Abstract
Droplet-based microfluidics has emerged as a powerful tool for single-cell screening with ultrahigh throughput, but its widespread application remains limited by the accessibility of a droplet microfluidic high-throughput screening (HTS) platform, especially to common laboratories having no background in microfluidics. Here, we first developed a microfluidic HTS platform based on fluorescence-activated droplet sorting technology. This platform allowed (i) encapsulation of single cells in monodisperse water-in-oil droplets; (ii) cell growth and protein production in droplets; and (iii) sorting of droplets based on their fluorescence intensities. To validate the platform, a model selection experiment of a binary mixture of Bacillus strains was performed, and a 45.6-fold enrichment was achieved at a sorting rate of 300 droplets per second. Furthermore, we used the platform for the selection of higher α-amylase-producing Bacillus licheniformis strains from a mutant library generated by atmospheric and room temperature plasma mutagenesis, and clones displaying over 50% improvement in α-amylase productivity were isolated. This droplet screening system could be applied to the engineering of other industrially valuable strains.
Keywords: Enzyme, High-throughput screening, Droplet microfluidic platform, Bacillus licheniformis
Introduction
Enzymes play an increasingly important role as biocatalysts in many industrial processes, such as the food, pharmaceutical, and detergent industries (Adrio & Demain, 2014; Choi et al., 2015; Singh et al., 2016). Enzymes exhibit several advantages compared with conventional chemical catalysts owing to their high substrate specificity, high catalytic efficiency, biodegradability, nontoxicity, and relatively mild operational conditions concerning pH and temperature. These enzyme traits make them receive more attention from both industry and academia. Microorganisms are the main source of industrial enzyme preparation due to their economical bulk production capacity, eco-friendly nature, easier handling, and ease of product modification (Chandra et al., 2020; Kuhad et al., 2011; Pandey et al., 2000).
To meet the growing demands in the industry, strategies involving genetic and process engineering have been used for strain development to increase the yield and productivity as well as stress tolerance to industrial production conditions (Davy et al., 2017; Meyer et al., 2015; Xiao et al., 2014). However, the high complexity of the metabolic processes and incomplete understanding of their underlying mechanisms limit the power of rational engineering approaches. Moreover, the use of recombinant DNA technology in the food industry is tightly regulated, and consumers’ responses to genetically modified food ingredients are largely negative (He et al., 2017).
Directed evolution employing iterative rounds of random mutagenesis and library screening can be used for strain improvement purposes (Markel et al., 2020; Turner, 2009). Random mutagenesis is one of the most commonly employed methods for diversity generation. This method does not require detailed knowledge about the strain and enzyme. Recently, a novel mutation machine named atmospheric and room temperature plasma (ARTP) has been extensively used for microbial mutation (Jin et al., 2018; Zhang et al., 2014, 2018). Compared with traditional mutagenesis strategies using nitrosoguanidine and ultraviolet treatment, ARTP mutagenesis is a more environmentally friendly, safe, and highly diverse mutagenesis method (Zhang et al., 2014).
In addition to the mutation efficiency, the success of directed evolution experiments hinges on an efficient high-throughput screening (HTS) strategy to isolate the desired mutants from a large mutant library (Gielen et al., 2018; Markel et al., 2020; Ruff et al., 2012). Conventional screening methods, such as halo and microtiter plate assays, are costly and low throughput. Fluorescence-activated cell sorting (FACS) is an ultrahigh-throughput method for single-cell sorting and analysis. However, FACS requires that fluorescent marker(s) must remain either inside or on the surface of the cells to be sorted (Griffiths & Tawfik, 2006; Ruff et al., 2012). In vitro compartmentalization (IVC) by compartmentalized cells and their extracellular products in water-in-oil-in-water double emulsion enables screening of 10,000 events per second using FACS (Mastrobattista et al., 2005). However, the IVC-FACS assay is limited by complex double emulsion generation and modification (He et al., 2019). In contrast, droplet-based microfluidic technology enables the generation and manipulation of highly monodisperse water-in-oil droplets at ultrahigh frequencies (Agresti et al., 2010; Beneyton et al., 2016, 2017; Huang et al., 2015; Ma et al., 2018; Mazutis et al., 2013; Qiao et al., 2017; Sjostrom et al., 2014; Wang et al. 2014). Single cells and their extracellular products were encapsulated in picoliter-sized droplets using a microfluidic droplet generation device at a rate of thousands per second. Each droplet acts as an individual microreactor to link the phenotype of the cell to its genotype. Following droplet collection and off-chip incubation, the droplets were reinjected into the sorting device and sorted based on their fluorescence signals at a rate of hundreds per second. In the past decade, fluorescence-activated droplet sorting (FADS) has been applied to the screening of many microorganisms that produce various enzymes and metabolites (Van Tatenhove-Pel et al., 2020). Although commercial microfluidic devices and reagents (oil, surfactant, etc.) are available from several companies, such as Dolomite, Bio-Rad, and RainDance Technologies, the lack of commercial droplet-based microfluidic platforms has limited their widespread use in common laboratories (Kaminski et al., 2016). In contrast to the rapid progress of flow cytometry, only limited research has been done on the development of microfluidic instruments for water-in-oil droplet sorting (Qiao et al., 2017).
In this study, we developed an integrated microfluidic droplet sorting platform based on FADS. The platform allowed droplet generation, long-term droplet incubation, and sorting of droplets with a throughput of up to 1 × 106 droplets per hour. A mixture of Bacillus strains with high and low fluorescence was successfully enriched to verify the feasibility of our platform. We further applied the platform to screen a mutant library of α-amylase-producing Bacillus licheniformis generated by ARTP, and mutants with higher α-amylase production capacity were successfully identified. This microfluidic droplet sorting platform should be extremely useful for accelerating the process of strain development.
Materials and Methods
Strains and Culture Media
The industrial strain B. licheniformis wild type (WT) provided by Shandong Longkete Enzyme Preparations Co., Ltd. (Linyi, China) was used as the original strain for α-amylase production. One Bacillus subtilis strain carrying a plasmid with kanamycin resistance as well as the other B. subtilis strain carrying a plasmid with kanamycin resistance and expressing green fluorescent protein (GFP) were stocked in our laboratory and used to model the selection experiment. The amylase-inducing medium in water-in-oil droplets contained (per liter) the following: beef extract powder 8 g, yeast extract powder 2 g, peptone 10 g, NaCl 2 g, and glucose 50 g. The flask fermentation medium consisted of (per liter) the following: corn steep liquor 30 g, soybean meal 14 g, CaCl2 0.315 g, (NH4)2SO4 3.5 g, K2HPO4 14 g, KH2PO4 6 g, sodium citrate 2 g, MgSO4 4 mg, FeSO4 0.4 mg, MnSO4 0.1 mg, and lactose 40 g, with an initial pH adjusted to 7.0 before autoclaving at 121°C for 15 min.
Microfluidic Devices and Experimental Setup
Two microfluidic devices were used for droplet generation and sorting, respectively. The devices were fabricated from polydimethyl siloxane (PDMS) using standard soft lithography techniques based on previously described procedures (Qiao et al., 2017). The PDMS slabs had channel heights between 18 and 25 μm and 0.5-mm-diameter holes on channel inlets and outlets. The PDMS slabs were sealed on glass microscope slides by exposing both parts to oxygen plasma. Electrode channels of the sorting device were filled with low-melting-temperature solder wire. Finally, the microfluidic channel walls were treated by injecting Aquapel and then flushing with pressurized air to generate a hydrophobic surface.
The microfluidic device was fixed on the stage of a Nikon Eclipse Ti-U inverted fluorescence microscope. All fluids were pumped to the microfluidic devices by syringe pumps (Harvard Apparatus, USA). A high-speed video camera (Fastec HiSpec 1, USA) was mounted on the microscope for the imaging of droplet formation, reinjection, and sorting. The 473-nm laser (MBL-III-473, China) was transmitted to the microscope and focused on the channel of the sorting device. The fluorescence emitted by each droplet was detected by a photomultiplier tube (PMT, H10722-20, Japan) and recorded by a data acquisition (DAQ) card (USB‐7855R, USA) controlled using a program written in LabVIEW software. When the PMT signal exceeds a defined threshold, the software provides a signal to a high-voltage amplifier (Trek 609E-6, USA), which delivers a high-voltage electric field to solder injected electrodes of the sorting device to deflect the droplets.
Droplet Generation and Microfluidic Screening
Monodisperse droplets were generated by a flow-focusing microfluidic device using a flow rate of 100 μl/hr for the aqueous solution and 300 μl/hr for the fluorinated oil (HFE-7500, 3M, USA) containing 2% (wt/wt) EA surfactant (Raindance Technologies, USA) (Holtze et al., 2008). The droplets were collected into a 1-ml syringe (BD Biosciences, USA) and incubated for the desired period at 37°C. Afterward, the droplets were reinjected into a sorting device at a flow rate of 10 μl/hr and spaced by fluorinated oil at 200 μl/hr. In the sorting device, the fluorescence signal of each droplet was measured using the microfluidic platform by the droplet microfluidic system. Droplets with fluorescence intensity exceeding the predefined threshold were sorted by dielectrophoresis.
Model Selection
Two model selection experiments of artificial premixed libraries were performed. First, a binary mixture of fluorescent and nonfluorescent droplets with a 1:1 mixing ratio was produced and collected in the same syringe. The fluorescent droplets contained 1 μM fluorescein in 0.1 M tris-HCl, pH 8.0, while the nonfluorescent droplets contained 1% wt/wt bromophenol blue. The mixed droplets were subsequently sorted using the FADS platform, and the trajectories of droplets were monitored by a high-speed camera. Second, a premixed library used two strains of B. subtilis: “active” cells expressing the GFP reporter gene and the control “inactive” cells without any fluorescent markers. In model selection experiments, the initial ratio of active to inactive cells can be set to 1:9, 1:99, or higher as previously reported (Beneyton, et al., 2014). As for the same starting cell density, the initial 1:9 ratio results in more droplets with active cells, which can be more easily detected under a microscope, thus reducing the sorting time. Besides, based on the Poisson distribution, the initial 1:9 ratio results in more coencapsulation events, especially under suboptimal cell preparation conditions (Mazutis et al., 2013). This could help to optimize cell preparation methods of minimizing cell sedimentation and aggregation. Thus, we used an initial 1:9 ratio of active cells to inactive cells. The strains were grown overnight in Luria-Bertani (LB) medium at 37°C with shaking at 250 rpm. The cell suspensions were washed 3 times by centrifuging at 5000 × g for 2 min and then resuspended in fresh LB medium. To minimize clumping, the suspensions were filtered with an 8-μm filter membrane (Nuclepore Track-Etched Membrane, Whatman, USA). The OD600 was measured, and the cell suspensions were subsequently mixed in a 1:9 initial ratio of active cells to inactive cells. The cell mixture was diluted to appropriate concentrations before encapsulation. After off-chip incubation for 12 hr at 37°C, the droplets were sorted according to their fluorescence intensities. The sorted droplets were spread on LB plates and recovered at 37°C for 16 hr. The recovered cells were subsequently harvested from agar plates and analyzed using MoFlo XDP flow cytometry (Beckman Coulter, Brea, CA, USA). The enrichment efficiency is defined as the ratio of ε after sorting (ε1) to ε before sorting (ε0), where ε is the ratio of “active” cells to “inactive” cells (Baret et al., 2009).
ARTP Mutagenesis
A single colony of B. licheniformis from a fresh plate was inoculated in LB medium and grown to the logarithmic phase. The cells were washed twice with sterile 0.9% NaCl and diluted to an OD600 of 1.0. Afterward, 10 μl of the culture was spread on a sterilized stainless steel plate and treated with the ARTP mutation breeding system (ARTP-II, China) by following the instruction manual (He et al., 2019). The main operating parameters were as follows: the radiofrequency power input was 100 W, the helium gas flow rate was 10 l/min, and the distance between the plasma torch nozzle exit and the sample plate was 2 mm. The treatment times were set to 0, 10, 12, 15, 20, 25, 30, and 40 s, respectively. After that, all the cells were washed from the plates, properly diluted, and spread on LB plates. Plates were incubated at 37°C until colonies formed. The lethality rates were calculated as follows:
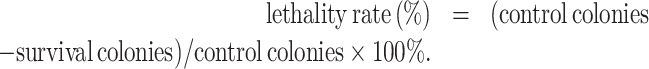 |
Library Screening
The mutant library on agar plates was carefully washed into 1 ml inducing medium. The cell suspension was treated with the same procedure as described in the model selection experiment. The suspension was diluted to 1.25 × 107 cells/ml to maintain a cell-to-droplet ratio of 0.1. The fluorogenic substrate was added to the treated suspension at a final concentration of 20 μM prior to emulsification. After incubation off-chip for the desired time, droplets with signals above the threshold were sorted and recovered for subsequent verification.
Analysis of Selected Clones
After sorting, cells were recovered from the sorted droplets by adding 200 μl of fresh media and 100 μl of 1H,1H,2H,2H-perfluoro-1-octanol (PFO, Sigma Aldrich, USA), gently mixed, spread on LB agar plates and incubated at 37°C until colonies formed (Mazutis et al., 2013). The clones from each of the plates were picked and cultivated in 250-ml flasks containing 30 ml of fermentation medium for 5 days at 37°C at 260 rpm. The WT strain was also treated with the same procedure as a control. The α-amylase activity was estimated using the starch–iodine color reaction (Xiao et al., 2006). One unit of α-amylase activity was defined as the amount of α-amylase required to produce a 1% reduction in the intensity of the blue color of the starch–iodine complex under conditions of the assay.
Results and Discussion
Development of Droplet-Based Microfluidic Screening Platform
Typically, a droplet-based microfluidic HTS pipeline was composed of the following three steps: encapsulation of single cells together with a fluorogenic substrate into picoliter droplets; off-chip incubation for cell growth, protein expression, and enzymatic reaction; and sorting of droplets based on their fluorescence intensities (Fig. 1). The pipeline incorporates two PDMS-glass microfluidic devices: a flow-focusing droplet-making device and a droplet-sorting device. To perform this pipeline, we developed a FADS microfluidic platform (Fig. 2a and b). This platform consisted of a fluid control module to transfer the fluids or droplets to the device, an optical setup to detect the fluorescence of droplets, a high-speed camera to record the movement of the droplets, a high-voltage amplifier connected to the electrodes of the sorting device, and control software to analyze the data. The platform can be used to generate monodispersed 8-pl droplets at a rate of over 6,000 droplets per second and sort droplets at 300 droplets per second. Additionally, droplets with different diameters (ranging from 20 to 100 μm) can be generated at rates of 1,000 to 10,000 droplets per second and sorted at rates of 10–400 droplets per second by adjusting the channel sizes of microfluidic devices in our platform (He et al., 2019; Tu et al., 2021). The performance of this platform is comparable to the previously reported droplet-based microfluidic platforms, in which droplets can be generated at rates of 1,000–30,000 droplets per second and sorting at rates of 1–2,000 droplets per second (Ma et al., 2018; Mazutis et al., 2013; Wang et al., 2014).
Fig. 1.
Droplet-based microfluidic high-throughput screening workflow. (a) Atmospheric and room temperature plasma (ARTP) mutagenesis. (b) Single cells were encapsulated into droplets with a fluorogenic substrate. (c) Droplets were incubated off-chip. (d) Droplets were reinjected into a sorting chip and sorted based on the fluorescence intensity.
Fig. 2.
Droplet-based microfluidic screening platform. (a) Schematic diagram of the platform. The 473-nm laser beam was aligned and focused through a 20× objective lens onto a specific point on the microfluidic chip, which was placed on the stage of an inverted microscope. The oil phase or droplets in the syringes controlled by syringe pumps were pumped into the microfluidic chip through Teflon tubing. A photomultiplier tube (PMT) and high-speed camera were connected to the side port of the inverted microscope to receive emission signals and image droplets, respectively. The droplet fluorescence signals were recorded by a data acquisition (DAQ) card and processed by a custom LabVIEW software. A high-voltage amplifier was connected to the electrodes of the microfluidic chip to sort the target droplets. (b) Photograph of the droplet-based microfluidics screening platform.
Analytical detection techniques play critical roles in developing a more versatile droplet-based microfluidic platform (Basova & Foret, 2015; Zhu & Fang, 2013). To date, almost all HTS implemented in microdroplet formats mainly relies on fluorescence. Absorbance-activated droplet sorting has been explored, but the limited sensitivity when working with droplets with small path lengths limits this approach (Gielen et al., 2016). In addition, other detection techniques, such as mass spectrometry, Raman spectroscopy, electrochemistry, and surface tension, have also been demonstrated in droplet analysis but have similarly not been widely employed to droplet-based HTS due to high noise levels and low throughput (Payne et al., 2020; Wang et al., 2017).
Measurement of Sorting Efficiency
To validate the feasibility of the FADS platform and optimize the sorting condition, we designed two model selection experiments. First, the fluorescence droplets were sorted from a mixture of fluorescent and nonfluorescent droplets with a 1:1 mixing ratio. The mixed droplets were sorted by the platform at a rate of 300 droplets per second. High-speed videos recorded during this process were analyzed to determine the error rates of the platform. The two kinds of droplets can be distinguished from the video due to the color difference (Fig. 3a). The false-positive error rate was found to be less than 1 in 104 droplets during the sorting process from video recording and counting. The second model selection experiment was performed by enriching active B. subtilis with a GFP reporter from abundant inactive B. subtilis without a fluorescent marker using an initial 1:9 mixture. The mixture of active and inactive cells was encapsulated in 8-pl, 25-μm-diameter droplets (Fig. 3b). The number of cells per droplet generally followed a Poisson distribution. Therefore, the cell suspension was diluted to 0.1 cells per droplet, which resulted in 91% of the droplets being empty, 9% of the droplets having a single cell, and less than 1% of the droplets having two or multiple cells. The droplets were off-chip incubated to allow cell growth and protein expression. Following incubation, the droplets were reinjected to a FADS chip and sorted at a throughput of 300 droplets per second. The sorted cells were then recovered and analyzed to determine the sorting efficiency. We obtained a 45.6-fold enrichment after one round of sorting, corresponding to 80.9% of active cells in the sorted population (Fig. 3c and d). This enrichment result verified the effectiveness of our screening platform. In the first experiment involving a binary mixture of fluorescent droplets, the sorting error rate was 0.01%. In the second sorting experiment involving a mixture of two strains, there was a slightly higher error rate, in which the proportion of active cells was enriched from 8.5% to 80.9% after one round of sorting. The main factor affecting the sorting efficiency involved in a mixture of two strains is the coencapsulation events of inactive and active cells (Baret et al., 2009). Therefore, the preparation of cell suspensions for encapsulation is an important procedure. In this study, we used a filter membrane strategy to prevent cell aggregation. Different methods can also be used to minimize cell sedimentation and aggregation, such as proper control over cell density, adding a density-matching solution to the medium, and ultrasonication of the cell suspension under proper power and time (Yang et al., 2021).
Fig. 3.
Validation of the droplet-based microfluidics screening platform. (a) Separate frame from the high-speed video during sorting of the mixed color droplets. The fluorescent droplets (light droplets) containing 1 μM fluorescein in 0.1 M tris-HCl were deflected into the upper channel by the sorting electrodes, while the nonfluorescent droplets (dark droplets) containing 1% wt/wt bromophenol blue flowed into the bottom channel. (b) Fluorescence microscopy images of droplets with the encapsulated mixture of active (GFP-positive) and inactive (nonfluorescent) cells mixed in a 1:9 ratio after 12 hr of incubation at 37°C. (c) Flow cytometry data from the sample mixture of active and inactive cells. (d) Flow cytometry data from the analysis of the sorted sample. Gated areas indicate percentages of fluorescent cells.
Development of a Fluorescence Assay for α-Amylase Activity
Droplets sorted on this platform relied on fluorescent readouts. To assay α-amylase activity in the droplets, fluorogenic substrate BODIPY fluorescein-labeled starch contained in the EnzChek Ultra Amylase Assay Kit was used (Beneyton et al., 2016; Sjostrom et al., 2014). When α-amylase hydrolyzes the starch backbone, the fluorophores are released, and the fluorescence intensity increases (Fig. 4a). The fluorogenic assay of α-amylase activity was performed in 96-well microtiter plates, and fluorescence was monitored over 24 hr at 37°C using a fluorescence spectrophotometer (SpectraMax M2, Molecular Devices) at a 473 nm excitation and 520 nm emission wavelength. As shown in Fig. 4b, the accompanying increase in fluorescence intensity was found to be proportional to the amylase activity (correlation coefficient R2 = 0.9376). In addition, the fluorogenic reaction in droplets was tested by encapsulating both single cells and the fluorogenic substrate in droplets, followed by fluorescence imaging over time of droplets incubated at 37°C. We imaged the droplets at generation, or after 4, 8, 12, 16, and 24 hr of incubation (Fig. 4c). The cells proliferated readily, and the fluorescence intensities gradually increased. This result demonstrated that the BODIPY FL-labeled starch substrate was suitable for the direct measurement of α-amylase activity in droplets. To allow secretion of enough enzyme for screening and avoid excess cell proliferation, we chose an 8-hr incubation time in the following screening experiments.
Fig. 4.
Assay of α-amylase from Bacillus licheniformis using the substrate BODIPY FL-labeled starch. (a) α-Amylase hydrolyzes the starch backbone to release the fluorophores simultaneously with an increase in fluorescence. (b) Reactions in 96-well microtiter plates were performed by mixing 196 μl of the fermentation supernatant of the B. licheniformis wild-type strain with different diluted concentrations and 4 μl of 1 mg/ml fluorogenic enzyme substrate BODIPY. Fluorescence was measured using 473 nm excitation and 520 nm emission. (c) Fluorescence microscopy images of microdroplets containing a single cell with a fluorogenic substrate after 0, 4, 8, 12, 16, and 24 hr of incubation at 37°C.
Screening of Mutant Library for Improved α-Amylase Production
The platform was used to screen a whole-genome-mutated B. licheniformis library, constructed by ARTP irradiation, to select for cells with high α-amylase production. It has been reported that the ARTP mutation method has the highest positive mutation probability, with a lethality between 90% and 95% (Zhang et al., 2018). As shown in Fig. 5, when the exposure time exceeded 20 s, the lethality rate was over 90%, an appropriate level to mutagenize the cells. Therefore, an exposure time of 20 s was chosen in subsequent experiments. To increase the genetic diversity, the mutation procedure was repeated 10 times, and all treatments were eluted altogether into the same sterile tube. After ARTP mutagenesis, the cells were recovered on agar plates, followed by droplet encapsulation, cultivation, and sorting using a similar procedure as mentioned earlier. After screening, a total of ∼648 (∼0.03%) droplets were sorted from 2.1 × 106 droplets within 2 hr. The sorted droplets were broken, and the released cells were recovered by spreading on LB agar plates.
Fig. 5.

The lethality curve of the Bacillus licheniformis wild-type strain after atmospheric and room temperature plasma (ARTP) treatment. Data represent the mean of triplicate samples, and the error bars represent standard deviations.
For a 106 mutant library, compared with conventional microtiter plate-based screening formats (1000 cells per hour and microliter per well), the FADS platform (300 droplets per second and picoliter per droplet) could shorten the entire screening process from weeks to hours while reducing the costs of reagents and consumables by millions of fold. Furthermore, this method would be possible to repeat for another round of selection for even better-producing strains by using the top strains as templates. In addition, the platform would be adapted to screen microbes for the production of other enzymes or metabolites as long as they can be linked to a fluorescence assay.
Validation of α-Amylase Production
Ninety individual colonies were randomly picked from the sorted plates and analyzed for their α-amylase activities in shake-flask fermentation. As shown in Fig. 6a, 64.4% of the 90 colonies exhibited higher enzyme activity levels than the WT strain, and the top-performing clone had a more than 50% increase in α-amylase activity. To examine the hereditary stability of the isolated strains, the 10 best performers were subcultured on fresh agar plates, and their enzyme activities were analyzed by fermentation again. Nine of the 10 strains still maintained the same level, with a more than 40–60% improvement in α-amylase activity relative to wild type, compared with the first round of testing (Fig. 6b).
Fig. 6.
Evaluation of α-amylase production by the sorted mutants. (a) Characterization of the sorted clones in the shake-flask fermentation assay. The red dashed line indicates the isolated strains with higher enzyme production compared with the wild type, and the red stars indicate the top 10 hyperproducers. (b) Ten of the most active strains were subcultured on fresh agar plates and again analyzed for their enzyme activities. The histogram displaying the α-amylase level of each clone was normalized to the mother strain.
In the past few years, droplet-based microfluidic platforms have been applied for screening different categories of enzymes, including horseradish peroxidase, α-amylase, cellulase, xylanase, aldolase, and esterase. Recently, Zhang et al. (2021) also applied this approach to isolate B. licheniformis mutants with high-yield amylase production. In addition, the understanding of the genetic basis might be valuable feedback to further development via rational design. With advances in genome sequencing technologies, whole-genome sequencing of these select clones with increased protein production can uncover the mutations responsible for improved phenotypes (Huang et al., 2015). These mutations can be applied to other products and cell types for the engineering of novel microbial cell factories.
Conclusion
In this study, a droplet-based microfluidic HTS platform was established, and the platform achieved a sorting rate of up to 300 droplets per second. Compared with the traditional microtiter plate screening system, this platform greatly accelerated the process of directed evolution while significantly reducing the costs. The combination of ARTP mutagenesis and the microfluidic HTS platform was shown to have successfully improved the α-amylase productivity of B. licheniformis, further demonstrating the ability of the platform. Furthermore, the platform can be widely applied to strain engineering of other microbial cell factories for extracellular enzymes or metabolites.
Acknowledgement
We are grateful to Prof. David A. Weitz Lab at Harvard University for valuable discussions and help in the construction of the microfluidic platform.
Contributor Information
Huiling Yuan, Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin 300308, China; National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Ran Tu, Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin 300308, China; National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China.
Xinwei Tong, Shandong Longkete Enzyme Preparations Co., Ltd, Linyi 276400, China.
Yuping Lin, Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin 300308, China; National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China.
Yuanyuan Zhang, Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin 300308, China; National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China.
Qinhong Wang, Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, 32 West 7th Avenue, Tianjin Airport Economic Area, Tianjin 300308, China; National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China.
Funding
This work was supported by the National Key Research and Development Program of China (nos. 2021YFC2100201 and 2021YFC2103300), the Instrument Developing Project of the Chinese Academy of Sciences (no. YJKYYQ20170023), and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (no. TSBICIP-PTJS-003). Q.W. and Y.L. were supported by the Tianjin High-Level Innovation and Entrepreneurship Team Program.
Conflict of Interest
The authors declare no conflict of interest.
References
- Adrio J. L., Demain A. L. (2014). Microbial enzymes: Tools for biotechnological processes. Biomolecules, 4, 117–139. 10.3390/biom4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J. C., Marquez M., Klibanov A. M., Griffiths A. D., Weitz D. A. (2010). Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proceedings of the National Academy of Sciences, 107, 4004–4009. 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J. C., Miller O. J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M. L., Hutchison J. B., Agresti J. J., Link D. R., Weitz D. A., Griffiths A. D. (2009). Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab on a Chip, 9, 1850–1858. 10.1039/B902504A. [DOI] [PubMed] [Google Scholar]
- Basova E. Y., Foret F. (2015). Droplet microfluidics in (bio)chemical analysis. The Analyst, 140, 22–38. 10.1039/C4AN01209G. [DOI] [PubMed] [Google Scholar]
- Beneyton T., Coldren F., Baret J. C., Griffiths A. D., Taly V. (2014). CotA laccase: High-throughput manipulation and analysis of recombinant enzyme libraries expressed in E. coli using droplet-based microfluidics. Analyst, 139(13), 3314–3323. [DOI] [PubMed] [Google Scholar]
- Beneyton T., Thomas S., Griffiths A. D., Nicaud J. M., Drevelle A., Rossignol T. (2017). Droplet-based microfluidic high-throughput screening of heterologous enzymes secreted by the yeast Yarrowia lipolytica. Microbial Cell Factories, 16, 18. 10.1186/s12934-017-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneyton T., Wijaya I. P., Postros P., Najah M., Leblond P., Couvent A., Mayot E., Griffiths A. D., Drevelle A. (2016). High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Scientific Reports, 6, 27223. 10.1038/srep27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra P., Enespa Singh R., Arora P. K. (2020). Microbial lipases and their industrial applications: A comprehensive review. Microbial Cell Factories, 19, 169. 10.1186/s12934-020-01428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. M., Han S. S., Kim H. S. (2015). Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnology Advances, 33, 1443–1454. 10.1016/j.biotechadv.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Davy A. M., Kildegaard H. F., Andersen M. R. (2017). Cell factory engineering. Cell Systems, 4, 262–275. 10.1016/j.cels.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Gielen F., Colin P. Y., Mair P., Hollfelder F. (2018). Ultrahigh-throughput screening of single-cell lysates for directed evolution and functional metagenomics. Methods in Molecular Biology (Clifton, N.J.), 1685, 297–309. [DOI] [PubMed] [Google Scholar]
- Gielen F., Hours R., Emond S., Fischlechner M., Schell U., Hollfelder F. (2016). Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proceedings of the National Academy of Sciences, 113, E7383–E7389. 10.1073/pnas.1606927113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A. D., Tawfik D. S. (2006). Miniaturising the laboratory in emulsion droplets. Trends in Biotechnology, 24, 395–402. 10.1016/j.tibtech.2006.06.009. [DOI] [PubMed] [Google Scholar]
- He P. H., Zhang Z. Y., Cai D. B., Chen Y. Z., Wang H., Wei X. T., Li S. Y., Chen S. W. (2017). High-level production of α-amylase by manipulating the expression of alanine racamase in Bacillus licheniformis. Biotechnology Letters, 39, 1389–1394. 10.1007/s10529-017-2359-5. [DOI] [PubMed] [Google Scholar]
- He R., Ding R., Heyman J. A., Zhang D., Tu R. (2019). Ultra-high-throughput picoliter-droplet microfluidics screening of the industrial cellulase-producing filamentous fungus Trichoderma reesei. Journal of Industrial Microbiology and Biotechnology, 46, 1603–1610. 10.1007/s10295-019-02221-2. [DOI] [PubMed] [Google Scholar]
- Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angile F. E., Schmitz C. H., Koster S., Duan H., Humphry K. J., Scanga R. A., Johnson J. S., Pisignano D., Weitz D. A. (2008). Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab on a Chip, 8, 1632–1639. 10.1039/B806706F. [DOI] [PubMed] [Google Scholar]
- Huang M. T., Bai Y. P., Sjostrom S. L., Hallstrom B. M., Liu Z. H., Petranovic D., Uhlen M., Joensson H. N., Andersson-Svahn H., Nielsen J. (2015). Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast. Proceedings of the National Academy of Sciences, 112, E4689–E4696. 10.1073/pnas.1506460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Wang Y., Yao M. D., Gu X. L., Li B., Liu H., Ding M. Z., Xiao W. H., Yuan Y. J. (2018). Astaxanthin overproduction in yeast by strain engineering and new gene target uncovering. Biotechnology for Biofuels, 11, 1, 230. 10.1186/s13068-018-1227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski T. S., Scheler O., Garstecki P. (2016). Droplet microfluidics for microbiology: techniques, applications and challenges. Lab on a Chip, 16, 2168–2187. 10.1039/C6LC00367B. [DOI] [PubMed] [Google Scholar]
- Kuhad R. C., Gupta R., Singh A. (2011). Microbial cellulases and their industrial applications. Enzyme Research, 2011, 280696. 10.4061/2011/280696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Chung M. T., Yao Y., Nidetz R., Lee L. M., Liu A. P., Feng Y., Kurabayashi K., Yang G. Y. (2018). Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nature Communications, 9, 1030. 10.1038/s41467-018-03492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel U., Essani K. D., Besirlioglu V., Schiffels J., Streit W. R., Schwaneberg U. (2020). Advances in ultrahigh-throughput screening for directed enzyme evolution. Chemical Society Reviews, 49, 233–262. [DOI] [PubMed] [Google Scholar]
- Mastrobattista E., Taly V., Chanudet E., Treacy P., Kelly B. T., Griffiths A. D. (2005). High-throughput screening of enzyme libraries: In vitro evolution of a beta-galactosidase by fluorescence-activated sorting of double emulsions. Chemistry & Biology, 12, 1291–1300. 10.1016/j.chembiol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Mazutis L., Gilbert J., Ung W. L., Weitz D. A., Griffiths A. D., Heyman J. A. (2013). Single-cell analysis and sorting using droplet-based microfluidics. Nature Protocols, 8, 870–891. 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A., Pellaux R., Potot S., Becker K., Hohmann H. P., Panke S., Held M. (2015). Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors. Nature Chemistry, 7, 673–678. 10.1038/nchem.2301. [DOI] [PubMed] [Google Scholar]
- Pandey A., Nigam P., Soccol C. R., Soccol V. T., Singh D., Mohan R. (2000). Advances in microbial amylases. Biotechnology and Applied Biochemistry, 31, 135–152. 10.1042/ba19990073. [DOI] [PubMed] [Google Scholar]
- Payne E. M., Holland-Moritz D. A., Sun S., Kennedy R. T. (2020). High-throughput screening by droplet microfluidics: Perspective into key challenges and future prospects. Lab on a Chip, 20, 2247–2262. 10.1039/D0LC00347F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y. X., Zhao X. Y., Zhu J., Tu R., Dong L. B., Wang L., Dong Z. Y., Wang Q. H., Du W. B. (2017). Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system. Lab on a Chip, 18, 190–196. 10.1039/C7LC00993C. [DOI] [PubMed] [Google Scholar]
- Ruff A. J., Dennig A., Wirtz G., Blanusa M., Schwaneberg U. (2012). Flow cytometer-based high-throughput screening system for accelerated directed evolution of P450 monooxygenases. ACS Catalysis, 2, 2724–2728. 10.1021/cs300115d. [DOI] [Google Scholar]
- Singh R., Kumar M., Mittal A., Mehta P. K. (2016). Microbial enzymes: Industrial progress in 21st century. 3 Biotech, 6, 174. 10.1007/s13205-016-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrom S. L., Bai Y. P., Huang M. T., Liu Z. H., Nielsen J., Joensson H. N., Svahn H. A. (2014). High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab on A Chip, 14, 806–813. 10.1039/C3LC51202A. [DOI] [PubMed] [Google Scholar]
- Turner N. J. (2009). Directed evolution drives the next generation of biocatalysts. Nature Chemical Biology, 5, 567–573. 10.1038/nchembio.203. [DOI] [PubMed] [Google Scholar]
- Tu R., Zhang Y., Hua E., Bai L. K., Huang H. M., Yun K. Y., Wang M. (2021). Droplet-based microfluidic platform for high-throughput screening of streptomyces. Communications Biology, 4, 647. 10.1038/s42003-021-02186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tatenhove-Pel R. J., Hernandez-Valdes J. A., Teusink B., Kuipers O. P., Fischlechner M., Bachmann H. (2020). Microdroplet screening and selection for improved microbial production of extracellular compounds. Current Opinion in Biotechnology, 61, 72–81. 10.1016/j.copbio.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Wang B. L., Ghaderi A., Zhou H., Agresti J., Weitz D. A., Fink G. R., Stephanopoulos G. (2014). Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nature Biotechnology, 32, 473–478. 10.1038/nbt.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. X., Ren L. H., Su Y. T., Ji Y. T., Liu Y. P., Li C. Y., Li X. R., Zhang Y., Wang W., Hu Q., Han D. X., Xu J., Ma B. (2017). Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells. Analytical Chemistry, 89, 12569–12577. 10.1021/acs.analchem.7b03884. [DOI] [PubMed] [Google Scholar]
- Xiao S., Shiloach J., Betenbaugh M. (2014). Engineering cells to improve protein expression. Current Opinion in Structural Biology, 26, 32–38. 10.1016/j.sbi.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z. Z., Storms R., Tsang A. (2006). A quantitative starch–iodine method for measuring alpha-amylase and glucoamylase activities. Analytical Biochemistry, 351, 146–148. 10.1016/j.ab.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Tu R., Yuan H. L., Wang Q. H., Zhu L. L. (2021). Recent advances in droplet microfluidics for enzyme and cell factory engineering. Critical Reviews in Biotechnology, 41, 1023–1045. 10.1080/07388551.2021.1898326. [DOI] [PubMed] [Google Scholar]
- Zhang G. Q., Chen Y. K., Li Q. H., Zhou J. W., Li J. H., Du G. C. (2021). Growth-coupled evolution and high-throughput screening assisted rapid enhancement for amylase-producing Bacillus licheniformis. Bioresource Technology, 337, 125467. 10.1016/j.biortech.2021.125467. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang X. F., Li H. P., Wang L. Y., Zhang C., Xing X. H., Bao C. Y. (2014). Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Applied Microbiology and Biotechnology, 98, 5387–5396. 10.1007/s00253-014-5755-y. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang X. M., Xu G. Q., Zhang X. J., Shi J. S., Xu Z.H. (2018). Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve l-serine yield in Corynebacterium glutamicum. Applied Microbiology and Biotechnology, 102, 5939–5951. 10.1007/s00253-018-9025-2. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Fang Q. (2013). Analytical detection techniques for droplet microfluidics—a review. Analytica Chimica Acta, 787, 24–35. 10.1016/j.aca.2013.04.064. [DOI] [PubMed] [Google Scholar]







