Abstract
Over the past year, the AI community has constructed several deep learning models for diagnosing COVID-19 based on the visual features of chest X-rays. While deep learning researchers have commonly focused much of their attention on designing deep learning classifiers, only a fraction of these same researchers have dedicated effort to including a segmentation module in their system. This is unfortunate since other applications in radiology typically require segmentation as a necessary prerequisite step in building truly deployable clinical models. Differentiating COVID-19 from other pulmonary diseases can be challenging as various lung diseases share common visual features with COVID-19. To help clarify the diagnosis of suspected COVID-19 patients, we have designed our deep learning pipeline with a segmentation module and ensemble classifier. Following a detailed description of our deep learning pipeline, we present the strengths and shortcomings of our approach and compare our model with other similarly constructed models. While doing so, we focus our attention on widely circulated public datasets and describe several fallacies we have noticed in the literature concerning them. After performing a thorough comparative analysis, we demonstrate that our best model can successfully obtain an accuracy of 91 percent and sensitivity of 92 percent.
Keywords: Coronavirus, COVID-19, Convolutional neural network, Deep learning, Chest X-ray, Computer vision
1. Introduction
The artificial intelligence (AI) research community has recently invested considerable time and resources into developing deep learning models based on chest radiographs for the purpose of diagnosing coronavirus disease 2019 (COVID-19). Many medical institutions are finding themselves in difficult positions when faced with countless numbers of patients presenting with symptoms of the illness. There is a need for new diagnostic models to alleviate this important need. Recently deep learning techniques have come to permeate “the entire field of medical image analysis” [1]. With deep learning methodologies, AI researchers have made considerable progress in improving the quality of automated diagnostic medical imaging systems. Because of their pioneering work, many promising directions are now opening up that could potentially help diagnose COVID-19.
There are several kinds of COVID-19 tests that are currently on the market. Molecular tests (polymerase chain reaction tests), Antigen tests (rapid tests), and antibody tests (blood tests) have seen widespread use. Of these three tests, the real-time reverse transcription-polymerase chain reaction (RT-PCR) test is considered the present gold standard for diagnosing COVID-19 [2]. RT-PCR tests are not perfect however and reports have been made considering problems with the tests overall sensitivity [3]. Luo et al. [4] in a study including 4653 participants found that RT-PCR tests have a sensitivity of around 71%. Kucirka et al. [5] in a Johns Hopkins study reported that an RT-PCR test’s sensitivity has wide variability over the 21 days after a patient is first exposed. They also noted that “although the false-negative rate is minimized 1 week after exposure, it remains high at 21%” [5]. Kucirka et al. [5] therefore ultimately found that it takes about a week from the time of symptom onset, for RT-PCR testing to deliver the lowest false-negative rate. This leaves room for other tests that may work better over the time that RT-PCR tests are less accurate. Radiological testing is a leading contender in the research community for such a scenario. Research has been shown it to be useful over the time that a patient has obtained a negative RT-PCR test [6]. It can therefore be used in conjunction with other tests and possibly give more clarity regarding a patients current diagnosis.
Many researchers have focused on using computerized tomographic (CT) scanners in diagnosing COVID-19 because of their ability to analyze three-dimensional information. As a modality for COVID-19 testing, however, CT scanners are expensive resources to employ. For a system to be practical during a pandemic, a cheaper and faster solution needs to be available to deal with the sheer number of patients waiting for a test. Chest X-rays (CXRs) are the other alternative modality typically employed by radiologists in imaging thoracic illnesses such as COVID-19. Some advantages of chest X-rays for this particular application include the portability of an X-ray scanner, the requirement of only cleaning a single surface when reusing it on patients, the speed of the diagnostic measurements required, and the overall expense of the procedure. Given these significant advantages, it is entirely practical for researchers to explore the use of X-ray technology in COVID-19 testing.
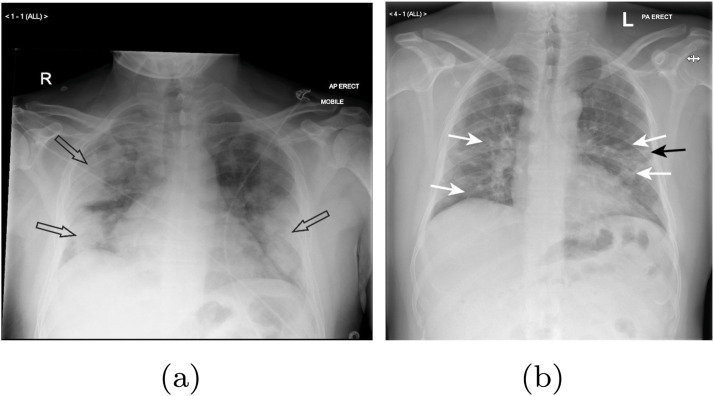
Before discussing how a proposed deep learning pipeline can diagnose COVID-19 in suspected patients, we first need to understand the features in a patients lungs that require imaging. Rousan et al.[7] in a study involving 88 patients, found that ground-glass opacities (GGO) were the most frequent finding in COVID-19 X-rays. The chest X-rays of normal patients generally show a black background within a patients lungs. In chest X-rays with GGOs, radiologists find lighter colored patches of haziness that are indicative of a possible pathology. Rousan et al.[7] also found that consolidation increases in severity in the X-rays of many COVID-19 patients up until approximately the second week of the illness. This aligns well with another study performed by Song et al. [8] who found that consolidations do indeed increase as the disease progresses. Consolidation in radiography represents areas of a patient’s lungs that are filled with extraneous liquids (pus, blood, and water) and solid materials (stomach contents or cells) that do not exist in healthy lungs. In comparing the number of COVID-19 X-rays with consolidation vs. GGOs, consolidation tends to occur less frequently. It is still, however, the second most frequent visual cue mentioned in the radiological literature. Fig. 1 shows the chest X-rays of two older patients with COVID-19 showing the aforementioned symptoms.
Fig. 1.
Lungs of 2 older COVID-19 patients revealing (a) bilateral consolidation and (b) ground glass opacities (white arrows) and linear opacity (black arrow) [7].
Many deep learning X-ray studies up until now have solely focused on classification in diagnosing COVID-19 in X-rays. While excellent research has occurred in this space, the number of articles dealing with COVID-19 X-ray segmentation has been quite limited. Segmentation is an important preprocessing technique that can shield a classifier from unnecessary pixel information when categorizing an image. In this way, many imaging-based studies in other computer vision applications have found that proper segmentation has increased the overall accuracies of their classifiers [9], [10], [11]. It is vital, therefore, to employ segmentation when training a COVID-19 classifier. The following lists the main contributions of our work:
-
•
Our pipeline employs an advanced segmentation network (ResUnet [12])
-
•
We have made available a COVID-19 X-ray classification dataset that is larger than all similar datasets we have found in the literature
-
•
Our overall pipeline makes use of majority voting and weighted average ensembles
-
•
We have included a thorough comparative analysis that benchmarks our model’s performance against other deep learning models in the literature
Our work begins in Section 2 with an overview of various research studies that have constructed segmentation-classification deep learning pipelines to diagnose COVID-19. In Section 3, we thereafter present our proposed deep learning pipeline’s architecture, showing the internal details of our segmentation and classification modules. Following a discussion of our pipeline’s architecture, in Section 4 we present the experimental results of our overall system. In Section 4, we additionally present a detailed comparative analysis of our pipeline versus other well-constructed models in the literature. Concluding in Section 5, we discuss potential future directions for this research.
2. Related works
There are many papers in the literature that use deep learning classification and segmentation for making medical predictions [13], [14], [15], [16], [17]. Our main focus in this review, however, is on COVID-19 X-ray articles that combine a segmentation unit and classifier [18], [19], [20], [21], [22], [23], [24], [25], [26]. We did so in order to see how our deep learning pipeline compares with the studies that are the most related to our own. There are several public datasets available in circulation for segmenting chest X-rays that have been cited in the articles below. There are also a number of public and private datasets mentioned in these articles that were prepared specifically for COVID-19 classification. The following works below are all studies that influenced how we ultimately implemented our final system.
Rajaraman et al. [18] created a segmentation – classification deep learning pipeline to diagnose COVID-19 that included an ensemble of iteratively pruned CNNs. The authors trained several CNN models (VGG-16/VGG-19 [27], Inception-V3 [28], Xception [29], DenseNet-201 [30], etc.) after their dataset had been preprocessed by a U-Net [31] segmentation module that included a Gaussian dropout layer [32]. The authors of this paper tried to employ many different ensemble strategies and, in the end, found that weighted averaging produced the best results. The authors of this paper unfortunately listed Kermany et al.’s [33] dataset as being contained in their dataset which likely contributed to exaggerated evaluation metrics. It is incorrect to bias a dataset with only certain categories of the dataset having images of children’s lungs.
Alom et al. [19] designed an X-ray-based system that diagnoses COVID-19 with a NABLA-N segmentation network [34] and an Inception Residual Recurrent Convolutional Neural Network (IRRCNN). Their X-ray model is initially trained on a normal vs. pneumonia dataset first as more images are in the public sphere for making such a comparison. After obtaining acceptable performance on this separate task, they fine-tune their model on a smaller COVID-19 dataset. This segmentation-classification pipeline ultimately achieves a final test accuracy of 84.67 percent. The authors of this paper, unfortunately, used Paul Mooney’s chest X-ray dataset on Kaggle [35] to obtain pneumonia and normal images for training their classifiers. This contains images from Kermany et al.’s dataset [33] of children’s lungs. Their classifier was intended for identifying COVID-19 in adult lungs. Training a classifier with children’s lungs that is intended for adult lungs is incorrect, however, and caused Alom et al.’s [19] classifier to be biased. They used normal images from children but COVID-19 images from adults in their dataset. Their normal vs. COVID-19 classifier, therefore, incorrectly could use the features of adult lungs to identify COVID-19.
Yeh et al. [20] combined several public datasets as well as datasets from several private medical institutions when training their segmentation-classification pipeline. Unlike the two previous studies, the authors of this work look like they have constructed an unbiased dataset. They do, however, reference several private datasets that are unavailable to the research community. It is therefore impossible to directly compare our pipeline against their work. They initially trained a U-Net segmentation model [31] as a preprocessing step to exclude non-informative regions of CXRs from their model. Yeh et al. [20] trained this segmentation unit on the Montgomery County X-ray Set and the Shenzhen Hospital X-ray Set [36]. After training their segmentation unit, they obtained a dice similarity coefficient (DSC) of 88 percent. Following this preprocessing step, they trained a DenseNet-121 [30] classifier on segmented images and obtained a COVID-19 sensitivity of 83.33% on their validation set. Their hold-out test set contained 306 COVID-19 images and their final COVID-19 sensitivity on this test set corrected to 81.8 percent.
Horry et al. [37] developed a segmentation–classification deep learning pipeline for diagnosing COVID-19 that was trained and tested on a relatively small preprocessed dataset. While Horry et al.’s [37] final curated dataset was not biased, it contained only 100 COVID-19 images, so it is difficult to ultimately know how well their work would translate to a larger number of images. Horry et al. [37] additionally removed images from their dataset which contained features they believed their model would have difficulty classifying. The authors’ segmentation model was not based on a deep learning model. They simply used OpenCV’s GrabCut function and reasoned that “that the lung area could be considered the foreground of the X-ray image” [37]. After preprocessing they trained five base models with their segmented images (VGG-16 [27], VGG-19 [27], Inception-V3 [28], Xception [29], and ResNet-50 [38]). Their best base model (VGG-19 [27]) ultimately achieved an F1-score of 81 percent.
Wehbe et al.’s [21] published deep learning pipeline that was trained on the largest COVID-19 X-ray dataset we have found reported in the literature. The authors developed their pipeline by working in collaboration with a private US medical institution. Their large classification dataset is therefore inaccessible to the public at this time. This dataset also appears to have not been improperly biased with the inclusion of incorrect data. The authors were aware of the need to divide their training and test sets by patient number. The authors chose to train their U-Net-based segmentation module [31] on the Montgomery [36] and JSRT [39] datasets. Wehbe et al. [21] in their study also created an ensemble model to detect COVID-19. Their final model contained a weighted average of 6 popular CNNs (Inception [28], Inception-ResNet [40] Xception [29], and ResNet-50 [38], DenseNet-121 [30], and EfficientNet-B2 [41]). An important reason to include this paper in our discussion is that the authors managed to perform an interesting study that up until now we have not seen reproduced elsewhere. The authors commissioned a study involving five radiologists to determine the effectiveness of experts in the field in differentiating COVID-19 from other illnesses. This is important when trying to approximate Bayes error prior to building a deep learning model. Wehbe et al.’s [21] compared the results of their model with the performance of expert radiologists and discovered their model to a minor extent outcompetes them. Their final binary weighted average model obtained a final accuracy of 82% on their test set. The expert radiologists manually obtained a consensus accuracy of 81% on the same images. These final results coincided very nicely with one another.
Tabik et al. [22] created a dataset dubbed the “COVID-GR-1.0” dataset which was used in training their “COVID-SDNet” model in diagnosing COVID-19. Their dataset was divided in a novel fashion whereby COVID-19 positive patients were subdivided into four risk categories (normal-PCR+, mild, moderate, and severe). The authors created this dataset to see how many of weak COVID-19 cases would be analyzed by a prospective classifier correctly. More often than not, in COVID-19 datasets, there is an unequal number of severe COVID-19 patients. Typically, patients who end up undergoing a radiological examination end up being patients experiencing increased complications. COVID-GR-1.0 is a small but well-curated dataset that has utility in that it can be employed to determine a classifier’s efficacy on weak COVID-19 images. Tabik et al.’s [22] pipeline consisted of a segmentation module and a classification module that performs “inference based on the fusion of CNN twins.” [22] The authors used a U-Net [31] segmentation module and trained it on the Montgomery County X-ray dataset [36], the Shenzhen Hospital X-ray datasets [36] and the RSNA Pneumonia CXR challenge dataset [42]. They calculated the smallest rectangle around each segmented image and added a border containing 2.5% of the pixels around each rectangle to obtain their final masked images. The X-rays they segmented were, therefore, never fully masked. The authors did not want to exclude relevant information in these images that could contain useful diagnostic information. After performing binary classification on their segmented COVID-GR-1.0 dataset, Tabik et al.’s [22] classifier obtained a COVID-19 sensitivity of 72.59%.
Teixeira et al. [23] designed a segmentation–classification pipeline used to diagnose COVID-19 that consisted of a U-Net [31] and InceptionV3 [28] CNN. Their U-Net [31] segmentation module was trained on images and masks that were hand-picked from a mixture of public datasets [36], [39], [43]. The number of images and mask pairings they chose in the Darwin V7 labs [43] segmentation dataset (489) was significantly lower than the total number of pairings available in that dataset (6504). This approach looks as though it allowed them to train their U-Net [31] to have a higher dice similarity coefficient (0.982) than other segmentation units we have seen in the literature for this task. For classification they otherwise used the RYDLS-20 dataset [44]. They had developed this dataset in a previous work and further added images to it to create a new “RYDLS-20-v2” dataset. They attempted to use several classifiers but ultimately found that using an InceptionV3 [28] CNN resulted in giving them their best overall multiclass performance metrics.
Oh et al. [24] published a novel “patch-based deep neural network architecture with random patch cropping” [24] for detecting COVID-19. Their model initially begins with a preprocessing step whereby a fully convolutional DenseNet-103 segments incoming chest X-rays. The authors thereafter use a ResNet-18 on the segmented images for classification. The authors generate 100 randomly cropped patches from the previously segmented chest X-rays and feed those patches through ResNet-18s as well. In this process, the authors have selected a sufficient number of lung patches to ensure that the entire surface area of the segmented lungs is covered. The authors of this paper unfortunately selected images from Kermany et al. [33] to include in their work and thereby biased their classifier.
Abdullah et al. [25] implemented a segmentation – classification pipeline that used a unique segmentation unit and ensemble model for classification. Their segmentation unit, the Res-CR-Net, is a new kind of segmentation model the authors introduced in a previous study [45] that does not contain the same encoder-decoder structure that the popular U-Net [31] contains. According to the authors, the Res-CR-Net [45] “combines residual blocks based on separable, atrous convolutions [46], [47] with residual blocks based on recurrent NNs [48] ” Abdallah et al. [45]. The authors trained their Res-CR-Net [45] on several open-source sets of masks and images [36], [39], [43]. They acquired their classification dataset from the Henry Ford Health System (HFHS) hospital in Detroit. This private dataset contained 1417 COVID-negative patients and 848 COVID-positive patients. The authors used this dataset to train a unique hybrid convnet called the “CXR-Net” that contains a Wavelet Scattering Transform (WST) block [49], [50], an attention block containing two MultiHeadAttention layers [51], [52], and several convolutional residual blocks. This segmentation-classification pipeline ultimately achieved an accuracy of 79.3% and an F1 Score of 72.3% on their test set.
Wang et al. [26] created a deep learning segmentation - classification pipeline for COVID-19 detection and severity assessment. After a CXR standardization module the authors included a common thoraic disease module that was used to determine whether a patient is suffering from pneumonia. This is followed by segmentation and classification modules. Wang et al.’s [26] lung segmentation network was trained on X-ray scans that were resized to images. They chose to use a DeepLabv3 segmentation architecture [47] after additionally training a U-Net [31] and Fully Convolutional Network [53]. Their DeepLabv3 segmentation architecture in the left lung field obtained a DSC of 0.873, in the right lung field obtained a DSC of 0.910, in the periphery of the left lung field obtained a DSC of 0.864, and in the periphery of the right lung field obtained a DSC of 0.893. Across all categories this averages out to a total DSC of 0.885. Following this segmentation operation the authors performed COVID-19 detection and severity assessments. During training their COVID-19 detection module was trained on 1407 COVID-19 X-rays, 5515 viral pneumonia X-rays and 10,961 “other” pneumonia X-rays. They evaluated their model on a test set with 164 COVID-19 CXRs and 630 other pneumonia CXRs. In the task of differentiating between COVID-19 and other X-rays they ultimately obtained an accuracy of 91% and a COVID-19 sensitivity of 92%.
3. Proposed network architecture
3.1. Segmentation dataset
To train our segmentation model, we looked at the datasets used in our literature review and decided to use the Darwin V7 Labs dataset[43]. We opted in favor of this dataset for three reasons. The first reason was its overall size. The Darwin V7 Labs dataset [43] is significantly larger (6504 images/masks) than most lung segmentation datasets. This being the case, we were able to train a robust segmentation unit that could accurately operate on a wide range of chest X-rays. Our second reason for using the dataset involved considerations involving the regions of the chest X-rays that its masks cover. Most masks in popular lung segmentation datasets include only the lungs. The Darwin V7 Labs [43] masks, however, included space next to the lungs. This left room for the heart to not be excluded. Initially, we did not give the heart and its size any consideration. Eventually, we came to realize, however, that cardiomegaly (an enlarged heart) is found in 29.9% of COVID-19 patients [54]. This symptom would not show up with most general-purpose lung segmentation masks. Our third reason for using the Darwin V7 Labs dataset [43] was that its masks were created for patients with a variety of conditions. Some masks were created for normal patients and others were created for patients exhibiting a variety of lung pathologies including COVID-19, bacterial pneumonia, viral pneumonia, Pneumocystis pneumonia, fungal pneumonia, and Chlamydophila pneumonia.
Some preprocessing was required on the Darwin V7 labs dataset [43] to create a model that operated correctly on the segmentation unit we later created. The segmentation unit we chose for this study was a ResUnet [12], and this segmentation unit was designed for images/masks. We needed to perform some data wrangling using the JSON files that were included with the dataset to ensure that images smaller than were excluded. The JSON files provided with the Darwin V7 Labs dataset [43] had a field indicating which kind of X-ray each image was. We, therefore, were able to automate a process whereby we removed all of the lateral X-rays that were sparsely hidden throughout the dataset. Our dataset, therefore, solely contained posteroanterior (PA) X-rays. After preprocessing, we were left with 6377 masks/image pairings. We finally divided this preprocessed Darwin V7 Lab dataset [43] into the 80% training / 20% validation split shown in Table 1 .
Table 1.
Number of images/masks in the preprocessed Darwin V7 labs dataset [43].
| Number of image/mask pairings | |
|---|---|
| V7 Labs preprocessed training set | 5102 |
| V7 Labs preprocessed test set | 1275 |
3.2. Classification datasets
In medical imaging, the ability of a model to generalize to new examples typically is limited by the size of the training set. Because research into imaging COVID-19 is relatively recent, there is only approximately a year’s worth of images that have been collected for classification purposes. For this reason, most published studies cannot present a model that can be deployed in a clinical setting. This study is no different, although in the work presented here we have taken significant steps forward in remediating several mistakes we have witnessed in the datasets of most papers.
When we first started gathering data, we initially realized that publicly available datasets generally have very little metadata available. That being the case, we decided to build a classifier that works on images alone. While doing so, we came to realize that the classification datasets in many studies have been incorrectly assembled. The majority of papers that have focused on differentiating COVID-19 from similar illnesses have cited using Kermany et al.’s [33] images in their dataset. As we have previously mentioned in our related works section, this dataset is composed of children that are suffering from various forms of bacterial and viral pneumonia. Since the lungs of small children have different features than adult lungs, we realized these images should not be included in our final classification dataset. This dataset likely poses more of a problem in biasing classifiers that are trained on nonsegmented images. The bones of adults are fused and the bones of children are not fused. This is feature can easily be picked up by a CNN. Kermany et al.’s [33] dataset, however, still would pose an issue even with a segmentation unit as the spatial features of adult lungs would differ from those of children’s lungs. The classifiers in studies that include this dataset, therefore, can pick up features both internal and external to the lungs that are inconsistent between adults’ and childrens’ lungs. This has, unfortunately, lead to the unfair biasing of several COVID-19 classifiers in the literature.
Another difficulty facing many studies is the lack of metadata accompanying images. At least some metadata is required alongside images to ensure that X-rays from individual patients do not get mixed in the training and test/validation sets. This problem of data leakage, we believe, is an issue in some studies we have reviewed. We find it disconcerting that most studies do not mention how they ensured the separation of patients’ X-ray scans between training and test sets. An enthusiasm surrounding finding the most images possible has resulted in a large number of images being harvested from medical research papers. Wang et al. [55] last year released a popular ‘COVIDx5’ dataset [55] that has been able to avoid this pitfall. They also did not include Kermany et al.’s dataset [33] in their COVIDX dataset [55], and improperly bias their classifier which many studies have done. We additionally used this dataset because it was larger than many existing datasets and included 14,258 CXR images. In total, this consisted of 617 COVID-19 images, 8066 normal images, and 5575 pneumonia images.
We added more COVID-19 images to the COVIDx5 dataset [55] because of the large COVID-19 class imbalance that existed within it. We hoped it would help to reduce overfitting in our classifier. We therefore added 922 COVID-19 images from the MIDRC-RICORD-1C database [56] and 2474 images from the BIMCV dataset [57]. In total, we constructed a dataset that contains 4013 COVID-19 images, 8066 normal images, and 5445 pneumonia images. The images from the COVIDx5 dataset [55] had the necessary metadata needed to allow us to split these images into three sets (80% training/ 10% validation/ 10%test) without creating data leakage. The MIDRC-RICORD-1C dataset [56] and BIMCV dataset [57] were released long after the COVIDx5 dataset [55], and none of these datasets had any relation with one another. It was therefore possible to split the COVID-19 images within these datasets into three sets without creating data leakage between them. The BIMCV [57] COVID-19 images were entirely used in the training set and the COVIDx5 [55] COVID-19 images were entirely split evenly between the validation and test set. The MIDRC-RICORD-1C [56] COVID-19 images were used in all three sets. The MIDRC-RICORD-1C [56] images came with metadata. Fortunately, the metadata allowed us to be able to divide the images from the MIDRC-RICORD-1C [56] dataset by patient between our training and validation/test sets. In this way we were able to create the datasets shown in Tables 2 and 3 . We created both multiclass (3-class) and binary datasets to later compare our segmentation-classification pipeline with models that are reported in various other papers. It was important to produce our large COVID-19 dataset with both validation and test sets to help mitigate concerns that have been brought up by Wehbe et al. [21] concerning overfitting.
Table 2.
Number of images in our multiclass training and test sets.
| COVID-19 | Normal | Pneumonia | |
|---|---|---|---|
| Multiclass Training Set | 3209 | 7262 | 4771 |
| Multiclass Validation Set | 402 | 402 | 402 |
| Multiclass Test Set | 402 | 402 | 402 |
Table 3.
Number of images in our binary training and test sets.
| COVID-19 | Non-COVID-19 | |
|---|---|---|
| Binary Training Set | 3209 | 12033 |
| Binary Validation Set | 402 | 402 |
| Binary Test Set | 402 | 402 |
In addition to the above dataset that we created, we also directly tested our model on another dataset that was used in Tabik et al.’s [22] study. We wanted to test our segmentation-classification against Tabik et al.’s [22] pipeline because their model worked on many of the same principles ours did. Their model used a segmentation algorithm that leaves more pixels surrounding the lungs in the images they segment. It has been difficult to find segmentation-classification pipelines like our own with unbiased and correctly constructed datasets. We were unable to find a study to directly compare ourselves against that uses a segmentation-classification pipeline and has a larger public dataset. Tabik et al.’s [22] study used a very conservative dataset that was meant to measure the performance of a deep learning model on weaker COVID-19 cases. Their “COVID-GR-1.0” binary dataset has 426 COVID-19 patients and 426 normal patients. The authors originally split this dataset into a 80% training / 20% test split. The dataset split in this format is shown in Table 4 .
Table 4.
Number of images in the COVID-GR-1.0 training and test sets [22].
| COVID-19 | Normal | |
|---|---|---|
| COVID-GR-1.0 Training Set | 340 | 340 |
| COVID-GR-1.0 Test Set | 86 | 86 |
3.3. System design
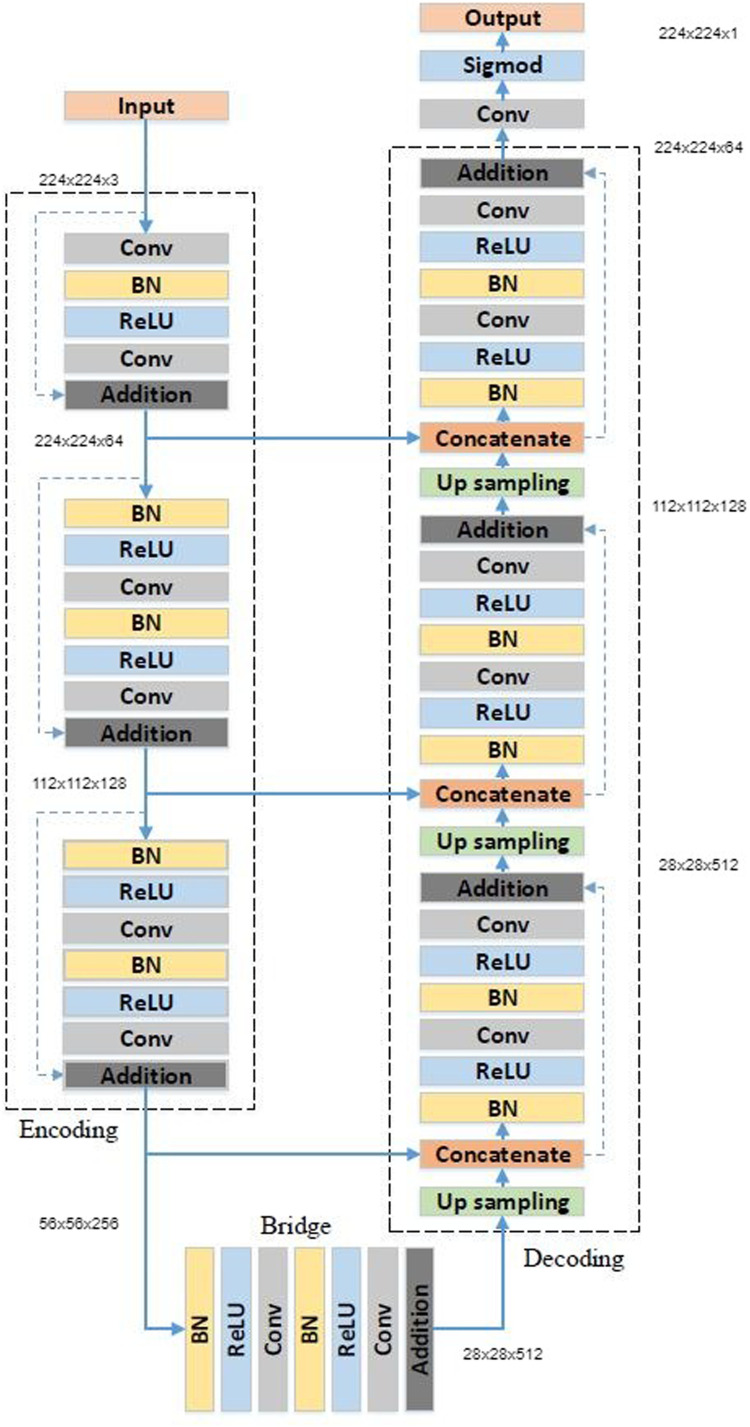
We set out to construct our deep learning segmentation-classification pipeline by first choosing an appropriate segmentation module to preprocess our classification dataset. We tested the preprocessed Darwin V7 Labs dataset [43] on a host of different segmentation modules including the popular U-Net [31], the ResUnet [12], the ResUNet-a [58], the TransResUNet [59] and U-Nets containing VGG and DenseNet backbones. Before training, we required the images in our preprocessed V7 Labs dataset [43] to undergo additional preprocessing in the form of image augmentation. During augmentation, we set the rotation range to 180 degrees, width/height shift ranges to 30%, shear range to 20%, zoom range to 20%, and set horizontal flips to true. We ultimately found that our best results on the preprocessed Darwin V7 Labs dataset [43] were obtained using Zhang et al’s ResUnet [12]. We therefore decided to move forward using this segmentation module in our pipeline. The ResUnet [12] on our preprocessed V7 Labs dataset ultimately obtained a dice similarity coefficient of 95.04% after 45 epochs. This segmentation module uses a 7-level architecture shown in Fig. 2 and Table 5 . Its architecture can be understood by dividing it conceptually into three main parts. The first part of the architecture is an encoder that fits the images input into the module into smaller and more compact representations. The last main segment of this architecture is the decoder which “recovers the representations to a pixel-wise categorization, i.e., semantic segmentation” [12]. The second middle part of the classifier serves as a bridge between the encoder at the ResUNet’s [12] input and the decoder at the ResUNet’s [12] output.
Fig. 2.
ResUnet architecture [12].
Table 5.
ResUnet architecture [12].
| Unit level | Conv layer | Filter | Stride | Output size |
|---|---|---|---|---|
| Input | ||||
| Encoder Lev 1 | Conv 1 | /64 | 1 | |
| Encoder Lev 1 | Conv 2 | /64 | 1 | |
| Encoder Lev 2 | Conv 3 | /128 | 2 | |
| Encoder Lev 2 | Conv 4 | /128 | 1 | |
| Encoder Lev 3 | Conv 5 | /256 | 2 | |
| Encoder Lev 3 | Conv 6 | /256 | 1 | |
| Bridge Lev 4 | Conv 7 | /512 | 2 | |
| Bridge Lev 4 | Conv 8 | /512 | 1 | |
| Decoder Lev 5 | Conv 9 | /256 | 1 | |
| Decoder Lev 5 | Conv 10 | /256 | 1 | |
| Decoder Lev 6 | Conv 11 | /128 | 1 | |
| Decoder Lev 6 | Conv 12 | /128 | 1 | |
| Decoder Lev 7 | Conv 13 | /64 | 1 | |
| Decoder Lev 7 | Conv 14 | /64 | 1 | |
| Output | Conv 15 | 1 |
Having discussed the segmentation portion of the deep learning pipeline, we now move on to discussing the models that we have constructed for classifying COVID-19 images. All of our models were trained in TensorFlow2.5. We ran our algorithms on an Intel Xeon CPU (2.30 GHz) using 26 GB RAM and a Tesla P100-PCIE-16GB GPU. We trained our preprocessed multiclass training set on a DenseNet-201 [30], a ResNet-152 [38], and a VGG-19 [27]. Each of these models was set to pretrained ImageNet weights. While designing each of these models we added an extra dense layer and dropout layer to the end of each model. The DenseNet-201’s [30] extra dense layer contained 128 neurons. The ResNet-152’s [38] extra dense layer contained 1024 neurons. The VGG-19’s [27] extra dense layer contained 4096 neurons. Each of the activation functions in these dense layers was set to a ReLU activation. The dropout layer added to the end of each model was set to a dropout rate of 10 percent. This helped each model to avoid overfitting and deal with the limited size of our dataset. We constructed both binary and multiclass versions of all of these classifiers. For the binary version of each classifier, we replaced the final softmax layer of each classifier with a single neuron containing a sigmoid activation function. For the multiclass version of each of these classifiers, our final layers contained three neurons each and had a softmax activation function.
Prior to training our DenseNet-201 [30], ResNet-152 [38], and VGG-19 [27] CNNs, we noticed that a class imbalance existed in our multiclass and binary datasets. There were lower amounts of COVID-19 images in comparison to the other categories of images in our datasets. We, therefore, needed to weigh the loss functions of our classifiers to correct for this imbalance. We did this because we wanted sure that all of our categories were evenly represented. Prior to training our classifiers, we additionally used image augmentation on the segmented images from our ResUNet [12] to prevent overfitting in our classifiers. There is often limited data in most medical imaging problems, and we noticed this helped us to improve the accuracy of our classifiers. Using Kera’s ImageDataGenerator class, we set the rotation range to 15%, the width/height range to 15%, the shear range to 15%, the zoom range to 15%, and horizontal flips to true. Our training and test set batch sizes were set to 32. In addition to segmenting and augmenting our classification datasets, we also normalized our data. In doing so, we ensured that the scaled data in each batch had a mean of zero and a standard deviation of one.
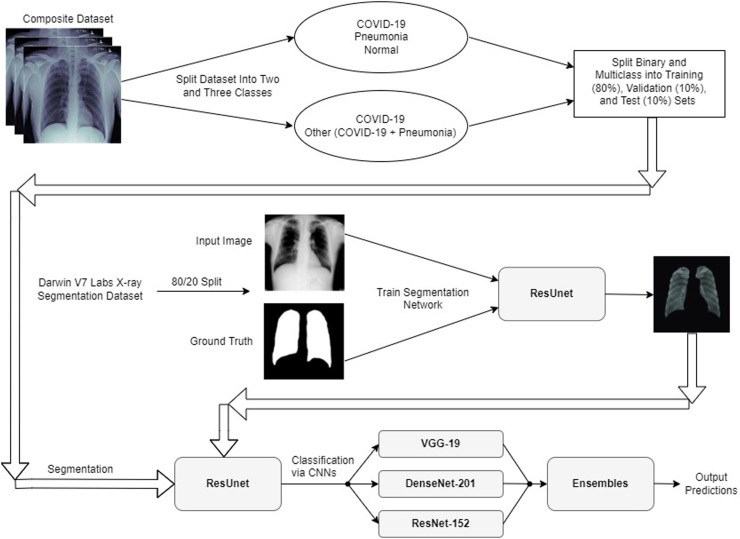
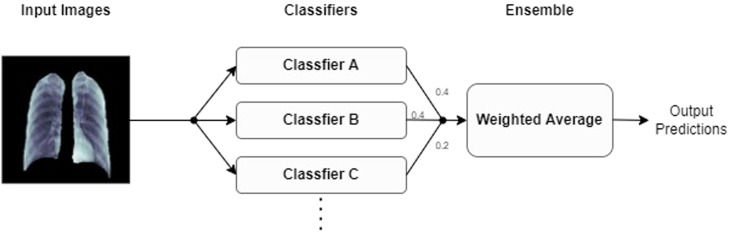
After our initial preprocessing steps, we trained the final fully-connected layers of each classifier alone for five epochs. We used the ADAM optimizer during this training and kept the ADAM optimizer set to its default settings. After performing this training, for each classifier we progressively unfroze each model’s layers and fine-tuned our models at a fixed learning rate of until each model hit its highest possible validation accuracy. Prior to unfreezing progressive layers in our models, we froze the moving mean and moving variance of the batches in our models’ batchnormalization layers to keep these parameters fixed to their pretrained ImageNet weights. After training each of our CNNs to their optimal validation accuracies, we constructed a majority voting ensemble and a weighted average ensemble that combined all of our classifiers together. We constructed both a binary version and a multiclass version of each type of ensemble classifier. An illustration showing our overall deep learning pipeline and can be observed in Fig. 5. The ensembles used in our deep learning pipeline are illustrated in Figs. 3 and 4 .
Fig. 5.
Proposed network architecture for COVID-19 classification.
Fig. 3.
Weighted average ensemble.
Fig. 4.
Majority voting ensemble.
4. Experimental results
4.1. Performance evaluation
Within the COVID-19 deep learning literature, we have found that most studies report common evaluation metrics. To compare our models against the literature we have reviewed, we have chosen to report the accuracy, sensitivity, specificity, F1-Score, precision, recall, negative predictive value (NPV), positive predictive value (PPV), and area under the receiver operating characteristic curve (AUC-ROC) of our deep learning pipeline.
We first set out to train our multiclass and binary DenseNet-201 [30], ResNet-152 [38], and VGG-19 [27] models for five epochs. On each model, we obtained a validation accuracy that ranged between 70 and 80 percent. This largely mirrored the performance of expert radiologists who had their expertise measured in a research study led by Wehbe et al. [21]. We performed this initial work using our multiclass and binary training sets before moving on to test ourselves against Tabik et al.’s [22] model (which was trained on the “COVID-GR-1.0” dataset). During this initial stage, we worked toward increasing the accuracy of all three of these classifiers by unfreezing each model during training progressively.
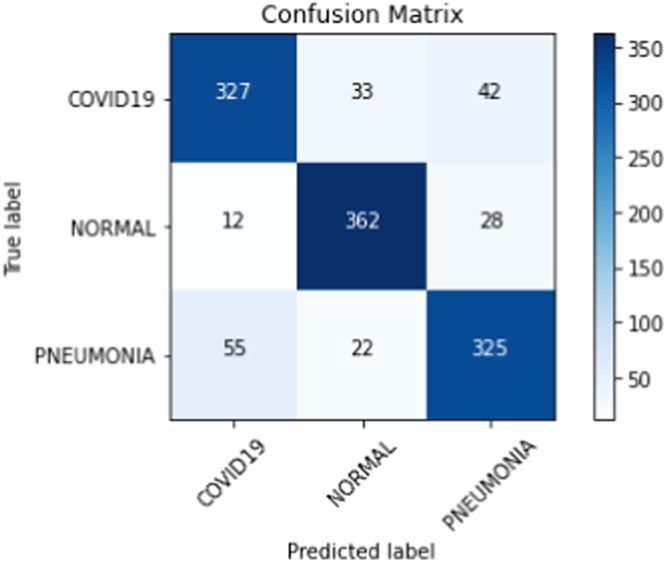
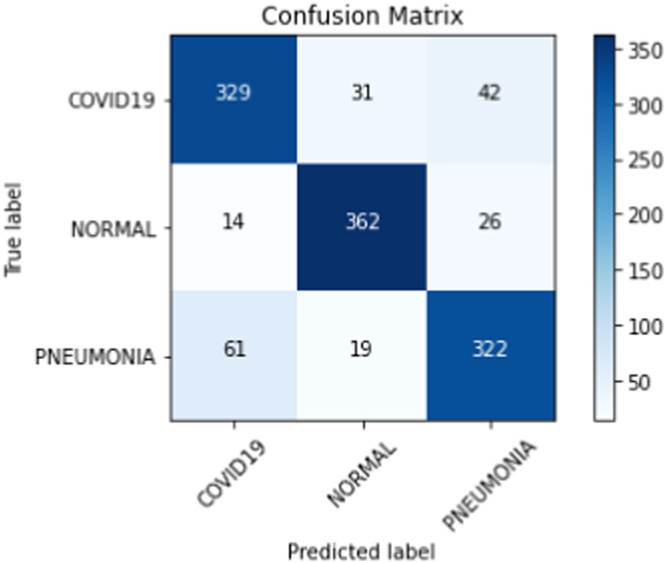
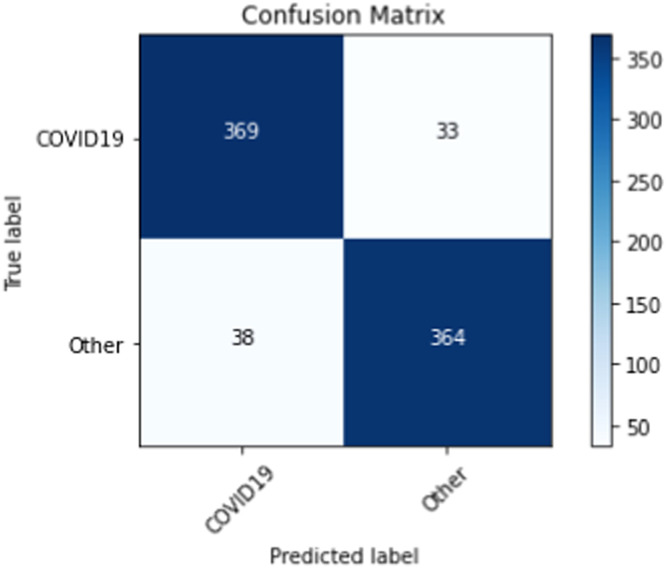
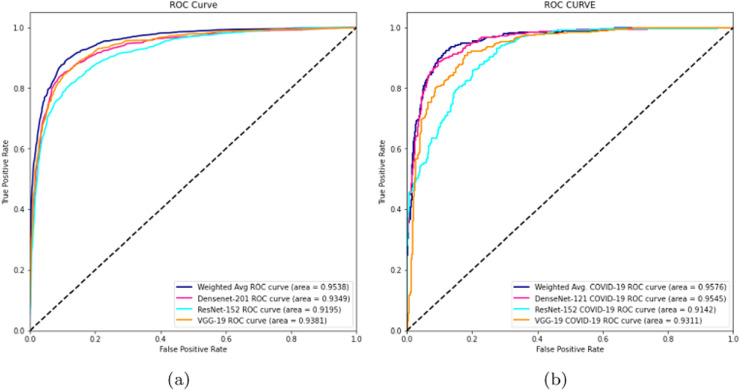
On our multiclass dataset set, we obtained final validation set accuracies of 82.16% on our DenseNet-201 [30], 84.25% on our ResNet-152 [38], and 81.09% on our VGG-19 [27]. Likewise, on our multiclass dataset set, we obtained final test set accuracies of 82.42% on our DenseNet-201 [30], 81.84% on our ResNet-152 [38], and 77.53% on our VGG-19 [27]. The test accuracies we obtained all saw a decrease of 2% - 4% from their corresponding validation set accuracies. When we ensembled all three classifiers into majority voting and weighted average ensembles, we saw an increase in performance on our validation and test sets. For our weighted average ensemble, we obtained a validation set accuracy of 87.40% and a test set accuracy of 84.07%. For our majority voting ensemble, we obtained a validation set accuracy of 87.14% and a test set accuracy of 84.00%. In both instances, we found that the test set accuracies of both ensembles outperformed our best individual classifier (DenseNet-201 [30]) by more than 1.5%. The overall performance of our three classifiers and our ensembles on our multiclass validation and test sets can be seen in Table 6 . Our binary classifiers were trained in the same way as our multiclass classifiers. The overall performance of our three classifiers and our ensembles on our binary validation and test sets can be seen in Table 7 . Table 8, Table 9, Table 10, Table 11 show a larger suite of statistics generated on the multiclass and binary test sets using both our weighted average and majority voting ensembles. Fig. 6, Fig. 7, Fig. 8, Fig. 9 show the corresponding confusion matrices generated by our weighted average and majority voting ensembles on our multiclass and binary test sets. Fig. 10 shows the AUC-ROC curves generated by our weighted average ensembles.
Table 6.
The performance of our classifiers on our multiclass dataset.
| Classifier | Val. Acc. | Test Acc. | Val. COV. Sen. | Test COV. Sen. |
|---|---|---|---|---|
| DenseNet-201 | 82.16% | 82.42% | 84.32% | 82.09% |
| ResNet-152 | 84.25% | 81.84% | 82.59% | 76.86% |
| VGG-19 | 81.09% | 77.53% | 81.34% | 75.62% |
| Weighted Avg. Ens. | 87.40% | 84.07% | 85.32% | 81.34% |
| Maj. Voting Ens. | 87.14% | 84.00% | 86.07% | 81.84% |
Table 7.
The performance of our classifiers on our binary dataset.
| Classifier | Val. Acc. | Test Acc. | Val. COV. Sen. | Test COV. Sen. |
|---|---|---|---|---|
| DenseNet-201 | 89.55% | 88.43% | 88.81% | 85.82% |
| ResNet-152 | 85.70% | 82.09% | 91.04% | 84.82% |
| VGG-19 | 89.55% | 84.55% | 89.30% | 83.08% |
| Weighted Avg. Ens. | 91.17% | 91.17% | 91.79% | 91.79% |
| Maj. Voting Ens. | 90.67% | 88.18% | 91.29% | 87.06% |
Table 8.
Weighted average ensemble performance metrics after training on our multiclass training set.
| TP | TN | FP | FN | Acc. | Sens. | Spec. | PPV | NPV | F1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 327 | 737 | 67 | 75 | 0.88 | 0.81 | 0.92 | 0.83 | 0.94 | 0.81 |
| Normal | 362 | 742 | 55 | 40 | 0.92 | 0.90 | 0.93 | 0.87 | 0.95 | 0.88 |
| Pneumonia | 325 | 734 | 70 | 77 | 0.88 | 0.81 | 0.91 | 0.82 | 0.91 | 0.81 |
Table 9.
Majority voting ensemble performance metrics after training on our multiclass training set.
| TP | TN | FP | FN | Acc. | Sens. | Spec. | PPV | NPV | F1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 329 | 729 | 75 | 73 | 0.88 | 0.82 | 0.91 | 0.81 | 0.91 | 0.82 |
| Normal | 362 | 754 | 50 | 40 | 0.93 | 0.90 | 0.94 | 0.88 | 0.95 | 0.89 |
| Pneumonia | 322 | 736 | 68 | 80 | 0.88 | 0.81 | 0.92 | 0.83 | 0.90 | 0.81 |
Table 10.
Weighted average ensemble performance metrics after training on our binary training set.
| TP | TN | FP | FN | Acc. | Sens. | Spec. | PPV | NPV | F1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 369 | 364 | 38 | 33 | 0.91 | 0.92 | 0.91 | 0.91 | 0.92 | 0.91 |
| Non-COVID-19 | 364 | 369 | 33 | 38 | 0.91 | 0.91 | 0.92 | 0.92 | 0.91 | 0.91 |
Table 11.
Majority voting ensemble performance metrics after training on our binary training set.
| TP | TN | FP | FN | Acc. | Sens. | Spec. | PPV | NPV | F1 | |
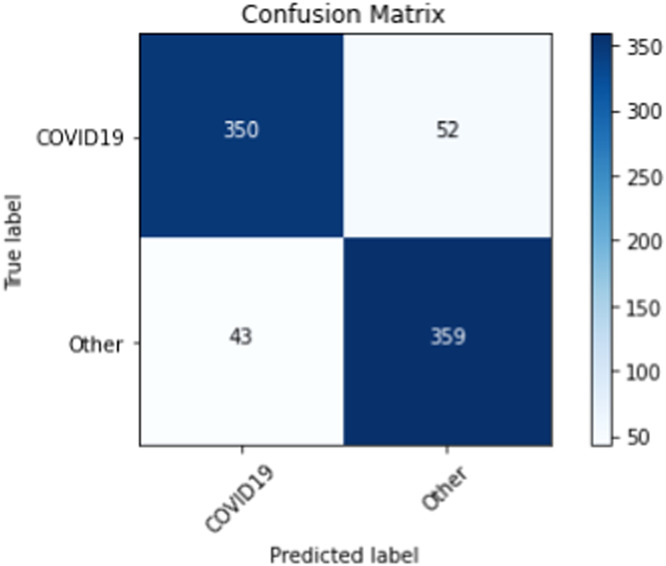
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | 350 | 359 | 43 | 52 | 0.88 | 0.87 | 0.89 | 0.89 | 0.87 | 0.88 |
| Non-COVID-19 | 359 | 350 | 52 | 43 | 0.88 | 0.89 | 0.87 | 0.87 | 0.89 | 0.88 |
Fig. 6.
Confusion matrix from weighted average ensemble after training on our multiclass training set.
Fig. 7.
Confusion matrix from majority voting ensemble after training on our multiclass training set.
Fig. 8.
Confusion matrix from weighted average ensemble after training on our binary training set.
Fig. 9.
Confusion matrix from majority voting ensemble after training on our binary training set.
Fig. 10.
AUC-ROC graphs of (a) Our multiclass weighted average ensemble trained on our multiclass training set and (b) Our binary weighted average ensemble trained on our binary training set.
After training and testing our segmentation-classification pipeline on our datasets, we also tested our binary pipeline directly against Tabik et al.’s [22] COVID-SDNet model. The details of their publicly available “COVID-GR-1.0” dataset [22] are provided in Section 3.2. It should be noted that Tabik et al.’s [22] dataset is smaller than ours and composed in a fashion whereby the authors collaborated with radiologists to intentionally incorporate weaker COVID-19 images into their dataset. This being the case, lower performance metrics should be expected out of this dataset. These two datasets have been designed to deal with separate problems and a detailed discussion concerning these differences is presented in the following section. Table 12 shows how our models compared against Tabik et al.’s [22] COVID-SDNet model.
Table 12.
Our binary models vs. COVID-SDNet on the COVID-GR-1.0 dataset [22].
| Classifier | Val. Acc. | Val. COV. Sen. |
|---|---|---|
| Weighted Avg. Ens. | 76.74% | 77.91% |
| Maj. Voting Ens. | 76.16% | 73.26% |
| COVID-SDNet | 76.18% | 72.59% |
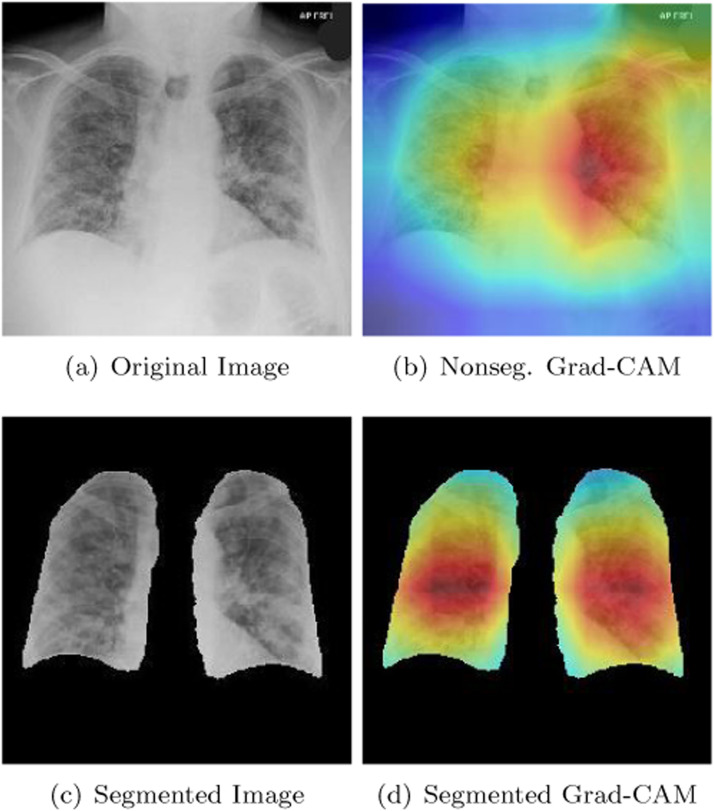
Every deep learning expert working in computer vision understands that it is necessary to validate the final version of a classifier after it has been trained. In medical imaging, saliency maps are widely employed on computer vision models to ensure that these models are correctly identifying important features in an image. In radiology, it is common for deep learning models to incorrectly focus on necklaces, medical devices, and the text within X-ray scans. The reason we included a segmentation unit in our study was to ensure that our model’s CNNs were rejecting unnecessary image details outside of the boundaries of the lungs. We used a Grad-CAM [60] in this study to ensure that our segmentation module was doing its job correctly in assisting our models to pick up the correct features of COVID-19. A Grad-CAM [60] functions by using the final feature maps in the last convolutional layer of a CNN to signal regions of importance within an image. We were interested in studying our CNNs that were trained on segmented images. We therefore devised a plan to compare them with CNNs that were trained on nonsegmented images. Fig. 11 shows the performance of our a DenseNet-201 [30] after being trained on segmented and nonsegmented X-rays. Our DenseNet-201 [30] was one of the three CNNs that we used in constructing our majority voting and weighted average ensembles. Part (b) of Fig. 11 shows the performance of our DenseNet-201 [30] on a test image after it was trained without a segmentation module. The red parts of the heatmap indicate the primary parts of the image that the DenseNet-201 [30] focused on when determining a patient has COVID-19. The orange/yellow portions of the heatmap represent areas of medium importance. The green/blue areas of the Grad-CAM [60] heatmap represented areas that were the least important diagnostically in determining that a patient is COVID-19 positive. Unfortunately, portions of the red and orange/yellow parts of the heatmap in part (b) of Fig. 11 are focused on areas outside of the lungs. The area that the Grad-CAM [60] partially focused on in the upper right-hand side of the image was a problem. This area should have been irrelevant to a COVID-19 diagnosis. When our DenseNet-201 [30] was trained on segmented images however, its behavior improved as is shown in part (d) of Fig. 11. We monitored the performance of our model in this way to ensure that our model was picking up the features of COVID-19 that we highlighted in Section 1.
Fig. 11.
Example of a segmented and non-segmented Grad-CAM heatmap produced by our DenseNet-201.
4.2. Discussion
Wehbe et al. [21] conducted an important study that measured the performance of practicing radiologists on a private COVID-19 vs. non-COVID-19 dataset. In our work, we took it upon ourselves to build a COVID-19 dataset of comparable size. We wanted to measure our pipeline’s ability to compete with the radiologists in their study and their model. We were more specifically interested in comparing our pipeline’s COVID-19 sensitivity with the radiologists in Wehbe et al.’s [21] study given the problems concerning RT-PCR test sensitivity we have read about in scientific journals. The radiologists’ consensus sensitivity in Wehbe et al.’s study [21] was 70%. All of our ensembles, including those trained on the weaker images in the “COVID-GR-1.0” dataset [22], obtained a higher COVID-19 sensitivity. The COVID-19 sensitivity of the five expert radiologists in Wehbe et al.’s [21] study versus that of our ensembles’ can be seen in Table 13 .
Table 13.
The COVID-19 sensitivity of five expert radiologists in Wehbe et al.’s study [21] vs. our classifiers.
| Group/Individual/Classifier | COV. Sens. |
|---|---|
| The Consensus of Expert Radiologists | 70% |
| The Best Radiologist | 76% |
| The Worst Radiologist | 60% |
| Weighted Avg. Ensemble (Our Binary dataset) | 91.79% |
| Weighted Avg. Ensemble (COVID-GR-1.0 dataset [22]) | 77.91% |
As can be seen in Table 13, when we compare our ensemble models with the performance of the radiologists in Wehbe et al.’s [21] study, we outperform even the best radiologist’s COVID-19 sensitivity. In Table 13, another item that stands out is the difference in sensitivity between the ensemble we trained on our binary dataset versus the ensemble we trained on the COVID-GR-1.0 dataset [22]. This discrepancy can be explained by the higher number of weak COVID-19 images that were intentionally placed by radiologists in the “COVID-GR-1.0” dataset [22]. Tabik et al. [22] created the “COVID-GR-1.0” dataset to measure the performance of their classifier on COVID-19 images that are more difficult to classify. Even after we trained our ensemble model on this extremely conservative dataset, we still managed to obtain a higher sensitivity than the radiologists in Wehbe et al.’s [21] study. This demonstrated the robustness of our technique. The COVID-GR-1.0 dataset intentionally contained a larger proportion of COVID-19 positive images that were difficult for radiologists to identify correctly. Many of the datasets currently available in the literature are constructed from the images of hospitalized patients. The COVID-19 severity of X-rays from patients who have been hospitalized is often worse than the severity seen in X-rays from patients who have not been hospitalized. Many COVID-19 X-ray datasets in the literature, therefore, have a larger proportion of severe COVID-19 images. These datasets may not always be representative of the population at large. That was an issue Tabik et al.’s [22] dataset was attempting to correct for. Our final results after training with Tabik et al.’s [22] dataset showed that our overall pipeline maintained good performance when working with a more conservative dataset.
When we constructed our binary dataset, we built our dataset so as to respond to a criticism that Wehbe et al. [21] mentioned in their paper concerning the size of public datasets. Wehbe et al.’s [21] study found that the consensus accuracy and sensitivity of expert radiologists are 81% and 70% respectively. After training their ensemble model, Wehbe et al. [21] found that their system achieved a test accuracy of 82% and test sensitivity of 75%. Many other studies however have reported performance metrics that are much higher than this. Wehbe et al. [21] explained this by showing how models with extremely high metrics often have very small COVID-19 datasets. They posited that if the number of COVID-19 images in these other studies increased, these models would see a correction. They believed that early COVID-19 deep learning models were overfitting on small COVID-19 datasets. We therefore set out to construct a larger COVID-19 dataset than any other public COVID-19 dataset we have seen in the literature thus far. We felt that it was additionally important to create separate validation and test sets in order to ensure that overfitting does not occur. To protect against overfitting, we also ensured that each of our CNNs in our pipeline had dropout layers in their second last layers.
Wehbe et al.’s [21] criticism of small public datasets was not the only concern we have ended up discovering when using public datasets. We later realized that many public datasets include images from Kermany et. al.’s [33] dataset which contains the chest X-rays of young children suffering from various forms of pneumonia. It is incorrect to take a model that was trained on children’s X-rays and deploy it on adult X-rays. When we attempted to use such a dataset for training one of our CNNs, we obtained extremely high-performance metrics (accuracy/sensitivity between 98% and 100%). We noticed that several deep learning segmentation-classification pipelines [18], [19], [24] made this mistake. In addition to this, we have come to discover that some authors may have unintentionally biased their classifiers by mixing multiple images from individual patients in their training and test sets. This ultimately results in an incorrect biasing of a deep learning model as the image in the test set often has similar features to the image in the training set that was derived from the same patient. If this biasing occurs, deep learning models often lock onto more closely related features than they would have otherwise been trained to recognize. To summarize, the following three main issues are, therefore, sometimes found with COVID-19 datasets in the literature:
-
1.
COVID-19 datasets have often been too small which has caused overfitting to occur in deep learning models
-
2.
Many datasets have been constructed with pneumonia X-rays collected from children. Models based on these datasets were later then deployed on adult lungs
-
3.
Some datasets may contain separate images from the same patients in both the training and test sets
In Table 14 we compare our work with other segmentation-classification pipelines that have not made the mistake of incorrectly biasing their datasets. Our best three-class and two-class ensemble models should only be compared against the first four classifiers in Table 14. Our three-class and two-class ensembles were trained on a dataset that we built after gathering as many COVID-19 images as possible. The authors of the first four papers in Table 14, composed their datasets in the same way. The COVID-GR-1.0 dataset [22], however, was trained intentionally on weak COVID-19 images resulting in a classifier that should be treated in isolation. In comparing our segmentation unit with Yeh et al.’s [20] U-Net [31] segmentation model, our ResUNet [12] achieved a dice similarity coefficient that was 7 percent higher. In terms of dataset size, our COVID-19 dataset contained over 3000 more COVID-19 images. Yeh et al. [20] had a smaller dataset, therefore, and were more likely to have overfit their model. Our model was, therefore, more likely to face downward pressure in our performance metrics. Our three-class model, however, was still capable of obtaining the same COVID-19 sensitivity as Yeh et al.’s [20] model. It likely was able to do so with the help of better segmentation and the use of a majority voting ensemble. This indicates that on datasets that are constructed with as many COVID-19 images as possible, a three-class model (COVID-19 vs. Normal vs. Pneumonia) can reasonably achieve a COVID-19 sensitivity of 82%. Our two-class weighted average ensemble outperformed Wehbe et al.’s [21] classifier by a substantial margin. This may have been caused by a difference in our approach to segmentation. Wehbe et al.’s [21] classifier was trained to crop out the smallest rectangle that a patient’s lungs can fit within. Our segmentation unit was trained on a set of masks that removed more pixels than Wehbe et al.’s [21] segmentation unit. We chose to not segment out the pixels showing the heart. Cardiomegaly (an enlarged heart) is a common symptom of COVID-19. Leaving the heart in our classified images allowed us to pick up this feature and likely assisted us to increase the performance metrics of our classifier. Our weighted average ensemble also outperformed Abdullah et al.’s [25] model despite our having a segmentation unit that under-performed Abdullah et al.’s Res-CR-Net [45] by one percent. We obtained a two-class accuracy that was 12 percent better than Abdullah et al.’s [45] classification model. We believe this is a result of our having constructed an extremely robust weighted average classification ensemble. Our best 2-class pipeline’s accuracy and sensitivity matched the accuracy and sensitivity of the Wang et al.’s [26] best classification model published in Nature. We should mention, however, that their pipeline was trained with half as many COVID-19 images. This made their model more vulnerable to possible overfitting.
Table 14.
Performance of similar segmentation-classification pipelines without dataset composition issues.
| Research paper | Seg. DSC | Acc. | COV. Sens. |
|---|---|---|---|
| Yeh et al. [20] | |||
| 3-class | 0.88 | – | 82% |
| Wehbe et al. [21] | |||
| 2-class | – | 82% | 75% |
| Abdullah et al. [25] | |||
| 2-class | 0.96 | 79% | – |
| Wang et al. [26] | |||
| 2-class | 0.89 | 91% | 92% |
| Tabik et al. [22] | |||
| 2-class (COVID-GR-1.0 dataset) | 0.885 | 76% | 73% |
| Ours | |||
| Best 3-class Ens. (Maj. Vot.) | 0.95 | 84% | 82% |
| Best 2-class Ens. (Wei. Avg.) | 0.95 | 91% | 92% |
| 2-class (COVID-GR-1.0 dataset) | 0.95 | 77% | 78% |
It should be noted that there are instances where using a segmentation unit can reduce a model’s accuracy. While segmentation units should generally always help a classifier’s accuracy, we have noticed in our work that classifiers without a segmentation unit can lock onto features of an image that are external to the lungs. Sometimes this helps to increase a CNN’s ability to classify particular images. For instance, if one category of images has more text than another you might notice the Grad-CAM [60] heatmaps for that category focusing on text. Our segmentation unit removed this possibility from happening and ultimately allowed us to boost our model’s accuracy in a more honest fashion. Our Grad-CAM [60] heatmaps in Fig. 11 additionally showed an improvement in discovering relevant COVID-19 features when we used our segmentation unit.
The approach to creating datasets that is followed by the vast majority of research papers is to obtain as many COVID-19 images as possible. During the early stages of the coronavirus pandemic, there was a lack of COVID19 images and many papers were being published that likely were overfitting on datasets containing only a couple of hundred COVID-19 images. Tabik et al. [22] published their paper when fewer COVID-19 images existed and therefore their paper only contained 426 COVID-19 images. The authors of this paper obtained the help of an expert radiologist. This radiologist located PCR positive images that did not have the visual features of COVID-19. They infused their dataset with such images and wanted to see the effect this would have. They eventually found that their classifier could identify COVID-19 in 85 to 97 percent of moderate to severe images. Mild COVID-19 images, however, could only be diagnosed correctly 46 percent of the time. They did not publish the accuracy of their classifier on Normal PCR positive images. We have to imagine that the accuracy for Normal PCR positive images was even lower. In total, their classifier had a final accuracy of 76 percent and COVID-19 sensitivity of 73 percent. When our binary weighted average ensemble was trained on their dataset, it achieved a 77 percent accuracy and a 78 percent COVID-19 sensitivity. We therefore achieved a COVID-19 sensitivity that was 5 percent better than Tabik et al’s [22] model on their dataset.
Tabik et al.’s [22] dataset was the only dataset that we could obtain that allowed us to directly compare our pipeline with another author’s segmentation-classification pipeline. It has been difficult to find publicly available datasets such Tabik et al.’s [22] where the authors have made clear how they segmented and classified their images. Tabik et al. [22] did not report a dice similarity coefficient because they segmented their images in such a way so as to create a small cropped rectangle around the lungs. This is similar in principle to how we segmented our images. We chose the Darwin V7 Labs dataset [43] for training our segmentation unit because the masks in this dataset left more room around the lungs to show the heart. We believe that if a segmentation unit were to remove these pixels, that COVID-19 symptoms like cardiomegaly could go unobserved by a classifier. We believe that our weighted average ensemble is ultimately what allowed us to achieve an improved accuracy and improved COVID-19 sensitivity when comparing our model with Tabik et al.’s [22] model. Our segmentation unit also likely helped as well, as it rejected a greater number of superfluous pixels around the lungs in comparison to Tabik et al.’s [22] segmentation methodology.
Unfortunately, at this time, public COVID-19 datasets that have been made available are somewhat incomplete. Public COVID-19 datasets are composed of images that previously came with corresponding positive RT-PCR tests. We know, however, that there are occasionally false-positive images, depending on when individual RT-PCR tests are performed. Sometimes, if a patient obtains a negative RT-PCR test, they will come back later and obtain a positive test. We, therefore, have datasets with RT-PCR-positive patients, but each image’s COVID-19 status has not been perfectly validated. There are occasional errors. This may have affected our work and the work of other papers we have reviewed. Our classifiers’ results, therefore, while promising, perhaps should not be clinically deployed until better external labeling processes have been followed in building COVID-19 datasets. Many deep learning models perform well in the lab before being deployed in a clinical setting. Our models would need to be tested alongside other administered COVID-19 tests in order to compare their efficacy against competing technologies.
5. Conclusion
The two-class and three-class datasets that we have constructed contain the largest number of publicly available COVID-19 images that we have found in the literature. In training our segmentation-classification pipeline we were ultimately able to design several ensembles that generated promising results. Our best two-class weighted average ensemble ultimately achieved a 91 percent COVID-19 accuracy and 92 percent COVID-19 sensitivity. We were also able to out-compete a segmentation-classification pipeline that we directly compared our pipeline against [22]. While our models show promising characteristics in terms of our Grad-CAM heatmaps and performance metrics, our models are still not ready to be implemented in a clinical setting.
For a deep learning pipeline such as ours to be advanced into a clinical setting, the medical community and AI experts require further collaboration. To the best of our knowledge, no study has been performed whereby every single incoming patient at a medical facility was tested for COVID-19 with an X-ray and RT-PCR test simultaneously. The COVID-19 images that can be found in public datasets tend to come from patients that were showing increased complications in relation to their illness. In private datasets, the same problem likely exists as well since radiological evaluations are typically reserved for patients showing a concerning trend in the development of their illness. It is important to find out the proportion of incoming patients at a medical clinic that are COVID-19 positive after blind X-rays get administered to every patient. Anyone wanting to clinically implement a deep learning system such as ours may also benefit from blindly administering competing molecular tests (RT-PCR tests), antigen tests, and antibody tests on the same patients during this data-gathering stage. In our future work, we aim to extend our pipeline with categorical and numerical data to improve the ability of our pipeline to diagnose COVID-19. This additional metadata concerning each patient’s age, sex, and relevant background details could really help to improve the performance metrics of our deep learning model. We also hope to eventually construct a deep learning pipeline capable of discovering the prognosis of COVID-19 patients. We believe that our pipeline is a promising step forward towards radiologically automating the detection of COVID-19. With a little more time and resources invested in these data-gathering processes, we believe that a clinically viable deep learning model is possible that allows for a truly better standard of care.
Dataset and code availability
We have made our dataset and scripts used in training our pipeline available at https://www.kaggle.com/roberthertel/covid-xray-dataset-with-segmentation-ensembles.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32101510
- 3.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32073353
- 4.L. Luo, D. Liu, X.-l. Liao, X.-b. Wu, Q.-l. Jing, J.-z. Zheng, F.-h. Liu, S.-g. Yang, B. Bi, Z.-h. Li, J.-p. Liu, W.-q. Song, W. Zhu, Z.-h. Wang, X.-r. Zhang, P.-l. Chen, H.-m. Liu, X. Cheng, M.-c. Cai, Q.-m. Huang, P. Yang, X.-f. Yang, Z.-g. Han, J.-l. Tang, Y. Ma, C. Mao, Modes of contact and risk of transmission in COVID-19among close contacts, medRxiv (2020). 10.1101/2020.03.24.20042606. [DOI]
- 5.Kucirka L., Lauer S., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 2020;173(4):262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32422057
- 6.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020;81(1):e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rousan L., Elobeid E., Karrar M., Khader Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20(1) doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., et al. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32027573
- 9.BenTaieb A., Kawahara J., Hamarneh G. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 2016. Multi-loss convolutional networks for gland analysis in microscopy; pp. 642–645. [DOI] [Google Scholar]
- 10.Arevalo J., González F.A., Ramos-Pollán R., Oliveira J.L., Guevara Lopez M.A. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput. Methods Programs Biomed. 2016;127:248–257. doi: 10.1016/j.cmpb.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Darwish A., Leukert K., Reinhardt W. IGARSS 2003. 2003 IEEE International Geoscience and Remote Sensing Symposium. Proceedings (IEEE Cat. No. 03CH37477) vol. 3. Citeseer; 2003. Image segmentation for the purpose of object-based classification; pp. 2039–2041. [Google Scholar]
- 12.Zhang Z., Liu Q., Wang Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018;15(5):749–753. doi: 10.1109/LGRS.2018.2802944. [DOI] [Google Scholar]
- 13.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam S.R., Maity S.P., Ray A.K., Mandal M. 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE) 2019. Automatic detection of pneumonia on compressed sensing images using deep learning; pp. 1–4. [DOI] [Google Scholar]
- 15.O’Quinn W., Haddad R.J., Moore D.L. 2019 IEEE 2nd International Conference on Electronic Information and Communication Technology (ICEICT) 2019. Pneumonia radiograph diagnosis utilizing deep learning network; pp. 763–767. [DOI] [Google Scholar]
- 16.Koppu S., Maddikunta P.K.R., Srivastava G. Deep learning disease prediction model for use with intelligent robots. Comput. Electr. Eng. 2020;87:106765. doi: 10.1016/j.compeleceng.2020.106765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., He J., Song L., Liu S., Srivastava G. Medical image classification based on an adaptive size deep learning model. ACM Trans. Multimed. Comput. Commun. Appl. 2021;17(3s) doi: 10.1145/3465220. [DOI] [Google Scholar]
- 18.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M.Z. Alom, M. Rahman, M.S. Nasrin, T.M. Taha, V.K. Asari, Covidmtnet: COVID-19 detection with multitask deep learning approaches, arXiv:2004.03747 (2020).
- 20.C.-F. Yeh, H.-T. Cheng, A. Wei, H.-M. Chen, P.-C. Kuo, K.-C. Liu et al. A cascaded learning strategy for robust COVID-19pneumonia chest X-ray screening, arXiv:2004.12786 (2020).
- 21.R.M. Wehbe, J. Sheng, S. Dutta, S. Chai, A. Dravid, S. Barutcu et al. DeepCOVID-XR: an artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large us clinical dataset, Radiology 0(0) (0) 203511. PMID: 33231531. 10.1148/radiol.2020203511. [DOI] [PMC free article] [PubMed]
- 22.Tabik S., Gómez-Ríos A., Martín-Rodríguez J.L., Sevillano-García I., Rey-Area M., Charte D., Guirado E., Suárez J.L., Luengo J., Valero-González M.A., García-Villanova P., Olmedo-Sánchez E., Herrera F. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images. IEEE J. Biomed. Health Inform. 2020;24(12):3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L.O. Teixeira, R.M. Pereira, D. Bertolini, L.S. Oliveira, L. Nanni, G.D.C. Cavalcanti, Y.M.G. Costa, Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images, arXiv:2009.09780 (2020). [DOI] [PMC free article] [PubMed]
- 24.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 25.H. Abdulah, B. Huber, S. Lal, H. Abdallah, L.L. Palese, H. Soltanian-Zadeh, D.L. Gatti, CXR-Net: an artificial intelligence pipeline for quick COVID-19 screening of chest X-rays, arXiv:2103.00087 (2021).
- 26.Wang G., Liu X., Shen J., Wang C., Li Z., Ye L., Wu X., Chen T., Wang K., Zhang X., Zhou Z., Yang J., Sang Y., Deng R., Liang W., Yu T., Gao M., Wang J., Yang Z., Lin T. A deep-learning pipeline for the diagnosis and discrimination of viral, non-viral and COVID-19 pneumonia from chest X-rayimages. Nat. Biomed. Eng. 2021;5:1–13. doi: 10.1038/s41551-021-00704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv:1409.1556 (2014).
- 28.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S.E. Reed, D. Anguelov, et al. Going deeper with convolutions, arXiv:1409.4842 (2014).
- 29.F. Chollet, Xception: deep learning with depthwise separable convolutions, arXiv:1610.02357 (2016).
- 30.G. Huang, Z. Liu, K.Q. Weinberger, Densely connected convolutional networks, arXiv:1608.06993 (2016).
- 31.O. Ronneberger, P. Fischer, T. Brox, U-Net: convolutional networks for biomedical image segmentation, arXiv:1505.04597 (2015).
- 32.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(56):1929–1958. [Google Scholar]; http://jmlr.org/papers/v15/srivastava14a.html
- 33.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 34.M.Z. Alom, T. Aspiras, T.M. Taha, V.K. Asari, Skin cancer segmentation and classification with NABLA-N and inception recurrent residual convolutional networks, arXiv:1904.11126 (2019).
- 35.Mooney P. Kaggle; 2018. Chest X-ray Images (Pneumonia) [Google Scholar]; https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 36.Jaeger S., Candemir S., Antani S., Wáng Y.-X.J., Lu P.-X., Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014;4(6):475–477. doi: 10.1016/j.cell.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Horry, S. Chakraborty, M. Paul, A. Ulhaq, B. Pradhan, M. Saha, N. Shukla, X-ray image based COVID-19 detection using pre-trained deep learning models, engrXiv (2020). 10.31224/osf.io/wx89s. [DOI]
- 38.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, arXiv:1512.03385 (2015).
- 39.Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K., Matsui M., Fujita H., Kodera Y., Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 2000;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 40.C. Szegedy, S. Ioffe, V. Vanhoucke, Inception-v4, inception-resnet and the impact of residual connections on learning, arXiv:1602.07261 (2016).
- 41.M. Tan, Q. Le, Efficientnet: rethinking model scaling for convolutional neural networks, arXiv (2019).
- 42.RSNA pneumonia detection challenge, 2018, (Kaggle). https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data.
- 43.COVID-19 chest X-ray dataset, 2020, https://darwin.v7labs.com/v7-labs/covid-19-chest-x-ray-dataset.
- 44.R.M. Pereira, D. Bertolini, L.O. Teixeira, C.N.S. Jr., Y.M.G. Costa, COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios, arXiv:2004.05835 (2020). [DOI] [PMC free article] [PubMed]
- 45.Abdallah H., Liyanaarachchi A., Saigh M., Silvers S., Arslanturk S., Taatjes D.J., Larsson L., Jena B.P., Gatti D.L. Res-CR-Net, a residual network with a novel architecture optimized for the semantic segmentation of microscopy images. Mach. Learn. 2020;1(1):045004. doi: 10.1088/2632-2153/aba8e8. [DOI] [Google Scholar]
- 46.L. Chen, G. Papandreou, I. Kokkinos, K. Murphy, A.L. Yuille, Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs, arXiv:1606.00915 (2016). [DOI] [PubMed]
- 47.L. Chen, G. Papandreou, F. Schroff, H. Adam, Rethinking atrous convolution for semantic image segmentation, arXiv:1706.05587 (2017).
- 48.Goodfellow I., Bengio Y., Courville A. MIT Press; 2016. Deep Learning. [Google Scholar]; http://www.deeplearningbook.org
- 49.E. Oyallon, E. Belilovsky, S. Zagoruyko, Scaling the scattering transform: deep hybrid networks, arXiv:1703.08961 (2017).
- 50.Oyallon E., Zagoruyko S., Huang G., Komodakis N., Lacoste-Julien S., Blaschko M., Belilovsky E. Scattering networks for hybrid representation learning. IEEE Trans. Pattern Anal. Mach. Intell. 2019;41(9):2208–2221. doi: 10.1109/tpami.2018.2855738. [DOI] [PubMed] [Google Scholar]
- 51.A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A.N. Gomez, L. Kaiser, I. Polosukhin, Attention is all you need, arXiv:1706.03762 (2017).
- 52.Parmar N., Vaswani A., Uszkoreit J., Kaiser L., Shazeer N., Ku A., Tran D. In: Proceedings of the 35th International Conference on Machine Learning. Dy J., Krause A., editors. vol. 80. 2018. Image transformer; pp. 4055–4064. (Proceedings of Machine Learning Research, PMLR). [Google Scholar]; http://proceedings.mlr.press/v80/parmar18a.html
- 53.J. Long, E. Shelhamer, T. Darrell, Fully convolutional networks for semantic segmentation, arXiv 79 (2014). [DOI] [PubMed]
- 54.Cozzi D., Albanesi M., Cavigli E., Moroni C., Bindi A., Luvará S., et al. Chest X-ray in new coronavirus disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol. Med. 2020;125 doi: 10.1007/s11547-020-01232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. Sci. Rep. 2020;10:19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.E. Bilello, Medical imaging data resource center (MIDRC) - RSNA international COVID-19 open radiology database (RICORD) release 1C - chest X-ray COVID+ (MIDRC-RICORD-1C), 2021, https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70230281.
- 57.M. de la Iglesia Vayá, J.M. Saborit, J.A. Montell, A. Pertusa, A. Bustos, M. Cazorla et al. BIMCV-COVID19, datasets related to COVID19’s pathology course, 2020, https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/.
- 58.Diakogiannis F.I., Waldner F., Caccetta P., Wu C. ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020;162:94–114. doi: 10.1016/j.isprsjprs.2020.01.013. [DOI] [Google Scholar]
- 59.Reza S., Amin O.B., Hashem M. 2020 IEEE Region 10 Symposium (TENSYMP) 2020. Transresunet: improving U-Net architecture for robust lungs segmentation in chest X-rays; pp. 1592–1595. [DOI] [Google Scholar]
- 60.R.R. Selvaraju, A. Das, R. Vedantam, M. Cogswell, D. Parikh, D. Batra, Grad-CAM: why did you say that? Visual explanations from deep networks via gradient-based localization, CoRRarXiv:1610.02391 (2016).