Abstract
G-quadruplex (G4)–prone structures are abundant in mammalian genomes, where they have been shown to influence DNA replication, transcription, and genome stability. In this article, we constructed cells with a single ectopic homopurine/homopyrimidine repeat tract derived from the polycystic kidney disease type 1 (PKD1) locus, which is capable of forming triplex (H3) and G4 DNA structures. We show that ligand stabilization of these G4 structures results in deletions of the G4 consensus sequence, as well as kilobase deletions spanning the G4 and ectopic sites. Furthermore, we show that DNA double-strand breaks at the ectopic site are dependent on the nuclease Mus81. Hypermutagenesis during sister chromatid repair extends several kilobases from the G4 site and breaks at the G4 site resulting in microhomology-mediated translocations. To determine whether H3 or G4 structures are responsible for homopurine/homopyrimidine tract instability, we derived constructs and cell lines from the PKD1 repeat, which can only form H3 or G4 structures. Under normal growth conditions, we found that G4 cell lines lost the G4 consensus sequence early during clonal outgrowth, whereas H3 cells showed DNA instability early during outgrowth but only lost reporter gene expression after prolonged growth. Thus, both the H3 and G4 non-B conformation DNAs exhibit genomic instability, but they respond differently to endogenous replication stress. Our results show that the outcomes of replication-dependent double-strand breaks at non–B-DNAs model the instability observed in microhomology-mediated break-induced replication (BIR). Marked variability in the frequency of mutagenesis during BIR suggests possible dynamic heterogeneity in the BIR replisome.
Keywords: DNA replication, DNA repair, DNA damage, G-quadruplex, DNA triplex, genomic instability
Abbreviations: ATR, ataxia telangiectasia and Rad3-related protein; BIR, break-induced replication; CHK1, checkpoint kinase 1; DDR, DNA damage response; DSB, double-strand break; eGFP, enhanced GFP; G4, G-quadruplex; H3, triplex; iPCR, inverse PCR; MMBIR, microhomology-mediated BIR; PKD1, polycystic kidney disease type 1; Pu/Py, homopurine/homopyrimidine; TMS, telomestatin
Microsatellite DNAs were originally identified as centrifugation peaks separate from bulk genomic DNA by virtue of their homogeneous base compositions (1). Most microsatellites comprise multiple tandem repeats of 1 to 9 nucleotide blocks. The tendency of microsatellite tracts to exhibit genomic instability by expansion, contraction, and recombination has been attributed to the predilection of these sequences to form non–B-DNA structures (2).
In yeast, expanded GAA (3, 4), CTG (5), and CGG (6) microsatellites are sites of DNA double-strand breaks (DSBs), chromosome fragility, and activation of the RAD9/ataxia telangiectasia and Rad3-related protein (ATR)/checkpoint kinase 1 (CHK1) DNA damage response (DDR) pathways (4, 7, 8, 9). Replication fork stalling at microsatellite non–B-DNA structures is one mechanism leading to single-ended DSBs and hypermutagenic break-induced replication (BIR) (10, 11, 12, 13, 14). An alternate mechanism for bypass of replication fork barriers by homologous recombination in Schizosaccharomyces pombe also leads to an error-prone pathway to circumvent replication barriers (15, 16).
In humans, microsatellite instability has been implicated in causing Huntington’s disease ((CAG)n) (17), the fragile X-related disorders, FXTAS, FXPOI, and FXS (18), myotonic dystrophy ((CTG)n and (CCTG)n) (19, 20), and more than 40 other neuromuscular and developmental diseases, and cancers. The presence of microsatellite repeats at translocation break points and junctions has implicated microhomology-mediated BIR (MMBIR) in gross chromosomal rearrangement and nonrecurrent copy number variation (21, 22, 23, 24). BIR is strongly indicated as an outcome of replication-dependent DSBs in human cells (25), and expanded CGG repeats have been shown to cause BIR in a murine cell culture system (11). Our laboratory has recently shown that Mus81 cleavage at the edge of an expanded CTG hairpin (26) causes replication-dependent BIR, hypermutation, and nonrandom chromosomal translocations (27).
Although it is not a strict tandem repeat, here we consider the asymmetric homopurine/homopyrimidine (Pu/Py) mirror repeat of the polycystic kidney disease type 1 (PKD1) gene intron 21 (IVS21) to behave as a microsatellite because of its homogeneous base composition, its replication polarity-dependent ability to stall replication forks through the formation of non–B-DNA structures, and its replication-dependent instability (27, 28). The Pu-rich strand of the PKD1 IVS21 (Pu/Py) repeat has the potential alternatively to form triplex (H3) DNA by Hoogsteen base pairing (29) or G-quadruplex (G4) DNA by Hoogsteen stabilization of G4 base stacking (30). The G4 consensus (G)>2(N)1–7(G)>2(N)1–7(G)>2(N)1–7(G)>2 has been used to identify potential sites of G4 formation (31), although later work indicates that longer loops ((N)≤30) can be readily accommodated into G4 structures (32).
Statistical calculations predict that approximately 200 G4 sequences with loops of (N)≤15 will occur in a random nucleotide collection of 3.5 × 109 bp, while algorithms that account for dinucleotide preferences in human DNA predict >370,000 potential G4 structures in the genome, and G4-targeted antibody binding has indicated >700,000 sites capable of stable G4 formation in human B-lymphocyte DNA (33, 34, 35, 36, 37, 38). G4 sequences have been implicated in the control of replication (39, 40, 41), transcription (30, 42), genomic looping (33), R-loop formation (43, 44), and genome instability (44). G4 structures are unwound by the FANCJ helicase in vitro (45), whereas depletion of FANCJ increases G4 formation in vivo (41, 46).
Our laboratory has shown that the PKD1 (Pu/Py)88 repeat stalls replication forks when the Pu strand is the lagging strand template downstream from the SV40 replication origin in vitro and when the Pu strand is replicated as the lagging strand template from an ectopic c-myc replication origin in vivo. At the same ectopic site, inversion of the Pu/Py tract stalls replication when the c-myc origin is inactivated, and the Pu lagging strand template is replicated from a neighboring downstream origin (28).
Cells containing the ectopic Pu/Py repeat in the Pu lagging strand orientation ((Pu)88) grow more slowly than cells containing the Pu/Py repeat at the same ectopic site in the Py lagging strand ((Py)88) orientation (27). (Pu)88 cells are also sensitive to the CHK1 inhibitor UCN-01 suggesting that the ATR/CHK1 DDR pathway is required for (Pu)88 cell viability. Precise excision of the Pu/Py repeat from the ectopic site of (Pu)88 cells using FLP recombinase eliminates the UCN-01 effect, demonstrating that the Pu/Py tract is directly responsible for UCN-01 sensitivity. Consistent with the requirement of the DDR for (Pu)88 cell viability, ATR and RAD9 bind preferentially to the (Pu)88 versus (Py)88 cell ectopic site (28).
We have constructed cells in which spontaneously shortened (Pu/Py)78 microsatellites are integrated by FLP recombinase at a single ectopic site alongside the core c-myc replication origin and flanked by dTomato and enhanced GFP (eGFP) reporter genes (26, 27, 28, 47). DNA DSBs occur with greater frequency in (Pu)78 cells than (Py)78 cells under nonperturbed conditions. To introduce replication stress in this system, we used the macrocyclic G4 ligand telomestatin (TMS) to stabilize G4s. TMS stacks tightly and specifically to the antiparallel basket conformation of G4s (48). Circular dichroism analyses show that TMS induces structural changes consistent with G4 formation in the PKD1 homoPu sequence but not homoPy, (CTG)n, or (CAG)n sequences (27). At elevated dosage, TMS also inhibits telomerase activity and reduces the growth of certain tumor cell types (49, 50, 51, 52). Here, we find that a low dose of TMS dramatically increases the frequency of ectopic site DSBs in (Pu)78 cells but not in (Py)78 cells.
Long-read sequencing of inverse PCR (iPCR) products show that TMS stabilization of the (Pu)78 G4 structure results in precise deletion of the repeat tract in a population of cells and hypermutation in the flanking DNA. This instability resembles the sister chromatid mutagenesis occurring during BIR. In other (Pu)78 cells, TMS treatment resulted in large deletions (>3–4 kb) of the ectopic site, hypermutagenesis, and nonallelic chromosomal translocations, similar to the effects of MMBIR. TMS also induced instability at G4 consensus sites flanking the ectopic Pu/Py repeat. Knockdown of the Mus81 resolvase in (Pu)78 cells significantly decreased replication-dependent DNA DSBs at the ectopic site.
To determine whether the G4 structure or the H3 structure of the Pu/Py microsatellite was responsible for instability under nonperturbed conditions, we constructed clonal cell lines containing versions of the Pu/Py tract that could only form H3 or G4 structures. Our results indicate that stable H3 and G4 DNA constructs are fragile in the absence of exogenous replication stress. We conclude that H3 and G4 DNA structures that are stable under perturbed and nonperturbed conditions lead to genomic instability with characteristics of BIR.
Results
TMS induces DSBs at the (Pu)78 ectopic site
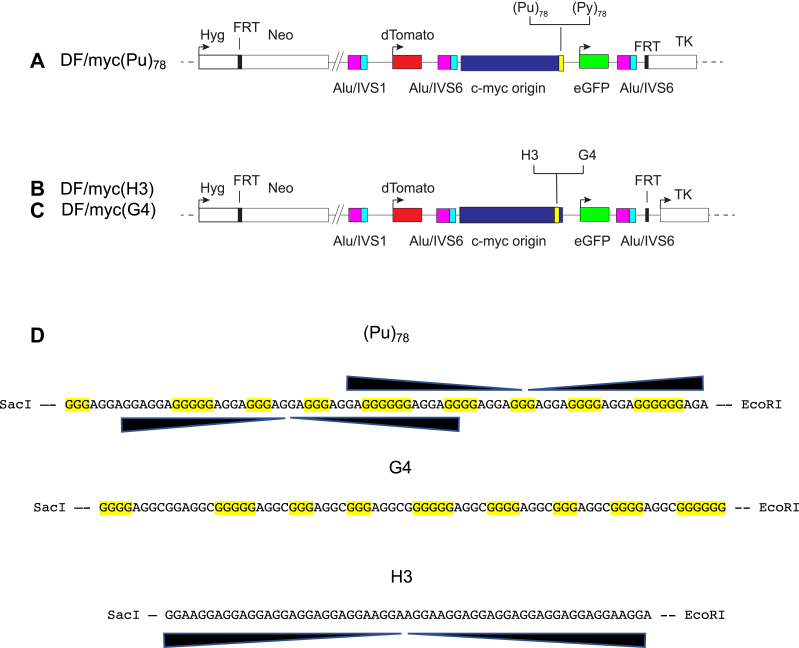
The ectopic site constructs integrated by FLP recombinase are shown in Figure 1. All constructs were placed at the same ectopic site, alongside a copy of the 2.4 kb c-myc core replication origin (53, 54, 55, 56, 57, 58). The ectopic site c-myc origin displays the chromatin structure, replication protein binding, and early S-phase replication firing pattern of the endogenous c-myc origin (26, 27, 28, 59, 60). The (Pu)78 repeat is in the lagging strand template for replication from the c-myc origin. dTomato and eGFP reporter genes are situated to allow ectopic site DSBs to be detected by flow cytometry. Three Alu/IVS elements derived from the UBE2T locus provide targets for homology-dependent recombination (61).
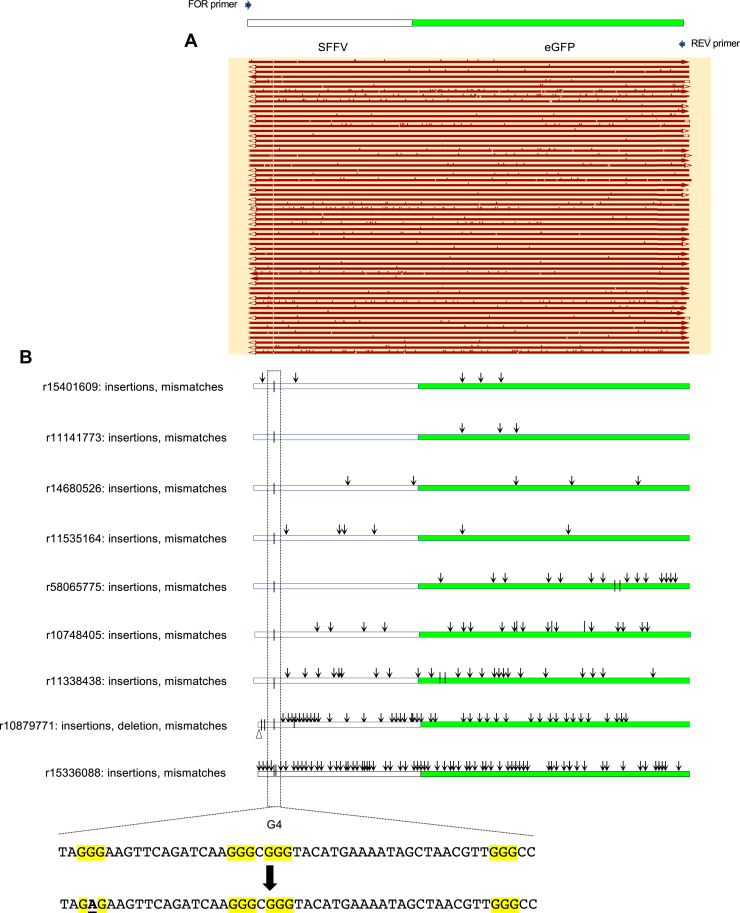
Figure 1.
Ectopic site maps.A, schematic map of the (Pu/Py)78 ectopic site integrant constructs. B and C, schematic maps of the H3 and G4 ectopic site integrant constructs. D, input DNA sequences. G4, G-quadruplex; H3, triplex; Pu/Py, homopurine/homopyrimidine.
(Pu)78 and (Py)78 cells (Fig. 1A) were untreated or treated with TMS and analyzed by flow cytometry (Fig. 2A). As measured by separation of the dTomato and eGFP reporter signals, TMS treatment had no visible effect on instability at the ectopic site in (Py)78 cells (Fig. 2, B and C). Consistent with previous results (27), TMS treatment reproducibly induced a dramatic loss of dTomato fluorescence in (Pu)78 cells (Fig. 2, D and E), in line with earlier circular dichroism analysis of the PKD1 tract treated with TMS, which confirmed the formation of G4 structures (27). We conclude that stabilization of G4 structures in the (Pu)78 replication orientation leads to DSBs at the ectopic site.
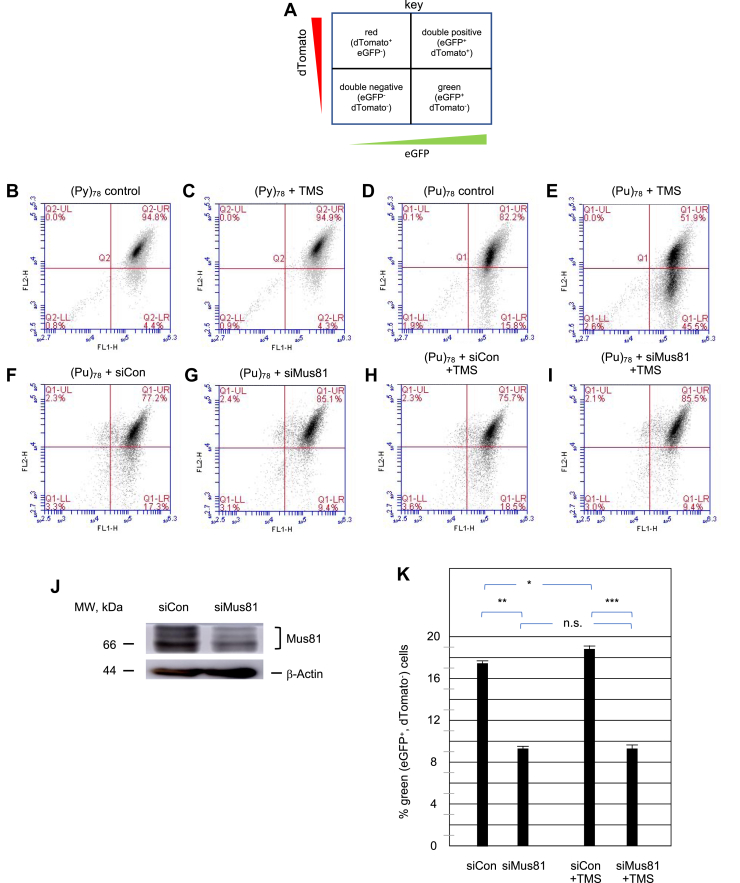
Figure 2.
Orientation-dependent sensitivity of the (Pu)78structure to telomestatin (TMS).A, flow cytometry key. Flow cytometry of (B) (Py)78 control cells. C, (Py)78 cells treated with TMS. D, (Pu)78 control cells. E, (Pu)78 cells treated with TMS. F, (Pu)78 cells treated with siCon. G, (Pu)78 cells treated with siMus81. H, (Pu)78 cells treated with siCon and TMS. I, (Pu)78 cells treated with siMus81 and TMS. J, Western blot. K, effect of siMus81 on the level of green (eGFP+, dTomato−) cells, +/− SD, n = 3, two-tailed Student’s t test. ∗p = 0.013; ∗∗p = 0.001; ∗∗∗p = 0.0001. ns, not significant; Pu, purine; Py, pyrimidine.
These results also show that initiation at the ectopic c-myc origin is not stochastic (62), since initiations upstream and downstream of the origin would eliminate the asymmetric effect of Pu/Py replication.
Treatment of (Pu)78 cells with TMS for approximately four cell division cycles (96 h) resulted in ca. 45% of cells losing the dTomato fluorescence. This suggested that not every cell division of the TMS-stabilized G4 resulted in the loss of the dTomato signal. When (Pu)78 cells were incubated with TMS for 48 h to allow 1 to 2 cell cycles in the presence of the drug, we reproducibly observed a smaller, but statistically significant, 1.2% increase in the percentage of eGFP+, dTomato− (“green”) cells (Figs. 2, F, H and K and S1, p ≤ 0.013). These results indicate that multiple recombinations at the ectopic site G4 occur before loss of the dTomato signal. Strikingly, as in the case of the ectopic (CTG)100 microsatellite (27), knockdown of Mus81 significantly reduced the percentage of green (eGFP+, dTomato−) cells in cultures treated or untreated with TMS (Fig. 2, G and I–K).
Single-molecule analysis of (Pu)78 DSBs
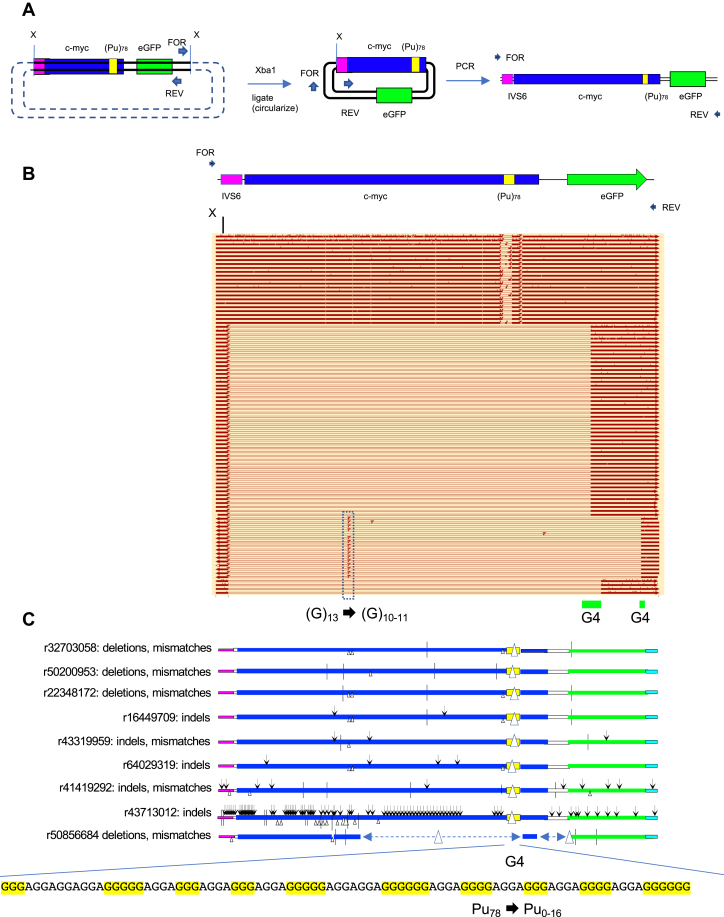
To characterize the effects of G4-mediated instability, DNA was isolated from TMS-treated (Pu)78 cells, digested with Xba1 and ligated at low DNA concentration to induce intramolecular circularization (63). Head-to-head PCR primers in the ectopic site were used to amplify the circles by iPCR (Fig. 3A), and the products were analyzed by PacBio long-read high-fidelity circular consensus sequencing (64).
Figure 3.
Inverse PCR (iPCR) analysis of (Pu)78instability.A, iPCR schematic. B, summary of PacBio sequencing results, aligned with an orientation map. Open bars, deletions; arrowheads, insertions; vertical ticks, mismatches; G4, G-quadruplex consensus sequences. C, expanded examples of mutagenesis results. Arrowheads, insertions; triangles, deletions; and vertical ticks, mismatches. The G4 sequence of the input (Pu)78 cassette is shown below. D, chromosome translocations at the (Pu)78 site. E, complex chromosome hopping at the ectopic site. Read designations (r) are indicated. Dashed arrows indicate the 5′ to 3′ polarity of the read. Pu, purine.
Sequencing of the iPCR products (iPCR-Seq) showed that a population of cells had precisely deleted all or part of the G4 consensus at the (Pu)78 insert (Fig. 3, B and C). In addition, a second larger population had deleted approximately 3 kb of DNA surrounding the G4 structure to break points at G4 consensus matches in the eGFP reporter gene (Fig. 3A). As in the case of cleavage by Mus81 alongside (CTG)100 hairpins (27), recurrent break points were largely confined to specific sites around the G4 consensus sequences, whereas small insertions, deletions, and single base substitutions were abundant but dispersed in the flanking DNA. The hypermutagenesis revealed by this single-molecule analysis is reminiscent of sister chromatid BIR initiated at (CTG)102 replication fork DSBs (27).
Should a translocation occur from the ectopic site, Xba1 cleavage of the acceptor chromosome would allow circularization, ligation, and identification of the break point junction (63). Thus, iPCR-Seq also revealed translocations to nonallelic sites (Fig. 3D) and complex chromosome rearrangements (Fig. 3E) in the TMS-treated (Pu)78 cells, characteristic of unstable D-loops formed during BIR (14, 65, 66, 67). Recombination on both the upstream and downstream sides of the (Pu)78 DSB supports the view that the (Pu)78 DSB is double ended, similar to the DSBs at the (CTG)100 ectopic site (27).
We note as well that the rate of mutation can change abruptly within a single translocation read (e.g., r62783876, r54198608, r27918549) and between reads covering the same sister chromatid sequence (e.g., r41419292, r43713012) suggesting that the composition of the BIR replisome may be dynamic. Consistent with MMBIR, 3 to 7 bp microhomology was detected at six of seven translocation junctions (Table 1).
Table 1.
Break point junction homology
| r27918549: mismatches, chromosome 5 (collagen alpha-1) | CCT |
| r53019448: chromosome 8 (POTE ankyrin domain family member A isoform 1) (SINE) | TCT |
| r36242316: chromosome 8 (DENN domain 3) | TGG |
| r62783876: chromosome 15 (DNAJ) | AGGCC |
| r27918549: chromosome 16 (SINE) | GCA |
| r5505505: chromosome 16 (Fox-1 homology region) | No homology |
| r54198608: chromosome 17 (chemokine 23 isoform CKb8) | AAAGCTT |
Mutagenesis of dTomato and eGFP flanking DNA
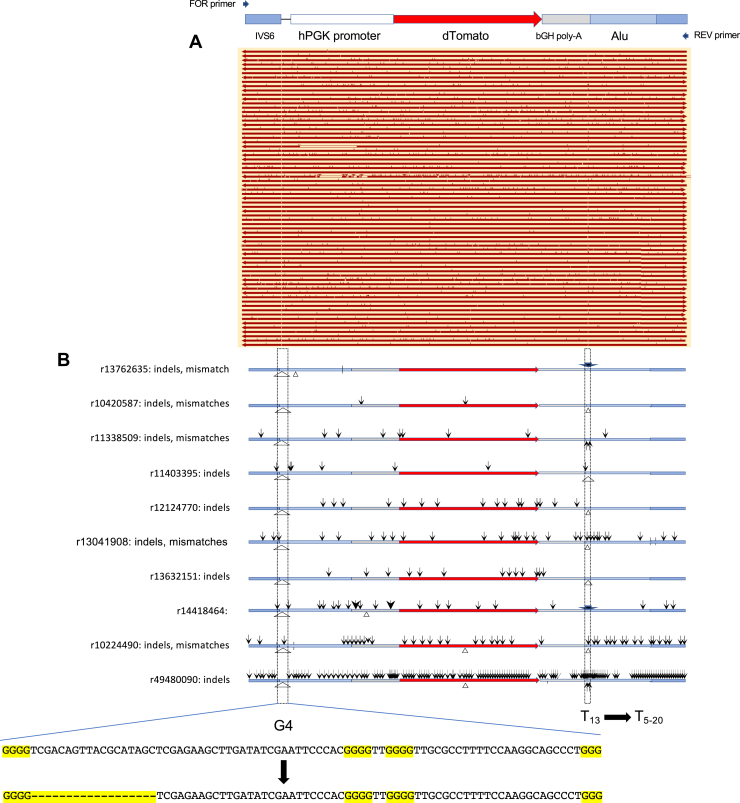
It is possible that loss of dTomato or eGFP signals could occur because of mutagenesis of the reporter genes. Sequencing of dTomato (Fig. 4, A and B) and eGFP (Fig. 5, A and B) (standard) PCR products showed that hypermutagenesis extended into these reporter genes over a distance of more than 5 to 6 kb. In both dTomato and eGFP genes, G4 and mononucleotide repeats were hotspots for mutation during homology-mediated sister chromatid exchange. Although these results imply that dTomato and eGFP fluorescence should be lost because of mutagenesis, the preferential appearance of green cells suggests that the acentric dTomato end of the chromosome is less frequently recombined or more often lost. A similar result has been observed at ectopic site unstable (CTG)100 repeats, where it was shown that BIR results in loss of dTomato fluorescence, whereas eGFP was lost by single-strand annealing repair of a clean I-Sce1 DSB (27). Again, the frequency of mutation varies substantially between individual reads.
Figure 4.
Mutagenesis of the dTomato region upstream of (Pu)78.A, summary of PCR sequencing results, aligned with an orientation map. B, expanded examples of mutagenesis results. Pu, purine.
Figure 5.
Mutagenesis of the eGFP region downstream of (Pu)78.A, summary of PCR sequencing results, aligned with an orientation map. B, expanded examples of mutagenesis results. eGFP, enhanced GFP; Pu, purine.
Instability of H3 and G4 repeats
The aforementioned results indicate that the PKD1 tract undergoes DSBs when the G4 forming sequence is stabilized by TMS. However, since both H3 and G4 structures are possible in this repeat under nonperturbed conditions, we decided to construct cell lines containing derivatives of the PKD1 repeat, which could only form H3 or G4 structures (Fig. 1, B and C). The H3 and G4 sequences were cloned in place of the G4 prone sequence (Pu27) (30) in the natural c-myc origin to avoid spreading of the G4 structure.
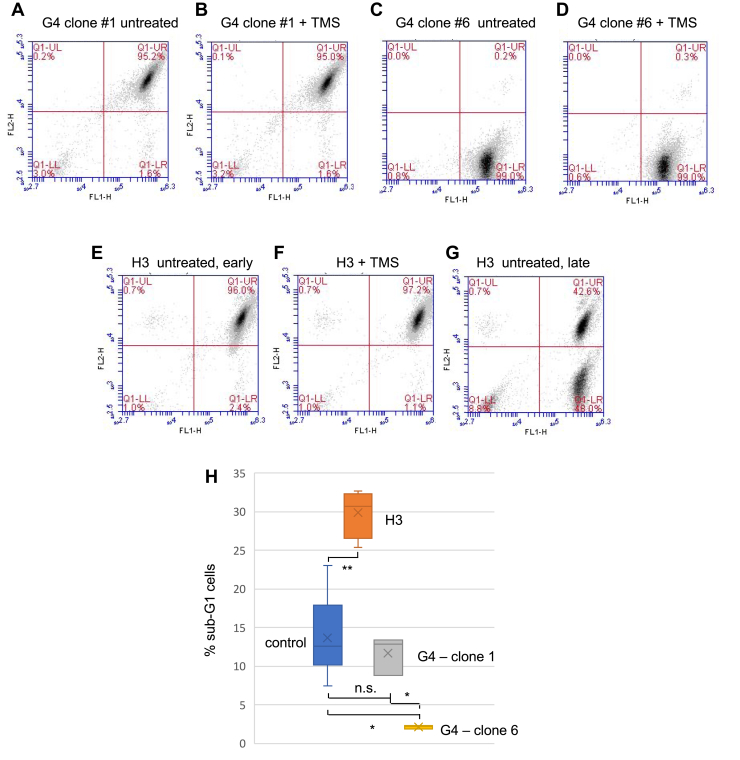
The H3 and G4 cell lines revealed multiple differences. Two clones of the G4 repeat cell lines (#1 and #6) displayed different flow cytometry patterns under nonstressed conditions (Fig. 6, A and C). Surprisingly, G4 clone #1 did not lose dTomato or eGFP to produce green or double-negative cells following TMS treatment (Fig. 6B); similarly, G4 clone #6 did not lose eGFP to produce double-negative cells (Fig. 6D). As expected, the H3 cells did not show sensitivity to TMS (Fig. 6, E and F) presumably because they do not contain an ectopic G4 consensus.
Figure 6.
Insensitivity of H3 and G4 clones to TMS. Flow cytometry of (A) G4 clone #1 untreated cells; (B) G4 clone #1 cells treated with TMS; (C) G4 clone #6 untreated cells; (D) G4 clone #6 cells treated with TMS; (E) H3 untreated cells, early passage; (F) H3 cells treated with TMS; (G) H3 untreated cells, late passage; (H) percent sub-G1 cells in control (minus ectopic site non–B-DNA), H3, and G4 cells. ∗p < 0.005; ∗∗p < 0.0002. n ≥ 3. G4, G-quadruplex; H3, triplex; TMS, telomestatin.
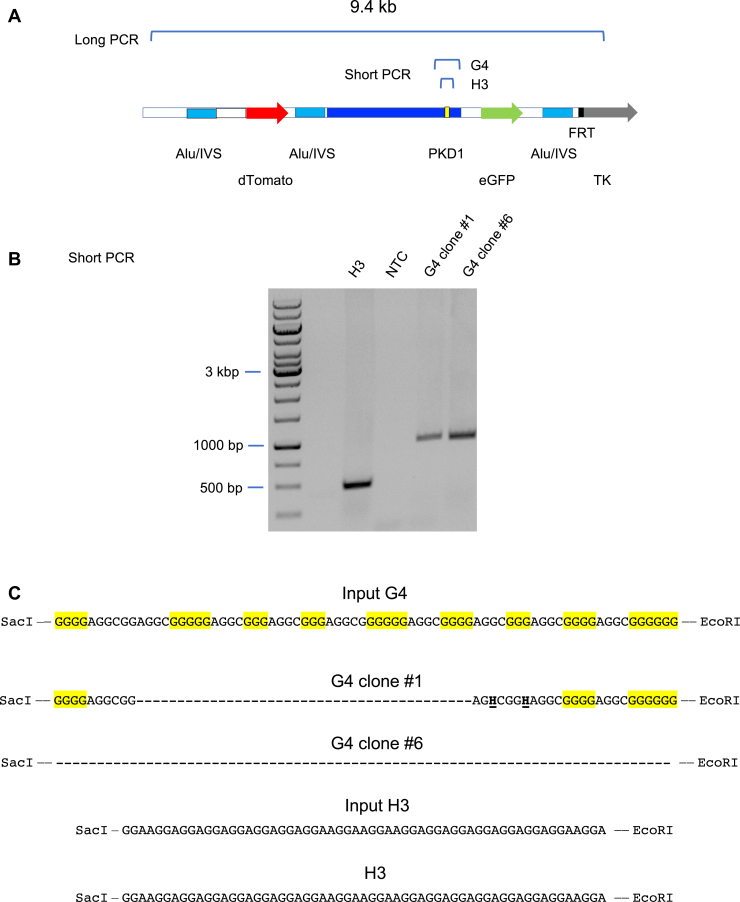
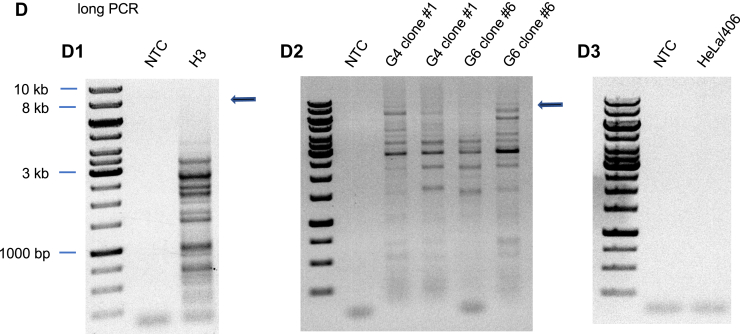
DNA sequencing of the short PCR products (Fig. 7A) across the H3 and G4 inserts revealed that G4 clone #1 had substituted and deleted G4 consensus nucleotides, and that the entire G4 cassette had been deleted in G4 clone #6, while no mutations were detected in the H3 repeat (Fig. 7B). The absence of perturbations in the flow cytometry patterns of G4 clone #1 and the H3 clone suggested that these inserts might not have caused ectopic site instability. However, PCR analysis of the H3 clone, and small-pool PCR analysis of G4 clone #1 and G4 clone #6 (long PCR, Fig. 7D) showed that significant deletions had occurred at both the H3 and G4 ectopic sites during clonal outgrowth under nonperturbed conditions.
Figure 7.
H3 cells and G4 ectopic sites are unstable under nonstressed growth conditions.A, map of short and long PCR templates. B, short PCR confirmation of H3 and G4 inserts. C, sequences of input and outcome H3 and G4 cassettes. H, non-G nucleotide. D, PCR analysis of H3, G4 recombination. Arrows, expected PCR product size (9.4 kb). G4, G-quadruplex; H3, triplex; NTC, no template control.
The long PCR results were confirmed to be specific for the ectopic site by the negative control amplification of genomic DNA of HeLa/406 acceptor cells containing an empty ectopic site (Fig. 7D.3). We conclude that G4 clones #1 and #6 had undergone distinct mutations prior to clonal growth and that the insensitivity of these clones to TMS is due to the loss of the G4 consensus repeat.
The retention of the unmutagenized H3 insert was surprising in view of the iPCR results demonstrating rearrangements in the H3 ectopic site. However, quantitation by flow cytometry (Fig. 6H) showed that the H3 cells generated more sub-G1 cells than either the G4 clones or control cells that did not contain non–B-DNA at the ectopic site. In addition, prolonged growth (3 months) of the H3 cells led to the appearance of >40% of green (eGFP+, dTomato−) cells (Fig. 6G), consistent with the view that multiple DSBs may be required before loss of either reporter gene signal. We conclude that both the H3 and G4 constructs lead to genomic instability and that different non–B-DNAs respond differently to replication stress.
Discussion
G4-prone sequences are abundant in mammalian DNA and have been implicated in having positive or negative effects on replication, transcription, and genome stability. In this study, we show that stabilization of G4 structures in the PKD1 IVS21 Pu/Py repeat by the TMS ligand leads to replication-dependent DSBs, hypermutagenesis, and gross chromosomal rearrangements associated with unstable D-loops characteristic of BIR. In cells that proliferated after TMS treatment, the PKD1 G4 consensus sequence was eliminated either by precise excision or kilobase length deletion. In either situation, deletion break points were nonrandomly located at G4-forming sequences. We note as well that G4 consensus sequences and mononucleotide repeats in the flanking DNA were also hotspots for mutation and that deletions at G4 upstream or downstream flanking sites were oriented at consensus sequences with Pus in the lagging strand template. Mutagenesis extended for >2 to 3 kb upstream and downstream of the ectopic site break points, suggesting that fork restart by homology-dependent sister chromatid exchange could initiate at both ends of the DSB.
(Pu)78 cells displayed greater instability than (Py)78 cells under nonperturbed conditions (cf. Fig. 2, B and D, and (27)). Cells containing a construct designed only to form G4 structures were also unstable under nonperturbed conditions, and G4 clones #1 and #6 grown out from these populations displayed deletions of the G4 consensus sequences similar to what was seen with TMS-treated (Pu)78 cells. We conclude that TMS ligand binding, or sequence optimization and spontaneous G4 formation, leads to stable G4 structures, and that mutation of the G4 consensus is responsible for the insensitivity of G4 clone #1 and G4 clone #6 cells to TMS and intrinsic replication stress. Similarly, an extended perfect mirror repeat H3-prone sequence appears to have increased the efficiency and instability of the H3 structure.
In contrast to the G4 cassette, we did not detect deletion of the H3 structure in early passage cells, despite dramatic recombinations at the H3 ectopic site. It is worth noting that the PCR results of Figure 7, B and D cannot be compared quantitively, since the short PCR is sufficiently sensitive to detect the H3 template retained in a small fraction of cells. Retention of the unaltered H3 cassette may occur because not all recombinations remove the ectopic microsatellite or because of structural differences in remodeling at G4 and H3 replication forks. The former explanation is supported by the observation that prolonged outgrowth is required to observe loss of the dTomato signal in H3 cells. Further next-generation sequencing analyses will address these questions.
In a broader sense, these results indicate that fork stalling at different non–B-DNAs can result in different forms of mutagenesis. Thus, different forms of replication stress may result in heterogeneous patterns of instability at a single non–B-DNA, and a single form of replication stress may induce multiple patterns of instability at different non–B-DNAs.
Experimental procedures
Cell lines
HeLa/406 cells containing the FLP recombinase target site and construction of ectopic cell lines have been described previously (26, 27, 47, 59, 68, 69, 70). The PKD1 insert in (Pu/Py) cells was cloned from the pJB4 plasmid (71). H3 and G4 inserts were designed to conform to the requirements of H3 mirror-repeat symmetry and G4 consensus matches and synthesized by Integrated DNA Technologies.
HeLa/406 acceptor cells contain a single FLP recombinase target site (69, 72). DF/myc cell lines were derived by cotransfecting HeLa/406 cells with dual fluorescence donor plasmids (27, 61) and the FLP recombinase expression vector pOG44 (73). Cells were maintained on Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 5% CO2 at 37 °C. For TMS treatment, DF/myc(Pu)78 cells were plated in 6-well plates at 30% confluency. Twenty-four hours later, they were treated with TMS at a final concentration of 0.5 μM. The treatment lasted for 4 days; this concentration of TMS did not significantly inhibit cell division, consistent with previous work (74). During the course of the treatment, TMS was replenished at 48 h. After the 4 day treatment ended, the cells were allowed to recover for 4 days. Following this, cells were harvested to extract genomic DNA for PCR, iPCR, or flow cytometry. Treatment and recovery of cells was in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 5% CO2 at 37 °C.
PCR and iPCR
iPCR was performed using genomic DNA from DF/myc(Pu)78 cells. About 1.5 μg DNA was digested with XbaI (New England Biolabs) for 1 h at 37 °C. After a 20 min heat inactivation step at 65 °C, 30 μl of the reaction (0.4–1 ng/μl) was used for ligation with T4 DNA ligase (New England Biolabs) overnight at 4 °C. The ligation reaction products were cleaned using E.Z.N.A. Cycle Pure kits (Omega). iPCR primers used for amplification were as follows: Nhe primer forward-BC-1 (5′-AGCTAAGCTTGCCTTGAGTGCTTC-3′) and eGFP primer reverse-BC-1 (5′-AGCTGTCCATGCCGAGAGTGATC-3′). Q5 HotStart polymerase (New England Biolabs) was used per manufacturer’s instructions with cycling conditions as follows: initial denaturation at 98 °C for 30 s. Touchdown PCR started with an annealing temperature of 66 °C for 15 s with a reduction of 0.5 °C for the first ten cycles. Subsequent cycles utilized 61 °C for 15 s. This was followed by an extension time of up to 5 min at each cycle. Final extension time was for 5 min followed by a hold at 4 °C.
Long PCR was performed using Lac—forward (5′-CTTCAAATCCGACCCGTAGA-3′) and TK—reverse-1 (5′-GGGTATCGACAGAGTGCCAG-3′) for H3, G4 clones #1 and #6, and HeLa/406 cells. Q5 HotStart polymerase was used per manufacturer’s instructions for 50 μl reactions using 1 to 200 ng template. Cycling conditions were as follows: initial denaturation at 98 °C for 30 s. Touchdown PCR started with an annealing temperature of 66 °C for 15 s with a reduction of 0.5 °C for the first ten cycles. Subsequent cycles utilized 61 °C for 15 s. This was followed by an extension time of up to 5 min at each cycle. Final extension time was for 5 min and then a hold temperature of 4 °C. PCR products were electrophoresed on a 0.4% agarose gel (Invitrogen) to verify the size of the recombination products. Appropriate no template controls were also run.
Short PCR was performed using multiple cloning site (MCS)—forward (5′-GGTTCCCAAAGCAGAGGG-3′) and MCS—reverse (5′-TGGAAACATCTGATGGGTCTT-3′) primers for H3 cells and BstZ17I NEBuilder—forward (5′-CATAACGCGCTCTCCAAGTA-3′) and MCS—reverse (5′-TGGAAACATCTGATGGGTCTT-3′) for G4 clones #1 and #6. Cycling conditions were as aforementioned.
DNA sequence analysis
Amplicon EZ sequencing was performed on standard PCR products less than 1 kb, whereas Pacific Biosciences HiFi sequencing was performed on longer amplicons (iPCR, eGFP, and dTomato) at GeneWiz. Circular consensus sequences were trimmed, and contaminants were removed by screening reads for sequences matching the amplification primers or sequences immediately adjacent to the primer binding sites. Exact sequence duplicates were removed, and the remaining unique sequences were used for mapping. Trimmed Amplicon EZ sequences were used directly for mapping. BWA-MEM (Burrows-Wheeler Aligner Maximal Exact Match) (75) was used to map reads against their respective ectopic site reference sequences or the human genome (hg38). Chimeric and nonchimeric reads were identified within the resulting .sam file and examined manually using BLAST or Muscle in SnapGene (Insightful Science).
Data availability
Data will be shared upon request to michael.leffak@wright.edu.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We gratefully acknowledge the support of the National Institutes of Health (grant no.: GM122976; to M. L.). We are grateful to Michael Bottomley of the WSU Statistics Center for the calculation of genomic G4 probabilities and to Dale Schiff for assistance with figure preparation.
Author contributions
S. D. R. Jr, R. Y. G., D. C. H., F. J. D. IV, N. Z., M. S., V. A., R. S., K. S.-y., and M. L. conceptualization; S. D. R. Jr, R. Y. G., D. C. H., F. J. D. IV, N. Z., M. S., V. A., R. S., K. S.-y., and M. L. methodology; S. D. R. Jr, R. Y. G., and D. C. H. software; S. D. R. Jr, R. Y. G., D. C. H., and M. L. validation; S. D. R. Jr, R. Y. G., D. C. H., F. J. D. IV, N. Z., M. S., V. A., R. S., K. S.-y., and M. L. writing–review and editing; M. L. project administration; M. L. funding acquisition.
Funding and additional information
V. A. and R. S. were supported by the Wright State University Biomedical Sciences PhD program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Patrick Sung
Supporting information
References
- 1.Kit S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J. Mol. Biol. 1961;3:711–716. doi: 10.1016/s0022-2836(61)80075-2. [DOI] [PubMed] [Google Scholar]
- 2.Khristich A.N., Mirkin S.M. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 2020;295:4134–4170. doi: 10.1074/jbc.REV119.007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krasilnikova M.M., Mirkin S.M. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spivakovsky-Gonzalez E., Polleys E.J., Masnovo C., Cebrian J., Molina-Vargas A.M., Freudenreich C.H., Mirkin S.M. Rad9-mediated checkpoint activation is responsible for elevated expansions of GAA repeats in CST-deficient yeast. Genetics. 2021;219 [Google Scholar]
- 5.Sundararajan R., Freudenreich C.H. Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in Saccharomyces cerevisiae. PLoS Genet. 2011;7 [Google Scholar]
- 6.Balakumaran B.S., Freudenreich C.H., Zakian V.A. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 7.Sundararajan R., Gellon L., Zunder R.M., Freudenreich C.H. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usdin K., House N.C., Freudenreich C.H. Repeat instability during DNA repair: Insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015;50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Freudenreich C.H. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393:110–115. doi: 10.1016/j.gene.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakofsky C.J., Malkova A. Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017;52:395–413. doi: 10.1080/10409238.2017.1314444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kononenko A.V., Ebersole T., Vasquez K.M., Mirkin S.M. Mechanisms of genetic instability caused by (CGG)n repeats in an experimental mammalian system. Nat. Struct. Mol. Biol. 2018;25:669–676. doi: 10.1038/s41594-018-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakofsky C.J., Roberts S.A., Malc E., Mieczkowski P.A., Resnick M.A., Gordenin D.A., Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malkova A., Haber J.E. Mutations arising during repair of chromosome breaks. Annu. Rev. Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 14.Donnianni R.A., Zhou Z.X., Lujan S.A., Al-Zain A., Garcia V., Glancy E., Burkholder A.B., Kunkel T.A., Symington L.S. DNA polymerase delta synthesizes both strands during break-induced replication. Mol. Cell. 2019;76:371–381.e374. doi: 10.1016/j.molcel.2019.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naiman K., Campillo-Funollet E., Watson A.T., Budden A., Miyabe I., Carr A.M. Replication dynamics of recombination-dependent replication forks. Nat. Commun. 2021;12:923. doi: 10.1038/s41467-021-21198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 18.Gazy I., Hayward B., Potapova S., Zhao X., Usdin K. Double-strand break repair plays a role in repeat instability in a fragile X mouse model. DNA Repair (Amst.) 2019;74:63–69. doi: 10.1016/j.dnarep.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., Sohn R., Zemelman B., Snell R.G., Rundle S.A., Crow S., et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 20.Cho D.H., Tapscott S.J. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Bahrambeigi V., Song X., Sperle K., Beck C.R., Hijazi H., Grochowski C.M., Gu S., Seeman P., Woodward K.J., Carvalho C.M.B., Hobson G.M., Lupski J.R. Distinct patterns of complex rearrangements and a mutational signature of microhomeology are frequently observed in PLP1 copy number gain structural variants. Genome Med. 2019;11:80. doi: 10.1186/s13073-019-0676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verdin H., D'Haene B., Beysen D., Novikova Y., Menten B., Sante T., Lapunzina P., Nevado J., Carvalho C.M., Lupski J.R., De Baere E. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 2013;9 [Google Scholar]
- 23.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bacolla A., Wells R.D. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009;48:273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 25.Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., Halazonetis T.D. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G., Chen X., Bissler J.J., Sinden R.R., Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadgil R.Y., Romer E.J., Goodman C.C., Rider S.D., Jr., Damewood F.J., Barthelemy J.R., Shin-Ya K., Hanenberg H., Leffak M. Replication stress at microsatellites causes DNA double-strand breaks and break-induced replication. J. Biol. Chem. 2020;295:15378–15397. doi: 10.1074/jbc.RA120.013495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G., Myers S., Chen X., Bissler J.J., Sinden R.R., Leffak M. Replication fork stalling and checkpoint activation by a PKD1 locus mirror repeat polypurine-polypyrimidine (Pu-Py) tract. J. Biol. Chem. 2012;287:33412–33423. doi: 10.1074/jbc.M112.402503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guedin A., Gros J., Alberti P., Mergny J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyu J., Shao R., Kwong Yung P.Y., Elsasser S.J. Genome-wide mapping of G-quadruplex structures with CUT&Tag. Nucleic Acids Res. 2022;50 [Google Scholar]
- 34.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Jr., Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 38.Hansel-Hertsch R., Di Antonio M., Balasubramanian S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 39.Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintome C., Riou J.F., Prioleau M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prorok P., Artufel M., Aze A., Coulombe P., Peiffer I., Lacroix L., Guedin A., Mergny J.L., Damaschke J., Schepers A., Cayrou C., Teulade-Fichou M.P., Ballester B., Mechali M. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun. 2019;10:3274. doi: 10.1038/s41467-019-11104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee W.T.C., Yin Y., Morten M.J., Tonzi P., Gwo P.P., Odermatt D.C., Modesti M., Cantor S.B., Gari K., Huang T.T., Rothenberg E. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021;12:2525. doi: 10.1038/s41467-021-22830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang D., Hurley L.H. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 43.Lokanga R.A., Kumari D., Usdin K. Common threads: Aphidicolin-inducible and folate-sensitive fragile sites in the human genome. Front. Genet. 2021;12:708860. doi: 10.3389/fgene.2021.708860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Magis A., Manzo S.G., Russo M., Marinello J., Morigi R., Sordet O., Capranico G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2019;116:816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta R., Sharma S., Sommers J.A., Jin Z., Cantor S.B., Brosh R.M., Jr. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 46.Brosh R.M., Jr., Cantor S.B. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014;5:372. doi: 10.3389/fgene.2014.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., Liu G., Leffak M. Activation of a human chromosomal replication origin by protein tethering. Nucleic Acids Res. 2013;41:6460–6474. doi: 10.1093/nar/gkt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim M.Y., Vankayalapati H., Shin-Ya K., Wierzba K., Hurley L.H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 49.Tauchi T., Shin-ya K., Sashida G., Sumi M., Okabe S., Ohyashiki J.H., Ohyashiki K. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: In vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 50.Shalaby T., von Bueren A.O., Hurlimann M.L., Fiaschetti G., Castelletti D., Masayuki T., Nagasawa K., Arcaro A., Jelesarov I., Shin-ya K., Grotzer M. Disabling c-Myc in childhood medulloblastoma and atypical teratoid/rhabdoid tumor cells by the potent G-quadruplex interactive agent S2T1-6OTD. Mol. Cancer Ther. 2010;9:167–179. doi: 10.1158/1535-7163.MCT-09-0586. [DOI] [PubMed] [Google Scholar]
- 51.Miyazaki T., Pan Y., Joshi K., Purohit D., Hu B., Demir H., Mazumder S., Okabe S., Yamori T., Viapiano M., Shin-ya K., Seimiya H., Nakano I. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin. Cancer Res. 2012;18:1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura T., Okabe S., Yoshida H., Iida K., Ma Y., Sasaki S., Yamori T., Shin-Ya K., Nakano I., Nagasawa K., Seimiya H. Targeting glioma stem cells in vivo by a G-quadruplex-stabilizing synthetic macrocyclic hexaoxazole. Sci. Rep. 2017;7:3605. doi: 10.1038/s41598-017-03785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McWhinney C., Leffak M. In: Cancer Cells. Stillman B., Kelly T., editors. CSH Laboratory Press; New York, NY: 1988. Episomal persistence of a plasmid containing human c-myc DNA; pp. 467–471. [Google Scholar]
- 54.McWhinney C., Leffak M. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 1990;18:1233–1242. doi: 10.1093/nar/18.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McWhinney C., Waltz S.E., Leffak M. Cis-acting effects of sequences within 2.4-kb upstream of the human c-myc gene on autonomous plasmid replication in HeLa cells. DNA Cell Biol. 1995;14:565–579. doi: 10.1089/dna.1995.14.565. [DOI] [PubMed] [Google Scholar]
- 56.Tao L., Dong Z., Leffak M., Zannis-Hadjopoulos M., Price G. Major DNA replication initiation sites in the c-myc locus in human cells. J. Cell. Biochem. 2000;78:442–457. doi: 10.1002/1097-4644(20000901)78:3<442::aid-jcb9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 57.Trivedi A., Waltz S.E., Kamath S., Leffak M. Multiple initiations in the c-myc replication origin independent of chromosomal location. DNA Cell Biol. 1998;17:885–896. doi: 10.1089/dna.1998.17.885. [DOI] [PubMed] [Google Scholar]
- 58.Waltz S.E., Trivedi A.A., Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;24:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu G., Bissler J.J., Sinden R.R., Leffak M. Unstable spinocerebellar ataxia type 10 (ATTCT)∗(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol. Cell. Biol. 2007;27:7828–7838. doi: 10.1128/MCB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu G., Chen X., Leffak M. Oligodeoxynucleotide binding to (CTG). (CAG) microsatellite repeats inhibits replication fork stalling, hairpin formation, and genome instability. Mol. Cell. Biol. 2013;33:571–581. doi: 10.1128/MCB.01265-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis T.W., Barthelemy J.R., Virts E.L., Kennedy F.M., Gadgil R.Y., Wiek C., Linka R.M., Zhang F., Andreassen P.R., Hanenberg H., Leffak M. Deficiency of the Fanconi anemia E2 ubiqitin conjugase UBE2T only partially abrogates Alu-mediated recombination in a new model of homology dependent recombination. Nucleic Acids Res. 2019;47:3503–3520. doi: 10.1093/nar/gkz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W., Klein K.N., Proesmans K., Yang H., Marchal C., Zhu X., Borrman T., Hastie A., Weng Z., Bechhoefer J., Chen C.L., Gilbert D.M., Rhind N. Genome-wide mapping of human DNA replication by optical replication mapping supports a stochastic model of eukaryotic replication. Mol. Cell. 2021;81:2975–2988.e6. doi: 10.1016/j.molcel.2021.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barthelemy J., Hanenberg H., Leffak M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016;44:6803–6816. doi: 10.1093/nar/gkw433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hiatt S.M., Lawlor J.M.J., Handley L.H., Ramaker R.C., Rogers B.B., Partridge E.C., Boston L.B., Williams M., Plott C.B., Jenkins J., Gray D.E., Holt J.M., Bowling K.M., Bebin E.M., Grimwood J., et al. Long-read genome sequencing for the molecular diagnosis of neurodevelopmental disorders. HGG Adv. 2021;2:100023. doi: 10.1016/j.xhgg.2021.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donnianni R.A., Symington L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kramara J., Osia B., Malkova A. Break-induced replication: An unhealthy choice for stress relief? Nat. Struct. Mol. Biol. 2017;24:11–12. doi: 10.1038/nsmb.3361. [DOI] [PubMed] [Google Scholar]
- 67.Kramara J., Osia B., Malkova A. Break-induced replication: The where, the why, and the how. Trends Genet. 2018;34:518–531. doi: 10.1016/j.tig.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu G., Leffak M. Instability of (CTG)n∗(CAG)n trinucleotide repeats and DNA synthesis. Cell Biosci. 2012;2:7. doi: 10.1186/2045-3701-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo J., Gu L., Leffak M., Li G.M. MutSbeta promotes trinucleotide repeat expansion by recruiting DNA polymerase beta to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 2016;26:775–786. doi: 10.1038/cr.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barthelemy J., Hanenberg H., Leffak M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2017;45:509–511. doi: 10.1093/nar/gkw1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel H.P., Lu L., Blaszak R.T., Bissler J.J. PKD1 intron 21: Triplex DNA formation and effect on replication. Nucleic Acids Res. 2004;32:1460–1468. doi: 10.1093/nar/gkh312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu G., Malott M., Leffak M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Gorman S., Fox D.T., Wahl G.M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 74.Tahara H., Shin-Ya K., Seimiya H., Yamada H., Tsuruo T., Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3' telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 75.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be shared upon request to michael.leffak@wright.edu.