Abstract
A 3-kb region containing the determinant for bacteriocin activity from Rhizobium leguminosarum 248 was isolated and characterized by Tn5 insertional mutagenesis and DNA sequencing. Southern hybridizations showed that this bacteriocin was encoded on the plasmid pRL1JI and that homologous loci were not found in other unrelated R. leguminosarum strains. Tn5 insertional mutagenesis showed that mutations in the C-terminal half of the bacteriocin open reading frame apparently did not abolish bacteriocin activity. Analysis of the deduced amino acid sequence revealed that, similarly to RTX proteins (such as hemolysin and leukotoxin), this protein contains a characteristic nonapeptide repeated up to 18 times within the protein. In addition, a novel 19- to 25-amino-acid motif that occurred every 130 amino acids was detected. Bacteriocin bioactivity was correlated with the presence of a protein of approximately 100 kDa in the culture supernatants, and the bacteriocin bioactivity demonstrated a calcium dependence in both R. leguminosarum and Sinorhizobium meliloti. A mutant of strain 248 unable to produce this bacteriocin was found to have a statistically significant reduction in competitiveness for nodule occupancy compared to two test strains in coinoculation assays. However, this strain was unable to compete any more successfully with a third test strain, 3841, than was wild-type 248.
Bacteriocins are often defined as narrow-spectrum antibiotics produced by bacteria and active against only closely related species or strains (43). Rhizobium leguminosarum strains have been shown to produce bacteriocins which have been characterized as small, medium, or large based on their assumed sizes and diffusion characteristics (19, 41). Large bacteriocins have been shown to resemble defective bacteriophages (19, 28, 41). Small bacteriocins were found to be chloroform soluble and heat labile and to have molecular masses of less than 2,000 daltons (19, 50). More recently, small bacteriocins were shown to be acylated homoserine lactone compounds related to quorum-sensing molecules (18, 39).
Very little is known about medium bacteriocins produced by R. leguminosarum. It has been shown that, whereas small bacteriocins appeared to be produced nearly ubiquitously, with all producers cross resistant to each other, relatively few strains produce medium bacteriocins. Cross-resistance patterns suggested that there may be several different bacteriocins within the medium bacteriocin family (19, 53).
R. leguminosarum 248 contains the symbiotic plasmid pRL1JI, which is one of the genetically best characterized nodulation plasmids. As well as containing genes necessary for nodulation and nitrogen fixation, this plasmid has been shown to carry determinants for medium bacteriocin production (20). In a study of field populations of R. leguminosarum, it was observed that pRL1JI or closely related nodulation plasmids were the most prevalent in strains isolated from nodules, suggesting that there must be a natural selective pressure to maintain this type of plasmid (55). However, the significance of bacteriocin production with respect to the prevalence of this family of plasmids has not been directly addressed.
In an effort to understand how medium bacteriocins may contribute to the prevalence of R. leguminosarum 248 in nodules, we have cloned and characterized a gene from pRL1JI capable of giving in vivo medium-bacteriocin activity. Nucleotide sequencing of this region suggests that medium bacteriocin is related to RTX-type proteins, which include calcium-dependent cytolysins, such as hemolysin and leukotoxin (52). Consistent with this, it was shown that the in vivo activity of this bacteriocin is enhanced by Ca2+. Nodulation competition experiments with other R. leguminosarum wild types and strain 248 show that the presence of bacteriocin activity in strain 248 may influence its competitiveness.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used or produced in this work are listed in Table 1. R. leguminosarum strains were grown on TY medium (4). Sinorhizobium meliloti strains were grown on either TY or Luria-Bertani (LB) medium (30), and E. coli was grown on LB medium. Antibiotics were used as necessary at the following concentrations: streptomycin, 200 μg/ml; neomycin, 100 μg/ml; tetracycline, 5 μg/ml; kanamycin, 20 μg/ml; chloramphenicol, 20 μg/ml; gentamicin, 50 μg/ml; and spectinomycin, 50 μg/ml.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| R. leguminosarum | ||
| 248 | Wild type | 19 |
| 306 | Wild type | 19 |
| 309 | Wild type | 19 |
| 336 | Wild type | 19 |
| VF39SM | Smr wild type | 34 |
| 3841 | Smr wild type | 33 |
| 6015 | Smr Rfrphe-1 trp-12 nod-6007 | 24 |
| GF160 | Smr wild type | 13 |
| W14-2 | Smr wild type | 2 |
| 162Y10 | Wild type | 31 |
| 8401/pJB5JI | rzc-1::Tn5 | A. Downie |
| Rl1003 | 248 rzc-1::Tn5 | This work |
| Rlv1031 | 336 Smr | This work |
| S. meliloti Rm1021 | SU47 str-21 | 29 |
| Plasmids | ||
| pPH1JI | IncP Gmr | 5 |
| pRK7813 | RK2 derivative carrying pUC9 polylinker; Tcr | 25 |
| pBluescript | Cloning vector; ColE1 Apr | Stratagene |
| pBAC1 | Tn5 from pJB5JI cloned into pBluescript as an EcoRI fragment | This work |
| pBAC4 | BamHI/EcoRI subclone of pBAC1 DNA flanking rzc248A-1 | This work |
| pBAC5 | BamHI subclone from pBAC1; Kmr DNA flanking rzc248A-1 | This work |
| pBAC8 | pRK7813 cosmid clone carrying rzc+ | This work |
| pBAC9 | pRK7813 cosmid clone carrying rzc+ | This work |
| pBAC10 | pRK7813 cosmid clone carrying rzc+ | This work |
| pBAC11 | pRK7813 cosmid clone carrying rzc+ | This work |
| pBAC12 | 5.3-kb EcoRI subclone from pBAC12 in pRK7813; rhizobiocin bioactivity | This work |
| pBAC17 | 3.8-kb BamHI/EcoRI subclone from pBAC12 in pRK7813; Tcr | This work |
| pBAC18 | 0.6-kb BamHI/EcoRI fragment from pBAC12 in pRK7813; Tcr | This work |
| pBAC19 | 1-kb BamHI fragment from pBAC12 in pRK7813; Tcr | This work |
| pBACp1 | pBAC12; rzc-3::TnphoA | This work |
| pBAC1p12 | pBAC12 1p12::TnphoA | This work |
| pBAC1p13 | pBAC12 1p13::TnphoA | This work |
| pBACL13 | pBAC12 rzc-8:::Tn5B20 | This work |
| pBACL14 | pBAC12 rzc-9::Tn5B20 | This work |
| pBAC2.13 | pBAC12 rzc-2::Tn5 | This work |
| pBAC5.8 | pBAC12 rzc-11::Tn5 | This work |
| pBAC5.13 | pBAC12 rzc-14::Tn5 | This work |
| pBACΔp1 | HindIII deletion from IS50 to polylinker of pBACp1 | This work |
| pBACΔL14 | HindIII deletion from IS50 to polylinker of pBACl14 | This work |
| pBACΔ2-13 | HindIII deletion from IS50 to polylinker of pBAC2.13 | This work |
| pBACΔL13 | HindIII deletion from IS50 to polylinker of pBACl13 | This work |
| Bacteriophage RL38 | General transducing phage for R. leguminosarum | 8 |
Abbreviations for antibiotics are as follows: Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Rf, rifampin; Sm, streptomycin; Tc, tetracycline.
Genetic techniques, DNA manipulations, and sequencing.
Bacterial matings, transposon mutageneses, and gene replacements were carried out essentially as described previously (9, 14, 15, 37, 42, 54). Standard procedures were used for plasmid isolation, restriction endonuclease digestions, ligations, transformations, agarose gel electrophoresis, and Southern transfers (38). Genomic DNA was isolated as described previously (29). Cosmid bank construction was carried out essentially as previously described (32). Briefly, 248 genomic DNA was partially digested with EcoRI and ligated to the costramid vector pRK7813 (25). The ligated DNA was packaged with a lambda packaging extract (Promega Corp., Madison, Wis.) as recommended by the manufacturer.
To move the rzc-1::Tn5 allele into 248 from 8401/pJB5JI, pJB5JI was first conjugally transferred into strain 6015, selecting for transfer of Nmr and counterselecting 8401 with rifampin. The rzc-1::Tn5 allele was subsequently transduced into strain 248 by using the generalized transducing phage RL38 and selecting for the transfer of Nmr. The resultant transductants were screened for loss of bacteriocin production, and one such colony was verified and designated Rl1003.
Nucleotide sequencing was carried out by using a combination of primer walking, subcloning, and sequencing of DNA flanking Tn5, Tn5B20, and TnphoA inserts. T3 and T7 primers were used to sequence the ends of the subclones, and an IS50 primer (5′TAGGAGGTCACATGGAAGTCAGAT 3′) was used to sequence the DNA flanking the IS50. Sequencing reactions were done with dye terminators and detected with an ABI automated sequencer at the University of Calgary Core DNA facilities.
Sequence data were analyzed with DNASIS (Hitachi Software Engineering Co., San Bruno, Calif.), and database searches were done with the BLASTX program (1).
Modified Eckhardt gel electrophoresis.
To verify plasmid profiles of R. leguminosarum strains or to isolate plasmid DNA, a modification of the Eckhardt (10) technique designed for horizontal gels was used, as previously described (22).
Bacteriocin assays.
To assay medium bacteriocin production, a saturated culture of the indicator strain (either 336 or VF39SM) grown in TY medium was diluted 10−2 and 1 ml was mixed with approximately 25 ml of soft TY agar (0.6% [wt/vol] agar) containing 5 mM Ca2+ (final concentration). Single colonies of strains to be tested for bacteriocin activity were stab inoculated into the soft agar within 2 h after the agar solidified. Halos were visible as cleared zones surrounding the stab-inoculated culture. Unless otherwise noted, the plates were scored approximately 48 h after stab inoculation.
Preparation of supernatant proteins.
To visualize bacteriocin protein, the method utilized by Hirsch (19) was modified. Rhizobium cultures were grown in 200 ml of TY medium for 5 days. The cultures were pelleted with a GSA rotor at 5,000 × g for 20 min. The supernatant was poured off and treated with chloroform (10% [vol/vol]) in a separatory funnel. The following steps were carried out on ice or at 5°C. The aqueous phase was collected, and the proteins were precipitated by the addition of ammonium sulfate to 80% (wt/vol) saturation. The precipitate was collected by spinning it at 10,000 × g for 30 min and was finally resuspended in approximately 3 ml of Tris · HCl (pH 6.8). Supernatants were routinely tested for bioactivity by testing dilutions of the extracts on TY agar plates seeded with the indicator strain 336. Proteins were separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (27) and stained with a Bio-Rad (Hercules, Calif.) silver stain kit as recommended by the manufacturer. The protein concentration was determined by a Bio-Rad protein determination protocol, with bovine serum albumin as a standard.
Plant tests.
Plant tests were carried out by the methods of Hynes et al. (21). Pea seeds (Pisum sativum cv. Trapper) were surface sterilized by 5 min of 70% ethanol treatment followed by 10 min in a 1/5 dilution of hypochlorite. The seeds were then rinsed in at least four changes of sterile distilled deionized water. The seeds were germinated on half-strength TY medium, and only those sterile seeds were aseptically planted into sterile vermiculite that had been presoaked with plant growth solution (51). Strains for inoculation were grown overnight in TY medium, pelleted, and resuspended to a uniform density in either sterile water or plant nutrient solution. The bacteria were then mixed in an approximate 1:1 ratio, the mixture was diluted, and 10 ml at approximately 105 CFU/ml was inoculated onto the seedlings. The inoculation mix was further diluted and plated to determine the precise inoculation ratio that had been used for each experiment.
Nodules were harvested after the plants had grown for 4 to 5 weeks in a growth chamber. The nodules were surface sterilized, crushed, and streaked out onto TY plates as described by Hynes and O’Connell (23). Four colonies from each crushed nodule were tested, and the identities of the strains in the nodules were determined by antibiotic resistance. The statistical significance of the data from nodule competition assays was analyzed by several techniques. The chi-square test was used to evaluate the significance of differences between the two strains used in a given experiment (i.e., whether deviations from the inoculation ratio were significant), and also as a preliminary means of determining whether strain Rl1003 was less competitive than 248 versus given test strains. In these tests, the observed data (pooled from three independent experiments) on the frequency of recovery of 248 when assayed against a given test strain were used as the expected frequency for Rl1003. To confirm the validity of this approach, analysis of variance and means model analyses of the data from the competition experiments were carried out with the SYSTAT package (version 5.2.1), with the null hypothesis being that the frequency of recovery of Rl1003 was no different from that of 248 when assayed against the same test strain. Adjustments to compensate for slight discrepancies in inoculation ratio did not change the results of these tests significantly.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in EMBL under accession no. AJ001518.
RESULTS
R. leguminosarum strains produce distinct bacteriocins.
R. leguminosarum medium bacteriocins have been previously described as compounds which produce a zone of inhibition against sensitive strains with a diameter between 2 and 10 mm (19). To identify strains capable of producing medium bacteriocin and to analyze cross-resistance patterns, 33 strains of R. leguminosarum (field isolates and laboratory strains) were tested with strain 248 as an indicator for small bacteriocin and 336 as an indicator for medium bacteriocin. It was found that only 4 of the 33 strains did not produce small bacteriocin and only 6 of the 33 strains produced a medium bacteriocin (data not shown). In addition to strains 248, 306, and 309, which were known to produce medium bacteriocin (19), strains GF160, 162Y10, and W14-2 were also found to produce medium-type bacteriocin (Table 2). Based on their production and resistance patterns, R. leguminosarum strains could be divided into at least four groups. Group I contained strain 248. The strains in group II (306 and 309) appeared to carry the group I determinant as well as a bacteriocin which distinguished strains 306 and 309 from 248. Group III contained strains GF160 and 162Y10, which had bacteriocin production patterns similar to that of W14-2 but were sensitive to the bacteriocin which is produced by strains 306 and 309. Group IV was defined by strain W14-2, which, based on its sensitivity patterns, was distinct from all other strains tested.
TABLE 2.
Comparison of bacteriocin production and resistance patterns for R. leguminosarum strainsa
| Strain | Presence of zone of inhibition with indicator strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| VF39SM | 336 | 248 | 306 | 309 | GF160 | 162Y10 | W14-2 | |
| VF39SM | − | − | + | + | + | + | + | + |
| 336 | − | − | + | + | + | + | + | + |
| 248 | + | + | − | − | − | − | − | + |
| 306 | + | + | + | − | − | + | + | + |
| 309 | + | + | + | − | − | + | + | + |
| GF160 | + | + | + | + | + | − | − | − |
| 162Y10 | + | + | + | + | + | − | − | − |
| W14-2 | + | + | + | + | + | − | − | − |
Strains were assayed for bacteriocin production by stab inoculating them into a soft TY agar plate that had been seeded with an indicator strain. +, presence of a zone of inhibition; −, absence of a zone of inhibition. VF39SM and 336 are small-bacteriocin-producing strains that are used as indicators for medium-bacteriocin production.
Based on the results from the cross-reactivity tests, it is clear that there are several different bacteriocins produced by R. leguminosarum strains which may or may not be similar in structure and function. These bacteriocins have traditionally been termed medium bacteriocins, presumably indicating molecular size (19). We intend to use the term rhizobiocins (rzc) to refer to these particular bacteriocins. This term does not presuppose any molecular characteristics and has already been used to refer to bacteriocins of rhizobial origin (36).
Cloning the region of 248.
It had previously been shown that a Tn5 insert localized to pRL1JI was unable to produce bacteriocin; this plasmid carrying the insert was renamed pJB5JI (24). Transduction of kanamycin resistance from pJB5JI back to the wild-type plasmid (pRL1JI) resulted in cotransduction of the bacteriocin phenotype, showing that the Tn5 insert (referred to here as rzc-1::Tn5) was responsible for the lack of bacteriocin production (24).
To generate a probe for the bacteriocin-encoding region on pRL1JI, a clone was made from the DNA flanking this Tn5 insert. Plasmid DNA from pJB5JI (containing rzc-1::Tn5) was isolated from preparative Eckhardt gels of strain 8401/pJB5JI. The DNA was restricted with EcoRI and ligated into pBlueScript. The Tn5 insert was in a 5.2-kb EcoRI fragment, and the clone containing this insert was designated pBAC1 (Table 1). The IS50R and flanking DNA from pBAC1 were subsequently subcloned to yield pBAC4 (Table 1), which was used as a probe to screen a genomic cosmid library of strain 248.
To find cosmids containing the wild-type bacteriocin region, 800 cosmid-containing colonies of Escherichia coli were screened by Southern blot analysis. Four independent cosmids were isolated and designated as pBAC8 to pBAC11 (Table 1). All four contained the 5.2-kb EcoRI fragment. Based on the complexity of the EcoRI restriction pattern, pBAC11 was chosen for further analysis.
To confirm that the cloned DNA originated from pRL1JI, the 5.2-kb EcoRI fragment from pBAC11 was subcloned, gel isolated, labelled, and used to probe Southern blotted Eckhardt gels containing strain 248 and other R. leguminosarum strains. The wild-type EcoRI fragment from pBAC11 hybridized to pRL1JI (data not shown). Moreover, Southern analysis of genomic DNA from strains carrying the rzc-1::Tn5 allele gave the expected hybridization pattern (data not shown). Together these data confirm that the 5.2-kb EcoRI fragment was not reiterated within the genome and originated from pRL1JI.
Group I and group II strains carry highly homologous bacteriocin production genes.
Based on the cross-resistance patterns of the various R. leguminosarum strains (Table 2), it was of interest to determine if any of the other strains carried the same bacteriocin determinants. Genomic DNA from these strains was probed with the labelled 5.2-kb EcoRI fragment. Consistent with the phenotypic data (Table 2) and what has been previously surmised (7, 19, 20), strains 306 and 309 gave hybridization patterns identical to that of strain 248, regardless of the restriction enzyme used to digest the DNA. The hybridization data suggest that 306, 309, and 248 all carry a highly homologous bacteriocin encoding region (data not shown). However, strains GF160, W14-2, and 162Y10 did not hybridize with this probe even at low stringencies, suggesting that genes coding for bacteriocins in these strains are not highly homologous to the bacteriocin gene encoded by pRL1JI (data not shown).
Genetic delineation of the bacteriocin region.
Mobilization of the cosmids pBAC8, pBAC9, pBAC10, and pBAC11 into strains that do not produce rhizobiocin, such as Rhizobium leguminosarum 3841, VF39, or S. meliloti Rm1021, was correlated with the ability of these strains to produce a rhizobiocin. Since the rhizobiocin was produced in S. meliloti, which is not sensitive to small or medium bacteriocins, constructs and inserts in the bacteriocin region were regularly screened for bioactivity in an Rm1021 background.
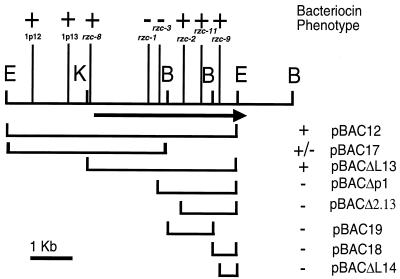
Introduction of pBAC12 into Rm1021, 3841, and Rl1003 showed that this subclone was sufficient for rhizobiocin bioactivity (Fig. 1 and data not shown). In an effort to delineate which regions of the insert contained the gene(s) necessary to confer bacteriocin production, a series of subclones and deletions was constructed. A subclone, pBAC17, containing the large BamHI/EcoRI fragment was shown to have partial activity (a smaller, less well-defined zone of inhibition) when tested for bacteriocin production (Fig. 1). Neither the smaller BamHI fragment (pBAC19) nor the 0.6-kb BamHI/EcoRI fragment (pBAC18) was sufficient to confer any activity. A series of directional deletions was constructed between the polylinker of pRK7813 and Tn5 inserts that were regularly spaced in pBAC12. The region necessary for bacteriocin production was deemed to be at most 3 kb, spanning from the KpnI site in pBAC12 to the distal EcoRI site (Fig. 1).
FIG. 1.
Delineation of bacteriocin-encoding region by transposon mutagenesis and deletion analysis. A partial restriction map of the bacteriocin region from pRL1JI is shown. The vertical lines indicate representative transposon inserts, with the bacteriocin phenotype marked above. Plasmids carrying deletions in the bacteriocin region are aligned below the map, with bacteriocin phenotypes marked along the right. The ORF and its direction of transcription are represented by the thick arrow. +, zone of inhibition like that of wild type; +/−, zone of inhibition smaller than that of wild type; −, absence of a zone of inhibition; E, EcoRI; B, BamHI; K, KpnI.
To delineate the extent of the bacteriocin gene more precisely, transposon mutageneses with Tn5, Tn5B20, and TnphoA were performed. The results from these mutageneses yielded many bacteriocin-positive inserts that flanked two definite bacteriocin knockouts. Based on the genetic data, this suggested a region of approximately 2 kb (Fig. 1).
Nucleotide sequencing and ORF determination.
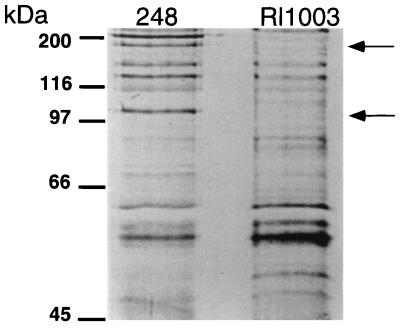
Nucleotide sequencing was carried out by a strategy that utilized subcloning, using IS50 elements from Tn5 inserts as priming sites, and primer walking. Analysis of the sequence data suggested that inserts giving an rzc mutant phenotype isolated from pBAC12 were within a 2,874-bp putative open reading frame (ORF) (Fig. 1). This ORF extends well beyond the first bacteriocin production-positive insert, rzc-2, as well as beyond the pBAC12 subclone itself (Fig. 1). To resolve this dilemma, supernatant preparations of secreted proteins were prepared from 248 and Rl1003 (carrying the rzc-1::Tn5) in an effort to determine the size of the wild-type bacteriocin protein. SDS-PAGE and Coomassie blue staining of ammonium sulfate-concentrated preparations did not reveal any differences between the supernatants of Rl1003 and 248. SDS-PAGE and silver staining of these preparations, however, demonstrated that the wild-type strain contained a low-abundance protein slightly larger than 97 kDa and another protein at less than 200 kDa, both of which were absent from the strain carrying the rzc-1 allele (Fig. 2).
FIG. 2.
Silver-stained SDS-PAGE analysis of the supernatant of 248 and Rl1003. The arrows on the right point to the positions of bands missing in the mutant strain. Molecular mass markers are shown on the left. Note that the Rl1003 lane has almost twice as much protein loaded as the 248 lane. All bands visible in Rl1003 have been shown to be present in 248.
The predicted molecular mass of the putative protein encoded by the ORF is 102,474 Da, which correlates well with the results of SDS-PAGE (Fig. 2). Consistent with this, rzc-9::Tn5B20 was mapped 696 bp from the end of the ORF, and it was shown to create a transcriptional fusion in the same orientation as the putative ORF. Expression studies with this fusion indicated that this ORF is constitutively expressed in TY medium and that transcriptional activity did not differ significantly with the culture growth phase (data not shown).
Together these data suggest that the rhizobiocin ORF codes for a low-abundance 102-kDa secreted protein that may form aggregates which were not separated by the denaturing conditions used (Fig. 2). Interestingly, the G+C content of this gene is 54%. This value is well below the typical 58 to 60% G+C content which is expected for Rhizobium strains (17).
248 rhizobiocin contains regions found in RTX proteins.
BLASTX searches of the databases with the bacteriocin ORF showed that the predicted translation product was related to FrpA from Neisseria meningitidis (44), HylA (12), and other related cytolysins and calcium-dependent epimerases. RTX (for repeat in toxin) proteins are characterized by the nonapeptide sequence L/I/F-X-G-G-X-G-N/D-D-X (52). Further analysis of the predicted amino acid sequence shows that the rhizobiocin protein contains only two such precise repeats (Fig. 3), but if positions 4 (G), and/or 7 (N/D) is relaxed, 18 additional repeat sequences can be found (Fig. 3). The predicted amino acid sequence of the bacteriocin also suggests that this is a glycine- and aspartate-rich protein. Together, these amino acids make up 27% of the total protein. The predicted pI of this protein is 3.7. The protein contains 3 cysteine residues, all of which are in the C-terminal 50 amino acids (Fig. 3).
FIG. 3.
Deduced amino acid sequence of the bacteriocin protein. Putative RTX repeats are underlined in the sequence, and the repeated motif is shown in boldface.
Transposon insertions that mapped within the C terminus-encoding half of the bacteriocin ORF did not abolish bacteriocin activity (Fig. 1 and data not shown). Using Rm1021 carrying pBAC12, or derivatives with inserts within the C terminus encoding half of the rhizobiocin ORF, we attempted to demonstrate a correlation between the presence of truncated proteins and bioactivity. These attempts were unsuccessful (data not shown). The predicted truncated protein products were not visualized by silver staining concentrated culture supernatants that were separated by SDS-PAGE. It appears that the partial rhizobiocin produced in these strains may be more unstable than the wild type, or perhaps it is not secreted efficiently, which complicates the analysis of these putative truncated proteins.
Bacteriocin ORF contains a second repeated motif.
Further analysis of the deduced amino acid sequence revealed the presence of a 19- to 25-amino-acid motif that is encoded six times within the bacteriocin ORF (Fig. 3). The motif is quite well conserved within the protein, and several amino acids appear to be very strongly conserved (Fig. 4). In addition, this sequence is periodic, occurring at consistent intervals of 127 to 132 amino acids within the protein. Database searches with various BLAST programs did not reveal any significant matches that might provide insight into the biological significance of this motif in any currently described protein or potential product of a sequenced gene. Interestingly, this motif was detected in another rhizobiocin that is currently being characterized (49) (see also accession no. AF141932).
FIG. 4.
Alignment of the repeated motif from the bacteriocin protein. The six repeated amino acid sequences were aligned with CLUSTALV. Positions that have amino acids that are conserved (∗) or have conservative substitutions (■) in all six repeats are shown below the alignment. The position of the repeat in the deduced protein is shown by the amino acid number immediately to the left of each sequence. Amino acids that are conserved in at least three of the six repeats are boxed. A possible signature sequence is shown below the alignment.
Bacteriocin bioactivity is affected by calcium concentration.
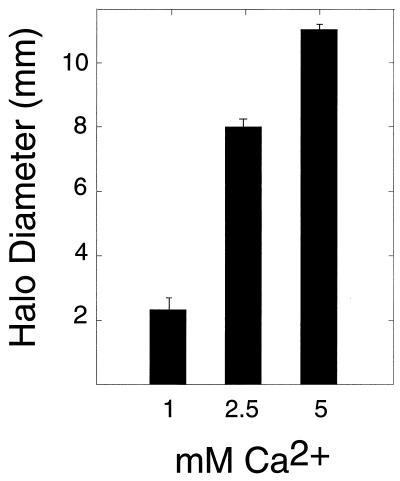
Proteins containing RTX nonapeptide repeats have been shown to bind calcium (3). Since the repeat sequence within the bacteriocin was not perfectly conserved, it was not clear whether calcium might have an effect on the bioactivity of the protein. To determine if calcium had an effect on the bioactivity of 248 bacteriocin, tests were carried out in which the concentration of calcium in the TY medium containing the indicator strain 336 was varied from 1 to 10 mM. It was found that the increase in halo size around strain 248 was positively correlated with the concentration of calcium up to 5 mM (Fig. 5). The halo sizes at 1, 2.5, and 5 mM were significantly different (Fig. 5). However, there were no discernible differences in the sizes of the halos formed around 248 with 5 and 10 mM calcium in the media. Moreover, the presence or absence of pBAC12 in strain 248 also did not alter the sizes of the halos.
FIG. 5.
Effect of calcium on zones of inhibition. Single colonies of R. leguminosarum 248 were stab inoculated into soft TY agar containing R. leguminosarum 336 as an indicator strain and either 1, 2.5, or 5 mM CaCl2. Zones of inhibition were scored after 48 h. n = 18 in each case. The error bars indicate standard deviations.
To determine if the calcium effect was a result of the differing sensitivity of 336 to the bacteriocin or a change in the potency of the 248 rhizobiocin, strain 248 was grown in medium containing 1 mM calcium. The supernatants were subsequently filter sterilized and treated with either 5 mM Ca2+, 5 mM EGTA, or a combination of the two. Bacteriocin potency was determined by serial twofold dilutions onto a plate seeded with the 336 indicator strain. The results showed that treatment of the supernatant with EGTA did not reduce the supernatant’s potency but treating it with 5 mM calcium resulted in a twofold increase in the potency of the rhizobiocin. The addition of EGTA and Ca2+ in combination also did not increase the potency of the rhizobiocin above that of untreated supernatant. This effect was seen with the supernatants of both overnight and 5-day-old cultures. Together, these data suggest that calcium, once bound to the rhizobiocin, is not removed by a simple EGTA treatment and that calcium affects the bioactivity of the rhizobiocin.
Bacteriocin plays a role in nodule competition.
To address the role that this bacteriocin may play in determining competition for nodulation, pBAC12 was introduced into VF39SM. This construct was then competed against VF39SM, which is sensitive to the bacteriocin produced by this subclone. These experiments were unsuccessful due to the instability of pBAC12 in the rhizosphere in the absence of antibiotic selection (data not shown). An alternate approach was undertaken in which the wild-type strain and the isogenic strain Rl1003 (containing rzc-1::Tn5) were each competed against VF39SM, 3841, and Rlv1031 under identical conditions. To validate this approach, we tested Rl1003 to ensure that it was immune to the 248 rhizobiocin and that it could compete equally well with the wild type under our assay conditions. The results showed that Rl1003 was resistant to the 248 rhizobiocin and that Rl1003 was equally competitive. These data were consistent with our expectations and allowed us to make comparisons between 248 and Rl1003 when assayed against other strains (Table 3).
TABLE 3.
Nodulation competition assays of bacteriocin-negative mutants with VF39SM, Rlv1031, and 3841 as indicator strains
| Competition | Inoculation ratio | Total no. of nodules scoreda | No. of nodules containing indicator straina |
|---|---|---|---|
| VF39SM/248 | 1.0 ± 0.1 | 216 | 10 |
| VF39SM/Rl1003 | 1.2 ± 0.2 | 223 | 35b |
| Rlv1031/248 | 0.5 ± 0.02 | 155 | 18 |
| Rlv1031/Rl1003 | 0.5 ± 0.04 | 142 | 52b |
| 3841/248 | 0.5 ± 0.02 | 144 | 76 |
| 3841/Rl1003 | 0.5 ± 0.03 | 148 | 76 |
| 248/Rl1003c | 1.0 | 98 | 49 |
Pooled data from three independent competition experiments. The data from all independent experiments showed identical trends.
Statistically significant result, showing reduction of competitiveness of Rl1003. Using analysis of variance (Systat package) on the independent replicates, the difference in recovery ratio for Rl1003 as opposed to 248 when assayed against VF39SM was significant at a P level of 0.05 (P value, 0.026). The results of the tests versus Rlv1031 were significant at a P level of 0.001 (P value, 0.2935 × 10−4). Fisher’s least significant difference was used as a means comparison test. As outlined in Materials and Methods, chi-square tests gave the same results.
Average of two independent competition experiments.
When strain 248 was competed against VF39SM, it was found to be more competitive than VF39, and under controlled conditions less than 5% of the nodules contained VF39SM (Table 3). However, when Rl1003 was competed against VF39SM under identical conditions, the data showed that VF39SM was significantly (99% confidence interval) more successful (Table 3). Similarly, when Rlv1031 was used as an indicator strain against the wild type, it typically occupied 11% of the nodules (at the given inoculation ratio), whereas when Rlv1031 was competed against Rl1003, the data show that Rlv1031 occupied 37% of the nodules analyzed (significant at the 99% confidence interval). Since the inoculation ratio in these experiments was 0.5, we would expect that if these strains were equally competitive, 33% of the nodules should be occupied by Rlv1031. This compares favorably with the observed value of 37% (52 of 142) (Table 3). Interestingly, competitions involving 3841 as an indicator strain appeared to be unaffected by the presence or absence of bacteriocin in strain 248 under our growth conditions, even though 3841 is sensitive to the bacteriocin produced by 248 in laboratory assays (Table 2). Taken together, these data show that, although the 248 bacteriocin appears to play a role in determining competitiveness for nodulation when assayed against some strains, it is probably only one of many factors affecting the outcome of competition experiments.
DISCUSSION
In her 1979 study, Hirsch (19) classified bacteriocins from R. leguminosarum as small or medium, based on apparent diffusion properties, while recognizing that other studies (28, 41) had revealed that even larger bacteriocins resembling defective phage particles existed in some strains. Our laboratory has elected to study medium bacteriocins, as the lack of cross-resistance between different medium bacteriocin-producing strains indicated that such compounds might potentially play a role in rhizosphere competition. The medium bacteriocins appear to have the properties commonly associated with “true” bacteriocins (i.e., they are proteinaceous and plasmid encoded, and a variety of different types are produced by different strains), and we propose to refer to them as rhizobiocins according to the suggestion of Roslycky (36). A further bacteriocin, which has been well characterized in R. leguminosarum, is the trifolitoxin produced by strain T24 (6, 40, 45–47), which has been shown to be a short peptide (6) somewhat similar to microcins and whose precise mode of action is unknown.
In the present study, the rhizobiocin-encoding region from plasmid pRL1JI was cloned and characterized. Evidence from DNA sequencing and Tn5 mutagenesis strongly indicates that there is only one ORF present at this locus which is necessary for rhizobiocin activity.
This ORF encodes a protein of 958 amino acids with a predicted molecular mass of 102.5 kDa. This predicted mass is in accordance with the masses of proteins seen in wild-type culture supernatants. The predicted protein has homology with various members of the RTX family of calcium binding proteins, many of which are toxins or hemolysins produced by pathogenic bacteria. The motif which gives rise to this homology is the repeated sequence believed to be involved in calcium binding (L/I/F-X-G-G-X-G-N/D-D-X), which is also present in some other Rhizobium proteins, such as NodO (11). It is noteworthy that calcium appears to be required for the activity of the rhizobiocin. The reading frame also encodes a second motif to which we have not been able to ascribe a biological function (Fig. 4). The regularity with which this motif is repeated (every 130 amino acids) makes it tempting to speculate that it may have a role in determining three-dimensional structure or functioning of the bacteriocin. Interestingly, this motif is also found in a rhizobiocin produced by 162Y10 (49).
The mechanism of secretion and transport of the rhizobiocin in strain 248 is also unresolved. Since the protein is produced in culture supernatants of a wide variety of host strains, and there is no specific transport system present in clones like pBAC12, it appears probable that the bacteriocin is secreted by some sort of general pathway or is released by spontaneous cell lysis.
Analysis of the C-terminal region of 248 bacteriocin did not reveal any secretory motifs which are used by other RTX proteins or by NodO. Interestingly, a R. leguminosarum prsD mutant which is unable to secrete NodO can still produce 248 bacteriocin (16). This suggests that if 248 bacteriocin is secreted, it is by a secretory system which is not related to the prsDE gene products, or that the bacteriocin may utilize more than one secretory pathway. Apparently the C terminus of rhizobiocin is not essential for biological activity or targeting of the protein, as several Tn5 inserts in the C-terminus-encoding portion of the rhizobiocin gene did not abolish activity (Fig. 1 and data not shown). Presumably, truncated proteins retaining activity are produced in these mutants. We were, however, unable to visualize these on silver-stained SDS-PAGE gels of ammonium sulfate-concentrated supernatants.
It is surprising, given that clones such as pBAC12 will confer bacteriocin production on a variety of different strains of R. leguminosarum, as well as S. meliloti, that there is no evidence for the presence of genes conferring immunity to the rhizobiocin or necessary for transport of the bacteriocin. Since pBAC12 and its various deletion derivatives (Fig. 1) can all be transferred to a variety of hosts, including sensitive strains, such as VF39, 3841, and 336, without any detrimental effects and strain Rl1003 (248; rzc::Tn5) is not sensitive to the bacteriocin, it must be assumed that pBAC12 carries a gene necessary for immunity; however, based on the locations of deletions, partial sequencing of upstream regions, and Tn5 inserts which we have constructed in pBAC12, there is very little room or evidence for an immunity gene.
The mode of action of the rhizobiocin remains to be resolved. Since many members of the RTX family of toxins are pore-forming cytolysins (52) and membrane depolarization is also a common mechanism of action for bacteriocins (26), it is tempting to speculate that the 248 rhizobiocin acts in a similar fashion. Hydrophobicity profiles, as well as algorithms used to predict transmembrane helices, have not detected any regions which have high probabilities of containing membrane-spanning domains. However, there is no experimental evidence that suggests that the bacteriocin does not interact with target membranes.
Evaluation of the role of other Rhizobium bacteriocins in the rhizosphere has been complicated by the fact that the best-studied medium-bacteriocin-producing strains were sensitive to small bacteriocin (19, 53), and thus most possible experiments examine mutually antagonistic strains (Table 3). Our nodule competition experiments under controlled conditions showed that the 248 rhizobiocin can play a statistically significant role in competition against certain test strains (Table 3). In these experiments the effect of bacteriocin in assays against VF39SM (which was radically less competitive than 248) was small, although statistically significant. No effect was seen with strain 3841, which was significantly more competitive than 248. The most pronounced effect was seen with strain Rlv1031 (a 336 derivative), which was intermediate in competitiveness with respect to 248 when compared to VF39SM and 3841. The bacteriocins may thus be most effective at tipping the balance when strains are otherwise of roughly equal competitiveness. It is also possible that a significant role of bacteriocins like the 248 rhizobiocin may be in plasmid maintenance in bacterial populations, analogous to that of plasmid addiction modules.
The presence of trifolitoxin genes in various rhizobia confers a selective advantage in the rhizosphere, both in soil and under more controlled conditions (48). It has been suggested that trifolitoxin could be used to enhance the competitiveness of inoculant strains, and there is strong evidence that this strategy can be successful (35). The cloning and molecular characterization of rhizobiocin genes, such as the 248 rhizobiocin, was undertaken to assess their ecological roles and to investigate the possibility of their use to enhance the rhizosphere competitiveness of inoculant strains. For this purpose, we have provided a molecular characterization of the rhizobiocin produced by R. leguminosarum 248 and some of its biological properties and provided evidence that the 248 rhizobiocin can in some cases confer an advantage with respect to competition for nodulation. The cloning and characterization of the rhizobiocin locus has, however, identified areas, such as partial bioactivity, transport, and immunity, that are at present unresolved. To understand how this rhizobiocin fulfills its biological role, we are addressing these questions.
ACKNOWLEDGMENTS
This work was funded by an Alberta Agricultural Research Institute Farming for the Future Grant and an NSERC operating grant to M.F.H. S.T. was supported by an Alberta Heritage Foundation for Medical Research studentship and an NSERC postgraduate scholarship.
REFERENCES
- 1.Altschul G S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baldani J I, Weaver R W, Hynes M F, Eardly B D. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured clover rhizobia. Appl Environ Microbiol. 1992;58:2308–2314. doi: 10.1128/aem.58.7.2308-2314.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann U. Crystal structure of the 50 kDa protease from Serratia marcescens. J Mol Biol. 1994;242:244–251. doi: 10.1006/jmbi.1994.1576. [DOI] [PubMed] [Google Scholar]
- 4.Beringer J E. R-factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 5.Beringer J E, Beynon J L, Buchanan-Wollaston A V, Johnston A W B. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature. 1978;276:633–634. [Google Scholar]
- 6.Breil B T, Ludden P W, Triplett E W. DNA sequence and mutational analysis of genes involved in the production and resistance of the antibiotic peptide trifolitoxin. J Bacteriol. 1993;175:3693–3702. doi: 10.1128/jb.175.12.3693-3702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewin N J, Beringer J E, Buchanan-Wollaston A V, Hirsch A W B, Hirsch P R. Transfer of symbiotic genes with bacteriocinogenic plasmids in Rhizobium leguminosarum. J Gen Microbiol. 1980;116:261–270. [Google Scholar]
- 8.Buchanan-Wollaston A V. Generalized transduction in Rhizobium leguminosarum. J Gen Microbiol. 1979;112:135–142. [Google Scholar]
- 9.Charles T C, Newcomb W, Finan T M. ndvF, a novel locus located on megaplasmid pRmeSu47b (pEXO) of R. meliloti, is required for normal nodule development. J Bacteriol. 1991;173:3981–3992. doi: 10.1128/jb.173.13.3981-3992.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckhardt T. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid. 1978;1:584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- 11.Economou A, Hamilton W D O, Johnston A W B, Downie J A. The rhizobium nodulation gene nodO encodes a Ca2+ binding protein that is exported without N terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 1990;9:349–354. doi: 10.1002/j.1460-2075.1990.tb08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Femlee T, Pellet S, Welch R A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finan T M, Wood J M, Jordan D C. Succinate transport in Rhizobium leguminosarum. J Bacteriol. 1981;148:193–202. doi: 10.1128/jb.148.1.193-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finan T M, Kunkel B, de Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finan T M, Oresnik I J, Bottacin A. Mutants of Rhizobium meliloti defective in succinate metabolism. J Bacteriol. 1988;170:3396–3403. doi: 10.1128/jb.170.8.3396-3403.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnie C, Hartley N M, Findlay K C, Downie J A. The Rhizobium leguminosarum prsDE genes are required for the secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation, and exopolysaccharide modification. Mol Microbiol. 1997;25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- 17.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 18.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenburg E P. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch P R. Plasmid-determined bacteriocin production by Rhizobium leguminosarum. J Gen Microbiol. 1979;113:219–228. [Google Scholar]
- 20.Hirsch P R, Van Montagu M, Johnston A W B, Brewin N J, Schell J. Physical identification of bacteriocinogenic, nodulation, and other plasmids in strains of Rhizobium leguminosarum. J Gen Microbiol. 1980;120:403–412. [Google Scholar]
- 21.Hynes M F, Brucksch K, Priefer U B. Melanin production encoded by a cryptic plasmid in a Rhizobium leguminosarum strain. Arch Microbiol. 1988;150:326–332. [Google Scholar]
- 22.Hynes M F, McGregor N F. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol Microbiol. 1990;4:567–574. doi: 10.1111/j.1365-2958.1990.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 23.Hynes M F, O’Connell M P. Host plant effect on competition among strains of Rhizobium leguminosarum. Can J Microbiol. 1990;36:864–890. [Google Scholar]
- 24.Johnston A W B, Beynon J L, Buchanan-Wollaston A V, Setchell S M, Hirsch P R, Beringer J E. High frequency transfer of nodulating ability between strains and species of Rhizobium. Nature. 1978;276:634–636. [Google Scholar]
- 25.Jones J D G, Gutterson N. An efficient mobilizable cosmid vector, and its use in a rapid marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 1987;61:299–306. doi: 10.1016/0378-1119(87)90193-4. [DOI] [PubMed] [Google Scholar]
- 26.Konisky J. Colicins and other bacteriocins with established modes of action. Annu Rev Microbiol. 1982;36:125–144. doi: 10.1146/annurev.mi.36.100182.001013. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lotz W, Mayer F. Isolation and characterization of a bacteriophage tail-like bacteriocin from a strain of Rhizobium. J Virol. 1982;9:160–173. doi: 10.1128/jvi.9.1.160-173.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Moënne-Loccoz Y, Sen D, Krause S E, Weaver R W. Plasmid profiles of rhizobia used in inoculants and isolated from clover fields. Agron J. 1994;86:117–121. [Google Scholar]
- 32.Oresnik I J, Charles T C, Finan T M. Second site mutations specifically suppress the Fix− phenotype of Rhizobium meliloti ndvF mutations on alfalfa: identification of a conditional ndvF-dependent mucoid colony phenotype. Genetics. 1994;136:1233–1244. doi: 10.1093/genetics/136.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole P S, Blyth A, Reid C J, Walters K. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology. 1994;140:2787–2795. [Google Scholar]
- 34.Priefer U B. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J Bacteriol. 1989;171:6161–6168. doi: 10.1128/jb.171.11.6161-6168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robleto E A, Scupham A J, Triplett E W. Trifolitoxin production in Rhizobium etli strain CE3 increases competitiveness for rhizosphere colonization and root nodulation of Phaseolous vulgaris in soil. Mol Plant-Microbe Interact. 1997;10:228–233. [Google Scholar]
- 36.Roslycky E B. Bacteriocin production in the Rhizobia bacteria. Can J Microbiol. 1967;13:431–433. doi: 10.1139/m67-057. [DOI] [PubMed] [Google Scholar]
- 37.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Schripsema J, DeRudder K E E, Van Vliet T B, Lankhorst P P, de Vroom E, Kijne J W, van Brussel A A N. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwinghamer E A, Belkengren R P. Inhibition of rhizobia by a strain of Rhizobium trifolii: some properties of the antibiotic and of the strain. Arch Mikrobiol. 1968;64:130–145. doi: 10.1007/BF00406972. [DOI] [PubMed] [Google Scholar]
- 41.Schwinghamer E A, Brockwell J. Competitive advantage of bacteriocin and phage-producing strains of Rhizobium trifolii in mixed culture. Soil Biol Biochem. 1978;10:383–387. [Google Scholar]
- 42.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicon, generation of operon fusions and induction of genes in Gram negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 43.Tagg J R, Dajani A S, Wannamaker L W. Bacteriocins of Gram positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson S A, Wong L L, West A, Sparling P F. Neisseria meningitidis produces iron regulated proteins related to the RTX family of exoproteins. J Bacteriol. 1993;175:811–818. doi: 10.1128/jb.175.3.811-818.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triplett E W. Isolation of genes involved in nodulation competitiveness from Rhizobium leguminosarum bv. trifolii T24. Proc Natl Acad Sci USA. 1988;85:3810–3814. doi: 10.1073/pnas.85.11.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triplett E W. Construction of a symbiotically effective strain of Rhizobium leguminosarum bv. trifolii with increased nodulation competitiveness. Appl Environ Microbiol. 1990;56:98–103. doi: 10.1128/aem.56.1.98-103.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Triplett E W, Barta T M. Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol. 1987;85:335–342. doi: 10.1104/pp.85.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triplett E W, Sadowsky M. Genetics of competition for nodulation of legumes. Annu Rev Microbiol. 1992;46:399–428. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 49.Twelker, S., I. J. Oresnik, and M. F. Hynes. Unpublished data.
- 50.Van Brussel A A N, Zaat S A J, Wijffelman C A, Pees E, Lugtenberg B J J. Bacteriocin small of fast growing rhizobia is chloroform soluble and is not required for effective nodulation. J Bacteriol. 1985;162:1079–1082. doi: 10.1128/jb.162.3.1079-1082.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent J M. A manual for the practical study of root-nodule bacteria, International Biological Programme Handbook 15. Oxford, United Kingdom: Blackwell Scientific Publications; 1970. [Google Scholar]
- 52.Welch R A. Pore-forming cytolysins of Gram negative bacteria. Mol Microbiol. 1990;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 53.Wijffelman C A, Pees E, Van Brussel A A N, Hooykaas P J J. Repression of small bacteriocin excretion in Rhizobium leguminosarum and Rhizobium trifolii by transmissible plasmids. Mol Gen Genet. 1983;192:171–176. [Google Scholar]
- 54.Yarosh O K, Charles T C, Finan T M. Analysis of C4 dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 55.Young J P W, Wexler M. Sym plasmid and chromosomal genotypes are correlated in field populations of Rhizobium leguminosarum. J Gen Microbiol. 1988;134:2371–2391. [Google Scholar]