Abstract
Verticillium wilt in cotton (Gossypium hirsutum) is primarily caused by Verticillium dahliae. Previous data suggest that prenylated RAB acceptors (PRAs) play essential roles in environmental plant adaptation, although the potential roles of PRA1 in cotton are unclear. Therefore, in this study, PRA1 family members were identified in G. hirsutum, and their roles in biotic and abiotic stresses were analyzed. Thirty-seven GhPRA1 family members were identified in upland cotton, which were divided into eight groups. Gene structure and domain analyses revealed that the sequences of GhPRA1 members in each group were highly conserved. Many environmental stress-related and hormone-response cis-acting elements were identified in the GhPRA1 promoter regions, indicating that they may respond to biotic and abiotic stresses. Expression analysis revealed that GhPRA1 members were widely expressed in upland cotton. The GhPRA1 genes responded to abiotic stress: drought, cold, salt, and heat stress. GhPRA1.B1-1A expression increased after V. dahliae infection. Furthermore, the functional role of GhPRA1.B1-1A was confirmed by overexpression in Arabidopsis thaliana, which enhanced the resistance to V. dahliae. In contrast, V. dahliae resistance was significantly weakened via virus-induced gene silencing of GhPRA1.B1-1A in upland cotton. Simultaneously, reactive oxygen species accumulation; the H2O2, salicylic acid, and jasmonic acid contents; and callose deposition were significantly decreased in cotton plants with GhPRA1.B1-1A silencing. These findings contribute to a better understanding of the biological roles of GhPRA1 proteins and provide candidate genes for cotton breeders for breeding V. dahliae-resistant cultivars.
Keywords: GhPRA1.B1-1A, PRA1, upland cotton, Verticillium dahliae, Verticillium wilt
1. Introduction
Cotton (Gossypium spp.) is the major crop for natural fiber quality and production and is an important source of edible oil and feed for livestock [1]. Upland cotton (G. hirsutum) is the most widely grown cotton species, accounting for approximately 90% of cotton production [2].
Verticillium wilt (V. wilt) is one of the most serious diseases affecting fiber production. V. wilt in cotton is primarily caused by Verticillium dahliae, and it can survive in the soil for long periods of time in the form of microsclerotia [3]. The leaves of cotton plants with V. wilt show symptoms of wilting and yellowing, browning occurs in the vascular bundles, and, in serious cases, the whole plant will wither and die. Blockage theory and toxin theory have been used to explain the pathogenesis of V. wilt [4]. According to blockage theory, when V. dahliae infects cotton roots, the hyphae pass through the roots of plants and colonize the vascular bundles [5]. The accumulation of mycelia blocks the vessels of xylem tissues, which affects the transportation of water and nutrients in plants. The toxin hypothesis holds that leaf necrosis occurs owing to toxins produced during fungal infection, which ultimately leads to the death of plants [6]. At present, barely effective strategies have been developed to protect cotton plants from V. dahliae infection. Thus, understanding the genetic basis of susceptibility to V. dahliae infection is crucial for cotton breeding [7]. In recent years, numbers of V. dahliae resistance genes have been detected in cotton. For example, laccase (GhLAC15) enhances V. dahliae resistance by increasing induced lignification and the abundance of lignin components in plant cell walls [8]. Hybrid proline-rich proteins (GbHyPRP1) negatively regulate V. wilt resistance through cell wall thickening and accumulation of reactive oxygen species (ROS) in cotton [9]. Wall-associated kinases (GhWAK7A) mediate the response to fungal pathogens by binding to chitin receptors [10]. The WAK-like kinase (GhWAKL) protein and DnaJ protein interact and participate in cotton plants against V. wilt infection [11]. Polygalacturonase inhibition protein (GhPGIP1) significantly improves cotton resistance to V. wilt [12]. U-boxE3 ubiquitin ligase (GhPUB17) interacts with the antifungal protein GhCyP3 to negatively regulate cotton’s resistance to V. wilt [13].
Rab GTPases are small GTPases [14] that play important roles in membrane recognition, vesicle movement and fusion, and intracellular vesicle transport [15]. In addition, Rab genes also respond to biotic and abiotic stress responses [16]. For example, the overexpression of the StRab gene can enhance resistance to potato late blight [17]. TaRab18 positively regulates resistance to wheat stripe rust by regulating allergic responses [18]. RabG3b participates in the death of immune-related allergic cells by activating autophagy [19]. Rab5 GTPase can activate and mediate plants’ immune responses to pathogen invasion [20]. In addition, an important feature of innate immunity is phagocytosis, and Rab GTPases can coordinate and transport phagocytic substances, which enhances immunity [21]. These findings show that Rab genes are involved in plant immunity and hypersensitive responses.
Prenylated Rab acceptor 1 (PRA1) is a group of small transmembrane proteins. As a receptor of Rab GTPases, PRA1 primarily regulates vesicular transport [22]. The first member of PRA1 was detected in yeast as a Ypt1-interacting protein [23]. Subsequently, many proteins similar to yeast Yip/PRA1 were found in mammals and higher plants [24]. The PRA1 protein can affect the membrane localization of Rab protein by interacting with the GDP dissociation inhibitor (GDI) [25], thereby promoting the transport of small GTPases through the intimal system [26]. AtPRA1.B6 is located in the Golgi apparatus and plays a role in transporting proteins [27]. OsPRA1 is located in the prevacuolar compartment and interacts with OsRab7 to participate in the transport of plant vacuoles [28]. In addition to their functions in vesicle transport, PRA1 proteins have recently been reported to be involved in plant immunity. For example, overexpression of the SlPRA1A gene reduces the expression of receptor-like protein (RLP-PRR) and regulates its transport and degradation together with AtRABA1e, thereby weakening the immune response [29]. As PRA1 is a receptor of Rab GTPases, PRA1–Rab interactions participate in the transport of plant-related proteins and attenuate the immune response by degrading LeEIX2 pattern-recognition receptors (PRRs) in SlPRA1A/Rab, indicating that they may have closely related functions [29]. To date, no reports have described the role of the PRA1 protein in cotton plants, particularly in defense responses to pathogens.
To explore the potential role of PRA1, we comprehensively analyzed GhPRA1 members in upland cotton. Additionally, GhPRA1.B1-1A was functionally analyzed by overexpression in Arabidopsis thaliana and virus-induced gene silencing (VIGS) in upland cotton. The results describe the characteristics of GhPRA1 members in upland cotton and highlight the potential role of GhPRA1.B1-1A in V. dahliae resistance.
2. Materials and Methods
2.1. Phylogenetic Analysis
Hidden Markov model (HMM) data for PRA1 (PF03208) were downloaded from the Pfam website (http://pfam.xfam.org/, accessed on 20 September 2021), and shared homologies with upland cotton proteins were determined using the threshold value of 1E-5. The PRA1 genes of G. arboreum [30], G. raimondii [31], G. barbadense [32], and G. hirsutum [33] (containing PRA1 domains) were detected by BLAST and regarded as PRA1 family members. The nomenclature of PRA1 genes was according to the corresponding orthologs in Arabidopsis thaliana and the chromosomal location in cotton. Meanwhile, members of PRA1 in tetraploid cotton species were named according to the homologous relationship in each subgenome, for which ‘A’ and ‘D’ at the end of each gene represent the homologs in the At and Dt subgenomes, respectively.
The protein sequences of PRA1 family members in cotton and A. thaliana were downloaded from the Cotton Gen [34] and TAR10 (https://www.arabidopsis.org/, accessed on 22 September 2021) databases, respectively. The amino acid sequences of the PRA1 proteins were aligned using the MUSCLE program. Next, a phylogenetic tree was constructed by using MEGA-X software via the neighbor-joining method.
2.2. Analysis of Gene Structures, Domains, and Motifs
The Gene Structure Display Server (GSDS) was used to display the exon–intron structure of the PRA1 genes according to their genome datasets [35]. The conserved domain of PRA1 protein was identified using the Simple Modular Architecture Research Tool [36] and was displayed using TBtools software [37]. The conserved motif of the PRA1 protein was identified using the MEME program [38].
2.3. Analysis of Cis-Acting Elements in the GhPRA1 Promoter Region
The sequences 2 kb upstream of the coding region of the GhPRA1 gene were extracted using TBtools software. The sequences were uploaded to the PlantCare website to predict cis-acting elements, which were graphically displayed using TBtools software.
2.4. Collinearity Analysis of the GhPRA1 Gene
The genomic location information for GhPRA1 was extracted from a downloaded genome annotation file, collinear gene analysis was conducted using MCScanX algorithm [39] software, and graphic visualization was conducted using Circos software [40].
2.5. Plant Materials and Growth Conditions
Upland cotton (G. hirsutum) cv. Zhongzhimian-2 seeds were soaked in distilled water for 12 h in a 28 °C incubator and then planted in soil. Cotton seedlings were grown in a greenhouse under 60% relative humidity with a dark/light cycle of 8 h in the dark (temperature: 23 °C and 16 h in the light (temperature: 28 °C).
The Colombian ecotype of A. thaliana was used in our experiment. Arabidopsis seeds were sterilized in 75% ethanol for 2 min, sterilized further in absolute ethanol for 2 min, and then sown on 1/2 MS medium for 10 days. The Arabidopsis seedlings were then transplanted to nutrient-supplemented soil and cultured under a 16/8 h light/dark cycle at 22/20 °C (light/dark) at 60% relative humidity.
2.6. Gene Cloning
Total RNA was extracted from the root tissues of Zhongzhimian 2 plants using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China). After quality testing, the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) was used for first-strand complementary DNA (cDNA) synthesis, according to the manufacturer’s instructions. The coding sequence (CDS) of GhPRA1.B1-1A was downloaded from the Cotton Gen Database, and specific primers were designed using SnapGene. The sequences of the specific primers are listed in Table S1. The full-length CDS of GhPRA1.B1-1A was cloned using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) and Zhongzhimian 2 cDNA as a template.
2.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
RT-qPCR analysis was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), according to the manufacturer’s recommended protocol, on a Light Cycler 480 system machine (Roche, Basel, Switzerland). The reactions were developed in a volume of 20 μL containing 1 μL cDNA, 1 μL forward primer (10 µM), 1 μL reverse primer (10 µM), 10 µL 2× ChamQ Universal SYBR Green Master Mix, and 7 μL nuclease-free water. The reaction conditions were as follows: 40 cycles of 95 °C for 30 s, 95 °C for 10 s, and 60 °C for 15 s, followed by 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. In the experiments, GhUBQ7 was used as an internal reference gene, and three biological replicates were performed. Relative expression levels were determined using the 2−∆∆T method [41].
2.8. Generating Transgenic Arabidopsis Plants
The full-length CDS of GhPRA1.B1-1A was amplified and inserted into the pCAMBIA2300 vector behind a CaMV 35S promoter to yield a recombinant vector, referred to as the p2300-GhPRA1.B1-1A vector. After transformation into the GV3101 Agrobacterium, genetic Arabidopsis transformants were obtained using the floral dip method [42]. Positive Arabidopsis seedlings were screened on 1/2 MS medium containing kanamycin and verified by PCR analysis. The T2 lines (in accordance with the 3:1 segregation ratio) were thought to contain single-copy insertions and were selected to observe GhPRA1.B1-1A expression levels by RT-qPCR. Transgenic Arabidopsis seedlings in the T3 generation were used for V. dahliae infection.
2.9. VIGS and V. dahliae Inoculation
Tobacco rattle virus was used in this study [43]. A specific 300 bp fragment of GhPRA1.B1-1A was inserted into the pTRV2 vector to generate the recombinant pTRV-GhPRA1.B1-1A vector, which was transformed into the A. tumefaciens strain GV3101. When both cotyledons of the cotton seedlings were fully expanded, Agrobacterium cultures containing pTRV1 and pTRV-GhPRA1.B1-1A at a 1:1 ratio (v/v) were transformed into cotton leaves using an injector. The chlorophyll gene CLA1 was used as a positive control, and cotton seedlings were injected with a mixture of Agrobacterium containing the empty pTRV1 and pTRV2 vectors as a negative control. The VIGS assay was performed three times, and at least 48 plants were used for each experiment.
Vd991, a highly aggressive defoliating V. dahliae strain, was inoculated on potato dextrose agar medium and incubated at 25 °C for 7 days. The hyphae were then inoculated in potato dextrose broth medium and cultured in a shaker at 25 °C and 180 rotations/min for 5 days. The cotton seedlings were immerged in a Vd991 spore suspension (1 × 107 spores mL−1) for 15 min. The method of A. thaliana infection by V. dahliae was according to the method of Zhao et al. [44]. The disease index and incidence rate were calculated 2 weeks after inoculation, based on the disease index (DI) classification [45].
2.10. Plant Disease Evaluation
According to the severity of the diseased phenotype in true cotton leaves, the DI was graded from 0 to 4, using the following formula: DI = (∑[incidence grade × number of plants at this level]/[total number of plants investigated × 4]) × 100 [46]. The lower the DI value, the lower the degree of disease in the plant. In addition, fungal restoration experiments and fungal biomass testing were performed to determine the degree of plant disease [47].
2.11. ROS and Callose Deposition Detection
After inoculation for 24 h, the true leaves were washed with distilled water, placed in a 50 mL centrifuge tube, and stained with a 3,3′-diaminobenzidine staining solution. After incubation at 25 °C for 8 h and destaining with 95% ethanol, the leaves were observed under a microscope and photographed. At least five seedlings were detected in each treatment.
For callose deposition, cotton leaves were sampled 2 weeks after inoculation. The leaves were decolorized in a 3:1 mixture of ethanol and acetic acid for 3 h, soaked sequentially in 70% and 50% ethanol for 2 h, and then soaked in water overnight. After treatment with 10% NaOH for 2 h, callose was stained with 0.01% aniline blue dye for 4 h. Next, the callose content was observed by ultraviolet excitation light under a fluorescence microscope.
2.12. Measuring Jasmonic Acid, Salicylic Acid, and H2O2 Content
The leaves of TRV:00 and TRV:GhPRA1.B1-1A plants were obtained at 48 h after inoculation, and the contents of JA and SA were measured by high-performance liquid chromatography. Three biological replicates were used for each experiment.
H2O2 was measured in the leaves of TRV:00 and TRV:GhPRA1.B1-1A plants inoculated with V. dahliae for 48 h using the H2O2 Kit (Solarbio; Beijing, China), according to the manufacturer’s instructions.
2.13. Statistical Analysis
GraphPad Prism8 software was used for constructing graphs, and the t-test was used to analyze statistical differences. “*” and “**” indicate significances at p-values of <0.05 and <0.01, respectively.
3. Results
3.1. Identification and Phylogenetic Analysis of PRA1 Family Members
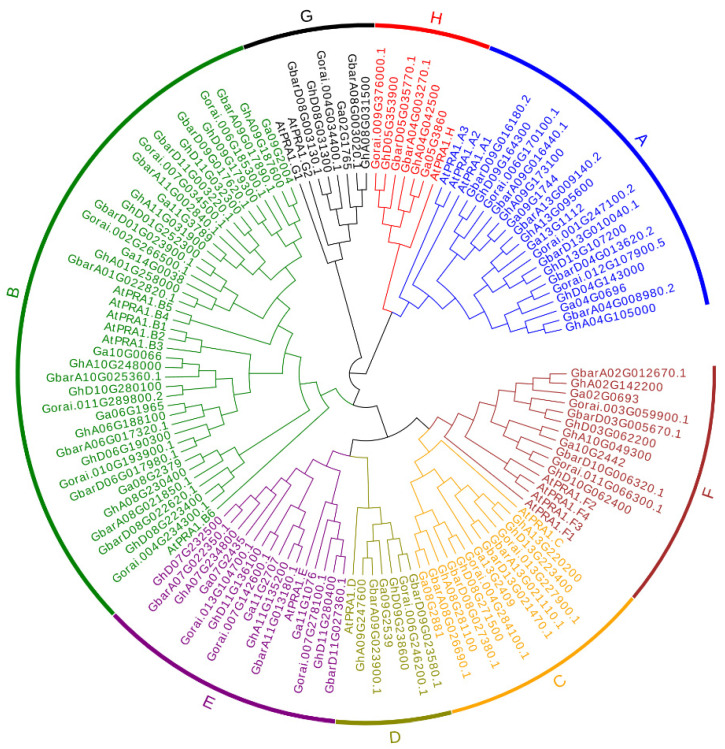
To understand the evolutionary relationship of the PRA1 gene family in G. hirsutum, G. raimondii, G. arboreum, and G. barbadense, the conserved domains of PRA1 in the four cotton species were detected by Pfam (PF03208). An evolutionary tree was constructed according to the amino acid sequence of PRA1 protein. (Figure 1). The PRA1 family can be divided into eight groups. Among these, the B subfamily of PRA1 contains the most members (41), has the farthest evolutionary relationship with the F subfamily, and has the closest evolutionary relationship with the E and G subfamilies. We observed that the number of PRA1 genes in tetraploid cotton species (G. hirsutum and G. barbadense) was approximately twice as much as that in diploid cotton (G. arboreum and G. raimondii). Meanwhile, PRA1 genes in the At subgenomes and A2 genome (G. arboreum) tended to form one clade, and PRA1 genes in the Dt subgenomes and D5 genome (G. raimondii) tended to form another clade. These results confirm that the tetraploid cotton species were a result of the hybridization of diploid cottons, followed by a genome doubling event during cotton’s evolution. Additionally, the phylogenetic tree showed that PRA1 members in each group were conserved in the four cotton species and A. thaliana, indicating that PRA1 family members exhibit conserved sequences in dicotyledons.
Figure 1.
Phylogenetic analysis of the PRA1 gene family. At, A. thaliana; Ga, G. arboreum; Gr, G. raimondii; Gb, G. barbadense; Gh, G. hirsutum.
Thirty-seven PRA1 members were identified in upland cotton, and detailed information is presented in Table 1. The CDSs of the 37 PRA1 genes were between 291 bp (GhPRA1.E-1D) and 801 bp (GhPRA1.G2-1A) in length. The molecular weight (MW) of PRA1 proteins ranged from 11.18 kDa (GhPRA1.E-1D) to 28.99 kDa (GhPRA1.G2-1A), and the isoelectric point ranged from 4.59 (GhPRA1.D-2D) to 10.84 (GhPRA1.E-1D). Subcellular localization analysis revealed that 14 of the PRA1 proteins were predicted to be located in the vacuoles, 12 PRA1 proteins were predicted to be located in the plasma membrane, nine PRA1 proteins were predicted to be located in the chloroplasts, and two PRA1 proteins were predicted to be located in the endoplasmic reticulum. The full-length 657 bp sequence of the GhPRA1.B1-1A gene encodes 218 amino acids, with an MW of 23.313 kDa and a theoretical isoelectric point of 9.566.
Table 1.
Identification of the GhPRA1 genes. Abbreviations: vacu, vacuole; plas, plasmalemma; golg, Golgi; chlo, chloroplast; ER, endoplasmic; extr, extracellular. The top three most possible subcellular localizations are shown, and the location numbers indicate the possibility of subcellular localizations.
| Gene Name | Gene Locus ID | Chr Location | CDS | Size | MW (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| GhPRA1.A1-1A | Gh_A04G105000.1 | A04: 70948155–70951205 | 630 | 209 | 24.19 | 10.58 | vacu: 5, plas: 4, extr: 2 |
| GhPRA1.A1-1D | Gh_D04G143000.1 | D04: 44330508–44333557 | 630 | 209 | 24.20 | 10.44 | vacu: 7, plas: 4, golg: 2, |
| GhPRA1.A1-2A | Gh_A09G173100.1 | A09: 74156483–74159609 | 630 | 209 | 24.17 | 10.58 | vacu: 12, plas: 1, golg: 1 |
| GhPRA1.A1-2D | Gh_D09G164300.1 | D09: 43855015–43858151 | 630 | 209 | 24.23 | 10.48 | vacu: 11, plas: 2, golg: 1 |
| GhPRA1.A1-3A | Gh_A13G095600.1 | A13: 40978102–40979417 | 630 | 209 | 23.98 | 10.41 | vacu: 6, plas: 4, golg: 2 |
| GhPRA1.A1-3D | Gh_D13G107200.1 | D13: 24914233–24915541 | 630 | 209 | 24.11 | 10.45 | plas: 5, vacu: 5, golg: 2 |
| GhPRA1.B1-1A | Gh_A06G188100.1 | A06: 117938223–117938879 | 657 | 218 | 23.31 | 9.44 | chlo: 5, ER: 4, plas: 3 |
| GhPRA1.B1-1D | Gh_D06G190300.1 | D06: 58492262–58492918 | 657 | 218 | 23.40 | 9.64 | chlo: 7, ER: 3, plas: 2 |
| GhPRA1.B1-2A | Gh_A10G248000.1 | A10: 114198294–114199248 | 648 | 215 | 23.49 | 9.64 | chlo: 6, ER: 3, plas: 2 |
| GhPRA1.B1-2D | Gh_D10G280100.1 | D10: 65324234–65325180 | 648 | 215 | 23.54 | 9.83 | chlo: 6, ER: 3, plas: 2 |
| GhPRA1.B4-1A | Gh_A01G258000.1 | A01: 115880792–115881439 | 648 | 215 | 23.32 | 7.82 | E.R.: 5, plas: 3, chlo: 2 |
| GhPRA1.B4-1D | Gh_D01G252300.1 | D01: 64514473–64515120 | 648 | 215 | 23.33 | 7.82 | E.R.: 4, plas: 3, chlo: 2, |
| GhPRA1.B4-2A | Gh_A08G230400.1 | A08: 119818691–119819320 | 630 | 209 | 22.74 | 8.68 | vacu: 7, ER: 3, plas: 2 |
| GhPRA1.B4-2D | Gh_D08G223400.1 | D08: 63336820–63337449 | 630 | 209 | 22.81 | 8.68 | vacu: 7, ER: 3, plas: 2 |
| GhPRA1.B4-3A | Gh_A09G187600.1 | A09: 75447349–75447999 | 651 | 216 | 23.70 | 9.03 | chlo: 5, plas: 5, golg: 2 |
| GhPRA1.B4-3D | Gh_D09G179300.1 | D09: 45181994–45182644 | 651 | 216 | 23.64 | 8.97 | chlo: 5, plas: 4, vacu: 2 |
| GhPRA1.B4-4A | Gh_A11G031900.1 | A11: 2693360–2694010 | 651 | 216 | 23.69 | 6.73 | vacu: 10, plas: 3, extr: 1 |
| GhPRA1.B4-4D | Gh_D11G032300.1 | D11: 2630603–2631253 | 651 | 216 | 23.69 | 6.10 | vacu: 12, plas: 1, extr: 1 |
| GhPRA1.C-1A | Gh_A08G281100.1 | A08: 124823067–124823489 | 423 | 140 | 16.10 | 6.71 | plas: 4.5, vacu: 3 |
| GhPRA1.C-1D | Gh_D08G271500.1 | D08: 67997394–67997933 | 540 | 179 | 20.29 | 6.06 | vacu: 9, golg: 3, plas: 1 |
| GhPRA1.C-2A | Gh_A13G220200.1 | A13: 104371813–104372518 | 606 | 201 | 22.70 | 5.11 | vacu: 11, plas: 1, extr: 1 |
| GhPRA1.C-2D | Gh_D13G223400.1 | D13: 59483095–59483754 | 591 | 196 | 22.08 | 8.05 | vacu: 7, plas: 2.5, golg: 2 |
| GhPRA1.D-2A | Gh_A09G247600.1 | A09: 81208181–81208732 | 552 | 183 | 20.15 | 4.71 | plas: 8, ER: 2, chlo: 1 |
| GhPRA1.D-2D | Gh_D09G238600.1 | D09: 50472807–50473358 | 552 | 183 | 20.20 | 4.59 | plas: 8, ER: 3, chlo: 1 |
| GhPRA1.E-1A | Gh_A11G135200.1 | A11: 14400779–14401303 | 525 | 174 | 19.28 | 7.95 | plas: 8, chlo: 3, ER: 3 |
| GhPRA1.E-1D | Gh_D11G136100.1 | D11: 12800664–12800954 | 291 | 96 | 11.19 | 10.84 | chlo: 7, plas: 6, ER: 1 |
| GhPRA1.E-2A | Gh_A07G234800.1 | A07: 93082823–93083368 | 546 | 181 | 20.04 | 9.68 | chlo: 7, plas: 6, ER: 1 |
| GhPRA1.E-2D | Gh_D07G232500.1 | D07: 55224484–55225535 | 546 | 181 | 20.17 | 9.68 | chlo: 7, plas: 6, ER: 1 |
| GhPRA1.F2-1A | Gh_A02G142200.1 | A02: 80712703–80713260 | 558 | 185 | 20.72 | 9.09 | plas: 10, golg: 2, vacu: 1 |
| GhPRA1.F2-1D | Gh_D03G062200.1 | D03: 10655558–10656115 | 558 | 185 | 20.76 | 8.07 | plas: 4, vacu: 3, ER: 3 |
| GhPRA1.F2-2A | Gh_A10G049300.1 | A10: 6090256–6090804 | 549 | 182 | 20.53 | 9.09 | plas: 5, golg: 3, chlo: 2 |
| GhPRA1.F2-2D | Gh_D10G062400.1 | D10: 6067187–6067735 | 549 | 182 | 20.47 | 7.93 | plas: 7, golg: 3, vacu: 2 |
| GhPRA1.F2-3D | Gh_D11G280400.1 | D11: 57861036–57861632 | 597 | 198 | 21.96 | 8.82 | plas: 7, ER: 3, chlo: 1 |
| GhPRA1.G2-1A | Gh_A08G031500.1 | A08: 3124048–3125076 | 801 | 266 | 28.99 | 8.76 | plas: 7, nucl: 2, vacu: 2 |
| GhPRA1.G2-1D | Gh_D08G031300.1 | D08: 2941239–2941814 | 576 | 191 | 20.78 | 7.09 | plas: 4.5, vacu: 3, ER: 3 |
| GhPRA1.H-1A | Gh_A04G042500.1 | A04: 7308990–7310582 | 717 | 238 | 26.79 | 9.14 | vacu: 8, plas: 2, extr: 2 |
| GhPRA1.H-1D | Gh_D05G353900.1 | D05: 56125424–56126730 | 717 | 238 | 26.84 | 9.30 | vacu: 9, plas: 2, extr: 1 |
3.2. Analysis of the Gene Organization and Conserved Domains of GhPRA1s
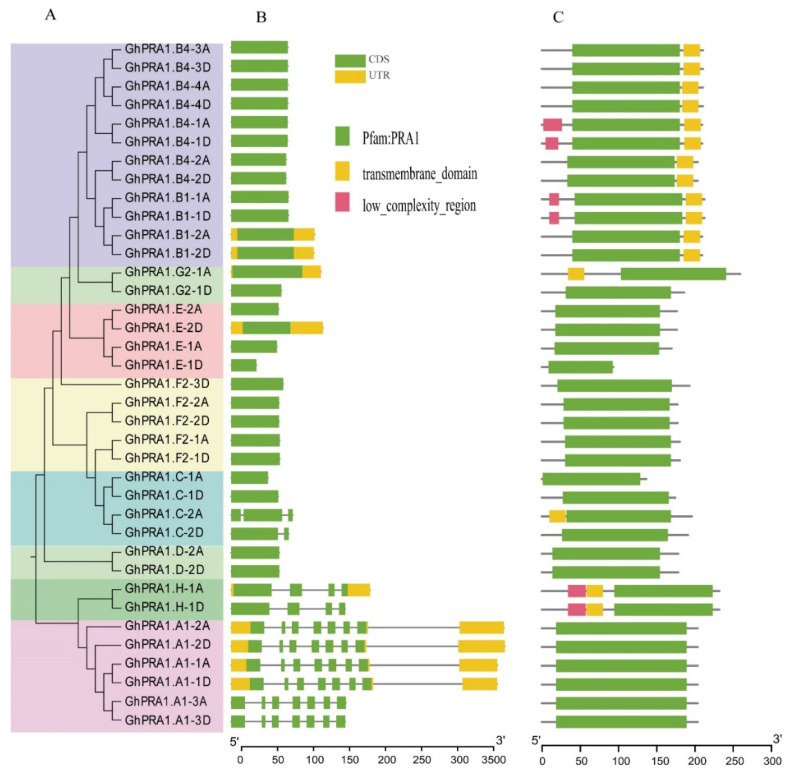
To understand the phylogenetic relationship of the PRA1 members in upland cotton, a phylogenetic tree was constructed using the 37 GhPRA1 amino acid sequences (Figure 2A). The results showed that the PRA1 proteins also can be divided into eight groups. PRA1 proteins in the same group were clustered together and shared high sequence identity. The exon–intron structure of PRA1 genes was analyzed using the online tool GSDS (http://gsds.gao-lab.org/, accessed on 27 September 2021). We observed that members of Subfamily A had the longest gene length and contained seven exons (Figure 2B). However, most GhPRA1 members in the other subfamilies were found to have only one exon, with a highly similar gene structure in the same subfamily.
Figure 2.
Gene structures and conserved domains in GhPRA1 family members. (A) Phylogenetic tree of GhPRA1 proteins. (B) The organization of exons and introns in GhPRA1 genes. (C) Conserved domains in GhPRA1 proteins.
In addition, the conserved domain of GhPRA1 was identified. All GhPRA1 members contained a PRA1 domain, and we also observed that GhPRA1.B1 group members, GhPRA1.B group members, GhPRA1.G2-1A, GhPRA1.C-2A, and GhPRA1.H-1A/D contain a transmembrane domain (Figure 2C). Furthermore, the PRA1 protein motif was analyzed using MEME software, and the top 10 most conserved motifs were presented. The results showed that all GhPRA1 proteins contain Motif 1. We also found that most homologous proteins have similar motifs and show highly conserved structures (Figure S1). Both GhPRA1.B1 and GhPRA1.B4 have Motifs 2 and 9. Except for GhPRA1.H and GhPRA1.A1, all the other GhPRA1 proteins were found to have Motifs 3 and 4. Only GhPRA1.A1 contains Motifs 5, 6, and 7. All GhPRA1 proteins contain Motif 8, except for GhPRA1.B1, GhPRA1.B4 and GhPRA1.A1. The motif organization of the GhPRA1 members was based on a phylogenetic tree, which indicated evolutionary relationships. The function of motifs was detected by the Pfam website, showing that Motif 1, Motif 3, Motif 4, Motif 7, and Motif 8 belong to the PRA1 domain, and Motif 6 was part of the Popeye protein’s conserved region (Table S2).
3.3. Analysis of Cis-Acting Elements of PRA1 Promoter
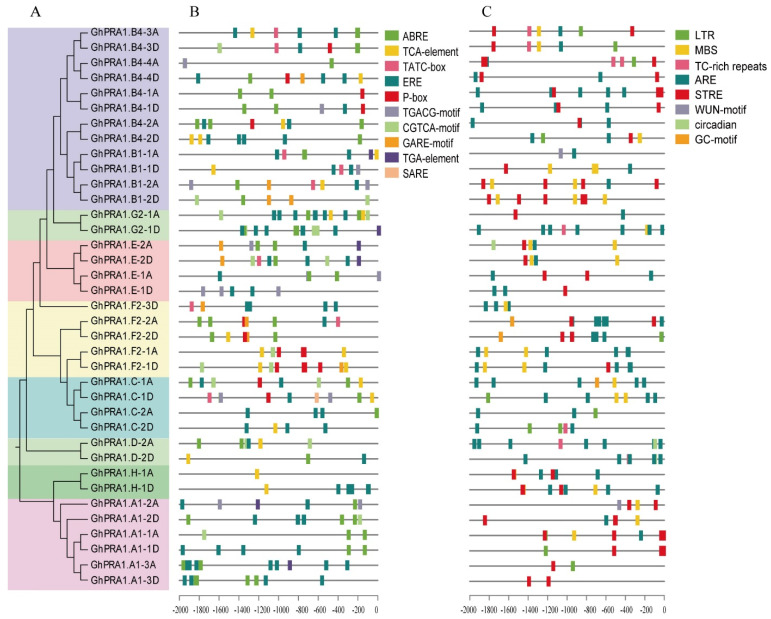
The phylogenetic tree showed that the 37 GhPRA1 members were divided to eight groups (Figure 3A). The 2 kb promoter regions upstream of the initiation codons of GhPRA1 genes were used to scan for cis-acting elements. Cis-acting elements (Table S3) in the GhPRA1 promoter regions were predicted to facilitate different functions, including anaerobic induction, meristem expression, hormone production, and stress defense. In particular, numbers of cis-elements related to hormones were predicted in the GhPRA1 promoter regions, such as ethylene (ET), abscisic acid (ABA), gibberellin (GA), salicylic acid (SA), methyl jasmonate (MeJA), and auxin (IAA), among others (Figure 3B). For instance, the cis-acting elements ERE, ABRE, TCA element, and CGTCA motif were predicted in the promoter region of GhPRA1.B1-1A, indicating that GhPRA1.B1-1A may be involved in ET, JA, and SA signal transduction pathways. In addition, cis-elements participated in environmental adaptation, such as ARE, STRE, MBS, LTR, the WUN-motif, the GC motif, and TC-rich repeats, and were also predicted in the GhPRA1 promoter regions (Figure 3C). The result suggest that the GhPRA1 family may play a role in drought, low temperature, defense, and stress responses.
Figure 3.
cis-elements in GhPRA1 promoter regions. (A) Phylogenetic tree of GhPRA1 proteins. (B) Hormone response elements. ABRE, abscisic acid response element; P-box; TATC box; GARE motif, gibberellin response element; TCA element; SARE, salicylic acid responsiveness element; CGTCA motif; TGACG motif. MeJA response cis-regulating elements: ERE, ethylene response element; TGA element, auxin response element. (C) Environmental stress related elements: STRE, stress response element; ARE, anaerobic induction element; MBS, MYB drought induction binding site; LTR, low temperature response element; GC motif, anoxic specific inducibility enhancer-like element; circadian, cis-acting regulatory element involved in circadian rhythm regulation; WUN motif, wound-responsive element; TC-rich repeats, defense and stress response cis-elements.
3.4. Chromosomal Location and Gene Synteny Analyses of GhPRA1 Genes
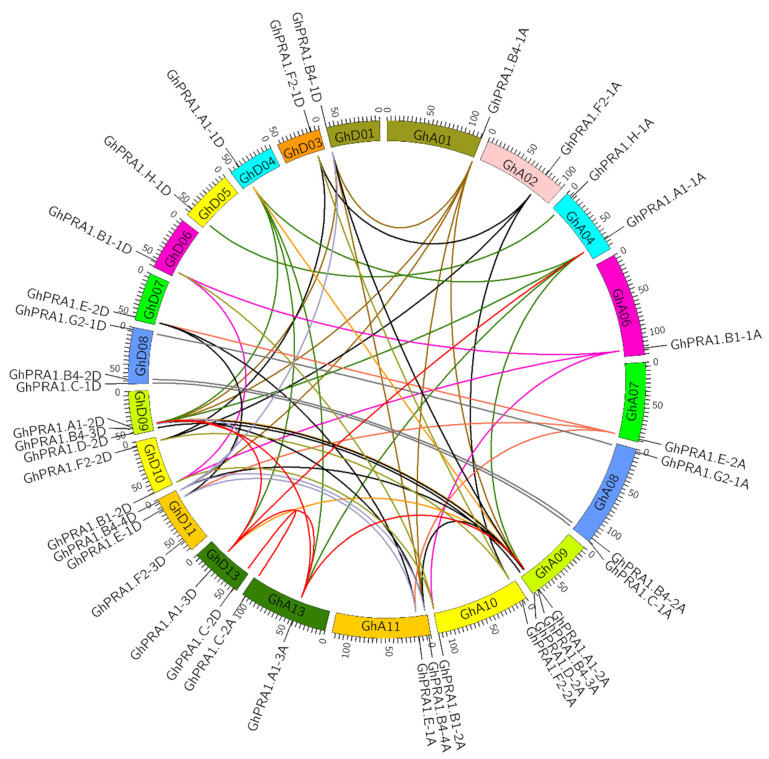
We conducted chromosomal location and gene synteny analyses of the GhPRA1 genes. The results showed that most of the GhPRA1 members were highly symmetrical between the At and Dt subgenomes. GhPRA1 genes were unevenly distributed on all chromosomes, except for chromosomes A03, A12, D02, and D12. However, GhPRA1.F2-1A and GhPRA1.H-1A were located on chromosomes A02 and A04, respectively, but the homologous genes GhPRA1.F2-1D and GhPRA1.H-1D were located on chromosomes D03 and D05, respectively (Figure 4).
Figure 4.
Syntenic relationships of GhPRA1 genes in upland cotton. Homologous chromosomes in the At and Dt subgenomes are displayed in the same color. The brown lines represent repetitive genes of GhPRA1.B4-1A/D; dark blue lines represent repetitive genes of GhPRA1.F2-1A/D; the green lines indicate repetitive genes of GhPRA1.A1-1A/D and homologous genes of GhPRA1.H-1A; the purple line indicates a repetitive gene of GhPRA1.A1-1A/D; the pink line indicates a repetitive gene of GhPRA1.E-2A; the gray line indicates a homologous gene of GhPRA1.G2-1A Magi GhPRA1.C-1A; the orange line represents a repetitive gene of GhPRA1.A1-2A/D; the black line represents a repetitive gene of GhPRA1.B4-3A/D; and the light brown line represents a repetitive gene of GhPRA1.C-1A. With respect to the repetitive genes in GhPRA1.B1-2A and GhPRA1.F2-2A, the light purple line indicates a repetitive gene of GhPRA1.B4-4A 3A/D, and the red line indicates a repetitive gene of GhPRA1.A1-3A/D and the homologous gene in GhPRA1.E-3A.
Duplicated GhPRA1 genes were analyzed in this study. We observed that GhPRA1.B4-1A/1D, GhPRA1.B4-3A/3D, and GhPRA1.B4-4A/4D; GhPRA1.F2-1A/1D and GhPRA1.F2-2A/2D; GhPRA1.A1-1A/1D, GhPRA1.A1-3A/3D, and GhPRA1.A1-2A/2D; GhPRA1.B1-1A/1D and GhPRA1.B1-2A/2D; and GhPRA1.E-2A/2D and GhPRA1.E-1A/1D may be duplicated genes. Based on the locations of the duplicated genes, these genes might have resulted from segmental duplication. The large number of duplicated genes suggested that the GhPRA1 genes duplicated frequently during upland cotton evolution.
3.5. Expression Profiles of GhPRA1 in Upland Cotton
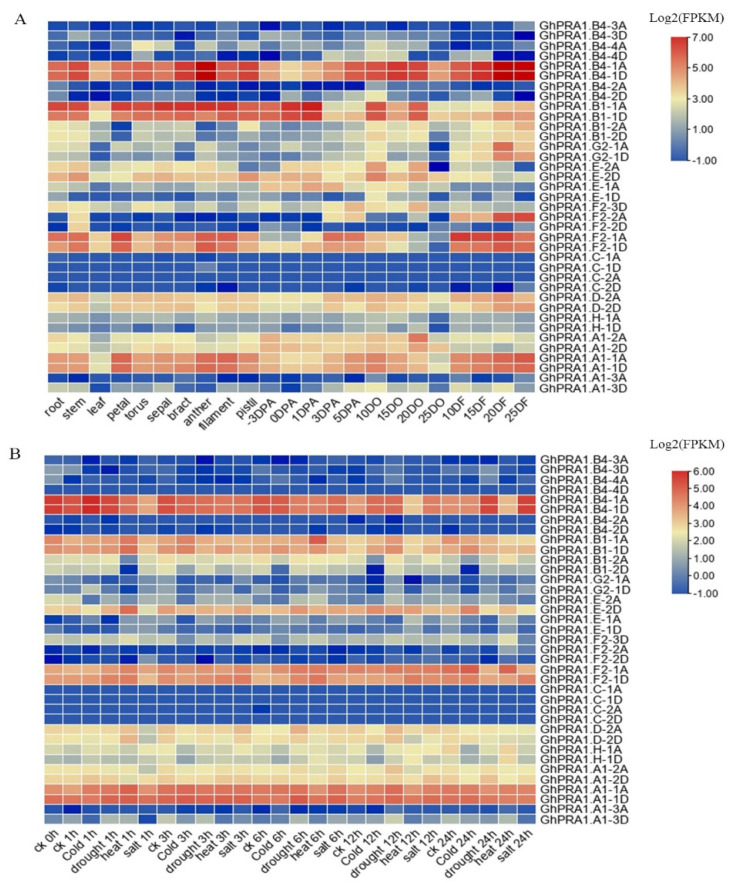
To have insights into the functions of GhPRA1 genes, transcriptome datasets [48] were used to analyze GhPRA1 expression patterns in different tissues during the development of upland cotton. We observed that GhPRA1.B4-3A/D, GhPRA1.B4-2A, GhPRA1.C-1A/D, GhPRA1.C-2A/D, and GhPRA1.A1-3A were barely expressed in upland cotton tissues. However, GhPRA1.B1-1A/D was highly expressed in roots, stems, petals, the torus, sepals, bracts, anthers, filaments, and early developmental ovules, indicating that GhPRA1.B1-1A/D might be involved in the growth and reproductive development of cotton. The finding that GhPRA1.B4-1A was abundantly expressed in anthers and fibers further suggested that GhPRA1.B4-1A may be related to reproductive development and fiber elongation. We also found that GhPRA1.F2-2A and GhPRA1.F2-1A/D were predominantly expressed in 25 DPA fibers, suggesting their roles in fiber growth (Figure 5A).
Figure 5.
Expression patterns of GhPRA1 genes in different tissues under abiotic stress in upland cotton. (A) The expression profiles of GhPRA1 genes in cotton tissue. DO and DF represent DPA ovules and fibers, respectively. (B) The expression profiles of GhPRA1 genes under abiotic stress.
We also investigated the expression profiles of GhPRA1 genes under abiotic stresses, including cold, heat, drought, and salt stress (Figure 5B). GhPRA1.B4-1A expression was elevated after 1 h and 12 h of cold stress, but was decreased after 3 h of cold stress. The GhPRA1.B4-1A gene was downregulated after 3 h and 6 h of drought stress, and was upregulated after 12 h and 24 h of exposure to drought conditions. GhPRA1.B4-1A exhibited decreased expression after 1 h, 3 h, 6 h, 12 h, and 24 h of heat stress. The GhPRA1.B4-1A gene was downregulated after 1 h, 3 h, and 6 h of salt stress, but was upregulated after 12 h and 24 h of exposure to saline conditions. The expression of GhPRA1.B1-1A was elevated after 12 h of cold and drought stress, but decreased after 24 h. The GhPRA1.B1-1A genes were abundantly expressed after 1 h and 6 h of exposure to heat conditions, but decreased after 24 h. The GhPRA1.B1-1A gene was downregulated after 3 h and 24 h of salt stress. The expression of GhPRA1.B1-1D was increased after 3 h, 6 h, 12 h, and 24 h of cold stress. The GhPRA1.B1-1D genes were abundantly expressed after 1 h and 6 h of exposure to heat conditions. The GhPRA1.E-2D gene was downregulated after 1 h and 3 h of cold stress, and upregulated after 6 h, 12 h, and 24 h of exposure to cold conditions. The GhPRA1.F2-1A genes were abundantly expressed after 6 h and 24 h of exposure to cold conditions. The GhPRA1.F2-1A gene showed increased expression after 1 h, 6 h, and 24 h of heat stress. The GhPRA1.F2-1D genes were abundantly expressed after 3 h, 6 h, and 12 h of exposure to salt conditions. These results suggest that GhPRA1 genes may participate widely in cotton’s adaptation to abiotic stresses.
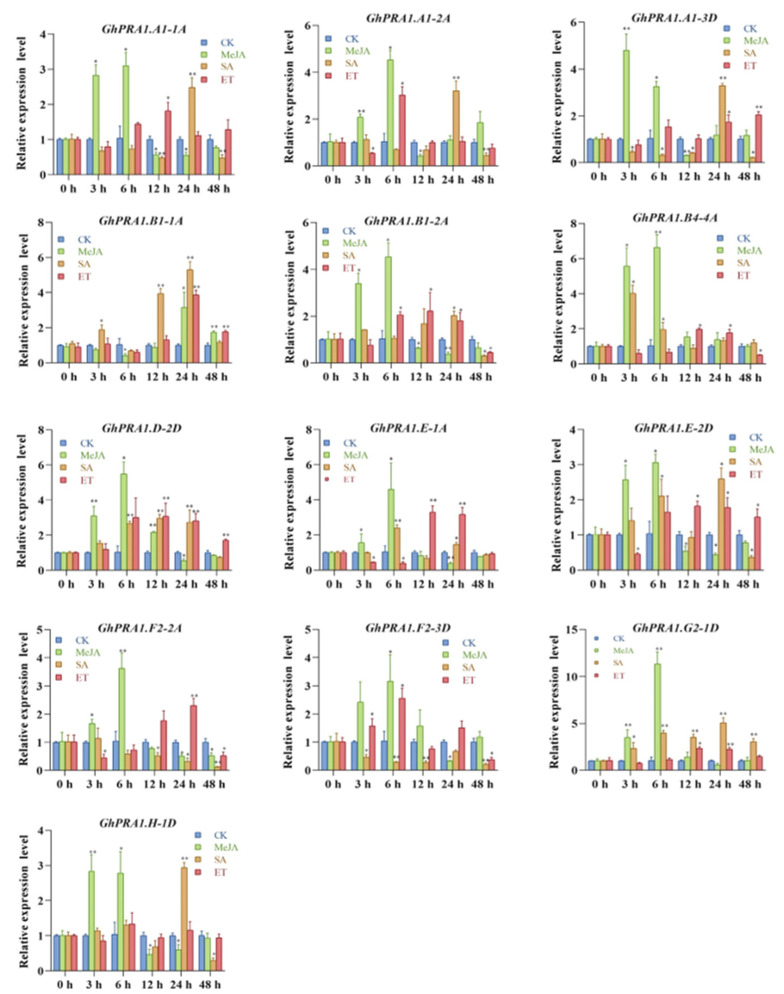
Plant hormone-mediated signal transduction is an important process in plant immune responses, and JA, SA, and ET have been found to play an important role in biological stress signals. Thus, we examined the effects of MeJA, MeSA, and ET treatments on GhPRA1 expression by the RT-qPCR approach (Figure 6). We observed that the transcriptional levels of the GhPRA1.B1-1A gene were upregulated at 24 h and 48 h after MeJA treatment. Most GhPRA1 genes were downregulated at 48 h after exposure to MeJA and SA. The expression levels of GhPRA1.F2-2A and GhPRA1.F2-3D decreased at multiple time points after SA and at 48 h after ET treatment, whereas those of GhPRA1.B1-1A and GhPRA1.D-2D increased. GhPRA1.D-2D, GhPRA1.E-1A GhPRA1.E-2D, and GhPRA1.G2-1D were induced at 12 h and 24 h after ET, and at 6 h and 24 h after SA treatment. These results show that the GhPRA1 genes responded to MeJA, SA, and ET treatments, and that GhPRA1 genes may be involved in plant immunity.
Figure 6.
Expression profiles of GhPRA1 genes under treatment with different plant hormones. Statistically significant differences are indicated as follows: * p < 0.05, ** p < 0.01.
3.6. Overexpression of GhPRA1.B1-1A Enhanced V. dahliae Resistance in Arabidopsis
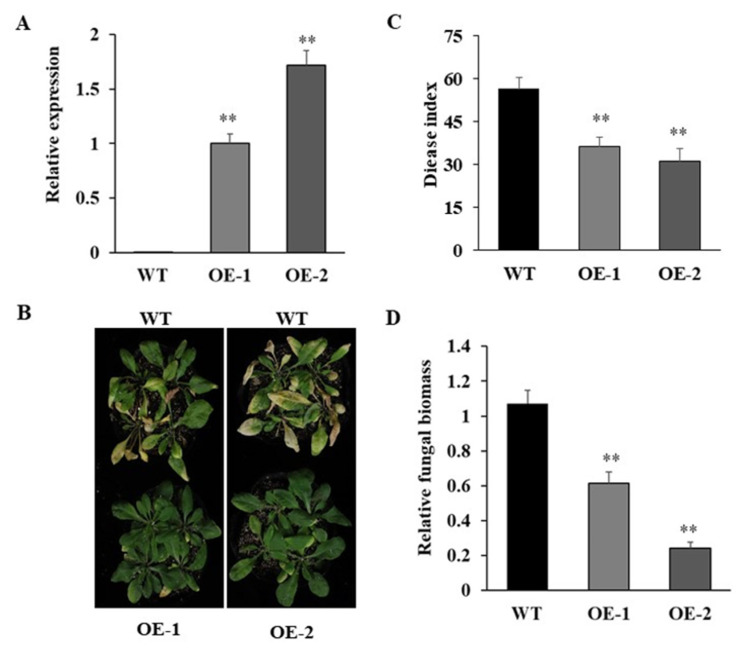
The expression patterns of GhPRA1 genes responding to V. dahliae infection were investigated by using public RNA-Seq data, in which only the expression of the homologous genes GhPRA1.B1-1A and GhPRA1.B1-1D were significantly induced (Figure S2). Meanwhile, the upregulated expression of GhPRA1.B1-1A upon Vd991 infection was confirmed by RT-qPCR (Figure S3). These results suggest that GhPRA1.B1-1A may play a defensive role in cotton against Vd991 infection. To further investigate the role of GhPRA1.B1-1A in V. dahliae resistance, the p2300-GhPRA1.B1-1A overexpression vector driven by the 35S promoter was transformed into A. thaliana by the flower dipping method. A single-copy insertion transgenic line with high GhPRA1.B1-1A expression in the T3 generation was used for V. dahliae infection. The results showed that V. dahliae resistance was enhanced in GhPAR1.B1-1A overexpressing (OE) lines (Figure 7B,C). In addition, the fungal biomass of the transgenic lines was significantly lower than that of the wild-type (Figure 7D). Therefore, these results indicate that GhPRA1.B1-1A overexpression improved V. dahliae resistance in A. thaliana.
Figure 7.
GhPRA1.B1-1A overexpression increased V. dahliae resistance in A. thaliana. (A) The transcriptional levels of GhPRA1.B1-1A in OE and wild-type lines were detected by RT-qPCR. (B) The V. dahliae-resistant phenotype of wild-type and transgenic plants after inoculation with Vd991. (C) Statistical analysis of the plants’ DI. (D) Fungal biomass in rosette leaves of A. thaliana, as detected by RT-qPCR. T-tests were performed to identify statistically significant differences (** p < 0.01). All experiments were repeated at least three times.
3.7. Silencing GhPRA1.B1-1A Reduced V. dahliae Resistance in Upland Cotton
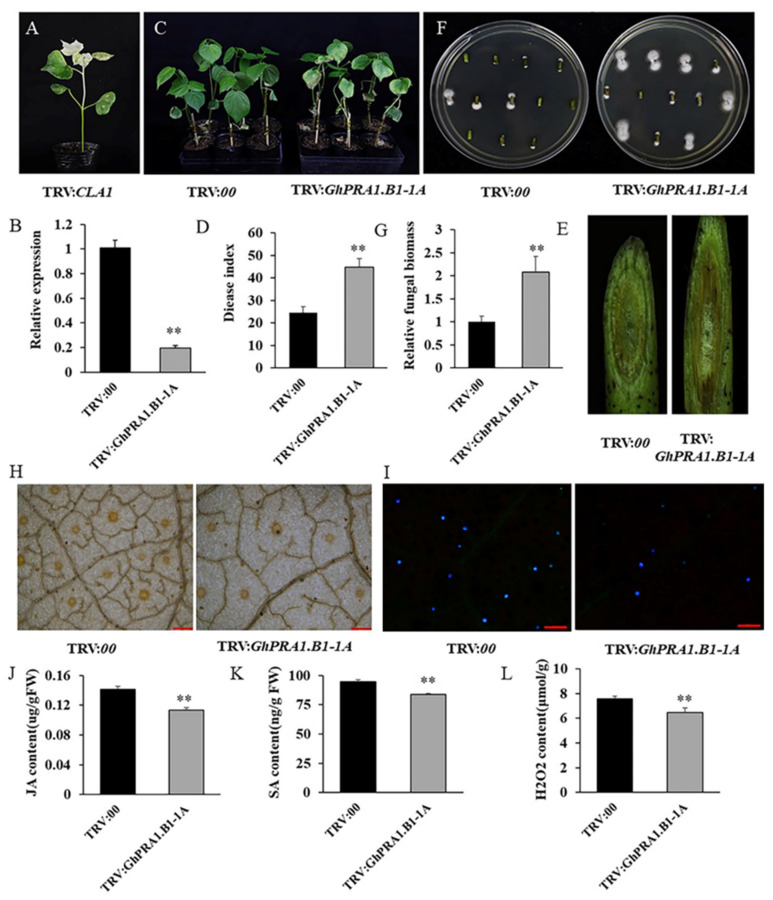
To determine the function of GhPRA1.B1-1A in V. dahliae resistance in upland cotton, the VIGS approach was used to downregulate GhPRA1.B1-1A expression. The pTRV2 vector (TRV:GhPRA1.B1-1A) was constructed with a specific fragment of GhPRA1.B1-1A, and the empty pTRV2 vector was used as a negative control (TRV:00). When photobleaching was observed in the positive control (TRV:CLA) (Figure 8A), RT-qPCR was used to detect GhPRA1.B expression in the silenced plants. The observation of GhPRA1.B1-1A downregulation suggested that GhPRA1.B1-1A was effectively silenced in cotton plants (Figure 8B). However, the expression of other members in GhPRA1.B subgroup were not affected by GhPRA1.B1-1A silencing, except for GhPRA1.B1-1D (the homologous gene of GhPRA1.B1-1A) (Figure S4). The plants were then infected with Vd991, and V. dahliae resistance was weakened in GhPRA1.B1-1A-silenced cotton plants, in which the leaves showed more serious yellowing and wilting phenotypes than the negative control (Figure 8C,D). In addition, the vascular bundles in GhPRA1.B1-1A-silenced plants were more browned than those in the negative control (Figure 8E). Fungal recovery experiments and fungal biomass detection showed that the fungal contents of TRV:GhPRA1.B1-1A samples were higher than those of TRV:00 (Figure 8F,G). Enhanced ROS production and callose deposition have been confirmed as important defense responses of plants to pathogen invasion [49]. Therefore, cotton leaves in the TRV:00 and TRV:GhPRA1.B1-1A plants were stained with DAB, and ROS levels were determined. We found that ROS accumulation and H2O2 levels in GhPRA1.B1-1A-silenced plants were significantly lower than in control plants (Figure 8H,L). In addition, less callose deposition was observed in GhPRA1.B1-1A-silenced cotton plants than in control plants (Figure 8I). These results indicated that V. dahliae resistance was weakened by downregulating the expression of GhPRA1.B1-1A in upland cotton.
Figure 8.
Silencing GhPRA1.B1-1A weakened V. dahliae resistance in upland cotton plants. (A) The photobleached phenotype after infection by TRV:GhCLA1. (B) Detection of the expression levels of the GhPRA1.B1-1A gene in GhPRA1.B1-1A-silenced plants. (C) Phenotypes of control (TRV:00) and TRV:GhPRA1.B1-1A plants. (D) DI after 21 days of Vd991 infection. (E) Observation of the browning degree in stem vascular bundles. (F,G) The degree of fungal colonization and relative fungal biomasses of TRV:00 and TRV:GhPRA1.B1-1A. (H) At 48 h after inoculation, the levels of ROS were detected by DAB; bar = 200 μm. (I) Callose deposition in TRV:00 and TRV:GhPRA1.B1-1A cotton leaves; bar = 200 μm. (J) JA contents of TRV:00 and TRV:GhPRA1.B1-1A. (K) SA contents of TRV:00 and TRV:GhPRA1.B1-1A. (L) H2O2 contents of TRV:00 and TRV:GhPRA1.B1-1A. The difference was statistically significant (** p < 0.01).
3.8. GhPRA1.B1-1A Affected PR Genes’ Expression and Regulated the SA and JA Signaling Pathways
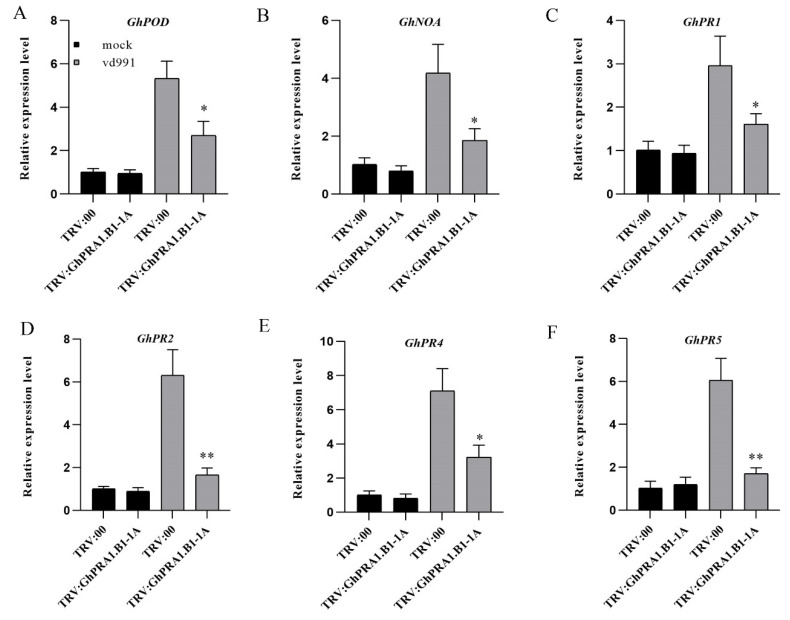
To study the molecular basis whereby GhPRA1.B1-1A participates in V. dahliae resistance, the expression patterns of several PR genes were detected in GhPRA1.B1-1A-silenced cotton plants. In particular, the expression levels of GhPR2 and GhPR5 decreased threefold in the silenced plants. The expression levels of many PR genes in TRV:GhPRA1.B1-1A plants were significantly downregulated compared with the control plants, suggesting that GhPRA1.B1-1A affects the expression of PR genes (Figure 9).
Figure 9.
Expression levels of PR genes in GhPRA1.B1-1A-silenced cotton. (A–F), RT-PCR analysis of the expression levels of PR genes in silenced plants and controls. All experiments were repeated at least three times (* p < 0.05, ** p < 0.01).
GhPRA1.B1-1A was induced by SA and MeJA treatments (Figure 6); thus, we determined the contents of SA and JA in TRV:00 and TRV:GhPRA1.B1-1A cotton leaves 48 h after inoculation with V. dahliae. We observed that the JA and SA contents in TRV:GhPRA1.B1-1A cotton seedlings were significantly lower than those in TRV:00 plants (Figure 8J,K). Therefore, our results indicated that GhPRA1.B1-1A promotes V. dahliae resistance via the SA and JA pathways.
4. Discussion
Pattern recognition receptors can trigger plant immune responses by recognizing microbe-related molecular patterns [50]. The recognition and transmission of pathogens by PRRs depends on transport to the plasma membrane [51]. Rab proteins play an important role in plant immunity [18,19,20,21], and the PRA1 protein regulates Rab proteins and mediates their transport by stabilizing their localization on the cell membrane [25]. The findings of this study expounded upon the role of GhPRA1 in cotton’s resistance to V. dahliae and provided a basis for functional analysis of the cotton PRA1 gene.
4.1. Evolution of GhPRA1 Genes
The PRA1 gene family has been identified in A. thaliana [22]. However, PRA1 gene members have not been identified or analyzed in cotton. In the present study, we analyzed the evolutionary relationship and expression profiles of PRA1 genes in upland cotton. The allotetraploid species of G. hirsutum and G. barbadense resulted from the hybridization of diploid cotton (G. arboreum and G. raimondii) [52]. We identified 108 PRA1 genes in G. arboreum, G. raimondii, G. hirsutum, and G. barbadense. Approximately twice as many PRA1 genes were identified in the tetraploid cotton species G. hirsutum (37 genes) and G. barbadense (33 genes) than in the diploid cotton species G. arboreum (19 genes) and G. raimondii (19 genes), which provides further evidence that the allotetraploid cotton species were derived from the hybridization of two diploid species.
PRA1 proteins can be divided into eight subfamilies according to phylogenetic analysis. The GhPRA1 proteins in the same subfamily clustered together and showed a high sequence identity. Furthermore, the GhPRA1 members in the same subfamily were found to have similar exon and intron structures. Furthermore, we found that most GhPRA1 members have only one exon.
In addition, PRA1 proteins in the same group have conserved protein motifs, which is in accordance with the results of our gene structure analysis. The results of synteny analysis suggested that a strong synteny relationship occurred with the PRA1 gene in upland cotton. Gene duplication analysis showed that GhPRA1.B4-3A/3D and GhPRA1.B4-4A/4D; GhPRA1.A1-1A/1D and GhPRA1.A1-3A/3D; GhPRA1.B1-1A/1D and GhPRA1.B1-2A/2D; and GhPRA1.E-2A/2D and GhPRA1.E-1A/1D have arisen through fragment duplications. Thus, fragment duplications may have promoted the evolution and expansion of GhPRA1 family members.
4.2. Expression of GhPRA1 in Tissue and under Abiotic Stress
PRA1 protein is a known receptor of Rab GTPases, which mediate vesicular transport; however, the role of PRA1 in abiotic stresses has not been extensively studied. Mutant atpr1.f4 and overexpressing AtPRA1.F4 plants were more sensitive to salt stress [53]. Analysis of the cis-acting elements showed that several STRE elements were found in the GhPRA1.B4-1A/D and GhPRA1.E-2D promoter regions. Wound-responsive elements were found in GhPRA1.A1-2A and GhPRA1.B1-1A. Additionally, the expression of GhPRA1.B4-1A increased under drought and salt stress after 24 h. The GhPRA1.E-2D gene was upregulated after exposure to heat and drought treatment at 1 h and 6 h. The expression of GhPRA1.F2-1A/D increased after 6 h under drought and heat stress conditions. Accordingly, MYB-binding site elements were predicted in their promoters. In summary, GhPRA1 family members may participate in environmental stress resistance in cotton plants.
4.3. GhPRA1.B1-1A Enhanced Cotton’s Resistance to V. dahliae through ROS Accumulation and the SA and JA Signaling Pathways
Plant hormones play vital roles in regulating plant defense responses. JA, SA, and ET are essential for transducing signals responsible for inhibiting pathogen infection [54,55]. SA is primarily involved in immunity against biological nutritional pathogens. JA plays roles in plants’ defenses against necrotizing pathogens and in the defense response to physical wounding [56]. GbMPK3 overexpression leads to cotton being more sensitive to V. dahliae by regulating SA-dependent signal transduction [57]. Silencing GbEDS1 significantly reduced SA and H2O2 accumulation and decreased the resistance of cotton to V. dahliae [46]. Gbvdr6 conferred V. dahliae resistance by regulating the JA and SA signaling pathways [58]. The ET response-related factor (GbERF1-like) participates in V. dahliae resistance by regulating lignin synthesis [59]. GaRPL18 participates in the SA signal transduction pathway and increases cotton’s resistance to V. dahliae [60]. In this study, many hormone-related elements were found in the GhPRA1 promoters (Figure 3B), such as the ethylene-response element (ERE), the TCA element, the salicylic acid response element (SARE), the CGTCA motif, and the TGACG motif (associated with the JA response element). In particular, we detected two EREs, two JA response elements, and one SA in the GhPRA1.B1-1A promoter region. These results imply that GhPRA1.B1-1A may be involved in cotton’s responses to pathogens by regulating SA and JA signals. To test this possibility, the VIGS approach was used to silence GhPRA1.B1-1A in upland cotton, which revealed that TRV:GhPRA1.B1-1A plants were more sensitive to V. dahliae infection. Furthermore, the JA and SA contents in TRV:GhPRA1.B1-1A seedlings were significantly lower than those in TRV:00 plants. Therefore, GhPRA1.B1-1A may be involved in SA and JA signaling pathways to enhance V. dahliae resistance in cotton.
ROS are cellular signaling molecules that can recognize pathogens and trigger immune and cell death responses in plants [3]. H2O2 production by cell wall peroxidases plays a vital role in the immune responses triggered by pathogen-associated molecular patterns. Previous findings showed that indicators of ROS accumulation were upregulated in cotton roots infected with V. dahliae, indicating the key role of ROS in defense responses [49,61,62]. HyPRP1 negatively regulates resistance in cotton through ROS accumulation and cell wall thickening [9], whereas GhGPA positively regulates V. dahliae resistance through H2O2 accumulation [7]. In this study, ROS accumulation and H2O2 levels were significantly lower in GhPRA1.B1-1A-silenced cotton than in the control (Figure 8L). These results suggest that GhPRA1.B1-1A protects cotton against V. dahliae infection through ROS accumulation. In this study, ROS and callose accumulation decreased, fungal biomasses increased, and vascular bundle browning was more serious in GhPRA1.B1-1A-silenced plants after V. dahliae inoculation, compared with the corresponding findings in the control plants. This result agrees with the toxin theory, and suggests that GhPRA1.B1-1A participates in V. dahliae resistance through the ROS pathway.
5. Conclusions
In the present study, we characterized PRA1 family genes in cotton, and the particular role of GhPRA1.B1-1A in V. dahliae resistance was highlighted. V. dahliae resistance was increased significantly by the overexpression of GhPRA1.B1-1A in A. thaliana, whereas V. dahliae resistance was decreased by silencing GhPRA1.B1-1A in cotton. Additionally, GhPRA1.B1-1A might participate in V. dahliae resistance through the SA and JA signal transduction pathways and ROS accumulation. The findings of this study help in understanding the potential role of GhPRA1.B1-1A in V. dahliae resistance and provide a theoretical basis for breeding cotton plants resistant to V. wilt.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13050765/s1, Figure S1: The conserved motifs identified in GhPRA1 proteins; (A) phylogenetic tree of GhPRA1 proteins; (B) the top ten conserved motifs in GhPRA1 proteins; (C) logos of the ten conserved motifs in GhPRA1 proteins, Figure S2: Expression patterns of GhPRA1 genes by Vd991 infection in RNA-Seq datasets, m, mock; v, Vd991 infection, Figure S3: Expression profiles of GhPRA1.B1-1A under Vd991 infection by RT-qPCR, Figure S4: Expression levels of GhPRA1.B group members in GhPRA1.B1-1A silencing cotton plants, Table S1: Primers used in this study, Table S2: The amino acid sequences and functional annotation of the conserved motifs, Table S3: Cis-elements detected in GhPRA1 promoter regions.
Author Contributions
N.W.: Formal analysis, experimentation, and writing. Y.-P.Z.: Formal analysis, review, and editing. W.-J.L.: Experimentation and data curation. C.C.: Methodology and data curation. Y.-X.H.: Supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31901578), the Science and Technologies R & D Program of Henan Province of China (222102110090), and the State Key Laboratory of Cotton Biology Open Fund (CB2022A06).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data analyzed in the current study are included in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ali F., Qanmber G., Li Y., Ma S., Lu L., Yang Z., Wang Z., Li F. Genome-wide identification of Gossypium INDETERMINATE DOMAIN genes and their expression profiles in ovule development and abiotic stress responses. J. Cotton Res. 2019;2:1–16. doi: 10.1186/s42397-019-0021-6. [DOI] [Google Scholar]
- 2.Wendel J.F. New world tetraploid cottons contain old-world cytoplasm. Proc. Natl. Acad. Sci. USA. 1989;86:4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaban M., Miao Y., Ullah A., Khan A.Q., Menghwar H., Khan A.H., Ahmed M.M., Tabassum M.A., Zhu L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018;125:193–204. doi: 10.1016/j.plaphy.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Daayf F. Verticillium wilts in crop plants: Pathogen invasion and host defence responses. Can. J. Plant Pathol. 2015;37:8–20. doi: 10.1080/07060661.2014.989908. [DOI] [Google Scholar]
- 5.Zhao P., Zhao Y.L., Jin Y., Zhang T., Guo H.S. Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell. 2014;5:94–98. doi: 10.1007/s13238-013-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talboys P.W. Association of tylosis and hyperplasia of the xylem with vascular invasion of the hop by Verticillium alboatrum. Trans. Br. Mycol. Soc. 1958;41:249–260. doi: 10.1016/S0007-1536(58)80037-6. [DOI] [Google Scholar]
- 7.Chen B., Zhang Y., Yang J., Zhang M., Ma Q., Wang X., Ma Z. The G-protein α subunit GhGPA positively regulates Gossypium hirsutum resistance to Verticillium dahliae via induction of SA and JA signaling pathways and ROS accumulation. Crop J. 2021;9:823–833. doi: 10.1016/j.cj.2020.09.008. [DOI] [Google Scholar]
- 8.Zhang Y., Wu L., Wang X., Chen B., Zhao J., Cui J., Li Z., Yang J., Wu L., Wu J. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2019;20:309–322. doi: 10.1111/mpp.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Zhang Y., Wang X., Wang W., Li Z., Wu J., Wang G., Wu L., Zhang G., Ma Z. HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 2018;18:1–18. doi: 10.1186/s12870-018-1565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P., Zhou L., Jamieson P., Zhang L., Zhao Z., Babilonia K., Shao W., Wu L., Mustafa R., Amin I. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell. 2020;32:3978–4001. doi: 10.1105/tpc.19.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng H., Li C., Zhou J., Yuan Y., Feng Z., Shi Y., Zhao L., Zhang Y., Wei F., Zhu H. A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 2021;167:633–643. doi: 10.1016/j.ijbiomac.2020.11.191. [DOI] [PubMed] [Google Scholar]
- 12.Liu N., Zhang X., Sun Y., Wang P., Li X., Pei Y., Li F., Hou Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017;7:1–18. doi: 10.1038/srep39840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin T., Liu S., Zhang Z., Sun L., He X., Lindsey K., Zhu L., Zhang X. GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17. Plant Mol. Biol. 2019;99:379–393. doi: 10.1007/s11103-019-00824-y. [DOI] [PubMed] [Google Scholar]
- 14.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 15.Stenmark H., Olkkonen V.M. The rab gtpase family. Genome Biol. 2001;2:1–7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal P., Reddy M., Sopory S., Agarwal P.K. Plant rabs: Characterization, functional diversity, and role in stress tolerance. Plant Mol. Biol. Rep. 2009;27:417–430. doi: 10.1007/s11105-009-0100-9. [DOI] [Google Scholar]
- 17.Gao W., Tian Z., Liu J., Lin Y., Xie C. Isolation, characterization and functional analysis of StRab, a cDNA clone from potato encoding a small GTP-binding protein. Sci. Hortic. 2012;135:80–86. doi: 10.1016/j.scienta.2011.12.019. [DOI] [Google Scholar]
- 18.Jiang Z., Wang H., Zhang G., Zhao R., Bie T., Zhang R., Gao D., Xing L., Cao A. Characterization of a small GTP-binding protein gene TaRab18 from wheat involved in the stripe rust resistance. Plant Physiol. Biochem. 2017;113:40–50. doi: 10.1016/j.plaphy.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Kwon S.I., Cho H.J., Kim S.R., Park O.K. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Pphysiol. 2013;161:1722–1736. doi: 10.1104/pp.112.208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen M.E., Jürgens G. Thordal-Christensen H: VPS9a activates the Rab5 GTPase ARA7 to confer distinct pre-and postinvasive plant innate immunity. Plant Cell. 2017;29:1927–1937. doi: 10.1105/tpc.16.00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taefehshokr N., Yin C., Heit B. Rab GTPases in the differential processing of phagocytosed pathogens versus efferocytosed apoptotic cells. Histol. Histopathol. 2021;36:123–135. doi: 10.14670/HH-18-252. [DOI] [PubMed] [Google Scholar]
- 22.Alvim Kamei C.L., Boruc J., Vandepoele K., Van den Daele H., Maes S., Russinova E., Inzé D., De Veylder L. The PRA1 gene family in Arabidopsis. Plant Physiol. 2008;147:1735–1749. doi: 10.1104/pp.108.122226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X., Matern H.T., Gallwitz D. Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 1998;17:4954–4963. doi: 10.1093/emboj/17.17.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calero M., Winand N.J., Collins R.N. Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 2002;515:89–98. doi: 10.1016/S0014-5793(02)02442-0. [DOI] [PubMed] [Google Scholar]
- 25.Hutt D.M., Da-Silva L.F., Chang L.H., Prosser D.C., Ngsee J.K. PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J. Biol. Chem. 2000;275:18511–18519. doi: 10.1074/jbc.M909309199. [DOI] [PubMed] [Google Scholar]
- 26.Figueroa C., Taylor J., Vojtek A.B. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 2001;276:28219–28225. doi: 10.1074/jbc.M101763200. [DOI] [PubMed] [Google Scholar]
- 27.Jung C.J., Hui Lee M., Ki Min M., Hwang I. Localization and trafficking of an isoform of the AtPRA1 family to the golgi apparatus depend on both N- and C- terminal sequence motifs. Traffic. 2011;12:185–200. doi: 10.1111/j.1600-0854.2010.01140.x. [DOI] [PubMed] [Google Scholar]
- 28.Rho H.S., Heo J.B., Bang W.Y., Hwang S.M., Nahm M.Y., Kwon H.J., Kim S.W., Lee B.H., Bahk J.D. The role of OsPRA1 in vacuolar trafficking by OsRab GTPases in plant system. Plant Sci. 2009;177:411–417. doi: 10.1016/j.plantsci.2009.07.003. [DOI] [Google Scholar]
- 29.Pizarro L., Leibman-Markus M., Schuster S., Bar M., Avni A. SlPRA1A/RAB attenuate EIX immune responses via degradation of LeEIX2 pattern recognition receptor. Plant Signal Behav. 2018;13:e1467689. doi: 10.1080/15592324.2018.1467689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 31.Du X., Huang G., He S., Yang Z., Sun G., Ma X., Li N., Zhang X., Sun J., Liu M., et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018;50:796–802. doi: 10.1038/s41588-018-0116-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G., et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., Ge X., Yang Z., Qin W., Sun G., Wang Z., Li Z., Liu J., Wu J., Wang Y., et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 2019;10:2989. doi: 10.1038/s41467-019-10820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J., Jung S., Cheng C.H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014;42:D1229–D1236. doi: 10.1093/nar/gkt1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I., Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.h., Jin H., Marler B., Guo H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2002;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 43.Gao X., Wheeler T., Li Z., Kenerley C.M., He P., Shan L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011;66:293–305. doi: 10.1111/j.1365-313X.2011.04491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y.P., Shen J.L., Li W.J., Wu N., Chen C., Hou Y.X. Evolutionary and characteristic analysis of RING-DUF1117 E3 ubiquitin ligase genes in Gossypium discerning the role of GhRDUF4D in Verticillium dahliae resistance. Biomolecules. 2021;11:1145. doi: 10.3390/biom11081145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Ma Q., Zhang Y., Wang X., Zhang G., Ma Z. Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene. 2016;575:687–694. doi: 10.1016/j.gene.2015.09.046. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Wang X., Rong W., Yang J., Ma Z. Island cotton enhanced disease susceptibility 1 gene encoding a lipase-like protein plays a crucial role in response to Verticillium dahliae by regulating the SA level and H2O2 accumulation. Front. Plant Sci. 2016;7:1830. doi: 10.3389/fpls.2016.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu S., Sun R., Zhang X., Feng Z., Wei F., Zhao L., Zhang Y., Zhu L., Feng H., Zhu H. Genome-wide analysis of OPR family genes in cotton identified a role for GhOPR9 in Verticillium dahliae resistance. Genes. 2020;11:1134. doi: 10.3390/genes11101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y., Chen J., Fang L., Zhang Z., Ma W., Niu Y., Ju L., Deng J., Zhao T., Lian J., et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019;51:739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 49.Jwa N.S., Hwang B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017;8:1687. doi: 10.3389/fpls.2017.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noman A., Aqeel M., Lou Y. PRRs and NB-LRRs: From signal perception to activation of plant innate immunity. Int. J. Mol. Sci. 2019;20:1882. doi: 10.3390/ijms20081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben Khaled S., Postma J., Robatzek S. A moving view: Subcellular trafficking processes in pattern recognition receptor–triggered plant immunity. Annu Rev. Phytopathol. 2015;53:379–402. doi: 10.1146/annurev-phyto-080614-120347. [DOI] [PubMed] [Google Scholar]
- 52.Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 53.Lee M.H., Yoo Y.J., Kim D.H., Hanh N.H., Kwon Y., Hwang I. The prenylated rab GTPase receptor PRA1. F4 contributes to protein exit from the Golgi apparatus. Plant Physiol. 2017;174:1576–1594. doi: 10.1104/pp.17.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spoel S.H., Johnson J.S., Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Nati. Acad. Sci. USA. 2007;104:18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bari R., Jones J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 56.Betsuyaku S., Katou S., Takebayashi Y., Sakakibara H., Nomura N., Fukuda H. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:8–16. doi: 10.1093/pcp/pcx181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long L., Xu F.C., Zhao J.R., Li B., Xu L., Gao W. GbMPK3 overexpression increases cotton sensitivity to Verticillium dahliae by regulating salicylic acid signaling. Plant Sci. 2020;292:110374. doi: 10.1016/j.plantsci.2019.110374. [DOI] [PubMed] [Google Scholar]
- 58.Yang Y., Chen T., Ling X., Ma Z. Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to Verticillium wilt in Arabidopsis and upland cotton. Front. Plant Sci. 2018;8:2272. doi: 10.3389/fpls.2017.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo W., Jin L., Miao Y., He X., Hu Q., Guo K., Zhu L., Zhang X. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016;91:305–318. doi: 10.1007/s11103-016-0467-6. [DOI] [PubMed] [Google Scholar]
- 60.Gong Q., Yang Z., Wang X., Butt H.I., Chen E., He S., Zhang C., Zhang X., Li F. Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 2017;17:1–15. doi: 10.1186/s12870-017-1007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ali M., Cheng Z., Ahmad H. Hayat S: Reactive oxygen species (ROS) as defenses against a broad range of plant fungal infections and case study on ROS employed by crops against Verticillium dahliae wilts. J. Plant Interact. 2018;13:353–363. doi: 10.1080/17429145.2018.1484188. [DOI] [Google Scholar]
- 62.Xie C., Wang C., Wang X., Yang X. Proteomics-based analysis reveals that Verticillium dahliae toxin induces cell death by modifying the synthesis of host proteins. J. Gen. Plant Pathol. 2013;79:335–345. doi: 10.1007/s10327-013-0467-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in the current study are included in this article.