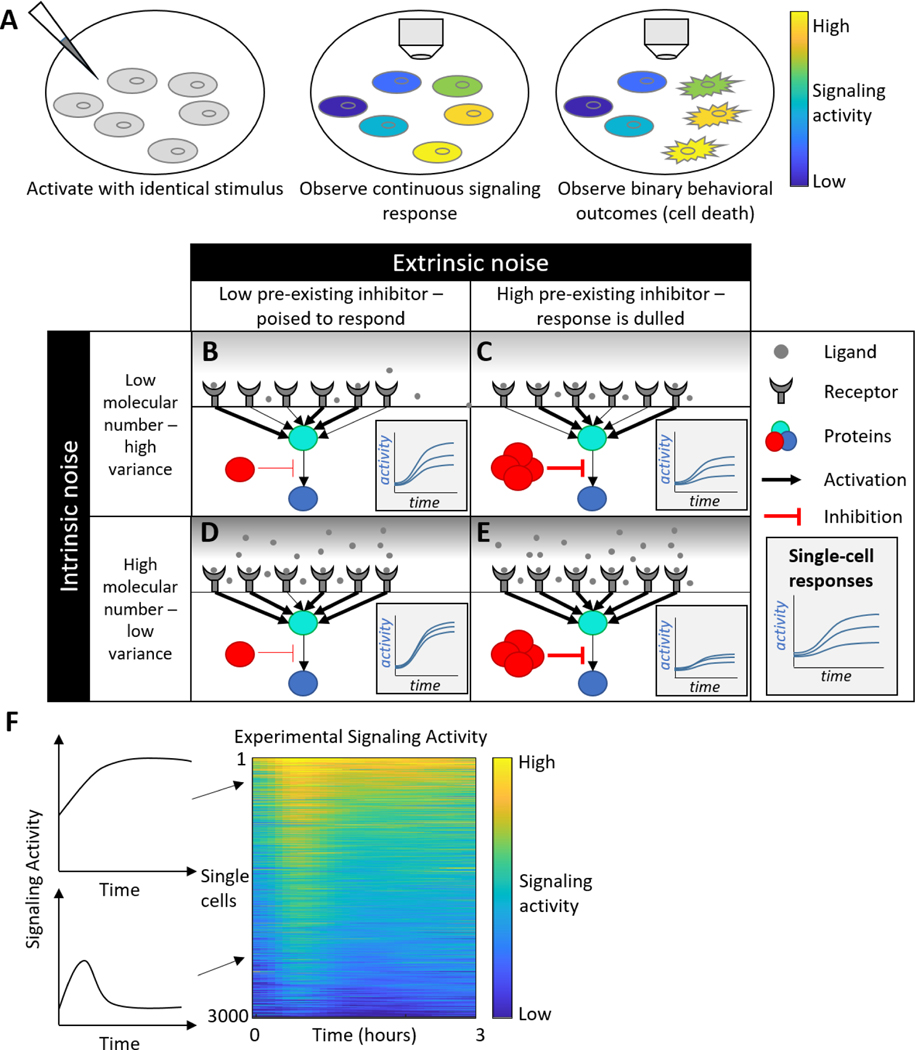
Sources of heterogeneity. A) Cells can exhibit continuous variation, for instance in signaling response (middle). This continuum may be converted into binary outcomes for phenotypes like cell death. B-E) Comparing the effects of intrinsic and extrinsic noise. Columns represent two cells in different pre-existing states based on the amount of an inhibitory protein in the cell. Rows represent differences in ligand concentration, with low ligand concentration leading to significant differences in response based on stochasticity of receptor-ligand binding. Insets represent signaling responses in 3 separate cells for each condition. F) A kymograph or colormap is commonly used to represent single-cell signaling data. Left: Two individual cell responses, one sustained and one transient. Right: A kymograph summarizing responses of 3000 cells.
In genetically identical cells, heterogeneity is thought to arise from both intrinsic and extrinsic sources [9–11]. Intrinsic heterogeneity occurs due to random fluctuations inherent to individual intermolecular collisions that underlie the chemical reactions that regulate biological processes. These fluctuations are present in any chemical reaction, but they decrease in influence as concentration increases and randomness in intermolecular collisions is smoothed out. However, intrinsic noise can be significant in some biological contexts, typically cellular processes involving low abundance molecules like gene transcripts (below 100 copies in mammalian cells, as a rough estimate) or rare events [12,13]. Intrinsic noise places fundamental limits on the ability of cells to sense their environment and control gene expression [14,15].
Extrinsic noise refers to pre-existing differences in cell state and are thus “extrinsic” from the fundamental fluctuations of chemical reactions. Cells that appear identical may in fact exhibit differences in cell-cycle timing, kinase pathway activation, or the concentration of receptors or other signaling pathway molecules [16,17]. When exposed to a stimulus, these cells will respond in various ways predicated on their pre-existing state. If cells were completely identical, extrinsic noise could be eliminated, but they would still respond differently due to intrinsic noise.
The clinical goals of controlling heterogeneity, e.g. to improve cancer treatment, require that researchers understand the sources of heterogeneity. Therefore, distinguishing between intrinsic and extrinsic noise is a key challenge, which can be addressed with computational and experimental tools. It may be possible to mitigate or alter heterogeneous populations of cells if extrinsic noise is driving their heterogeneity [18,19]. However, if intrinsic stochasticity is the root cause of heterogeneous phenotypes, interventions may not be possible or may need to target a downstream effector that is less impacted by intrinsic noise.
In this mini-review, we outline several methods used to analyze live-cell, time-lapse, fluorescence microscopy data. Given the key role that dynamic processes can play in controlling cellular behaviors, we focus here on live-cell, time-lapse methods, as opposed to endpoint methods including cyclic immunofluorescence, flow cytometry, or single-cell RNA-seq, all of which have been reviewed elsewhere e.g. [20,21]. Dynamic experimental methods can quickly generate hundreds of observations for thousands of cells, necessitating the use of computational methods to answer even simple questions about the data relating to the population level behaviors and variance among cells. Furthermore, watching cells in real time leads to new questions about the dynamics of cellular processes in single cells and their synchronization, which can be answered using dynamic computational models. We discuss data-driven methods that extract patterns from data, which can be quickly and easily applied to quantify and compare heterogeneous signaling behaviors. We then discuss model-driven methods, where researchers construct mechanistic models with varying degrees of complexity to explain heterogeneous behaviors in their data. Throughout this review, we provide examples of specific biological questions that are addressed.
Single-cell Signaling Data Acquisition
Single-cell time domain data can be obtained from any live single-cell imaging modality. Using bright-field microscopy, data about morphology, movement, lineage, and cell division can be measured for individual cells over time [22]. Using fluorescence to monitor biochemical processes including cell signaling can be accomplished with dyes or endogenously expressed fluorescent reporter proteins [23,24]. Ideal fluorescent measurement systems will feature a high quantum yield to minimize light exposure and rapid activation and deactivation kinetics to accurately track the underlying cellular process of interest. Other key factors are the photostability of the reporter protein and maximizing the overlap between reporter emission spectra and detector absorption spectra while minimizing overlap in emission spectra among different reporter proteins. Balancing these considerations is difficult and has been reviewed at length elsewhere [25].
Automated image processing is another key aspect of the experimental pipeline. Although ImageJ [26] can be used to extract intensities and calculate reporter activities, manually processing images will typically not scale effectively to larger datasets. Automated methods such as CellProfiler [27] have been developed to identify individual cells, extract reporter activities, and track cells from frame to frame [28,29]. Ideally, tracking will be consistent even in dense, confluent cultures, and cell division and lineage will be tracked. These programs generate trajectories for each cell tracked based on the activity of the reporters in that cell. These trajectories can be summarized quickly in kymographs, which display individual cell trajectories as rows in a colormap, with the intensity in each column corresponding to the reporter value at that time (Figure 1F).
Data-Driven Inference
Given a heterogeneous distribution of cell trajectories (Figure 2A), the simplest analytical approach is to mitigate the heterogeneity using summary statistics such as averages, medians, and percentiles (Figure 2B). This approach is quick and readily understood, providing a useful start to any analysis of heterogeneous data. However, there are two main problems with applying summary statistics to heterogeneous signaling data. Firstly, they blur the heterogeneity present in the data, leaving the researcher with aggregate behaviors that can be measured with other methods. One solution to this problem is to look not just at means or medians, but instead at various percentiles or outliers, which are only accessible from single cell data [30]. The second issue with mean or median behaviors is that they may not represent any real cells. For instance, if cell responses are binary (either high or low), the average of all cells will be a medium response, behavior that is not exhibited by any actual cells observed (Figure 2B) [31]. It can be difficult to know when aggregate data can provide insight or when aggregation obscures meaningful single-cell behaviors. For instance, Goglia et. al. found meaningful differences in drug responses by aggregating cells and looking at mean population behaviors [32]. However, when deciding what types of aggregation to use, researchers should consider if they are interested in studying collective or individual behaviors and should also balance the potential for greater explanatory power of single-cell data against the increased noise that can come from single cell trajectories. The methods discussed below can provide researchers with more flexibility when considering some of these issues.
Figure 2:
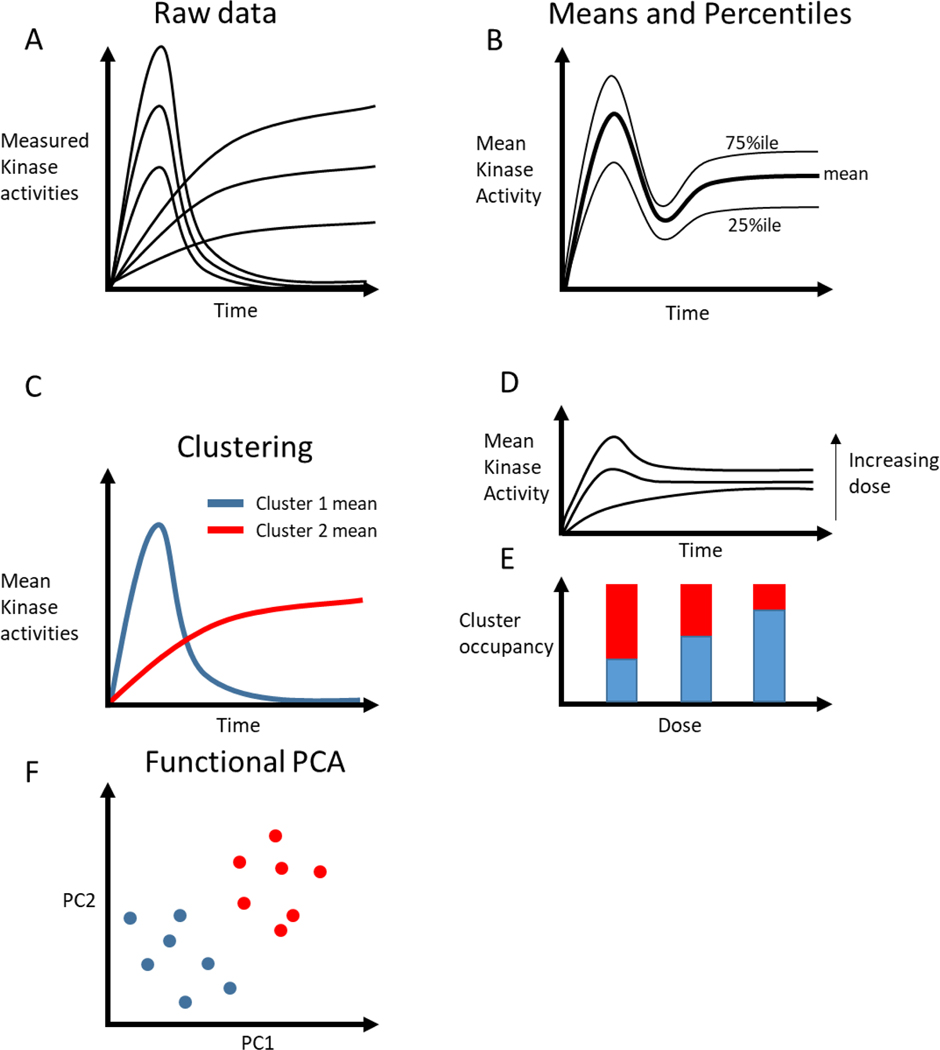
Analysis of dynamic, single-cell data. A) Each line indicates a cellular response from a single-cell experiment, showing two families of responses with variation in each family. B) Mean and percentile measures can capture some dynamics, but no experimental cells behave like the mean. C) Taking the mean of cells assigned to clusters can yield representative dynamics. D, E) Hypothetical data from a dose response experiment. D) The mean increases with dose, but the dynamics are not captured. E) Clustering the data reveals that increasing dose leads to more cells adopting fast, transient responses (blue cluster). F) Functional PCA can identify clusters of behavior while capturing variation within each cluster.
Clustering algorithms (Table 1) can summarize signaling trajectories while ensuring that the aggregation is representative of real behaviors. Clustering finds families of distinct trajectories within the data by first calculating the distance between individual trajectories and then identifying clusters of trajectories that have low within-cluster distances and high between-cluster distances. This allows data aggregation within each cluster (Figure 2C). Euclidean distance is often used as a distance metric to calculate the similarity of each cell with every other cell. However, cells may display dynamics at different times, or for different durations, despite undergoing the same biological processes. For instance, clustering dividing and non-dividing cells would require a distance metric that is sensitive to asynchronous divisions in cells and differences in the duration of cell division. Therefore, distance metrics using dynamic time warping or other algorithms may do a better job of describing similarities in time series data [33]. After defining a distance metric, commonly used clustering algorithms such as hierarchical clustering or k-means are used to label individual observations based on cluster occupancy. Using clustering, researchers can ensure that the aggregate behaviors that compose each cluster are representative of real cell behaviors. Blum et. al. wanted to determine if different growth factors induced distinct ERK activation dynamics in single cells [34,35]. They clustered ERK trajectories from cells activated by various growth factors, allowing them to represent over 1000 signaling trajectories using 6 clusters, drastically simplifying the analysis and presentation of their data. They found that the clustered behaviors were growth-factor dependent. Interestingly, their analysis also showed that for even low doses of growth factors, some cells would respond strongly, and at high doses, the occupancy of this cluster increased (Figure 2D, E). In this work, clustering was useful because it separated cells that had qualitatively different dynamics (e.g. transient vs. sustained activation). However, this may be an inappropriate constraint when cells occupy a continuum of responses.
Table 1:
Computational techniques for dimensionality reduction and mechanistic understanding of single-cell heterogeneous signaling data
| Method | Output | Benefits | Shortcomings | Software | Applications |
|---|---|---|---|---|---|
|
| |||||
| Clustering | Labels for each cell corresponding to cluster belonging. | Aggregates based on real cell behavior, relatively fast, interpretable. | Inappropriate or unhelpful for continuous responses | Time course inspector R package and web application [35] |
[22], [34], [47] |
| FPCA | Low-dimensional projection of each cell along axes of variation in the data. | Enables easy visualization of heterogeneity to identify patterns in data. | Primary axes of variation may not be physiologically meaningful. | FDA package [36,37] | [38] |
| Information Theory | Numerical quantification of mutual information between input (dose or ligand) and output (reporter activity) | Summarizes heterogeneous outcomes in a single number with a physical meaning. | Mutual information may be of limited interest for particular study. | SLEMI R package [59], EstCC Scala package [60] | [11,39–42,44] |
| Stochastic Dynamic modeling | A distribution of species dynamics corresponding to each parameter set used by modeler. | Can explicitly model the effects of intrinsic stochasticity and low molecular number. | Requires knowledge of rate constants and species interactions, along with simplifying assumptions to make system tractable. | Simbiology MATLAB package, Hy3S [61], PySB Python package [43] | [43,44,46] |
| ODE modeling | A set of deterministic species dynamics corresponding to each parameter set used by modeler. | Can vary parameters to test hypotheses about sources of heterogeneity, suggest mechanistic drivers. | Requires knowledge of rate constants and species interactions, along with simplifying assumptions to make system tractable. | Simbiology MATLAB package, PySB Python package [43] | [11,16,43,47,48] |
Another method that can summarize an arbitrarily large number of trajectories is functional principal component analysis (fPCA, Table 1) [36,37]. fPCA decomposes a set of trajectories into two different parts, harmonics and principal component scores. The harmonics of fPCA are orthogonal time-dependent trajectories representing times when there is the most variance in the data The key difference between fPCA and PCA is that fPCA captures time-dependence in trajectories, as opposed to principal components that ignore the time-dependence of the data. fPCA will also calculate a numerical score for each individual trajectory provided, based on how much each harmonic represents a specific trajectory. For instance, if some cells exhibited a transient increase in signal, while others had a sustained increase, these would be represented as different functional principal components, with transient cells scoring highly along one axis and sustained cells scoring highly along the other. By representing trajectories in lower-dimensional space, they can be visualized and compared more easily (Figure 2F). Sampattavanich et. al. used fPCA to identify differences in transcription factor FOXO3a dynamics in response to a variety of growth factors [38]. fPCA identified that key variation in trajectories existed through variation in the basal level, the post-stimulus steady-state, and the transient response. Furthermore, the authors were able to cluster responses to ligands based on their dynamics as represented by fPCA. A potential shortcoming of clustering is that it labels each cell discretely, implicitly erasing heterogeneity that may exist within clusters. Since fPCA relaxes this constraint and projects cells along a spectrum of PC scores, this heterogeneity is preserved, which may provide a clearer picture of the data. Furthermore, fPCA can serve as a pre-processing step for later clustering.
Clustering and fPCA focus on revealing structure within heterogeneous data by reducing dimensionality in a discrete or continuous manner. These are related to unsupervised machine learning methods, but supervised learning, where different observations are labeled, can also be applied to heterogeneous cell signaling. For example, by extending experimental acquisition times to several hours or days, cells can be labeled based on cell division, death, or migration. This gives researchers another way to categorize signaling data based on material observations and has the added benefit of reflecting phenotypes that are oftentimes of interest. However, this approach has attendant difficulties, including increased photodamage from long-term microscopy and the increased difficulty of tracking cells over long times. Miura et. al. showed that in cells exposed to UV light, p38 and JNK respond heterogeneously, with p38 suppressing JNK in some cells [2]. They extended imaging to 6–12 hours, and were able to identify JNK-high cells as apoptotic, while p38-high cells survived. This behavioral classification allowed them to associate signaling patterns with functional outputs.
Given that signaling pathways communicate the presence and amount of a ligand in the extracellular space to the cell, key questions are: How effective is this communication, and what tradeoffs do cells make to control effectiveness? Information theory answers these questions by quantifying how severely noise (both extrinsic and intrinsic) corrupts the ability of a signal sender to communicate with a receiver (Table 1). Information theory measures communication in units of bits, which correspond to the amount of information gained about an input from measurement of an output. In a system with high information transfer, measuring an output will give a high-confidence estimate of the input signal, while in a low information transfer system, noise will corrupt the output, making it difficult to determine a specific input based on a measured output. Most commonly, information theory has been used to quantify the ability of cells to differentiate between different doses of a ligand, given that each dose provokes a distribution of responses across different cells. Early work using this approach from Cheong et al. calculated information transfer for experimental measurements of NF- κB activation by TNF and compared them with theoretical results from statistical models incorporating different signaling pathway architectures and noise sources, finding that signaling architecture and the integration of a signal over time by cells can mitigate information loss [39]. In an expansive study, Selimkhanov et. al. quantified information transfer in ERK, calcium, and NF-kB signaling pathways [40]. They showed that information transfer can be much higher if signaling responses at multiple time points are considered, offering one explanation of the previous, static measurements of information transfer, which were surprisingly low. They also used a combination of information theory and experimental perturbation of the ERK pathway to quantify intrinsic and extrinsic sources of noise, finding that extrinsic sources of heterogeneity dominated. More recent work has built on these foundations, measuring information transfer by stimulating the same cells repeatedly [41,42]. These experiments and analyses have revealed that different cells in a population may have very different information capacities. Information theory facilitates the quantification of intrinsic and extrinsic contributions to heterogeneity, and summarizes heterogeneous data in a way that is consistent with signaling pathway function - to transmit information and enable cells to respond accordingly.
Mechanistic Models of Heterogeneity
The data-driven methods described above detail ways of extracting patterns or meaning from within a dataset. However, a separate class of methods involves constructing mechanistic models of cellular behavior, and synthesizing model and experimental data to draw conclusions about observed heterogeneity. Mechanistic modeling requires numerical parameters and information about mechanisms that are oftentimes difficult or impossible to obtain, which can make modeling significantly more difficult and time-consuming than data-driven methods. However, mechanistic models provide unique opportunities to computationally control the relative effects of biological mechanisms and parameters, which can yield deeper insights into the causes and consequences of heterogeneity. Models can be stochastic or deterministic, and can range from simple and qualitative to sprawling and numerically precise. In general, model building should be directed by specific questions that can dictate the specific interactions that need to be included in the model.
To model stochastic sources of intrinsic noise, specific frameworks have been developed that simulate individual molecular events (Table 1) [43]. The framework for these simulations uses the Gillespie algorithm, which calculates the probability of different events occurring based on their pre-defined interaction rate constants and species number. To run stochastic simulations, the researcher must define the species involved, the stoichiometry of each interaction between species, and probabilistic rate constants of each interaction. After these parameters are defined, simulations can be run, which produce individual realization of the random process being modeled. This approach was utilized to study heterogeneity in the ERK response to hormone stimulation by Garner et. al. They constructed a simple model of ERK signaling that incorporated multiple sources of feedback control [44]. They combined this model with calculations of information transfer carried out on their experimental and simulated data, and varied different sources of feedback to study how different regulatory mechanisms could influence information transfer. Their model enabled them to make experimentally testable predictions about the factors that influence information transfer in the presence of intrinsic noise. Notably, stochastic simulations were also used by Iwamoto et. al. to explore variability in EGFR signaling to ERK, as measured by flow cytometry [45]. They found that stochasticity caused by intrinsic noise was insufficient to explain the variability in their data because they measured sufficiently high concentrations of molecules for all components. This highlights the potential complexity of modeling intrinsic and extrinsic factors underlying heterogeneity, and shows how modeling can be used to differentiate between the two. A similar approach was used by Wang et. al. in analyzing p53 dynamics, where they found that in mammalian cells, p53 variability was consistent with extrinsic cell-to-cell differences [46]. These two examples illustrate how computational models can help identify extrinsic, and potentially controllable sources of noise, compared to intrinsic noise.
As consciousness of the role of pre-existing differences in seemingly identical cells grew, other work began to focus on using deterministic mechanistic models based on ordinary differential equations to understand which cellular components were different from cell to cell (Table 1) [11,16,43,47,48]. Yao et. al. observed heterogeneous calcium signaling dynamics (Figure 3A) [48]. They used summary statistics to summarize and cluster their data, but they followed up on those observations by constructing a differential equation model of calcium signaling (Figure 3B). They derived parameter sets that provided good fits to individual cellular responses, and recorded the parameter distributions that were extracted from the population of single-cell fits (Figure 3C, D). They found clusters of parameter distributions that corresponded to clusters of cellular responses. Observed heterogeneity of responses arose from differences in interactions between IP3 import to the endoplasmic reticulum and related regulation of this process by calcium. This approach required the construction of a mechanistic model, which has intrinsic difficulties and complications, but it uniquely facilitated a granular, mechanistic understanding of calcium signaling heterogeneity.
Figure 3:
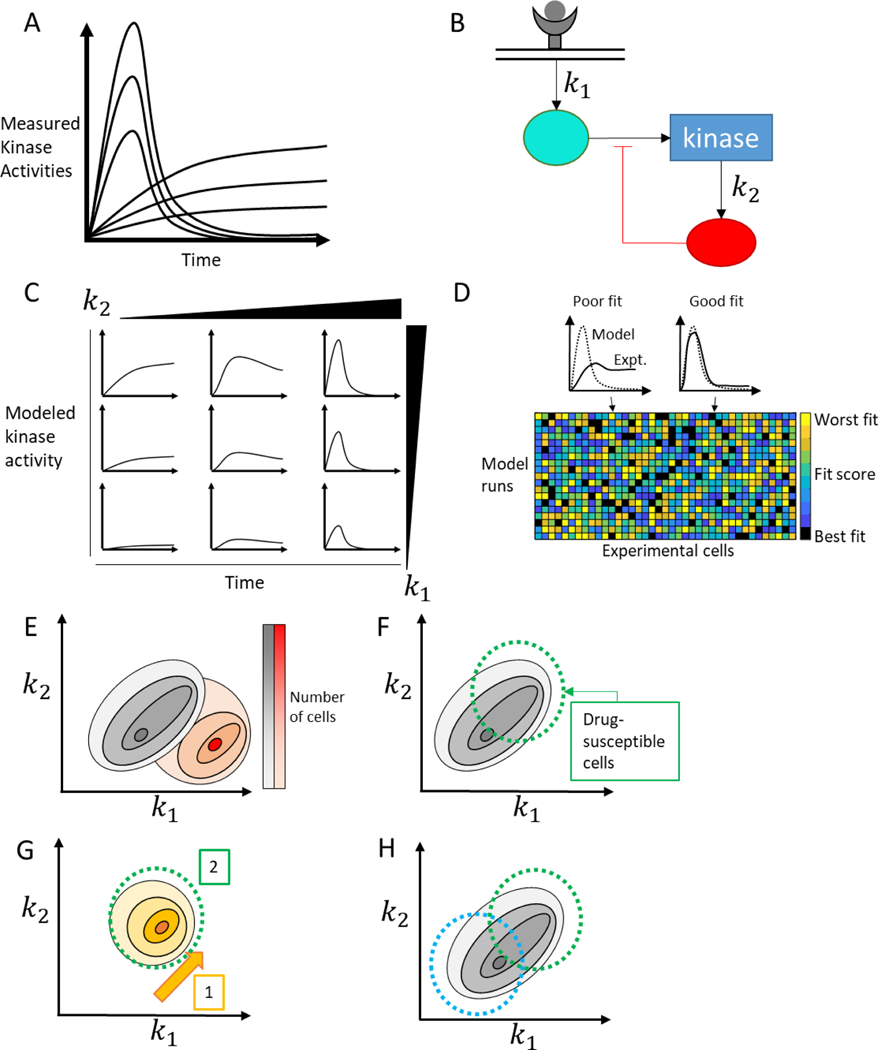
Mechanistic modeling of heterogeneous signaling data. A) Heterogeneous single cell responses can differ in both magnitude and shape, with some cells exhibiting a sustained peak and others a transient response. B) A simplified schematic of an ODE model which can be used to study the heterogeneity shown in A. Two rate constants, k1 and k2, out of many are identified. C) Systemic variation of parameters in the model shows that k1 increases the magnitude of the response, while changes in k2 tune the duration of the response. D) Model and experimental outputs can be compared (top) with different cells fitting well with different model outputs. This procedure can be completed for all cell/model combinations (bottom) to extract a parameter set which represents the pre-existing cell state. E) Model-based inference of parameter values – and therefore cell state - enables population of cells to be placed on a spectrum of heterogeneous mechanistic processes, and compared between two different experimental conditions (black distribution vs. red distribution). F) Understanding cell state can provide mechanistic explanations for why some subpopulations of cells are sensitive to a drug and others aren’t. Treating with a drug that only targets some of the cells (green circle) will leave behind an insensitive population of cell. G) Using this knowledge, we can combine drugs temporally, giving one drug (orange) to sensitize the entire population to a second drug (green). H) We can also combine drugs (blue and green) to target two subpopulations simultaneously, eliminating the whole population of cells.
The approach taken by Yao et. al., in varying every model parameter, implies a particular assumption about pre-existing cellular states - that cell state occupancy is pseudo-random, and that variability is equally likely in all components [49]. However, some pathway components are affected by many different inputs, while others are more confined to individual pathways [50]. Furthermore, different components are subject to different regulatory mechanisms which may be better or worse at suppressing variation [51]. Therefore, it is plausible that variability in kinase activation could be due to a small number of highly variable species. Spinosa et. al. explored this hypothesis when studying ERK and Akt signaling in response to CXCL12 (Figure 3B) [16]. They built a differential equation model including both pathways and accounted for the variability observed by varying just three parameters, corresponding to extrinsic noise leading to variable activation in specific pathway components Ras, PI3K, and mTORC1 (Figure 3C,D). This approach allowed them to locate each cell in an experiment to a specific point in 3D space corresponding to pre-existing cell state, and to make measurable predictions about the effects of various inhibitors in moving the population of cells around in state space (Figure 3E). Identifying key nodes of heterogeneity may be useful for inferring treatable therapeutic targets facilitating the control of heterogeneous cell states, or understanding why some cells are resistant to targeted inhibitors (Figure 3F).
Continuing Challenges and Promising Approaches
As experimental methods advance, we will have access to ever-growing forms of data, including multiplexed reporters that can be deployed in new ways and observed at new timescales. Making meaning from all of this data may be a daunting task, but it will yield unprecedented insights into the sources of cellular heterogeneity and means by which it can be controlled. Fortunately, extracting meaning from heterogeneous time-series data is central to the field of signal processing, and many challenges associated with single-cell signaling data are present in other fields. Some techniques, including information theory and functional data analysis, were borrowed from signal processing and creatively applied to single-cell signaling data. Researchers in many fields are studying heterogeneous, dynamic data to answer questions ranging from the geographic differences in bird calls to fault diagnosis in mechanical systems [52,53]. Novel machine learning techniques to extract features from time-series data are an open field of study [54]. All of this suggests that interdisciplinary collaboration inspired by work in many different fields may yield creative methods of analysis for heterogeneous cell signaling data.
There are also key biological challenges to address in order to fully realize the potential of single-cell imaging modalities. Specific challenges, which will require realizing novel computational and experimental methods, include the incorporation of disparate timescales, cellular systems, and streams of data. Meaningful signaling dynamics can occur on the seconds to minutes time scale, as shown by transient ERK pulses that can drive proliferation, on the minutes to hours timescale associated with ligand-receptor signaling, and on the several hour scale associated with cell division [55,56]. Connecting these timescales in a multi-scale model that explains variation in single-cell behaviors represents a major challenge, but also holds the key to understanding how cells integrate information to make decisions. Furthermore, signaling pathways are affected by numerous other cellular processes, including metabolism and cell cycle progression, all of which may help determine a cell’s pre-existing state. Understanding how cell-to-cell variation in these other systems, such as variations in glucose utilization, affect heterogeneity in signaling, and vice versa, will require merging computational models for each individual system [57]. Finally, heterogeneity is understood in a variety of ways, and single-cell sequencing technologies have exploded in recent years. Understanding how observable signaling dynamics shape heterogeneity in RNA transcript number is another major challenge, which again requires merging mechanistic signaling models with data-driven models of heterogeneity in RNA expression [58]. By extending our understanding of the situational balance between various sources of heterogeneity, we will be able to identify targets that will enable control over cellular populations, and possible dead-ends where intrinsic noise dominates over any plausible intervention. Clinically, identifying sources of heterogeneity may enable temporal treatment strategies that first shift heterogeneous cells to a more homogeneous set of states, making the whole population vulnerable to a second drug (Figure 3G) [18]. Another approach might identify treatments that affect specific subpopulations based on an identified axis of variation and combine those treatments to kill all populations simultaneously (Figure 3H). Creative combinations of experimental and computational methods are the key to making progress on all of these fronts.
Highlights:
Multiplexed fluorescent reporters offer unprecedented measurements of single-cell data.
Data-driven methods can reveal subpopulations with distinct dynamics or outcomes.
Mechanistic models enable discovery of drivers of heterogeneity.
Acknowledgements
The authors acknowledge funding from United States National Institutes of Health Grants R01CA196018, R01CA238042, R01CA238023, R33CA225549, R37CA222563, R50CA221807, and U01CA210152.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Min M, Rong Y, Tian C, Spencer SL: Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science 2020, 368:1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miura H, Kondo Y, Matsuda M, Aoki K: Cell-to-Cell Heterogeneity in p38-Mediated Cross-Inhibition of JNK Causes Stochastic Cell Death. Cell Rep 2018, 24:2658–2668. By measuring p38 and JNK responses in single cells exposed to UV light, the authors revealed that a spectrum of JNK activity was controlled by p38. They extended their analysis time to capture downstream cell death, and found that p38 inhibition of JNK protected cells from apoptosis.
- 3.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK: Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 2009, 459:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolly MK: Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front Oncol 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jolly MK, Ware KE, Gilja S, Somarelli JA, Levine H: EMT and MET: necessary or permissive for metastasis? Mol Oncol 2017, 11:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okabe S, Tauchi T, Ohyashiki K: Characteristics of Dasatinib- and Imatinib-Resistant Chronic Myelogenous Leukemia Cells. Clin Cancer Res 2008, 14:6181–6186. [DOI] [PubMed] [Google Scholar]
- 7.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. : A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 2010, 141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedard PL, Hansen AR, Ratain MJ, Siu LL: Tumour heterogeneity in the clinic. Nature 2013, 501:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elowitz MB: Stochastic Gene Expression in a Single Cell. Science 2002, 297:1183–1186. [DOI] [PubMed] [Google Scholar]
- 10.Swain PS, Elowitz MB, Siggia ED: Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci 2002, 99:12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sumit M, Jovic A, Neubig RR, Takayama S, Linderman JJ: A Two-Pulse Cellular Stimulation Test Elucidates Variability and Mechanisms in Signaling Pathways. Biophys J 2019, 116:962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JJY, Low-Nam ST, Alfieri KN, McAffee DB, Fay NC, Groves JT: Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci Signal 2019, 12:eaat8715. [Google Scholar]
- 13.Soltani M, Vargas-Garcia CA, Antunes D, Singh A: Intercellular Variability in Protein Levels from Stochastic Expression and Noisy Cell Cycle Processes. PLOS Comput Biol 2016, 12:e1004972. [Google Scholar]
- 14.Bialek W, Setayeshgar S: Physical limits to biochemical signaling. Proc Natl Acad Sci 2005, 102:10040–10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lestas I, Vinnicombe G, Paulsson J: Fundamental limits on the suppression of molecular fluctuations. Nature 2010, 467:174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spinosa PC, Humphries BA, Lewin Mejia D, Buschhaus JM, Linderman JJ, Luker GD, Luker KE: Short-term cellular memory tunes the signaling responses of the chemokine receptor CXCR4. Sci Signal 2019, 12:eaaw4204. Using multiplexed reporters for ERK and Akt, the authors demonstrated that heterogeneous responses in both pathways to CXCR4 stimulus could be explained by pre-existing differences in cell state. Their ordinary differential equation model of pre-existing state enabled them to predict the effects of specific clinical inhibitors used to treat breast cancer and in particular explained why some targeted inhibitors can activate other signaling pathways.
- 17.Yuan TL, Wulf G, Burga L, Cantley LC: Cell-to-Cell Variability in PI3K Protein Level Regulates PI3K-AKT Pathway Activity in Cell Populations. Curr Biol 2011, 21:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB: Sequential Application of Anticancer Drugs Enhances Cell Death by Rewiring Apoptotic Signaling Networks. Cell 2012, 149:780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morton SW, Lee MJ, Deng ZJ, Dreaden EC, Siouve E, Shopsowitz KE, Shah NJ, Yaffe MB, Hammond PT: A nanoparticle-based combination chemotherapy delivery system for enhanced tumor killing by dynamic rewiring of signaling pathways. Sci Signal 2014, 7:ra44–ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JP, Rajwa B, Patsekin V, Davisson VJ: Computational analysis of high-throughput flow cytometry data. Expert Opin Drug Discov 2012, 7:679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Ning B, Shi T: Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front Genet 2019, 10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordonov S, Hwang MK, Wells A, Gertler FB, Lauffenburger DA, Bathe M: Time series modeling of live-cell shape dynamics for image-based phenotypic profiling. Integr Biol 2016, 8:73–90. [Google Scholar]
- 23.Zhou X, Herbst-Robinson KJ, Zhang J: Visualizing Dynamic Activities of Signaling Enzymes Using Genetically Encodable Fret-Based Biosensors. In Methods in Enzymology. . Elsevier; 2012:317–340. [Google Scholar]
- 24.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW: High-Sensitivity Measurements of Multiple Kinase Activities in Live Single Cells. Cell 2014, 157:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pargett M, Albeck JG: Live Cell Imaging and Analysis with Multiple Genetically Encoded Reporters. Curr Protoc Cell Biol 2018, 78. [Google Scholar]
- 26.Abramoff MD, Magalhães PJ, Ram SJ: Image processing with ImageJ. Biophotonics Int 2004, 11:36–42. [Google Scholar]
- 27.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. : CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 2018, 16:e2005970. [Google Scholar]
- 28.Tian C, Yang C, Spencer SL: EllipTrack: A Global-Local Cell-Tracking Pipeline for 2D Fluorescence Time-Lapse Microscopy. Cell Rep 2020, 32:107984. [Google Scholar]
- 29.Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW: TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115:80–90. [DOI] [PubMed] [Google Scholar]
- 30.Gerosa L, Chidley C, Fröhlich F, Sanchez G, Lim SK, Muhlich J, Chen J-Y, Vallabhaneni S, Baker GJ, Schapiro D, et al. : Receptor-Driven ERK Pulses Reconfigure MAPK Signaling and Enable Persistence of Drug-Adapted BRAF-Mutant Melanoma Cells. Cell Syst 2020, 11:478–494.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birtwistle MR, Rauch J, Kiyatkin A, Aksamitiene E, Dobrzyński M, Hoek JB, Kolch W, Ogunnaike BA, Kholodenko BN: Emergence of bimodal cell population responses from the interplay between analog single-cell signaling and protein expression noise. BMC Syst Biol 2012, 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goglia AG, Wilson MZ, Jena SG, Silbert J, Basta LP, Devenport D, Toettcher JE: A Live-Cell Screen for Altered Erk Dynamics Reveals Principles of Proliferative Control. Cell Syst 2020, 10:240–253.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aghabozorgi S, Shirkhorshidi AS, Wah TY: Time-series clustering –A decade review. Inf Syst 2015, 53:16–38. [Google Scholar]
- 34. Blum Y, Mikelson J, Dobrzyński M, Ryu H, Jacques M, Jeon NL, Khammash M, Pertz O: Temporal perturbation of ERK dynamics reveals network architecture of FGF2/MAPK signaling. Mol Syst Biol 2019, 15. The authors used a combination of microfluidics and live cell imaging to study how the dynamics and dosage of three growth factors affected ERK signaling dynamics. They used clustering to identify growth-factor specific signaling dynamics, and developed a mechanistic model to explain complex FGF-ERK signaling dynamics.
- 35.Dobrzyński M, Jacques M-A, Pertz O: Mining single-cell time-series datasets with Time Course Inspector. Bioinformatics 2020, 36:1968–1969. [Google Scholar]
- 36.Ramsay J, Silverman BW: Functional Data Analysis. Springer-Verlag; 2005. [Google Scholar]
- 37.Chen K, Zhang X, Petersen A, Müller H-G: Quantifying Infinite-Dimensional Data: Functional Data Analysis in Action. Stat Biosci 2017, 9:582–604. [Google Scholar]
- 38. Sampattavanich S, Steiert B, Kramer BA, Gyori BM, Albeck JG, Sorger PK: Encoding Growth Factor Identity in the Temporal Dynamics of FOXO3 under the Combinatorial Control of ERK and AKT Kinases. Cell Syst 2018, 6:664–678.e9. These authors were the first to apply functional principal component analysis to heterogeneous cell signaling data. They found that immediate responses to different growth factors dynamics could be identified through fPCA.
- 39.Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A: Information Transduction Capacity of Noisy Biochemical Signaling Networks. Science 2011, 334:354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L, Wollman R: Accurate information transmission through dynamic biochemical signaling networks. Science 2014, 346:1370–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keshelava A, Solis GP, Hersch M, Koval A, Kryuchkov M, Bergmann S, Katanaev VL: High capacity in G protein-coupled receptor signaling. Nat Commun 2018, 9:876. This work demonstrates some of the first information-theoretic analysis of cells stimulated repeatedly. Their novel approach enabled them to quantify information transfer in single cells, and revealed that some cells had much higher information capacity than previously suggested.
- 42.Gross SM, Dane MA, Bucher E, Heiser LM: Individual Cells Can Resolve Variations in Stimulus Intensity along the IGF-PI3 K-AKT Signaling Axis. Cell Syst 2019, 9:580588.e4. [Google Scholar]
- 43.Lopez CF, Muhlich JL, Bachman JA, Sorger PK: Programming biological models in Python using PySB. Mol Syst Biol 2013, 9:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garner KL, Perrett RM, Voliotis M, Bowsher C, Pope GR, Pham T, Caunt CJ, Tsaneva-Atanasova K, McArdle CA: Information Transfer in Gonadotropin-releasing Hormone (GnRH) Signaling. J Biol Chem 2016, 291:2246–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwamoto K, Shindo Y, Takahashi K: Modeling Cellular Noise Underlying Heterogeneous Cell Responses in the Epidermal Growth Factor Signaling Pathway. PLOS Comput Biol 2016, 12:e1005222. [Google Scholar]
- 46.Wang D-G, Wang S, Huang B, Liu F: Roles of cellular heterogeneity, intrinsic and extrinsic noise in variability of p53 oscillation. Sci Rep 2019, 9:5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomida T, Takekawa M, Saito H: Oscillation of p38 activity controls efficient pro-inflammatory gene expression. Nat Commun 2015, 6. [Google Scholar]
- 48.Yao J, Pilko A, Wollman R: Distinct cellular states determine calcium signaling response. Mol Syst Biol 2016, 12:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahama PA, Linderman JJ: Calcium signaling in individual BC3H1 cells: Speed of calcium mobilization and heterogeneity. Biotechnol Prog 1994, 10:45–54. [Google Scholar]
- 50.Hase T, Tanaka H, Suzuki Y, Nakagawa S, Kitano H: Structure of Protein Interaction Networks and Their Implications on Drug Design. PLoS Comput Biol 2009, 5:e1000550. [Google Scholar]
- 51.Jeschke M, Baumgärtner S, Legewie S: Determinants of Cell-to-Cell Variability in Protein Kinase Signaling. PLoS Comput Biol 2013, 9:e1003357. [Google Scholar]
- 52.Lin J, Qu L: FEATURE EXTRACTION BASED ON MORLET WAVELET AND ITS APPLICATION FOR MECHANICAL FAULT DIAGNOSIS. J Sound Vib 2000, 234:135–148. [Google Scholar]
- 53.Salamon J, Bello JP, Farnsworth A, Robbins M, Keen S, Klinck H, Kelling S: Towards the Automatic Classification of Avian Flight Calls for Bioacoustic Monitoring. PLOS ONE 2016, 11:e0166866. [Google Scholar]
- 54.Längkvist M, Karlsson L, Loutfi A: A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit Lett 2014, 42:11–24. [Google Scholar]
- 55.Albeck JG, Mills GB, Brugge JS: Frequency-Modulated Pulses of ERK Activity Transmit Quantitative Proliferation Signals. Mol Cell 2013, 49:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benary M, Bohn S, Lüthen M, Nolis IK, Blüthgen N, Loewer A: Disentangling Pro-mitotic Signaling during Cell Cycle Progression using Time-Resolved Single-Cell Imaging. Cell Rep 2020, 31:107514. [Google Scholar]
- 57.Hung YP, Teragawa C, Kosaisawe N, Gillies TE, Pargett M, Minguet M, Distor K, Rocha-Gregg BL, Coloff JL, Keibler MA, et al. : Akt regulation of glycolysis mediates bioenergetic stability in epithelial cells. eLife 2017, 6:e27293. [Google Scholar]
- 58.Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, Jaimovich A, Carr A, Meyer T, Pe’er D, Boutet SC, et al. : Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-κB Activation. Cell Syst 2017, 4:458–469.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jetka T, Nienałtowski K, Winarski T, Błoński S, Komorowski M: Information-theoretic analysis of multivariate single-cell signaling responses. PLoS Comput Biol 2019, 15:e1007132. [Google Scholar]
- 60.Suderman R, Bachman JA, Smith A, Sorger PK, Deeds EJ: Fundamental trade-offs between information flow in single cells and cellular populations. Proc Natl Acad Sci 2017, 114:5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salis H, Sotiropoulos V, Kaznessis Y: Multiscale Hy3S: Hybrid stochastic simulation for supercomputers. BMC Bioinformatics 2006, 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]