Abstract
Background
Serological assays are being used to monitor antibody responses in individuals who had SARS-CoV-2 infection and those who received a COVID-19 vaccine. We aimed to determine whether such assays can predict neutralising antibody titres as antibody levels wane and viral variants emerge.
Methods
We measured antibody levels in serum samples from a cohort of 112 participants with SARS-CoV-2 infection using ten high-throughput serological tests and functional neutralisation assays. Serum samples were taken at baseline and at up to four subsequent visits. We assessed the effects of time and spike protein sequence variation on the performance and predictive value of the various assays. We did correlation analyses for individual timepoints using non-parametric Spearman correlation, and differences between timepoints were determined by use of a two-tailed Wilcoxon matched-pairs signed rank test.
Findings
Neutralising antibody titres decreased over the first few months post-infection but stabilised thereafter, at about 30% of the level observed shortly after infection. Serological assays commonly used to measure antibodies against SARS-CoV-2 displayed a range of sensitivities that declined to varying extents over time. Quantitative measurements generated by serological assays based on the spike protein were better at predicting neutralising antibody titres than those based on nucleocapsid, but performance was variable, and manufacturer positivity thresholds were not able to predict the presence or absence of detectable neutralising activity. Although we observed some deterioration in correlation between serological measurements and functional neutralisation activity, some assays maintained an ability to predict neutralising titres, even against variants of concern.
Interpretation
The ability of high-throughput serological assays to predict neutralising antibody titres is likely to be crucial for evaluation of immunity at the population scale. These data can facilitate the selection of the most suitable assays as surrogates of functional neutralising activity and suggest that such measurements might be useful in clinical practice.
Funding
US National Institutes of Health and National Health Service Research Scotland BioResource.
Introduction
The world has experienced an unprecedented pandemic after the emergence of SARS-CoV-2. Millions have died, and the repercussions have affected every aspect of life. The remarkable mobilisation of the scientific community in response to the COVID-19 pandemic has led to the rapid development of safe and effective vaccines, as well as reagents and assays to aid in the detection and mitigation of virus spread.
An early prominent issue in the pandemic was the accurate identification at a large scale of individuals who were infected. Although PCR-based assays remain a reliable and sensitive test for infection, they are not amenable to mass population screening. Therefore, serological assays, despite limitations,1, 2, 3 have been instrumental for surveillance and providing selection criteria to recruit participants for vaccine trials and convalescent plasma donors. Monitoring antibody titres is necessary to measure the magnitude and longevity of immune responses induced by natural infection or vaccination. As immune responses to SARS-CoV-2 antigens are increasingly elicited by infection, vaccination, or both, the measurement of antibody titres and the ability of such measurements to predict protection from infection or disease will be of great importance. Whether simple serological tests will be able to predict neutralising antibody titres and immunity to SARS-CoV-2 is yet to be determined. Moreover, as antibodies mature and acquire greater affinity while their total levels decline4, 5, 6 and new SARS-CoV-2 variants emerge, the predictive value of serological tests based on the prototype viral strain will need to be evaluated.
Several high-throughput serological assays are routinely used to detect antibodies against nucleocapsid or spike viral antigens. These assays were initially designed to provide a positive or negative test result, but they also generate quantitative measurements of antibody levels. Previous studies of how these quantitative serological values correlate with neutralising antibody titres have yielded variable results.7, 8, 9 Importantly, the sensitivity of the assays as diagnostic tools and their predictive value for immune parameters several months after infection and against variants of concern have not been assessed. Here we expand the study of our previously reported cohort of patients who recovered from COVID-19 to include additional longitudinal samples reaching over 6 months post-infection, and we evaluate the diagnostic sensitivity of ten serological assays for their ability to predict neutralising antibody activity against SARS-CoV-2 and its variants.
Research in context.
Evidence before this study
We searched PubMed during May, 2021, using serological assays, comparison, and evaluation terms, with no language restrictions. Although several studies have correlated serological measurements with neutralisation activity in SARS-CoV-2 infection, at the time of submission, only three papers in peer-reviewed journals have directly assessed the ability of commercial, high-throughput serological assays used in routine clinical practice to predict functional neutralising activity. The first study was done in 190 patients in New York (NY, USA) and compared the ability of two commercial serological assays with that of a lateral flow assay and three ELISA assays to predict neutralising titres in serum samples obtained shortly after infection (about 30 days). This study concluded that the variation in neutralising antibody titres was variably predicted by the commercial assays. The second study was done in a cohort overlapping with the current study (97 participants) from the National Health Service hospitals in Scotland and evaluated the performance of four serological assays with serum samples collected over a period of up to about 84 days post-infection. The study showed that neutralising activity declined over time and that declining neutralising titres were not predicted by serological assays. The third study was done in a cohort of 40 patients in St Louis (MO, USA) and compared values obtained by use of three commercial serological assays with neutralisation titres obtained shortly post-infection (about 30 days). The study reported that the assays evaluated were not optimal for predicting neutralisation. At present, several high-throughput serological assays are routinely used to measure SARS-CoV-2 antibodies, but their ability to predict immunity has not been evaluated. Importantly, none of the previous studies extended beyond about 80 days post-infection, and none evaluated the ability of the serological assays to predict functional antibody activity against variants of concern.
Added value of this study
To our knowledge, this is the first study to evaluate ten different high-throughput serological assay platforms over a period longer than 140 days post-SARS-CoV-2 infection and assess their ability to predict functional antibody activity against variants of concern. Our analysis substantially expands our previous study in extending the time of sample acquisition from 80 days to 140 days, increasing the number of serological assays evaluated from four to ten and including variants of concern. Our study showed that neutralising antibody titres declined shortly after infection but stabilised thereafter. The serological assays did not accurately reflect this initial decline. Diagnostic sensitivity of each assay declined to widely varying extents over time. Manufacturer assay diagnostic cutoffs were not suitable for accurate prediction of the presence or absence of serum neutralising activity. Nevertheless, quantitative values measured with some high-throughput serological assays based on the spike protein correlated well with serum neutralising titres over time, even when neutralisation activity was measured against variants of concern.
Implications of all the available evidence
Serological assays are playing an important role in efforts to manage the SARS-CoV-2 pandemic, including the identification of correlates of immune protection after infection or vaccination, and the selection of individuals recommended for monoclonal antibody prophylaxis or additional vaccinations. These data provide a detailed analysis and comparison of the ability of multiple commercial high-throughput serological assays to predict functional neutralising antibody activity, a key component of immune protection, and reveal parameters that need to be taken into consideration when evaluating serology results, such as assay cutoff, presence of SARS-CoV-2 variants, time post-infection, or vaccination. In summary, our study provides guidance for the use of serological assays for various applications that can inform clinical practice and policy.
Methods
Participants
112 participants with a history of SARS-CoV-2 infection diagnosed by RT-PCR, who developed mild symptoms, were recruited (more cohort details in appendix pp 2, 11–14).8 Participants were surveyed to determine the date of the positive PCR test, the date of onset of symptoms, and if their symptoms required hospitalisation. Serum samples were taken at a baseline visit (visit 1; about 3·5 to 8·5 weeks after the PCR test) and 2 weeks (visit 2), 4 weeks (visit 3), 8 weeks (visit 4), and 22 weeks (visit 5) later. In total, 101 participants completed at least three visits and 58 participants completed the fifth visit and were included in the neutralisation assays. Four of 101 patients were admitted to hospital but none required intensive care. 23 additional participants attended for a single visit a mean of 171 days (range 44–202) after the PCR test. The mean age of the participants was 45 years (21–65), and 72 (71%) were women and 29 (29%) were men. At visit 1 (baseline), the average number of days between PCR test and the visit was 41 days (24–64); at visit 2, the average number of days post-PCR test was 55 days (40–79); at visit 3, the average number of days post-PCR test was 70 days (55–95); at visit 4, the average number of days post-PCR test was 98 days (85–110); and at visit 5, the average number of days post-PCR test was 194 days (160–216). Ethical approval was granted through the NHS Lothian BioResource (SR1407) and London-Brent Research Ethics Committee (20/HRA/3764 IRAS:28653). All participants gave written and informed consent for serial blood sample collection. Deidentified samples were shipped to the Rockefeller University (New York, NY, USA) under a protocol reviewed and approved by an institutional review board.
Serological assays
In this study, we assessed the following ten serological assays. The Abbott SARS-CoV-2 IgG (N) (Abbott, Chicago, IL, USA; used at National Health Service [NHS] Lothian, UK) and SARS-CoV-2 IgGII Quant (Abbott; used at NHS Greater Glasgow and Clyde, UK) assays are two-step chemiluminescent microparticle immunoassays (CMIA) designed to detect IgG antibodies against nucleocapsid and the receptor binding domain (RBD), respectively. The LIASON SARS-CoV-2 S1/S2 IgG (DiaSorin, Saluggia, Italy; used at NHS Lothian), LIASON Trimeric S IgG (DiaSorin; used at NHS Highland, UK), and sCOVG RBD IgG (Siemens, Erlangen, Germany; used at NHS Tayside, UK) assays are also two-step CMIA designed to detect IgG antibodies. The Euroimmun SARS-CoV-2 IgG assay (Euroimmun, Lübeck, Germany; used at Scottish National Blood Transfusion Service, UK) is an indirect ELISA that uses the S1 domain of the spike protein as the antigen. The Anti-SARS-CoV-2 N and S assays (Roche, Basel, Switzerland; used at NHS Lanarkshire, UK) and the COV2T assay (Siemens; used at NHS Tayside) are two-step bridging electrochemiluminescent immunoassays that use nucleocapsid or the RBD of the spike protein as antigens. The cPass assay (Genscript, Piscataway, NJ, USA) detects antibodies that block binding of a soluble RBD to an immobilised cellular receptor protein.
SARS-CoV-2 pseudotyped reporter virus
SARS-CoV-2 pseudotyped particles were generated as previously described.10 Briefly, 293Tcells were transfected with pNL4–3ΔEnv-nanoluc and pSARS-CoV-2-SΔ19. 48 h later, particles were harvested, filtered, and stored at –80°C.
The amino acid deletions or substitutions corresponding to SARS-CoV-2 variants of concern were incorporated into a spike expression plasmid with use of synthetic gene fragments or overlap extension PCR-mediated mutagenesis and Gibson assembly. Specifically, the variant-specific deletions and substitutions introduced are detailed in the panel .
Panel. Variant-specific deletions and substitutions.
Alpha variant (B.1.1.7)
His69_Val70del, Tyr144del, Asn501Tyr, Ala570Asp, Asp614Gly, Pro681His, Thr761Ile, Ser982Ala, and Asp1118His
Alpha variant version with an additional Glu484Lys mutation
His69_Val70del, Tyr144del, Asn501Tyr, Ala570Asp, Glu484Lys, Asp614Gly, Pro681His, Thr761Ile, Ser982Ala, and Asp1118His
Beta variant (B.1.351) version 1
Leu18Phe, Asp80Ala, Asp215Gly, 242_4del, Lys417Asn, Glu484Lys, Asn501Tyr, Asp614Gly, and Ala701Val
Beta variant version 2
Asp80Ala, Asp215Gly, Leu242His, Arg246Ile, Lys417Asn, Glu484Lys, Asn501Tyr, Asp614Gly, and Ala701Val
Delta variant (B.1.617.2)
Thr19Arg, 156_8del, Leu452Arg, Thr478Lys, Asp614Gly, Pro681Arg, and Asp950Asn
The variant spike proteins included the Arg683Gly substitution, which disrupts the furin cleavage site and increases particle infectivity and neutralisation sensitivity. Therefore, in these neutralisation assays, we used a wild type SARS-CoV-2 spike (NC_045512), carrying Arg683Gly for comparative purposes (appendix pp 8–9).
Pseudotyped virus neutralisation assay
We incubated five-fold serially diluted serum samples from patients who recovered from COVID-19 with SARS-CoV-2 pseudotyped virus for 1 h at 37°C. The mixture was subsequently added to 293TAce2 cl22 cells (for analyses using SARS-CoV-2 Wuhan-Hu-1 [NC_045512] pseudovirus) or HT1080Ace2 cl14 cells (for analyses involving variant pseudovirus panels and the respective Wuhan-Hu-1 Arg683Gly controls).10 The starting serum dilution on cells was 1:50. We measured NanoLuc luciferase (Promega, Madison, WI, USA) activity in lysates 48 h post-inoculation using the Nano-Glo Luciferase Assay System (Promega) with the Glomax Navigator (Promega). Relative luminescence units were normalised to those derived from cells infected with SARS-CoV-2 pseudotyped virus in the absence of serum. We determined the half-maximal neutralisation titres for serum samples (NT50) using four-parameter non-linear regression using the least squares regression method without weighting.
We considered samples from visit 1 and visit 5 independently, and we evaluated selected cutoff values for each serological assay scale for sensitivity, specificity, and predictive value. More details on the assessment and selection of cutoffs are presented in the appendix (p 2).
Statistical analysis
Analyses and the statistics used are described in the appendix (pp 2–3). To determine whether assay values differed significantly between visit 1 and 5, we applied the two-tailed Wilcoxon matched-pairs signed rank test with a 95% CI, and we report the p values. We did correlation analyses by calculating the non-parametric Spearman correlation coefficient; we report here Spearman's r with 95% CIs, as well as two-tailed p values. Correlation analyses were always done for the individual timepoints, as indicated. We used GraphPad Prism for statistical analyses.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
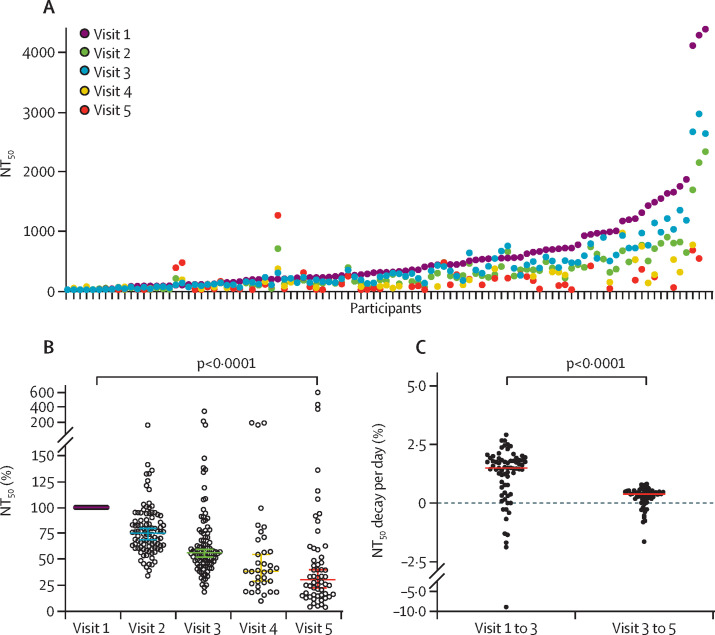
A previously reported8 cohort of participants who developed mild symptoms after SARS-CoV-2 infection was repeatedly sampled, up to 7·2 months post-infection, to evaluate how neutralising antibody levels correlate over time with antibodies measured with ten high-throughput serological assays. Neutralising antibody titres were measured at up to five visits for each participant by use of a pseudovirus neutralisation assay. We have previously shown that NT50 values determined with this assay correlate well with those obtained with use of authentic SARS-CoV-2.10 Consistent with previous studies,4, 6, 8, 11, 12 NT50 values declined over time in the majority of patients (figure 1 A, B). The most significant rate of decrease, approximately 25%, was observed between the early visits, reaching a 45% decrease in NT50 values by visit 3, approximately 70 days post-infection (figure 1C). Thereafter, the rate of decrease became less pronounced, and NT50 values at visits 4 and 5 (approximately 3–7 months post-infection) appeared to stabilise at about 30% of the levels observed within the first 2 months post-infection (figure 1B, C). The overall rate of NT50 decline from the first to the last visit was greater for male participants than for female participants (appendix p 4). The sex difference gradually diminished and was not discernible by visit 4. We did not observe any correlation between NT50 values and age at any timepoint (appendix p 4).
Figure 1.
Neutralisation activity in longitudinal serum samples from patients with COVID-19
(A) NT50s for each sample collected at the visit indicated. (B) Relative NT50 values in serum samples obtained at visits 1 to 5, normalised to visit 1; coloured horizontal bars indicate median values with 95% CIs; statistical significance was determined with the two-tailed Wilcoxon matched-pairs signed rank test. (C) Relative decay of NT50 per day between visits 1 and 3 and relative decay of NT50 per day between visits 3 and 5; red horizontal bars indicate the median; statistical significance was assessed with the Wilcoxon test. NT50=half-maximal neutralisation titre.
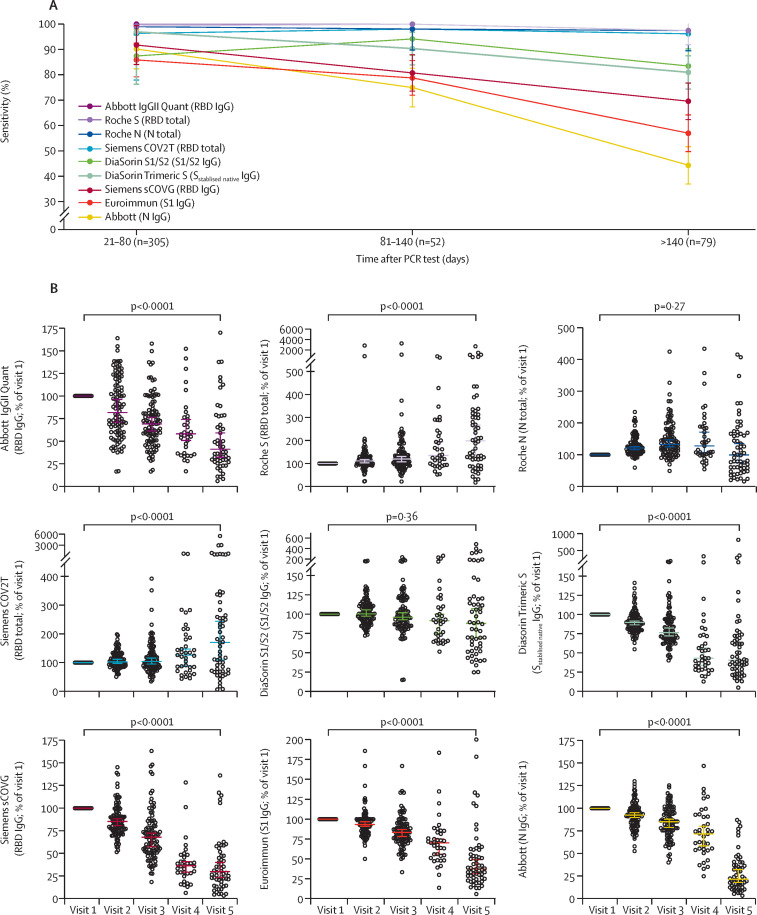
We analysed the same serum samples using nine different serological assays that detect antibodies against either the viral nucleocapsid protein, or various forms of the spike protein that included trimeric spike, S1/S2 subunits, or the RBD. First, we determined the sensitivity of each assay for three time windows over the course of the study (appendix p 2). All assays were sensitive at the first window of 21–80 days post-infection: Abbott IgGII Quant, Roche S, and Roche N had the highest sensitivities (100%); Siemens COV2T and DiaSorin Trimeric S were 95% sensitive; and DiaSorin S1/S2, Euroimmun, and Abbott (N) ranged from 85% to 90%. Whereas Abbott IgGII Quant, Roche N, and DiaSorin S1/S2 maintained their sensitivity over time, the sensitivity of other assays declined to varying degrees, ranging from 45% to 85% at more than 140 days post-infection (figure 2 A). Therefore, the performance of the assays for serosurveillance applications at more than 140 days after SARS-CoV-2 infection was extremely variable.
Figure 2.
Serological analysis of longitudinal serum samples from patients with COVID-19
(A) Sensitivity of the indicated serological assays in samples collected at three different time intervals after the PCR test; mean and 95% CIs are shown. (B) Relative serological results at visits 1 to 5, normalised to visit 1 for the indicated serological assays; horizontal bars indicate median with 95% CIs, and statistical significance was determined with the two-tailed Wilcoxon matched-pairs signed rank test. N=nucleocapsid. RBD=receptor binding domain. S=spike protein.
In addition to indicating whether a serum sample is negative or positive for antibodies against viral antigens, each assay indicates quantitative antibody levels within assay-specific scales. Analysis of antibody levels over time showed assay-dependent differences in trajectory that were not dependent on whether nucleocapsid or spike antigens were used (figure 2B, appendix p 5). Median antibody levels measured by the Roche S and Siemens COV2T assays increased slightly over time, those measured by the Roche N and DiaSorin S1/S2 assays remained approximately constant, whereas levels measured by the Siemens sCOVG, DiaSorin Trimeric S, Euroimmun, and both Abbott assays decreased over time. The deviation from the mean of individual participant antibody levels increased over time in all assays except Abbott IgGII Quant, which showed high deviations from the first timepoint.
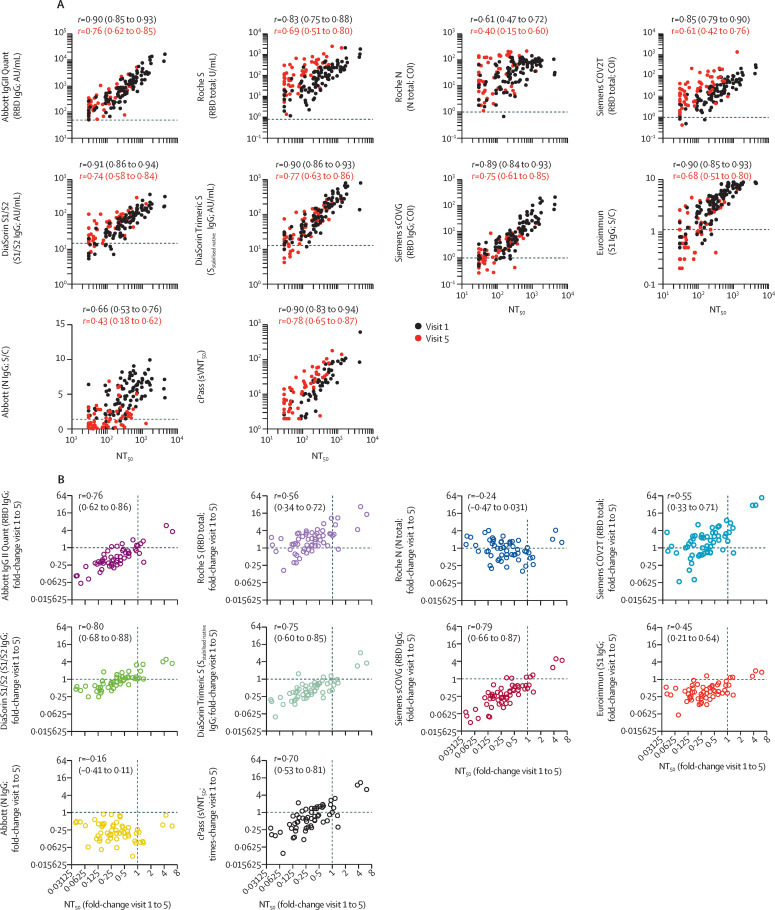
We determined the ability of each serological assay to predict pseudotype virus neutralising antibody titres over time (figure 3 , appendix p 6). For each assay, the correlation with NT50 values was closest at early timepoints and deteriorated over time, with the poorest correlation observed at visit 5 in each case. We included in this analysis an additional assay (cPass), which measures antibodies that block the interaction between the RBD and the virus receptor, using samples only from visits 1 and 5. At visit 1, antibody levels measured with the DiaSorin S1/S2 and Trimeric S assays had the highest correlation with NT50, followed by Abbott IgGII Quant, Siemens sCOVG, Euroimmun, and cPass (figure 3A, appendix p 6). Antibody levels measured with the remaining spike antigen-based assays (Roche S and Siemens COV2T), which are designed to detect total antibody levels against the spike antigen regardless of antibody class, had a lower correlation with NT50 titres, whereas the nucleocapsid-based assays had the poorest correlation. For all serological assays, the correlation with neutralising antibody titres decreased over time, but for all spike antigen-based IgG assays (DiaSorin S1/S2 and Trimeric S assays, Abbott IgGII Quant, and Siemens sCOVG) and cPass, the correlation coefficient r remained higher than 0·75 even at visit 5 (figure 3A, appendix p 6).
Figure 3.
Correlation of neutralisation titres and serology assays
(A) Correlation of NT50 (x-axis) with serological assay values (y-axis) obtained at visit 1 and visit 5 for each participant; statistical significance was determined with the Spearman correlation test for samples obtained at visit 1 and visit 5 independently, with Spearman's r and respective 95% CIs as indicated; dotted lines indicate serological assay thresholds. (B) Correlation of fold change (visit 1 to visit 5) of NT50s with corresponding fold change in serological assay values for indicated serology assays. Statistical significance was determined using the Spearman correlation test, with Spearman's r and respective 95% CIs as indicated. Dotted lines at x=1 and y=1 indicate unchanged assay results over time. For the cPass assay, the serum dilution at which 50% signal inhibition was achieved was defined as 50% surrogate virus neutralisation titre (sVNT50). AU=arbitrary unit. COI=confidence interval. N=nucleocapsid. NT50=half-maximal neutralisation titre. RBD=receptor binding domain. S=spike protein. S/C=ratio over threshold value. U=unit.
In general, the decrease in neutralising antibody titres over time was proportionately greater than the corresponding decrease in levels measured with serological assays (figure 3B), particularly between early timepoints (appendix p 6). Therefore, declines in antibody levels over time measured with serological assays did not, in some cases, accurately reflect the decrease in neutralisation activity. This was particularly the case for the nucleocapsid-based assays, where the magnitude of the decrease in antibody measurements did not correlate with the decrease in NT50. Nevertheless, for assays that correlated best with NT50 titres at early timepoints, declining antibody levels measured with the serological assays predicted declining NT50 (figure 3B, appendix p 6).
We estimated the ability of serological assays to qualitatively identify serum samples that did or did not have detectable neutralising activity (appendix pp 7, 10). None of the assays were effective in qualitatively discriminating neutralising versus non-neutralising serum samples when manufacturer-recommended cutoffs were used, with specificity ranging from 3% for the Roche S and N total antibody assays to 72% for the Euroimmun assay. By selecting different cutoff values, the sensitivity, specificity, and predictive values could be improved for some assays (appendix p 10).
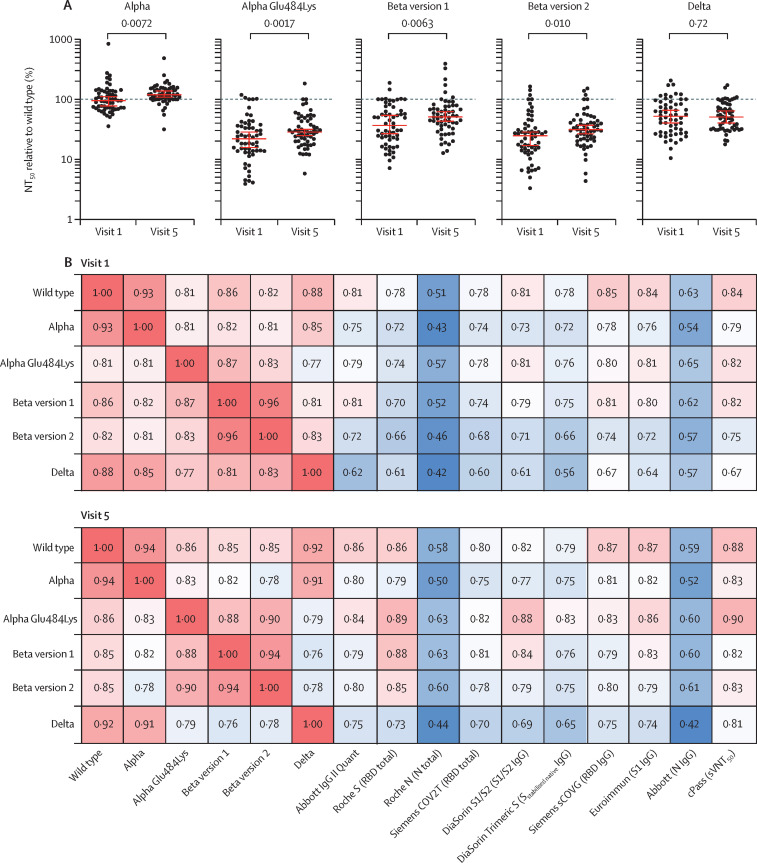
The occurrence of SARS-CoV-2 variants compromises the ability of antibodies resulting from first wave infections or vaccination to neutralise contemporaneous viruses and might further erode the ability of serological assays based on proteins derived from the prototype (Wuhan-hu-1) virus to predict neutralising antibody titres. We determined the ability of 58 serum samples obtained at visits 1 and 5 to neutralise selected variants of concern of the alpha, beta, and delta lineages that have been associated with partial resistance to neutralisation.13, 14, 15 Serum samples from both visits were able to neutralise viruses carrying the alpha spike, with potencies similar to those observed with the Wuhan-hu-1 spike protein (figure 4 A, appendix p 8). By contrast, titres against viruses with the alpha variant with Glu484Lys amino acid substitution, delta variant, or either of the two beta spike variants were decreased by approximately 2·5–5·0-times at visit 1 (figure 4A, appendix p 8). Differences between the ability to neutralise Wuhan-hu-1 and variant spike-bearing viruses became somewhat less pronounced at visit 5 (figure 4A). Therefore, the inclusion of amino acid substitution Glu484Lys in the context of the alpha variant or substitutions found in the spikes from other variants of concern appeared to substantially reduce neutralisation titres, with the largest effect seen with substitutions found in the beta variants, consistent with previous reports.6, 13, 14, 15, 16, 17, 18
Figure 4.
Neutralisation of variants of concern
(A) NT50 values for variant pseudoviruses alpha (B.1.1.7), alpha with a Glu484Lys, beta (B.1.351) version 1, beta version 2, or delta (B.1.617.2) measured at indicated timepoints, normalised to wild type NT50; analysis done in a cohort of 58 participants; statistical significance was determined with Wilcoxon test; dotted line indicates a ratio of 1 (equal NT50). (B) Correlation coefficients (Spearman's r values) of NT50 values between original and variant viruses and each serological assay, measured at visit 1 and visit 5; analysis done in 58 participants for all groups shown, except for DiaSorin S1/S2 (n=54). N=nucleocapsid. NT50=half-maximal neutralisation titre. RBD=receptor binding domain. S=spike protein. sVNT50=50% surrogate virus neutralisation titer.
Overall, the correlation of antibody levels, as measured by serological assays, with neutralising activity was marginally weaker for variant pseudotyped viruses than for the Wuhan-hu-1 variant (figure 4B, appendix p 9). The weakest correlations were observed for the delta variant.
Discussion
Tracking transmission dynamics, spread, and prevalence of viral infections, is crucial in mitigating viral epidemics, particularly when many cases remain asymptomatic during infection, as is the case with individuals infected with SARS-CoV-2. Moreover, the widespread use of vaccines necessitates the accurate determination of the vaccination and immune status of individuals. High-throughput serological assays address these needs, but their usefulness obviously depends on their accuracy and reliability. In this study, we compared the results provided by several SARS-CoV-2 serological assays, with an emphasis on their ability to predict neutralisation activity as antibodies both wane and evolve and SARS-CoV-2 variants emerge.4, 8
The study limitations include the sample size of 112 individuals, of which a small proportion were lost to follow-up during the study. Participants predominantly had mild disease, with few being admitted to hospital, and the findings might not be applicable to individuals with severe COVID-19 disease. Nevertheless, our cohort is representative of a large proportion of the infected population, who have only mild symptoms. This study was done during the first wave of the COVID-19 pandemic, with study visits completed between March and November, 2020, a time at which individuals would have been infected with Wuhan-hu-1-like variants. Although this scenario does not allow for comparison with participants infected afterwards with other variants, it allows for comparisons of neutralisation titres between participants and for correlations of neutralisation and serological data by use of assays based on antigens derived from closely related sequences. And although the levels of neutralising antibodies required for robust protection against currently circulating variants are unknown, neutralising antibodies are recognised as a correlate of protection. We used a pseudotyped virus assay to measure neutralising titres, but we note that titres obtained with the assay used herein correlate very well with those obtained against live SARS-CoV-2.10
Serological assays are generally optimised for increased sensitivity, so they can reliably diagnose the presence or absence of antibodies against viral antigens.1, 19 Most of the assays we used accomplished this goal with serum samples obtained shortly after infection; however, their sensitivity was not always maintained over time, and some assays showed a sharp decline in sensitivity at later timepoints after infection. The reason for the difference in assay trajectories over time is unclear, but it might reflect differences in assay dependence on antibody concentrations versus affinity. This loss of sensitivity was not related to the antigen on which the assays were based.
Serological assays also provide a quantitative result that could enable their use for estimation of antibody levels and prediction of immunity, especially if antibody levels correlate with functional neutralising antibody titres. Assays that detect spike-binding antibodies can use various protein subdomains or conformations (eg, isolated RBD, S1 subunit, or a stabilised trimeric spike) as their antigens. Moreover, some assays only detect specific antibody classes such as IgG, or those that directly interfere with RBD receptor binding. By contrast, neutralisation assays detect all antibodies capable of inhibiting spike-mediated virus entry into cells. Although neutralising antibodies are sometimes dominated by those targeting RBD,20, 21 including those that block ACE2 binding, antibodies targeting the N-terminal domain of the S1 subunit can substantially contribute to the overall serum neutralisation activity in plasma.22, 23, 24 Weak neutralising activity has also been ascribed to antibodies targeting the region of the S2 subunit involved in fusion.25
Multiple studies have shown that neutralising antibody titres after natural infection or vaccination wane over time,4, 8, 11, 12, 26 a decline that is not always accurately reflected by serological assays. Nevertheless, some of the assays we used that detect spike-specific antibodies maintained good levels of correlation with neutralising titres over time. Assays measuring spike-specific IgG antibodies predicted neutralising antibody titres more accurately than those measuring total antibodies against spike or those against the nucleocapsid protein. The DiaSorin assays, Abbott IgGII Quant, Siemens sCOVG, Euroimmun, and cPass had the highest correlation with neutralising antibody titres across all comparisons, and changes in quantitative values over time for these assays were most closely correlated with changes in neutralising antibody levels within individuals. Although the cPass assay is designed to measure antibodies that block the interaction between RBD and ACE2, it correlated with neutralising antibody titres as well as the best of the serological assays that measure antibodies to the entire spike protein. Therefore, although the cPass assay does not measure all neutralising antibodies, this finding suggests that antibodies that block RBD–ACE2 interaction either dominate the overall population of neutralising antibodies or correlate with overall neutralising antibody levels.
The quantitative results from the aforementioned assays are thus best suited for estimating neutralising antibody levels at a population level. By contrast, qualitative assay results, when based solely on the manufacturer-recommended cutoffs, were poorly specific for detecting the presence of neutralising antibodies and might lead to a substantial over-estimation of antibody-related immunity. For some assays, it might be possible to improve specificity for the presence of neutralising antibodies by selecting a higher quantitative cutoff value, thus improving the positive predictive value of these assays for neutralising antibody detection.
Most of the naturally infected population included in our study were infected with viral variants closely related to a prototype variant (Wuhan-hu-1). The antigens used by all serological assays rely on protein sequences derived from that prototype. However, over the past several months, new variants have emerged that encode multiple amino acid substitutions in their spike proteins, some of which affect neutralisation by antibodies resulting from previous infections or vaccination.6, 13, 15, 27, 28 Our data indicate that antibody levels measured with several serological assays maintained good correlation with neutralisation titres against some of the most important variants that have emerged thus far. However, this property will need to be monitored in the future if serological assays are to be used to predict immunity, particularly as antibodies diversify in response to variant virus infection and, potentially, variant booster vaccination.
The need for serological assays in monitoring natural infection at the population level remains. Moreover, the introduction of vaccines raises new requirements for serological assays. These requirements include the distinction between individuals who were vaccinated and those who were naturally infected, the prognostication of levels of protection against infection and disease afforded by vaccines, and the identification of individuals for whom a boosting immunisation or monoclonal antibody therapy is indicated. Furthermore, in instances where countries are considering deploying so-called immunity passports to allow, for example, travel, the selection of assays and the diagnostic cutoff used can have important ramifications. Finally, since the measurement of antibody function (eg, neutralisation) at a population level is not practical, establishing the ability of serological assays to predict neutralisation and immunity will help determine correlates of protection that can be applied at a large scale and, perhaps, as part of routine clinical practice.
Data sharing
De-identified data will be available for sharing after manuscript publication upon request to the corresponding author.
Declaration of interests
SJ and EF received honoraria from Siemens for an online webinar in October, 2020, which was paid to their institutions. EF received sCOVG reagent from Siemens to support this study. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
We acknowledge the support of NRS BioResource and NHS Lothian Outpatients with the provision of this research study. We are grateful to Linfa Wang and GenScript for providing the cPass assay kits. This study received funding from the National Institutes of Health (R01AI78788 to TH, R01 AI50111 to PDB) and NRS BioResource (SR1407 to SJ).
Contributors
HW, SJ, TH, and PDB conceived and designed the study. FM did and analysed the neutralisation and cPASS assays and did cross-assay data analyses and data visualisation. HW, KT, and SJ acquired and analysed data using the serological assay platforms. HW and SJ did receiver operating characteristic curve and sensitivity analyses and respective visualisation. HW, SJ, and MS provided Abbott N assay data. EF provided data from Siemens assays. SM provided data from DiaSorin assays. IG and AB provided Abbott S IgGII Quant data. LJ and KMa provided EuroImmun assay data. CR and JM provided data from Roche assays. BB and KMc assisted with collection of research clinic samples. FZ and FS provided spike plasmids. TH wrote the manuscript with help from FM, PDB, and SJ, with input from all authors. All authors had access to all the data in the study and accept responsibility for publication. HW and FM verified the data.
Supplementary Material
References
- 1.GeurtsvanKessel CH, Okba NMA, Igloi Z, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Marca A, Capuzzo M, Paglia T, Roli L, Trenti T, Nelson SM. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online. 2020;41:483–499. doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Özçürümez MK, Ambrosch A, Frey O, et al. SARS-CoV-2 antibody testing-questions to be asked. J Allergy Clin Immunol. 2020;146:35–43. doi: 10.1016/j.jaci.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaebler C, Wang Z, Lorenzi JCC, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muecksch F, Weisblum Y, Barnes CO, et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853. doi: 10.1016/j.immuni.2021.07.008. 68.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Muecksch F, Schaefer-Babajew D, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luchsinger LL, Ransegnola BP, Jin DK, et al. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID-19 patients. J Clin Microbiol. 2020;58:e02005–e02020. doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muecksch F, Wise H, Batchelor B, et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. 2021;223:389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang MS, Case JB, Franks CE, et al. Association between SARS-CoV-2 neutralizing antibodies and commercial serological assays. Clin Chem. 2020;66:1538–1547. doi: 10.1093/clinchem/hvaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford KHD, Dingens AS, Eguia R, et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;223:197–205. doi: 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seow J, Graham C, Merrick B, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 16.Beaudoin-Bussières G, Laumaea A, Anand SP, et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. MBio. 2020;11:e02590–e02620. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tada T, Dcosta BM, Samanovic MI, et al. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. MBio. 2021;12 doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Y, Liu F, Xu X, et al. Durability of neutralizing antibodies and T-cell response post SARS-CoV-2 infection. Front Med. 2020;14:746–751. doi: 10.1007/s11684-020-0822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galipeau Y, Greig M, Liu G, Driedger M, Langlois MA. Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaney AJ, Starr TN, Gilchuk P, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 23.McCallum M, De Marco A, Lempp FA, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332. doi: 10.1016/j.cell.2021.03.028. 47.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerutti G, Guo Y, Zhou T, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819. doi: 10.1016/j.chom.2021.03.005. 33.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer MM, Tortorici MA, Park YJ, et al. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- 26.Widge AT, Rouphael NG, Jackson LA, et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N Engl J Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified data will be available for sharing after manuscript publication upon request to the corresponding author.