Abstract
In this work, a brief electrochemical aptasensor was developed for highly sensitive detection of SARS-CoV-2 antigen utilizing an aptamer-binding induced multiple hairpin assembly strategy for signal amplification. In the presence of SARS-CoV-2, a pair of aptamers was brought in a close proximity according to the aptamer-protein antigen binding, which initiated strand displacement reaction thereby triggering a multiple hairpin assembly to obtain long linear DNA concatemers on the electrode surface. As the fabricated hairpin probes were labeled with biotin, massive streptavidin-alkaline phosphatases (ST-ALP) could be further introduced on the electrode interface via biotin-streptavidin interaction thus generating strong electrochemical signal in electrolyte solution containing 1-naphthol phosphate. Benefiting from the non-enzymatic multiple hairpin assembly signal amplification strategy, the designed aptasensor for SARS-CoV-2 spike protein detection exhibited the wide linear range from 50 fg·mL−1 to 50 ng·mL−1 and low detection limit of 9.79 fg·mL−1. Meaningfully, this proposed electrochemical assay provided a potential application for the point of care analysis of viral diseases under ambient temperature.
Keywords: Aptamer/protein proximity binding, Multiple hairpin assembly, SARS-CoV-2, Electrochemical aptasensor, Spike protein
Graphical abstract
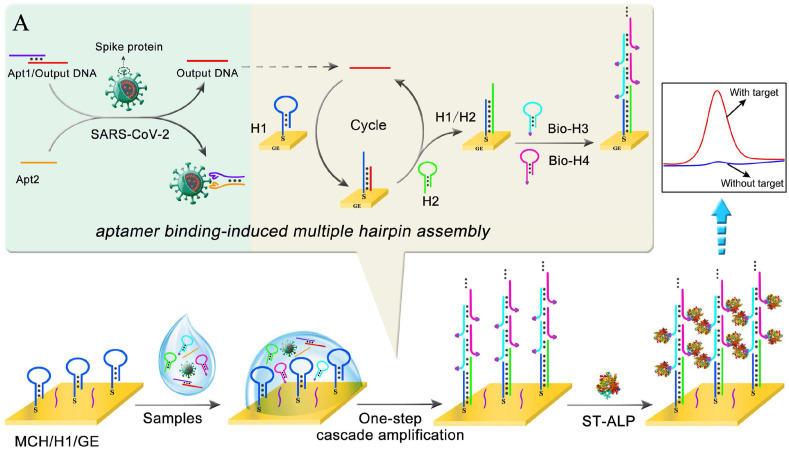
A highly sensitive electrochemical aptasensor for rapid determination of SARS-CoV-2 spike protein based on aptamer-binding induced multiple hairpin assembly as the efficient signal amplification.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as the causative agent of the COVID-19 global pandemic, is highly infectious and spreads through human-to-human interaction primarily by the airborne route [1]. Interventions including the quarantine [2], wearing surgical mask [3] and vaccination were identified as effective measures to interrupt the viral transmission [4,5]. Among these interventions, vaccination was considered as the ideal way to reduce the morbidity and mortality caused by SARS-CoV-2, which brought about less impact on economy and society. However, vaccination was an absolute protection, as COVID-19 epidemic had broken through the protection of vaccination time after time, even in the high vaccination coverage areas [[6], [7], [8]]. Moreover, the vaccines were scarce in most of the world, where the epidemic control was an arduous task. From this perspective, timely quarantine and wearing surgical mask were still the key measure to deal with epidemic outbreak. Therefore, rapid and extensive detection of the SARS-CoV-2 was of significance in the foreseeable future.
Currently, reverse transcription real-time quantitative PCR (RT-qPCR) assay is the gold standard to diagnose SARS-CoV-2 infection by targeting for SARS-CoV-2 nucleic acids [9]. However, this assay takes hours in detection procedure and requires expensive instruments and trained personnel, which is adverse to extensive screening in the case of epidemic outbreak. Antibody detection-based serum immunological examination is low-cost and rapid in detection procedure while not sensitive enough to early diagnosis of SARS-CoV-2 infection. Meanwhile, the specificity of serological examination is barely satisfactory, because the positive antibody test may indicate past infection, or just vaccinated [10]. Conversely, antigen detection-based etiological examination presents specificity comparable to nucleic acid detection but needn't rigorous laboratory condition, expensive instruments and professional operation [11], which has promising application to on-site diagnosis of SARS-CoV-2 infection. However, limited by the affinity of antibody, classical sandwich immunoassays present significant deficiency in detection sensitivity.
Herein, an aptamer-binding induced multiple hairpin assembly was proposed to construct an electrochemical biosensor for rapid and sensitive detection of SARS-CoV-2 spike protein. As shown in Scheme 1 , a pair of aptamers was employed to probe the target SARS-CoV-2 spike protein [12]. The sandwich aptamers/spike protein proximity binding increased the local concentration of the DNA strands, thus inducing DNA strand displacement that transformed SARS-CoV-2 spike protein to output DNA. Then, the output DNA induced a multiple hairpin assembly to fabricate large amount of alkaline phosphatase (ALP) labeled probes on the electrode surface. With the addition of 1-naphthol phosphate (1-NPP) in the electrolyte solution, the electrochemical oxidation signal could be obtained corresponding to the concentration of target SARS-CoV-2 spike protein, as 1-NPP would be hydrolyzed into oxidizable 1-naphthol in the presence of ALP. As the signal amplification of the aptamer-binding induced multiple hairpin assembly, the electrochemical aptasensor achieved sensitive detection of SARS-CoV-2 spike protein with the linear range from 50 fg·mL−1 to 50 ng·mL−1 and detection limit down to 9.79 fg·mL−1. Moreover, the whole detection procedure could be finished in 1.5 h, which presented good application potential in screening of SARS-CoV-2 infection.
Scheme 1.
Schematic representation of stepwise electrochemical aptasensor fabrication process for SARS-CoV-2 detection. (A) The aptamer-binding induced multiple hairpin assembly signal amplification.
2. Experimental methods
2.1. Reagents and chemicals
Streptavidin-alkaline phosphatase (ST-ALP) was purchased from Solarbio Science & Technology Co. (Beijing, China). 6-Mercapto-1-hexanol (MCH) and 1-naphthyl phosphate (1-NPP) were purchased from Sigma-Aldrich (St. Louis, USA). SARS-CoV spike protein, MERS-CoV spike protein, SARS-CoV-2 spike protein, HCoV-HKU1 spike protein and HCoV-OC43 spike protein were purchased from Sino Biological Inc. (Beijing, China). The HPLC-purified DNA oligonucleotides were purchased from Sangon Biotechnology Co. (Shanghai, China). The DNA sequences were presented in Table 1 .
Table 1.
Sequences of the used oligonucleotides in this work.
| Name | Sequence (5’→3′) |
|---|---|
| Output DNA | TAG CTT ATC AGA CTG ATG TTG ACT AGC |
| Apt1 | CAG CAC CGA CCT TGT GCT TTG GGA GTG CTG GTC CAA GGG CGT TAA TGG ACA TTT TTT TTT TTC AAC ATC AGT CTG |
| Apt2 | GAC TGA TGT TGA TTT TTT TTT TAT CCA GAG TGA CGC AGC ATT TCA TCG GGT CCA AAA GGG GCT GCT CGG GAT TGC GGA TAT GGA CAC GT |
| H1 | TCA ACA TCA GTC TGA TAA GCT ACT ACA ACT GGC GGG GTA GCT TAT CAG ACT–(CH2)6–SH |
| H2 | TAA GCT ACC CCG CCA GTT GTA GTA GCT TAT CAG ACT CTA CAA CTG GCG GGG |
| Bio-H3 | Biotin-CTG GCG GGG AGG AAG CCC CGC CAG TTG TAG |
| Bio-H4 | Biotin-CTT CCT CCC CGC CAG CTA CAA CTG GCG GGG |
The buffers involved in this work were as follows: The annealing buffer was sodium chloride-Tris-EDTA (STE) buffer (10 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA, pH 8.0). The washing buffer was 20 mM Tris-HCl. Diethanolamine (DEA) buffer included 100 mM DEA, 100 mM KCl and 1 mM MgCl2 (pH 9.6). The detection buffer for the differential pulse voltammetry (DPV) measurement was 100 mM DEA buffer (pH 9.6) containing 1 mg·mL−1 1-NPP. The detection buffer for cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) was 0.1 M PBS (pH 7.4) containing 0.1 M KCl and 5 mM [Fe(CN)6]3-/4-. All other reagents were of analytical grade and used without further purification. Ultrapure water (18.2 MΩ/cm, Millipore System Inc) was employed throughout the entire experiment.
2.2. Apparatus
Electrochemical measurements, including EIS, CV and DPV were implemented on an AUTOLAB PGSTAT302 N electrochemical workstation (Metrohm Technology Co. Ltd., Switzerland). The electrochemical workstation was used with a conventional three-electrode system consisting of a saturated calomel electrode (reference electrode), a platinum wire (auxiliary electrode) and a 3-mm diameter gold electrode (working electrode). A JY600C electrophoresis analyzer (Beijing Junyi Electrophoresis Co. Ltd., China) was used to perform gel electrophoresis experiments, and gel images were imaged on a G:BOX F3 Gel Documentation System (Gene Co. Ltd., China).
2.3. Preparation of hairpin probes and Apt1/output DNA complex
All the hairpin probes (2 μM) containing H1, H2, Bio-H3 and Bio-H4 were dissolved in STE buffer and then heating to 95 °C for 5 min and later slowly cooled to ambient temperature to form the stem-loop structure. Next, the solutions were stored at 4 °C for further use.
Apt1/output DNA complex for proximity binding-induced strand displacement was prepared by hybridizing the predesigned output DNA with aptamer 1 (Apt1) of spike protein at a final concentration of 400 nM by mixing 9 μL of 10 μM Apt1 with 6 μL of 10 μM output DNA in 7.5 μL ultrapure water, heating to 95 °C for 5 min and slowly cooled down to ambient temperature. Finally, the Apt1/output DNA complex was further purified by the polyacrylamide gel DNA recovery kit.
2.4. Fabrication of the proposed electrochemical aptasensor
Firstly, the gold electrode (GE) was cleaned following procedures reported in a previous work [13]. Next, 10 μL of 1 μM thiolated H1 was dropped on the GE surface at 4 °C for 12 h through Au-thiol interaction. After washed with the washing buffer, the electrode was covered with 1 mM MCH at room temperature for 1 h to block the nonspecific area. Finally, the fabricated electrochemical aptasensor was stored at 4 °C when not in use.
2.5. Electrochemical measurement procedure
Initially, 1 μL of 100 nM Apt1/output DNA complex, 1 μL of 100 nM aptamer 2 (Apt2), 1 μL of 10 μM H2, 1 μL of 10 μM biotin-modified H3 (Bio-H3), 1 μL of 10 μM Bio-H4 and 5 μL of different concentrations of SARS-CoV-2 spike protein were mixed (the final volume was 10 μL) and then reacted for 45 min at 37 °C. After washed with the washing buffer, 10 μL of DEA buffer including 2.5 μg·mL−1 ST-ALP solution was dropped onto the surface of the electrode for 30 min at 37 °C. The ST-ALP would be immobilized on the electrode interface due to the streptavidin-biotin affinity reaction. Finally, the electrochemical signal was measured in DEA buffer containing 1 mg·mL−1 1-NPP substrate by DPV. The DPV measurements were carried out within the voltage range from −0.05 to 0.55 V, modulation amplitude of 0.07 V, modulation time of 0.05 s and interval time of 0.2 s.
3. Results and discussion
3.1. Working mechanism of the electrochemical aptasensor
The construction of this biosensing platform was based on the cascade signal amplification strategy of programmed catalytic hairpin assembly/hybridization chain reaction (CHA/HCR) triggered by the output DNA released by the proximity binding-induced DNA strand displacement. As shown in Scheme 1A, when the SARS-CoV-2 spike protein appeared, the Apt2 was brought in close proximity to the Apt1/output DNA complex due to the specific aptamer-spike protein binding, resulting in the output DNA releasing. The released output DNA continuously hybridized with H1 immobilized on the electrode surface and unfolded H1 to form a DNA duplex H1/output DNA. Then the opened toehold of H1 further hybridized with H2 to form the hybridized complex H1/H2, followed by the displacement of output DNA from the complex H1/H2 to realize the CHA target recycling amplification. Meanwhile, the CHA products (H1/H2) could unfold Bio-H3, then the opened Bio-H3 hybridized with Bio-H4. The HCR generated the long linear DNA concatemers by alternating hybridization between Bio-H3 and Bio-H4. Based on the numerous biotins modified on the Bio-H3 and Bio-H4, lots of ST-ALP were captured on the electrode surface via biotin-streptavidin interaction. Immediately, the cascade non-enzymatic signal amplification strategy integrating the CHA and HCR was achieved. Finally, the DPV signal response was measured under the enzymatic catalysis of ST-ALP with the substrate 1-NPP to an electroactive phenol for producing an amplified electrochemical signal. The obtained electrochemical signals were positively related to SARS-CoV-2 spike protein concentrations.
3.2. PAGE characterization of aptamer binding-induced multiple hairpin assembly
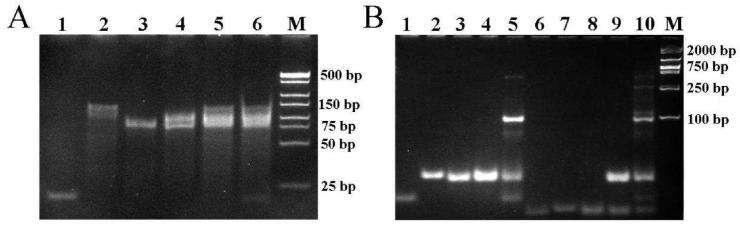
The aptamer binding-induced multiple hairpin assembly process was confirmed by polyacrylamide gel electrophoresis (PAGE). As depicted in Fig. 1 A, output DNA, Apt2 and Apt1 presented three distinct bands in lane 1, 2 and 3, respectively. The Apt1/output DNA complex was stable with no extra bands (lane 4). In the absence of the SARS-CoV-2 spike protein, the band of output DNA could not be observed, indicating that the Apt2 could not displace output DNA from Apt1/output DNA complex (lane 5). Conversely, in the presence of SARS-CoV-2 spike protein, the band of output DNA was clearly observed, suggesting that the Apt1-spike protein-Apt2 complex was formed along with the release of output DNA (lane 6). These results suggested that the proximity binding between SARS-CoV-2 spike protein and its aptamers induced output DNA released from the Apt1/output DNA complex.
Fig. 1.
(A) Native PAGE analysis of oligonucleotides from the proximity binding-induced DNA strand displacement. Lane 1, 2 μM output DNA; lane 2, 2 μM Apt2; lane 3, 2 μM Apt1; lane 4, 2 μM Apt1/output DNA; lane 5, the mixture of 2 μM Apt1/output DNA and 2 μM Apt2; lane 6, the mixture of 2 μM Apt1/output DNA, 2 μM Apt2, and 1 μg·mL−1 SARS-CoV-2 spike protein; lane 7, DL500 Marker. (B) Native PAGE displaying the assembly of programmed CHA and HCR. Lane 1, output DNA; lane 2, H1; lane 3, H2; lane 4, the mixture of H1 and H2; lane 5, the mixture of output DNA, H1, and H2; lane 6, H3; lane 7, H4; lane 8, the mixture of H3 and H4; lane 9, the mixture of H1, H2, H3, and H4; lane 10, the mixture of output DNA, H1, H2, H3, and H4.
As exhibited in Fig. 1B, the output DNA, H1 and H2 corresponded to lane 1, lane 2 and lane 3, respectively. Further, the mixture of H1 and H2 presented the specific bands of H1 and H2 in lane 4. When output DNA was added into the mixture of H1 and H2, a new band with lower migration appeared, indicating the output DNA-triggered the assembly of H1 and H2 (lane 5). It could be seen that H3 and H4 in lane 6 and lane 7 exhibited distinct bands, respectively. When H3 and H4 were mixed, the specific bands of H3 and H4 were separately observed in lane 8. When H1, H2, H3 and H4 were mixed, the multiple hairpin assembly process had not initiated (lane 9), as no new band with lower migration could be observed. Compared to lane 9, ladder-like bands appeared in the presence of output DNA, indicating the output DNA-induced cascading assembly of the hairpin probes H1, H2, H3 and H4 (lane 10). Therefore, the PAGE results demonstrated the feasibility of the designed multiple hairpin assembly strategy.
3.3. Characterization of electrode assembly process
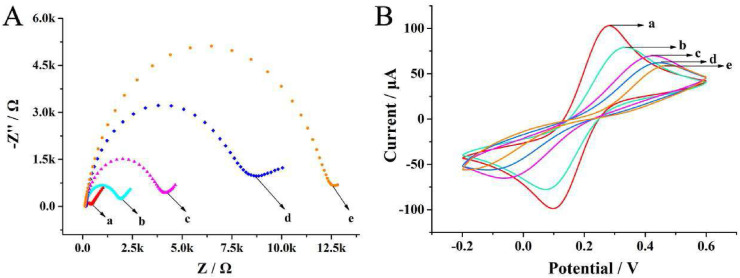
To investigate the fabrication process of the proposed electrochemical aptasensor, the different stepwise modifications were investigated using CV and EIS. In EIS, the semicircle diameter is equivalent to the electron transfer resistance (Ret), which controls the electron transfer kinetics of the redox probes at the electrode interface [14]. As displayed in Fig. 2 A, the cleaned bare gold electrode (GE) showed a narrow semicircle (curve a), indicating a good charge transfer ability. After thiolated H1 assembly on the gold electrode through Au–S interaction, the Ret was increased (curve b) owing to the resistance of negative DNA phosphoric backbone towards the access and redox of [Fe(CN)6]3-/4- on the electrode surface. The semicircle diameter was further increased with the nonconductive MCH immobilized on the electrode (curve c). After adding the mixed solution of SARS-CoV-2 spike protein, Apt1/output DNA, Apt2, H2, Bio-H3 and Bio-H4 on the gold electrode surface, the Ret increased significantly (curve d), which demonstrated the successful proximity binding-triggered DNA strand displacement reaction and the CHA/HCR cascade enzyme-free signal amplification process. Furthermore, with the incubation of ST-ALP, the Ret was further increased (curve e), because multiple streptavidin-alkaline phosphatases were assembled on the electrode via biotin-streptavidin interaction. As described in Fig. 2B, the results of CV measurements were consistent with the EIS, where the peak currents varied with the stepwise assembly process. These results indicated that the aptasensor functioned well, as expected.
Fig. 2.
(A) EIS and (B) CV characterization of electrodes at various modification stages in 5 mM [Fe(CN)6]3-/4- solution: (a) bare GE, (b) H1/GE, (c) MCH/H1/GE, (d) multiple hairpin assembly/MCH/H1/GE, (e) ST-ALP/multiple hairpin assembly/MCH/H1/GE. The EIS measurement was recorded with a frequency sweep range from 10−1 to 105 Hz and 10 mV amplitude. The CV measurement was recorded with a potential window from −0.2 to 0.6 V and a scan rate of 100 mV s−1.
3.4. Analytical performance of the electrochemical aptasensor
To achieve the best analytical performance of the biosensor, the reaction time of the mixture and H1 was optimized. The result was described in Supplementary Information and Fig. S1. Under the optimal condition, the current responses toward SARS-CoV-2 antigen in different concentrations were recorded through DPV measurements. As shown in Fig. 3 A, the electrochemical signals increased proportionally with the SARS-CoV-2 concentrations increased from 50 fg ·mL−1 to 50 ng·mL−1. The current intensity displayed a strong linear relationship to the logarithm value of target SARS-CoV-2 antigen concentration ranging from 50 fg·mL−1 to 50 ng·mL−1 (Fig. 3B). The linear regression equation was i = 2.2864 lg c + 12.0861 with r = 0.9987 (i and c represent the current intensity and the concentration of target SARS-CoV-2 antigen, respectively). The detection limit was calculated to be 9.79 fg·mL−1 (S/N = 3). In addition, compared with other analytical methods for SARS-CoV-2 spike protein detection (Table 2 ), it could be found that the prepared electrochemical aptasensor possessed a broader linear range and a lower detection limit due to the non-enzymatic multiple hairpin assembly signal amplification strategy.
Fig. 3.
(A) DPV curves response of the electrochemical aptasensor upon the increase in SARS-CoV-2 spike protein concentration (from bottom to top: 0, 50 fg·mL−1, 500 fg·mL−1, 5 pg·mL−1, 50 pg·mL−1, 500 pg·mL−1, 5 ng·mL−1 and 50 ng·mL−1, separately). (B) Calibration curve of current intensity and logarithm of concentration of SARS-CoV-2 spike protein.
Table 2.
Comparison of other reported methods for the SARS-CoV-2 spike protein analysis.
| Methods | linear rang | LOD | Ref. |
|---|---|---|---|
| Carbon magnetic beads-based electrochemical immunosensor | 40 ng·mL−1–10 μg·mL−1 | 14 ng·mL−1 | [15] |
| Nanozyme-based chemiluminescence immunoassay | 0.2 ng·mL−1–100 ng·mL−1 | 0.1 ng·mL−1 | [16] |
| Fiber-optic label-free biosensor | 1 pg·mL−1–100 μg·mL−1 | _ | [17] |
| Field-effect transistor-based electrical immunosensor | _ | 100 fg·mL−1 | [18] |
| Aptamer binding-induced multiple hairpin assembly-based electrochemical aptasensor | 50 fg mL−1∼ 50 ng mL−1 | 9.79 fg mL−1 | This work |
3.5. Specificity and repeatability of the electrochemical aptasensor
Previous research showed that SARS-CoV-2 shared more than 60% similarities with SARS-CoV, MERS-CoV, HCoV-HKU1 as well as HCoV-OC43 identified by genome sequence analysis [19]. Thus, the above-mentioned four human coronaviruses were selected as interferences to investigate the specificity of the fabricated electrochemical aptasensor for SARS-CoV-2 detection. Concretely, specificity of the aptasensor was evaluated by detecting SARS-CoV-2 antigen (50 ng mL−1), four human coronaviruses interferences (50 ng mL−1) and the mixture (composed of the 50 ng mL−1 interference coronavirus antigens and 0.5 ng∙mL−1 SARS-CoV-2 spike protein) at the same experimental conditions (Fig. 4 A). The current responses of other human coronavirus antigens were nearly the same as the blank even in the presence of 100-fold higher concentrations (50 ng mL−1) compared to SARS-CoV-2 spike protein (0.5 ng mL−1). When other human coronavirus antigens coexisted with SARS-CoV-2 spike protein, the current response was nearly the same as when only SARS-CoV-2 spike protein existed even if the concentrations of other human coronavirus antigens (50 ng mL−1) were 100-fold higher than that of SARS-CoV-2 (0.5 ng·mL−1). Therefore, the proposed electrochemical aptasensor exhibited high specificity for SARS-CoV-2 detection in complex interference environment.
Fig. 4.
(A) Specificity of the aptasensor for the detection of SARS-CoV-2 against the interference substances: Blank, 50 ng·mL−1 MERS-CoV spike protein, 50 ng·mL−1 SARS-CoV spike protein, 50 ng·mL−1 HCoV-OC43 spike protein, 50 ng·mL−1 HCoV-HKU1 spike protein, 0.5 ng·mL−1 SARS-CoV-2 spike protein, and the mixture of 50 ng·mL−1 MERS-CoV spike protein, 50 ng·mL−1 SARS-CoV spike protein, 50 ng·mL−1 HCoV-OC43 spike protein, 50 ng·mL−1 HCoV-HKU1 spike protein, 0.5 ng·mL−1 SARS-CoV-2 spike protein. (B) Reproducibility of five independent electrodes modified with 50 ng·mL−1, 0.5 ng·mL−1 and 5 pg·mL−1 of SARS-CoV-2 spike protein.
The repeatability of the aptasensor was also examined by independently measuring the SARS-CoV-2 antigen at different concentrations: 50 ng·mL−1, 0.5 ng·mL−1 and 5.0 pg·mL−1. As shown in Fig. 4B, after five independent measurements, these electrodes exhibited similar current responses, and the relative standard deviations (RSD) for SARS-CoV-2 antigen detection at different concentrations were 3.55%, 1.80%, and 3.46%, respectively. These results revealed that the proposed aptasensor showed acceptable repeatability.
3.6. The practical application of the electrochemical aptasensor
To evaluate the application of the aptasensor in practical complicated matrices, different concentrations of SARS-CoV-2 spike protein were added into the saline solution, which soaked nasopharyngeal swab from healthy people to simulate positive clinical samples. As shown in Table 3 , the RSD ranged from 0.42% to 2.88%, and the recovery rate ranged from 95.01% to 106.52%. These results demonstrated that the designed aptasensor was applicable for detection of SARS-CoV-2 antigen in clinical analysis.
Table 3.
Determination of SARS-CoV-2 spike protein in saline solution soaked nasopharyngeal swab from healthy people with the proposed aptasensor.
| Sample | Added (ng·mL−1) | Found (ng·mL−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 5.00 × 10 | 5.33 × 10 | 106.52 | 2.88 |
| 2 | 5.00 | 4.75 | 95.01 | 0.42 |
| 3 | 5.00 × 10−1 | 5.18 × 10−1 | 103.64 | 0.50 |
| 4 | 5.00 × 10−2 | 4.78 × 10−2 | 95.59 | 2.30 |
| 5 | 5.00 × 10−3 | 4.88 × 10−3 | 97.51 | 2.55 |
| 6 | 5.00 × 10−4 | 5.14 × 10−4 | 102.86 | 1.26 |
4. Conclusion
In summary, an efficient aptamer binding-induced multiple hairpin assembly signal amplification strategy was designed and applied to construct an electrochemical aptasensor for sensitive detection of SARS-CoV-2 spike protein. The aptamer binding-induced strand displacement converted the SARS-CoV-2 spike protein to DNA stand output, which was further amplified by the multiple hairpin assembly. The aptamer binding-induced multiple hairpin assembly strategy exhibited specific recognition as well as efficient signal amplification to SARS-CoV-2 spike protein. As a result, the proposed electrochemical aptasensor presented good specificity, satisfactory repeatability, and high sensitivity for SARS-CoV-2 spike protein detection under ambient temperature. Moreover, good performance was achieved with the aptasensor in spiking test, indicating the great application potential for the point of care analysis of SARS-CoV-2 antigen in practice.
Credit author statement
Jian Xue: Conceptualization, Methodology, Investigation, Data curation, Writing – original draft. Ying Li: Methodology, Investigation, Data curation, Formal analysis. Jie Liu: Visualization, Conceptualization, Resources. Zixuan Zhang: Software, Formal analysis. Rongjun Yu: Conceptualization, Visualization. Yaling Huang: Methodology, Software. Anyi Chen: Methodology, Data curation, Supervision, Writing – review & editing, Funding acquisition, Project administration. Jingfu Qiu: Project administration, Formal analysis, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was sponsored by Natural Science Foundation of Chongqing (cstc2020jcyj-zdxm0015), China Postdoctoral Science Foundation (2020M683259), the Science and Technology Support Program of Guizhou Science and Technology Department ((2020)4Y183) and the Science and Technology Support Program of Zunyi Science and Technology Bureau [NS(2020)22].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123605.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Morawska L., Milton D.K. It is time to address airborne transmission of coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020;71(9):2311–2313. doi: 10.1093/cid/ciaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soltesz K., Gustafsson F., Timpka T., Jalden J., Jidling C., Heimerson A., Schon T.B., Spreco A., Ekberg J., Dahlstrom O., Bagge Carlson F., Joud A., Bernhardsson B. The effect of interventions on COVID-19. Nature. 2020;588(7839):E26–E28. doi: 10.1038/s41586-020-3025-y. [DOI] [PubMed] [Google Scholar]
- 3.Abboah-Offei M., Salifu Y., Adewale B., Bayuo J., Ofosu-Poku R., Opare-Lokko E.B.A. A rapid review of the use of face mask in preventing the spread of COVID-19. Int. J. Nurs. Stud. Adv. 2021;3:100013. doi: 10.1016/j.ijnsa.2020.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasileiou E., Simpson C.R., Shi T., Kerr S., Agrawal U., Akbari A., Bedston S., Beggs J., Bradley D., Chuter A., de Lusignan S., Docherty A.B., Ford D., Hobbs F.D.R., Joy M., Katikireddi S.V., Marple J., McCowan C., McGagh D., McMenamin J., Moore E., Murray J.L.K., Pan J., Ritchie L., Shah S.A., Stock S., Torabi F., Tsang R.S.M., Wood R., Woolhouse M., Robertson C., Sheikh A. Interim findings from first-dose mass COVID-19 vaccination roll-out and COVID-19 hospital admissions in Scotland: a national prospective cohort study. Lancet. 2021;397(10285):1646–1657. doi: 10.1016/S0140-6736(21)00677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K., Southern J., Swerdlow D.L., Jodar L., Levy Y., Alroy-Preis S. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397(10287):1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. N. Engl. J. Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., Oikawa M.K., Salomon T., Cucunuba Z.M., Castro M.C., de Souza Santos A.A., Nascimento V.H., Pereira H.S., Ferguson N.M., Pybus O.G., Kucharski A., Busch M.P., Dye C., Faria N.R. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 9.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z.W., Chen H.L., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman V.M., Haage V.C., Bleicker T., Schmidt M.L., Muhlemann B., Zuchowski M., Jo W.K., Tscheak P., Moncke-Buchner E., Muller M.A., Krumbholz A., Drexler J.F., Drosten C. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. The Lancet Microbe. 2021;2(7):e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92(14):9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 13.H.B. Sun, J.L. Liu, Y.L. Qiu, J.M. Kong, X.J. Zhang, High sensitive electrochemical methamphetamine detection in serum and urine via atom transfer radical polymerization signal amplification. Talanta, 238 (2022), 123026. [DOI] [PubMed]
- 14.Zhang B., Zhang Y., Liang W.B., Cui B., Li J.B., Yu X.J., Huang L. Nanogold-penetrated poly(amidoamine) dendrimer for enzyme-free electrochemical immunoassay of cardiac biomarker using cathodic stripping voltammetric method. Anal. Chim. Acta. 2016;904:51–57. doi: 10.1016/j.aca.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2020;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S.L., Kim J., Choi S., Han J., Seoc G., Lee Y.W. Fiber-optic label-free biosensor for SARS‐CoV‐2 spike protein detection using biofunctionalized long-period fiber grating. Talanta. 2021;235(1):122801. doi: 10.1016/j.talanta.2021.122801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Liu B., Cui J., Wang Z., Shen Y., Xu Y., Yao K., Guan Y. 2020. Similarities and Evolutionary Relationships of COVID-19 and Related Viruses. arXiv preprint arXiv: 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.