Abstract
Sphingomonas paucimobilis SYK-6 has the ability to transform a lignin-related biphenyl compound, 2,2′-dihydroxy-3,3′-dimethoxy-5,5′-dicarboxybiphenyl (DDVA), to 5-carboxyvanillic acid (5CVA) via 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA). In the 4.9-kb HindIII fragment containing the OH-DDVA meta-cleavage dioxygenase gene (ligZ), we found a novel hydrolase gene (ligY) responsible for the conversion of the meta-cleavage compound of OH-DDVA to 5CVA. Incorporation of 18O from H218O into 5CVA indicated there was a hydrolytic conversion of the OH-DDVA meta-cleavage compound to 5CVA. LigY exhibited hydrolase activity only toward the meta-cleavage compound of OH-DDVA, suggesting its restricted substrate specificity.
The complex aromatic polymer lignin comprises about 25% of the land-based biomass on earth, and its recycling is a vital component of the earth’s carbon cycle. The study of the biochemical and enzymatic processes involved in lignin biotransformation can supply a variety of catalytic reactions useful for the production of valuable aromatic chemicals. Lignin is composed of various intermolecular linkages between phenylpropanes, including guaiacyl, syringyl, and p-hydroxyphenyl, and contains biphenyl nuclei (2). Biphenyl linkages are one of the key connections between phenylpropane units. The biphenyl structure is so stable that its decomposition should be the rate-limiting step in lignin degradation.
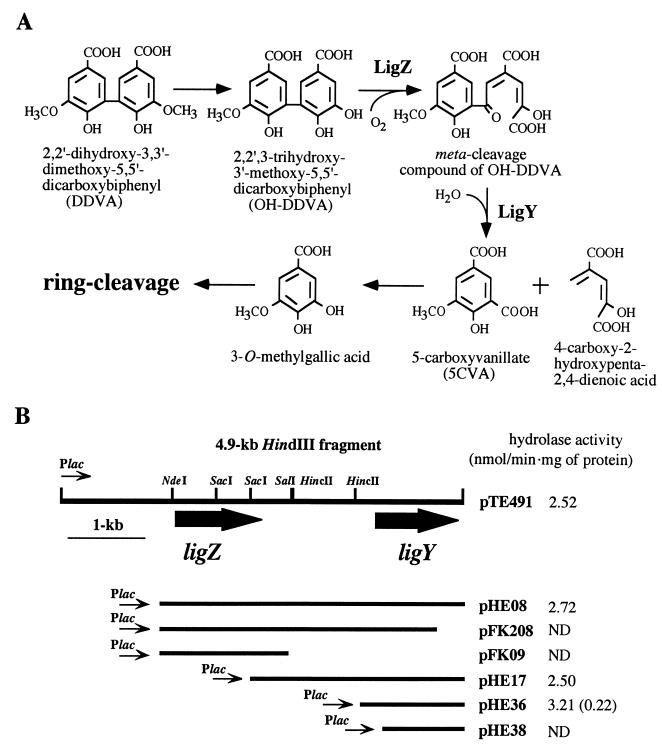
Sphingomonas paucimobilis SYK-6 was isolated from a kraft pulp effluent and degrades and grows on a lignin-related biphenyl compound, 2,2′-dihydroxy-3,3′-dimethoxy-5,5′-dicarboxybiphenyl (DDVA) (8). DDVA is transformed to a diol com-pound, 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA), by demethylation (7) (Fig. 1). A meta-ring cleavage of OH-DDVA is catalyzed by a meta-cleavage dioxygenase encoded by the ligZ gene, which resides in a 4.9-kb HindIII fragment of SYK-6 (16). A metabolite, 5-carboxyvanillic acid (5CVA), was observed during the degradation of DDVA by SYK-6, and is expected to be generated from a meta-ring cleavage compound of OH-DDVA.
FIG. 1.
(A) Proposed metabolic pathway for DDVA in S. paucimobilis SYK-6. LigZ, OH-DDVA dioxygenase; LigY, OH-DDVA meta-cleavage compound hydrolase. (B) Deletion analysis to locate the meta-cleavage compound hydrolase gene (ligY). The direction of transcription from the vector-located promoter (Plac) is depicted by a thin arrow. The large arrows represent the coding regions of ligZ and ligY genes. The hydrolase activities of E. coli strains containing each plasmid are presented on the right. ND, no product detected. The value in parentheses represents the activity obtained in the reaction in which the crude LigY was added 1 min after the incubation of LigZ with OH-DDVA.
In this study, we focused on the gene involved in the transformation of a meta-ring cleavage compound of OH-DDVA to elucidate the DDVA metabolic pathway in SYK-6. Hydrolysis of a meta-cleavage compound following meta-ring cleavage by a dioxygenase is a well-known metabolic sequence in aromatic compound metabolism, including those of toluene, xylene, naphthalene, and biphenyl (6, 9, 12, 13, 20). Here we characterized the ligY gene whose product catalyzes the hydrolysis of a meta-cleavage compound of OH-DDVA.
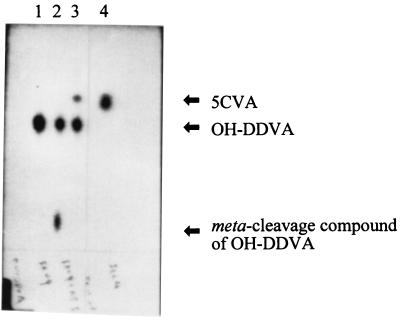
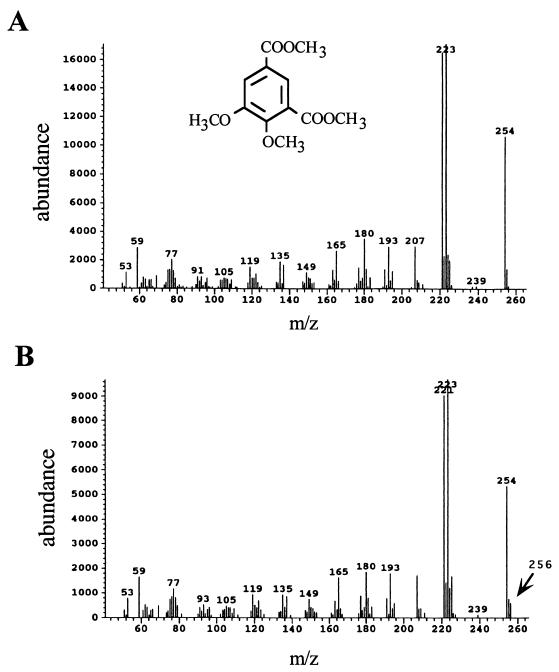
When OH-DDVA was incubated with the crude extract of Escherichia coli MV1190 cells containing plasmid pFK09 carrying the ligZ gene, a meta-cleavage compound of OH-DDVA accumulated (Fig. 2, lane 2), which showed an absorption maximum at 455 nm and presented the yellow color common to meta-cleavage compounds (3). When OH-DDVA was incubated with the crude extract of E. coli cells, which harbored pTE491 carrying the 4.9-kb SYK-6 HindIII fragment containing the ligZ gene and its adjacent region, OH-DDVA was converted to 5CVA. A 20-μl portion of 50 mM OH-DDVA was added to the crude extract (0.8 mg of protein) in 1 ml of 20 mM Tris-HCl (pH 7.5), and this reaction mixture was incubated for 10 min at 25°C. It was acidified with hydrochloric acid and extracted with 400 μl of ethyl acetate. The organic phase was dried in vacuo and dissolved in 20 μl of ethyl acetate. The resulting sample was analyzed by thin-layer chromatography (TLC) by using silica gel 60 F254 (E. Merck, Darmstadt, Germany). The developing solvent was chloroform-ethyl acetate-formic acid (10:8:3 [vol/vol/vol]). Compounds were visualized under UV light (at 254 nm). The metabolite 5CVA was provisionally identified by comparing the Rf value on TLC with that of authentic 5CVA (Fig. 2, lanes 3 and 4). Authentic 5CVA was synthesized according to the method of Profft and Krause (18). Its identification was confirmed by gas chromatography-mass spectrometry (GC-MS) analysis. The metabolite was methylated by using trimethylsilyldiazomethane (Wako Chemical Industries, Ltd., Tokyo, Japan). The resultant methyl ester was analyzed by GC-MS (model 5971A; Hewlett-Packard Co., Palo Alto, Calif.) by using an Ultra-2 capillary column (50 m by 0.2 mm; Hewlett-Packard Co.). The injection and detector temperatures were 250 and 280°C, respectively. The sample was chromatographed by using a temperature program which began at 60°C, was raised to 150°C at 20°C/min, and then was raised to 300°C at 3°C/min. The retention time and mass spectrum of the methyl ester are fully equivalent to those of authentic 5CVA (see Fig. 5). The mass spectrum of the methyl ester derivative of 5CVA had a molecular ion (M) at an m/z of 254 and a major fragment ion at an m/z of 223. Since the hydrolysis products of a meta-cleavage compound of OH-DDVA are presumed to be 5CVA and 4-carboxy-2-hydroxypenta-2,4-dienoic acid (Fig. 1A), these results indicated that the 4.9-kb SYK-6 HindIII fragment carrying the ligZ gene contains a meta-cleavage compound hydrolase gene. In these experiments, 4-carboxy-2-hydroxypenta-2,4-dienoic acid was not observed. It might have been metabolized in E. coli cells.
FIG. 2.
Thin-layer chromatogram of the reaction products of OH-DDVA with E. coli crude extracts. Lanes: 1, synthetic OH-DDVA; 2, E. coli (pFK09); 3, E. coli (pTE491); 4, synthetic 5CVA.
FIG. 5.
(A) The mass spectrum of the methyl ester derivative of the product 5CVA. The inset shows the chemical structure of the 5CVA methyl ester derivative. (B) The mass spectrum of the methyl ester derivative of 5CVA produced in the presence of H218O. The molecular ion that originated from 18O-containing 5CVA is indicated by an arrow.
A series of deletion derivatives of the 4.9-kb HindIII fragment of pTE491 were constructed to limit the region encoding an OH-DDVA meta-cleavage compound hydrolase by using restriction enzymes and E. coli exonuclease III (Takara Shuzo Co., Ltd., Kyoto, Japan). The production of 5CVA from OH-DDVA was then catalyzed by sequential actions of LigZ, and an OH-DDVA meta-cleavage compound hydrolase was examined. The crude extracts of deletion clones and a ligZ recombinant clone (0.8 mg of protein each) were incubated with OH-DDVA, and the 5CVA formed from OH-DDVA was evaluated by GC-MS, as described above. A deletion clone, pHE36, contained the minimum fragment which conferred hydrolase activity toward the meta-cleavage compound of OH-DDVA (Fig. 1B). The 1.3-kb insert in pHE36 was subjected to nucleotide sequencing. The determination of the nucleotide sequence was performed by the dideoxy termination method (19) with an ALFred DNA sequencer (Pharmacia, Milwaukee, Wis.). Analysis of nucleotide sequence was done with GeneWorks software (Intelligenetics, Inc., Mountain View, Calif.).
A 996-bp open reading frame (ORF) was found in the 1.3-kb insert (Fig. 3). The 5′- and 3′-terminal parts of this ORF were deleted in pHE38 and pFK208, respectively, both of which lacked the hydrolase activity (Fig. 1B). These results indicated that this ORF encodes an OH-DDVA meta-cleavage compound hydrolase. This ORF was designated ligY. The ligY gene encoded a protein of 332 amino acid residues, whose molecular mass was estimated to be 37,280 Da. Its G+C content was 63%, which was almost equivalent to those of the proteins encoded by the other lignin-degradative genes of SYK-6 (11, 14–16). A putative ribosome binding sequence, ′AAGGGGA′, was present in the upstream region of the start codon for ligY (Fig. 3). The deduced amino acid sequence of ligY showed no similarity to those of other aromatic compound hydrolases involved in benzene, toluene, xylene, and biphenyl metabolism, and there was little identity with previously reported enzymes, indicating that ligY encodes a novel aromatic compound hydrolase. These aromatic hydrolases contain a Gly-X-Ser-X-Gly motif constituting an active site, which is shared by serine hydrolases (1, 5). The deduced LigY amino acid sequence did not contain this motif, suggesting that LigY does not belong to serine hydrolase family. In addition, LigZ has little similarity to the deduced amino acid sequences of meta-ring cleavage dioxygenases for benzene, toluene, xylene, and biphenyl degradation. The ligZ and ligY genes seem to have evolved from separate ancestors of the genes coding for meta-ring cleavage dioxygenases and meta-cleavage compound hydrolases for benzene, toluene, xylene, and biphenyl degradation.
FIG. 3.
Nucleotide and deduced amino acid sequences of the hydrolase gene (ligY) from S. paucimobilis SYK-6. The putative ribosome binding sequence (RBS) for ligY is underlined. A stop codon is indicated by an asterisk. The deduced amino acid sequence of ligY is presented below the nucleotide sequence.
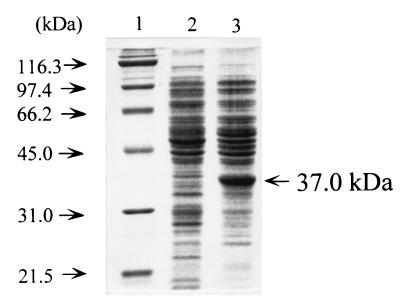
The LigY hydrolase was overproduced in E. coli MV1190 under the control of the lac promoter in pHE36. A 37-kDa polypeptide was found by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10) with 12% (wt/vol) polyacrylamide, and its molecular mass is in good agreement with that estimated from the deduced amino acid sequence of LigY (Fig. 4).
FIG. 4.
SDS-PAGE of LigY hydrolase produced in E. coli. Proteins (10 μg each) were separated on an SDS–12% PAGE gel and stained with Coomassie brilliant blue. Lanes: 1, molecular mass standard proteins; 2, E. coli MV1190(pBluescript II KS+); 3, E. coli MV1190(pHE36).
When the crude LigY prepared from the E. coli cells harboring pHE36 was added together with the crude LigZ to the reaction mixture containing OH-DDVA, OH-DDVA was transformed to 5CVA. However, the crude LigY was added 1 min after the reaction of LigZ with OH-DDVA, meta-cleavage compound of OH-DDVA remained, and a small amount of 5CVA was produced (Fig. 1B). These results suggested that the meta-cleavage compound of OH-DDVA would be so unstable that the sequential actions of LigZ and LigY are required. The close interaction between LigZ and LigY might be needed for the efficient transformation of OH-DDVA to 5CVA. The close physical association between meta-cleavage pathway enzymes has been reported. Aldehyde dehydrogenase (acylating) is associated with the preceding enzyme, 4-hydroxy-2-ketovalerate aldolase, in the pathway from Pseudomonas sp. strain CF600 (17), and 2-oxopent-4-enoate hydratase is associated with the preceding enzyme, 4-oxalocrotonate decarboxylase, from Pseudomonas putida (4). In the latter case, the close association between two enzymes is supposed to ensure efficient transformation of the unstable intermediate by avoiding the conversion of the enol form to the keto form. Assuming a conversion between the enol and keto forms of the meta-cleavage compound of OH-DDVA, the association between LigZ and LigY seems to be advantageous. Further investigations are needed to address this notion.
We examined the activity of LigY on the meta-cleavage compounds formed during benzene, toluene, and biphenyl degradation. The substrates were enzymatically produced from catechol, 3-methylcatechol, and 2,3-dihydroxybiphenyl by using 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) of Pseudomonas sp. strain KKS102 (9). The crude enzyme preparations of BphC, LigZ, and LigY were added to the reaction mixture containing 100 μM substrate in 1 ml of 20 mM Tris-HCl (pH 7.5), and this mixture was then incubated for 30 min at 25°C. The absorbance spectrum of this reaction mixture was measured with a DU-640 spectrophotometer (Beckman Instruments, Inc., Fullerton, Calif.) to evaluate the transformation of a meta-cleavage compound. LigY did not exhibit hydrolase activity toward any of these meta-cleavage compounds, suggesting that the substrate specificity of LigY was restricted.
To confirm the hydrolysis reaction of a meta-cleavage compound of OH-DDVA catalyzed by LigY hydrolase, the incorporation of 18O into 5CVA from H218O was examined. The reaction mixture of crude LigZ and LigY containing 9% H218O (Aldrich Chemical Company, Milwaukee, Wis.) was incubated with OH-DDVA for 3 h at 25°C, and the methyl ester derivative of metabolite 5CVA was analyzed by GC-MS. A molecular ion peak at an m/z of 256 was observed and is specific to the reaction with H218O (Fig. 5). Its abundance is about 8% of the molecular ion at an m/z of 254, which is mostly equivalent to the proportion of H218O in the reaction mixture. These results indicate that this molecular ion originated from 5CVA, in which 18O was incorporated from H218O, and prove the hydrolysis of a meta-cleavage compound of OH-DDVA catalyzed by LigY.
Nucleotide sequence accession number.
The nucleotide sequence of ligY has been deposited in the DDBJ, EMBL, and GenBank sequence databases under accession no. AB018415.
REFERENCES
- 1.Ahmad D, Fraser J, Sylvestre M, Larose A, Khan A, Bergeron J, Juteau J M, Sondossi M. Sequence of the bphD gene encoding 2-hydroxy-6-oxo-(phenyl/chlorophenyl)hexa-2,4-dienoic acid (HOP/cPDA) hydrolase involved in the biphenyl/polychlorinated biphenyl degradation pathway in Comamonas testosteroni: evidence suggesting involvement of Ser112 in catalytic activity. Gene. 1995;156:69–74. doi: 10.1016/0378-1119(95)00073-f. [DOI] [PubMed] [Google Scholar]
- 2.Freudenberg K. The constitution and biosynthesis of lignin. In: Neish A C, Freudenberg K, editors. Constitution and biosynthesis of lignin. New York, N.Y: Springer-Verlag; 1968. pp. 47–122. [Google Scholar]
- 3.Furukawa K, Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harayama S, Rekik M, Ngai K-L, Ornston L N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofer B, Eltis L D, Dowling D N, Timmis K N. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/chlorinated biphenyl degradation. Gene. 1993;130:47–55. doi: 10.1016/0378-1119(93)90345-4. [DOI] [PubMed] [Google Scholar]
- 6.Horn J M, Harayama S, Timmis K N. DNA sequence determination of the TOL plasmid (pWW0) xylGFJ genes of Pseudomonas putida: implications for the evolution of the aromatic catabolism. Mol Microbiol. 1991;5:2459–2474. doi: 10.1111/j.1365-2958.1991.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 7.Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, Haraguchi T. The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6) FEBS Lett. 1988;233:129–133. [Google Scholar]
- 8.Katayama Y, Nishikawa S, Nakamura M, Yano K, Yamasaki M, Morohoshi N, Haraguchi T. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi. 1987;33:77–79. [Google Scholar]
- 9.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masai E, Sugiyama K, Iwasita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukuda M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 13.Menn F-M, Zylstra G J, Gibson D T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase in Pseudomonas putida F1. Gene. 1991;104:91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 14.Nishikawa S, Sonoki T, Kasahara T, Obi T, Kubota S, Kawai S, Morohoshi N, Katayama Y. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl Environ Microbiol. 1988;64:836–842. doi: 10.1128/aem.64.3.836-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda Y, Nishikawa S, Shiozuka K-I, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng X, Egashira T, Hanashiro K, Masai E, Nishikawa S, Katayama Y, Kimbara K, Fukuda M. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol. 1998;64:2520–2527. doi: 10.1128/aem.64.7.2520-2527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powlowski J, Sahlman L, Shingler V. Purification and properties of the physically associated meta-cleavage pathway enzymes 4-hydro-2-ketovalerate aldolase and aldehyde dehydrogenase (acylating) from Pseudomonas sp. strain CF600. J Bacteriol. 1993;175:377–385. doi: 10.1128/jb.175.2.377-385.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Profft E, Krause W. Über die chlormethylierung des o- und novo-vanillins und die gewinnung von 4-hydroxy-5-alkoxyisophthalaldehyden. Arch Pharm. 1964;298:148–162. doi: 10.1002/ardp.19652980303. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schell M A. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983;153:822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]