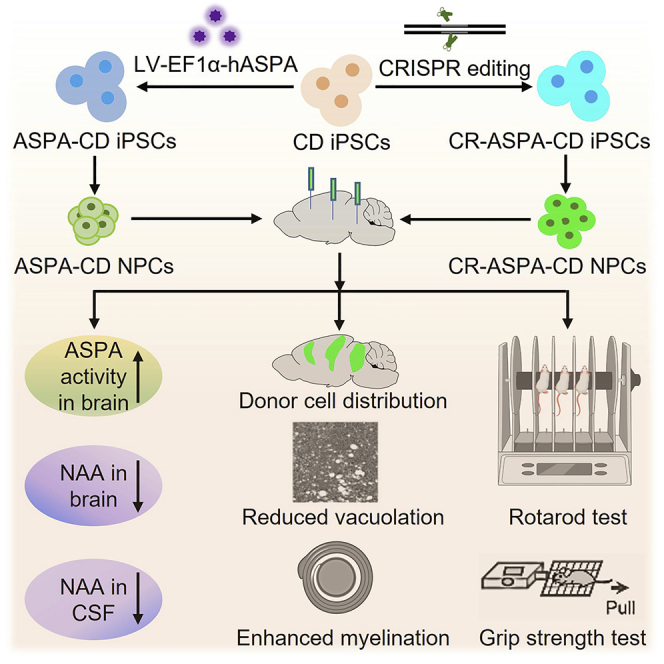
Summary
Canavan disease (CD) is a devastating neurological disease that lacks effective therapy. Because CD is caused by mutations of the aspartoacylase (ASPA) gene, we introduced the wild-type (WT) ASPA gene into patient iPSCs through lentiviral transduction or CRISPR/Cas9-mediated gene editing. We then differentiated the WT ASPA-expressing patient iPSCs (ASPA-CD iPSCs) into NPCs and showed that the resultant ASPA-CD NPCs exhibited potent ASPA enzymatic activity. The ASPA-CD NPCs were able to survive in brains of transplanted CD mice. The engrafted ASPA-CD NPCs reconstituted ASPA activity in CD mouse brains, reduced the abnormally elevated level of NAA in both brain tissues and cerebrospinal fluid (CSF), and rescued hallmark pathological phenotypes of the disease, including spongy degeneration, myelination defects, and motor function impairment in transplanted CD mice. These genetically modified patient iPSC-derived NPCs represent a promising cell therapy candidate for CD, a disease that has neither a cure nor a standard treatment.
Subject areas: Neuroscience, Biotechnology, Biotechnology of human disorders
Graphical abstract
Highlights
-
•
The wild-type ASPA gene was introduced into CD patient iPSCs to make ASPA-CD iPSCs
-
•
ASPA-CD iPSCs were differentiated into ASPA-CD NPCs with potent ASPA activity
-
•
Engrafted ASPA-CD NPCs could rescue major disease phenotypes in CD mice
-
•
CSF NAA level can be used as a biomarker to monitor the treatment outcome for CD
Neuroscience; Biotechnology; Biotechnology of human disorders
Introduction
Stem cell-based therapy provides great hope for the treatment of neurological disorders. However, the lack of expandable sources of stem cells is a critical issue in moving stem cell technology to the bedside. Human iPSCs derived by reprogramming human fibroblasts could provide a continuous and autologous cell source for the generation of specific somatic cell types and tissues from individual patients (Takahashi et al., 2007; Yu et al., 2007; Lowry et al., 2008; Park et al., 2008). Furthermore, autologous patient iPSCs could address the limitation of immune rejection associated with allogeneic donor cells. For diseases with known genetic mutations, patient iPSCs can be genetically modified by viral transduction or gene editing to introduce the wild-type (WT) gene into patient cells in order to overcome functional deficits caused by genetic mutations. These genetically engineered autologous iPSCs could provide exciting prospects for cell therapy.
Canavan disease (CD) is a devastating neurological disease and myelin disorder. The most prevalent disease onset form is infantile onset, symptoms of which appear in early infancy and progress rapidly. Clinical symptoms include mental retardation, loss of acquired motor skills, feeding difficulties, abnormal muscle tone, unusually large head, paralysis, blindness, and early death (Matalon et al., 1995). It has been shown that mutations in the aspartoacylase (ASPA) gene cause CD. ASPA is an enzyme expressed in oligodendrocytes of the brain (Matalon et al., 1988). It catalyzes the conversion of N-acetyl-aspartate (NAA), a highly abundant amino acid derivative produced by neurons, into aspartate and acetate in oligodendrocytes (Baslow, 2003). ASPA gene mutations lead to ASPA activity loss, which in turn results in elevated NAA level, spongy degeneration and demyelination of the brain, and motor function deficits (Canavan, 1931; van Bogaert and Bertrand, 1949; Matalon et al., 1988; Matalon and Michals-Matalon, 2000; Traka et al., 2008).
Studies have been designed to test potential therapies for CD. Directly targeting the mutant ASPA gene through gene therapy using functional human ASPA-expressing non-viral vectors or AAV vectors have been reported in both CD animal models and patients with CD in clinical trials (Janson et al., 2002; Matalon et al., 2003; McPhee et al., 2005; McPhee et al., 2006; Leone et al., 2012; Gessler et al., 2017; von Jonquieres et al., 2018; Lotun et al., 2021). These studies have demonstrated that the ASPA vector is well tolerated in both animals and human subjects (Leone et al., 2000; Janson et al., 2002; Matalon et al., 2003; McPhee et al., 2005; McPhee et al., 2006; Leone et al., 2012; Ahmed et al., 2013; Ahmed et al., 2016; Francis et al., 2016; Gessler et al., 2017; von Jonquieres et al., 2018). While substantial phenotypical improvements have been observed in rodent studies (Janson et al., 2002; Matalon et al., 2003; McPhee et al., 2005; McPhee et al., 2006; Ahmed et al., 2013; Ahmed et al., 2016; Francis et al., 2016; Gessler et al., 2017; von Jonquieres et al., 2018; Francis et al., 2021), the clinical benefits to patients remain to be demonstrated (Leone et al., 2000, 2012; Hoshino and Kubota, 2014; Bradbury and Ream, 2021; Mendell et al., 2021).
In addition to targeting the root course of the disease, the mutant ASPA gene, efforts have been devoted to reducing brain NAA level by suppressing the expression of the neuronal NAA-synthesizing enzyme N-acetyltransferase 8-like (Nat8l) (Guo et al., 2015; Sohn et al., 2017; Bannerman et al., 2018; Hull et al., 2020; Pleasure et al., 2020; Nesuta et al., 2021), or increasing acetate level by dietary supplementation with glyceryl triacetate or triheptanoin (Madhavarao et al., 2009; Arun et al., 2010; Segel et al., 2011; Francis et al., 2014). Both approaches have demonstrated efficacy in rodent studies (Arun et al., 2010; Francis et al., 2014; Guo et al., 2015; Sohn et al., 2017; Bannerman et al., 2018; Hull et al., 2020). While the former is waiting to be tested in patients, measurable neurological improvements in patients with CD remain to be demonstrated for the latter (Madhavarao et al., 2009; Segel et al., 2011; Pleasure et al., 2020). Therefore, developing an effective therapy for CD remains an unmet medical need.
This study is built on the knowledge we have gained through the previous preclinical and clinical studies, with an aim to provide a sustainable cellular source for the WT ASPA through a combined cell and gene therapy approach. We combined human iPSC technology with gene therapy to generate the WT ASPA gene-expressing CD patient iPSCs (ASPA-CD iPSCs) by introducing the WT ASPA gene into CD patient iPSCs through lentiviral transduction or CRISPR/Cas9-mediated gene editing. We further differentiated the ASPA-CD iPSCs into neural progenitor cells (NPCs) and demonstrated the therapeutic efficacy and preliminary safety of the ASPA-CD NPCs in a CD mouse model. We also developed a method to detect NAA level in the CSF as a biomarker for monitoring the therapeutic efficacy after ASPA-CD NPC transplantation.
Results
Generation of the WT ASPA-expressing ASPA-CD iPSCs
We have obtained primary dermal fibroblasts from a clinically affected patient with CD (STAR Methods) that has heterozygous mutations of G176E and A305E. Of note, A305E is the most common mutation (60%) in non-Jewish patients with CD (Kaul et al., 1994). The CD patient fibroblasts were reprogrammed to generate CD patient iPSCs (CD iPSCs), which expressed the key human pluripotency genes and the human embryonic stem cell (ESC)-specific surface markers (Figure S1A). Sequence analysis confirmed that the CD patient iPSCs harbored the ASPA mutation of the corresponding patient with CD (Figure S1B). Bisulfite sequencing confirmed epigenetic reprogramming as revealed by demethylation of the endogenous OCT4 and NANOG promoters in CD iPSCs (Figures S1C and S1D). The in vivo developmental potential of CD iPSCs was demonstrated by teratoma formation (Figure S1E). Cytogenetic analysis confirmed normal karyotype in all iPSC clones tested (Figure S1F). Together, these results demonstrate that the CD iPSCs we derived are characteristic pluripotent stem cells containing CD patient ASPA mutations.
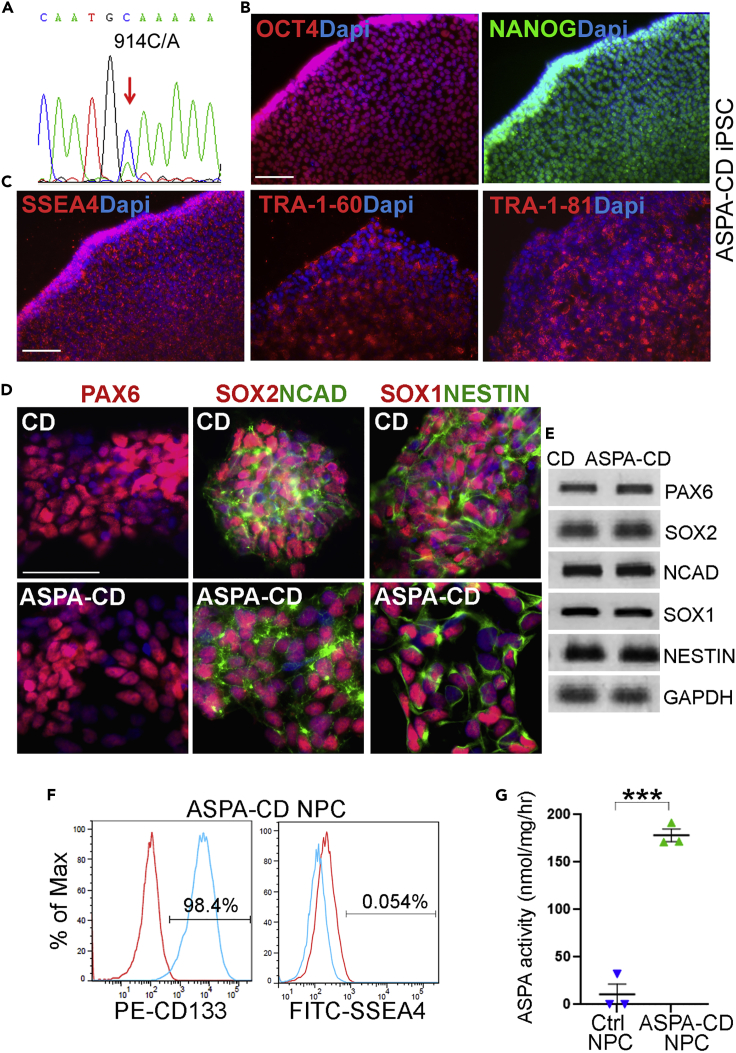
Because CD is caused by genetic mutations in the ASPA gene, in order to restore the WT ASPA gene expression, we transduced CD patient iPSCs with lentivirus expressing the human WT ASPA gene under the constitutive human EF1α promoter. The WT ASPA-expressing CD patient iPSCs were termed ASPA-CD iPSCs. The presence of the WT ASPA gene sequence in ASPA-CD iPSCs was confirmed by genomic DNA sequencing (Figure 1A). Immunostaining revealed that ASPA-CD iPSCs continued to express the pluripotency factors OCT4 and NANOG and the human ESC surface markers SSEA4, TRA-1-60, and TRA-1-81 (Figures 1B and 1C).
Figure 1.
Characterization of ASPA-CD iPSCs and ASPA-CD NPCs
(A) Genomic DNA sequencing confirmed the presence of the WT ASPA sequence in ASPA-CD iPSCs.
(B) Expression of the human pluripotency factors OCT4 and NANOG in the ASPA-CD iPSCs. Nuclei Dapi staining is shown in blue. Scale bar: 100 μm.
(C) Expression of the human ESC cell surface markers SSEA4, TRA-1-60, and TRA-1-8 in ASPA-CD iPSCs. Nuclear Dapi staining is shown in blue. Scale bar: 100 μm.
(D) Immunostaining for the NPC markers PAX6, SOX2, NCAD, SOX1, and NESTIN in NPCs derived from CD and ASPA-CD iPSCs. Nuclear Dapi staining is shown in blue. Scale bar: 50 μm.
(E) Expression of the NPC markers in ASPA-CD NPCs revealed by RT-PCR. CD NPCs were included as a control. GAPDH was included as a loading control.
(F) FACS analysis of ASPA-CD NPCs.
(G) ASPA-CD NPCs displayed potent ASPA enzymatic activity, compared to IMR90 control NPCs that were not transduced with lentivirus encoding the wild-type ASPA gene.
Error bars are SE of the mean (n = 3 biological repeats). ∗∗∗p < 0.001 by Student’s t test. See also Figure S1.
Generation and characterization of ASPA-CD NPCs
Next, we differentiated ASPA-CD iPSCs into NPCs following a published method (Liu et al., 2012). The resultant ASPA-CD NPCs expressed typical NPC markers, including PAX6, SOX2, N-cadherin (NCAD), SOX1, and NESTIN, as revealed by immunostaining and RT-PCR (Figures 1D and 1E). Fluorescence-activated cell sorting (FACS) revealed that vast majority of the ASPA-CD NPCs were CD133-positive NPCs, with minimal contamination of undifferentiated iPSCs as revealed by the negligible fraction of SSEA4-positive cells (Figure 1F). Importantly, the ASPA-CD NPCs exhibited potent ASPA enzymatic activity, compared to control NPCs that were not transduced with lentivirus encoding the wild-type ASPA gene, which exhibited no detectable ASPA activity (Figure 1G). Together, these results demonstrate the identity, purity, and potency of the ASPA-CD NPCs.
The bio-distribution and cell fate of ASPA-CD NPCs in transplanted CD mouse brains
The Aspanur7/nur7 mouse that contains a nonsense mutation (Q193X) in the ASPA gene (Traka et al., 2008) manifests major pathological phenotypes of CD, therefore have been used as a CD mouse model. We bred Aspanur7/nur7 mice with immunodeficient Rag2−/− mice to generate an immunodeficient CD mouse model to allow engraftment of human cells. These mice, called CD mice for short, exhibit characteristic pathological features of CD (Feng et al., 2020), and were thus used as a preclinical model for testing the effect of ASPA-CD NPCs in vivo in the following study.
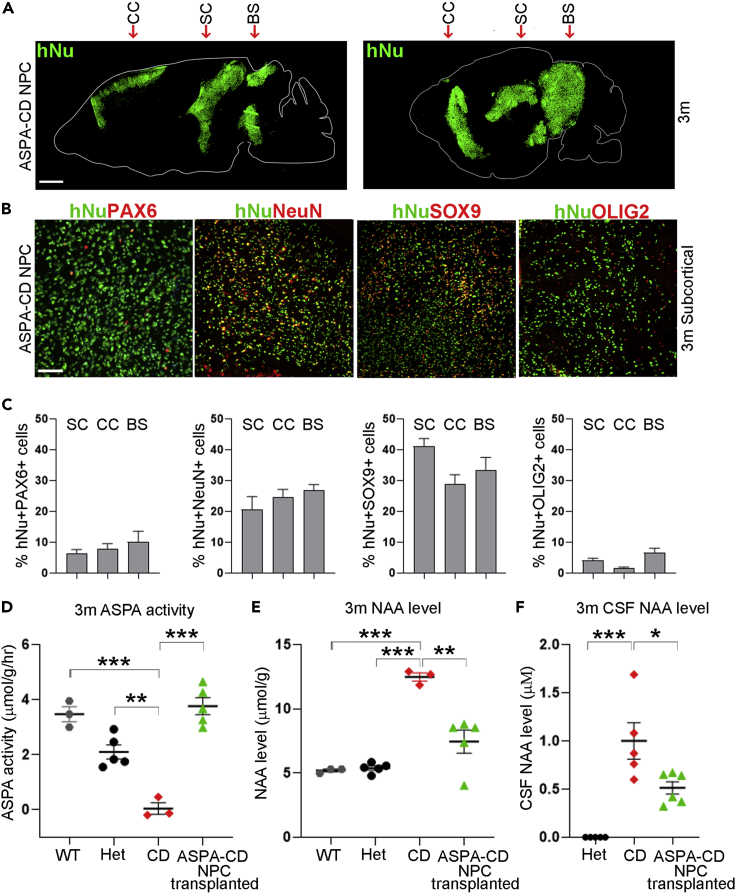
We transplanted ASPA-CD NPCs derived from ASPA-CD iPSCs into brains of postnatal day 1–4 CD pups stereotactically as we described previously (Feng et al., 2020). Specifically, ASPA-CD NPCs were transplanted bilaterally into the corpus callosum, the subcortical region, and the brain stem. Three months after transplantation, brains of transplanted mice were harvested and sectioned. The engraftment of the ASPA-CD NPCs was confirmed by immunostaining for the human nuclear antigen (hNu) in transplanted CD mice. The transplanted cells were detected in regions around the injection sites, including the corpus callosum, the subcortical, and the brain stem regions, with limited migration (Figure 2A). While a small portion of the engrafted ASPA-CD NPCs was maintained as NPCs, as revealed by double-positive staining for hNu and the NPC marker PAX6 (Figures 2B and 2C), more ASPA-CD NPCs differentiated into neurons or astrocytes as revealed by positive staining for hNu together with the neuronal lineage marker NeuN or the astroglial lineage marker SOX9 (Figures 2B and 2C). A small fraction of ASPA-CD NPCs became oligodendroglial lineage cells as revealed by co-staining for hNu and the oligodendroglial lineage marker OLIG2 (Figures 2B and 2C).
Figure 2.
Transplantation with ASPA-CD NPCs leads to increased ASPA activity and decreased NAA level in CD mouse brains
(A) Tiling images of human nuclear antigen (hNu) staining are shown by dot map. ASPA-CD NPCs were distributed around the injection track in transplanted CD mouse brains 3 months after transplantation. The arrows indicate the injection sites. CC: corpus callosum; SC: subcortical; BS: brain stem. Scale bar: 1 mm.
(B and C) Transplanted ASPA-CD NPCs differentiated into neuronal and glial lineage cells in CD mouse brains, as revealed by immunostaining for hNu and PAX6, NeuN, SOX9, and OLIG2, respectively. The images of the subcortical region are shown in panel B. Scale bar: 50 μm. Quantification of transplanted cells in the subcortical (SC), corpus callosum (CC), and brain stem (BS) regions is shown in panel C. n = 9 fields from three mice for each group.
(D–F) The ASPA activity was elevated (D) and the NAA level was reduced in the brain tissues (E) and the CSF (F) of ASPA-CD NPC-transplanted CD mice. Each dot corresponds to the result of one mouse. n = 3 mice for WT or CD mice, five for Het mice or ASPA-CD NPC-transplanted CD mice for panels (D and E). n = 5 mice for Het or CD mice, and n = 6 mice for ASPA-CD NPC-transplanted CD mice for panel (F).
Error bars are SE of the mean. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by one-way ANOVA followed by Tukey’s multiple comparisons test.
Engrafted ASPA-CD NPCs can increase ASPA activity and reduce NAA level in CD mice
The reduced ASPA enzymatic activity and elevated NAA level are characteristic phenotypes in both CD patients and animal models (Canavan, 1931; van Bogaert and Bertrand, 1949; Matalon et al., 1988; Matalon and Michals-Matalon, 2000). Three months after transplantation, ASPA-CD NPC-transplanted CD brains were subjected to ASPA activity and NAA level measurement. The ASPA-CD NPC-transplanted CD brains exhibited substantially higher ASPA activity than the control CD brains without transplantation. The ASPA activity in ASPA-CD NPC-transplanted CD mouse brains was similar to that in WT or heterozygous (Het) mouse brains (Figure 2D). In line with the elevated ASPA enzymatic activity, the NAA level was reduced in ASPA-CD NPC-transplanted CD mouse brains (Figure 2E). Importantly, we also detected reduced NAA level in the cerebrospinal fluid (CSF) of ASPA-CD NPC-transplanted CD mice (Figure 2F). Therefore, CSF NAA level could be a potential biomarker for evaluating the therapeutic effect of treatments, such as ASPA-CD NPC transplantation.
Engrafted ASPA-CD NPCs can reduce vacuolation in CD mouse brains
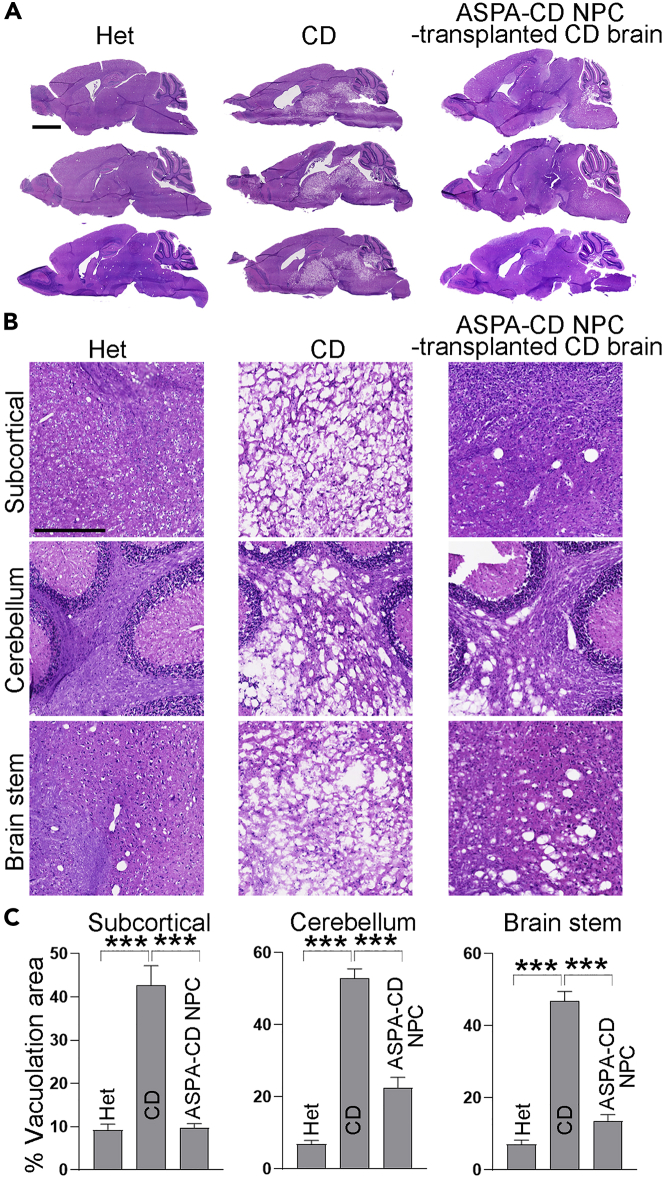
Extensive spongy degeneration is a key pathological feature of CD patients and mouse models, which is revealed by vacuolation in various brain regions (Matalon et al., 1988, 2000; Traka et al., 2008; Mersmann et al., 2011). We indeed found extensive vacuolation in the subcortical, brain stem, and cerebellum regions of CD mouse brains, which was not seen in the corresponding regions of Het mouse brains. To our excitement, we detected substantially reduced extent of vacuolation in various brain regions of ASPA-CD NPC-transplanted CD mice, including the subcortical, brain stem, and cerebellum regions (Figures 3A–3C). These results indicate that transplantation with ASPA-CD NPCs was able to rescue the spongy degeneration phenotype in CD mouse brains.
Figure 3.
Improved vacuolation in ASPA-CD NPC-transplanted CD mouse brains
(A and B) H&E staining of the subcortical, cerebellum, and brain stem region and whole sagittal section in Het, CD mice, and ASPA-CD NPC-transplanted CD mice. Scale bar: 2000 μm in panel (A), 500 μm in panel (B).
(C) The percentage of vacuolation area is shown. n = 3 mice in each group.
Error bars are SE of the mean. ∗∗∗p < 0.001 by one-way ANOVA followed by Tukey’s multiple comparisons test.
Engrafted ASPA-CD NPCs can rescue myelination deficits in CD mouse brains
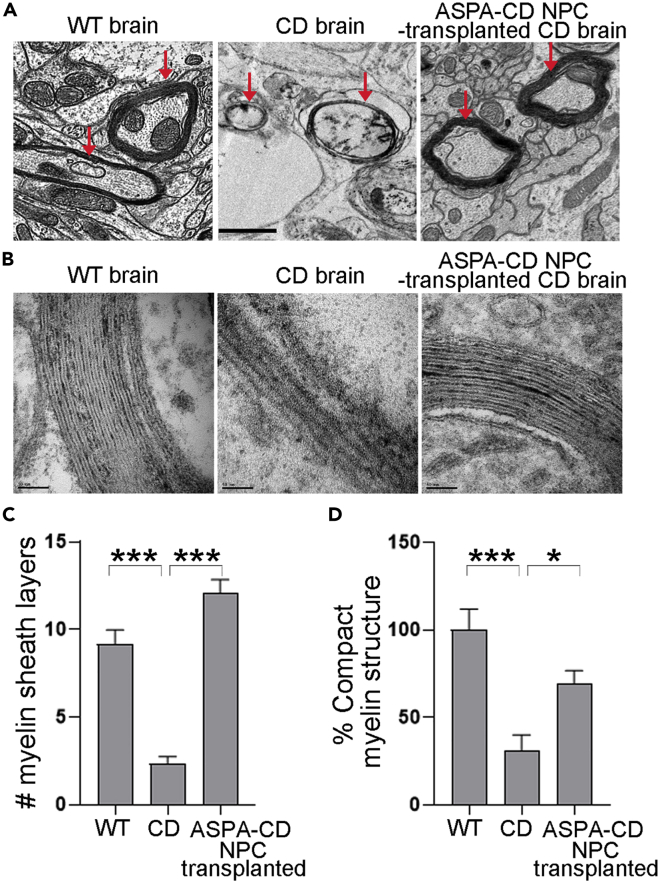
In keeping with extensive vacuolation in brains of CD mice, substantially reduced thickness of myelin sheaths and myelinated areas were detected in brains of CD mice, compared to that from WT mice (Figures 4A–4C). Myelin sheaths in ASPA-CD NPC-transplanted CD brains were much thicker than that of untreated CD brains, instead resembled more to that of WT brains (Figures 4A–4C). The percentage of compact myelin structure was also improved in transplanted CD mouse brains (Figure 4D). These results further support the therapeutic potential of ASPA-CD NPCs for their ability to ameliorate the pathological phenotypes of CD.
Figure 4.
ASPA-CD NPCs rescue myelination in transplanted CD mice
(A) The myelination was rescued in brain of ASPA-CD NPC-transplanted CD mice. Intact and thick myelin sheaths were detected by EM in brains of 3-month-old WT mice, whereas splitting and thinner myelin sheaths were seen in brains of littermate CD mice. Myelin sheaths in CD mice transplanted with ASPA-CD NPCs for three months appeared thicker. Images of brain stem region are shown. Scale bar: 1 μm. The arrows point to the myelin sheaths.
(B) Enlarged EM images of the brain stem region in WT, CD, and ASPA-CD NPC-transplanted mice. Scale bar: 50 nm.
(C) The number (#) of myelin sheath layers in WT, CD, and ASPA-CD NPC-transplanted CD mice. n = 12 axons from one WT and one CD mouse brain, respectively. n = 15 axons from two ASPA-CD NPC-transplanted CD mouse brains. Error bars are SE of the mean. ∗∗∗p < 0.001 by one-way ANOVA followed by Dunnett’s multiple comparisons test.
(D) The percentage of compact myelin structure in WT, CD, and ASPA-CD NPC-transplanted CD mice. n = 8 imaging fields from one mouse for each group.
Error bars are SE of the mean. ∗p < 0.05 and ∗∗∗p < 0.001 by one-way ANOVA followed by Dunnett’s multiple comparisons test.
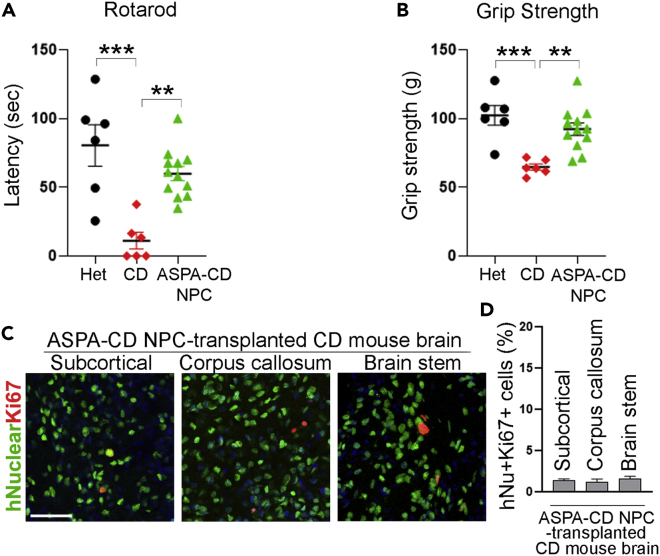
Engrafted ASPA-CD NPCs can improve motor function in CD mice
In addition to the improvement of CD phenotypes at the cellular level, transplantation with ASPA-CD NPCs was also associated with systemic effect on CD mice. Defective motor function is characteristic of CD patients and mouse models (Matalon et al., 1988, 2000; Traka et al., 2008; Mersmann et al., 2011). To investigate if ASPA-CD NPCs could rescue the defect in motor performance in CD mice, we subjected CD mice transplanted with ASPA-CD NPCs to two motor skill behavioral tests, the accelerating rotarod test, and the grip strength test. ASPA-CD NPCs improved rotarod performance in transplanted CD mice substantially three months after transplantation, compared to control CD mice without transplantation (Figure 5A). In addition, substantial improvement of the forepaw grip strength was detected in ASPA-CD NPC-transplanted CD mice, compared to that in control CD mice in the grip strength test (Figure 5B). The results from the two behavioral tests together indicate that transplantation with ASPA-CD NPCs can improve motor functions in a CD mouse model dramatically. Collectively, these results provide compelling evidence that ASPA-CD NPCs that carry a WT ASPA gene exhibit substantial therapeutic potential for rescuing the pathological phenotypes of CD.
Figure 5.
Transplantation with ASPA-CD NPCs improved motor function in CD mice
(A and B) Transplantation with ASPA-CD NPCs improved motor functions in CD mice as revealed in rotarod or grip strength test. n = 6 mice for the Het and the CD mouse group, n = 12 mice for the ASPA-CD NPC-transplanted CD mouse group. Each dot corresponds to the result of one mouse.
(C) The transplanted ASPA-CD NPCs exhibited low mitotic index in CD mouse brains as revealed by human nuclear antigen and Ki67 double staining. Scale bar: 50 μm.
(D) The percentage of human nuclear antigen (hNu)+Ki67 + cells out of total hNu + cells in transplanted brains. n = 9 fields from three mice for each group.
Error bars are SE of the mean. ∗∗p < 0.01, and ∗∗∗p < 0.001 by one-way ANOVA followed by Dunnett’s multiple comparisons test for panels (A and B). See also Figure S2.
No tumor formation in ASPA-CD NPC-transplanted CD mice
For preliminary safety study, mice transplanted with ASPA-CD NPCs were closely observed for 3 months. We detected no sign of tumor formation or other adverse events. By the end of 3 months, transplanted mice were euthanized. Their brains were collected and analyzed by H&E staining. No typical tumor tissue was detected in brain sections of ASPA-CD NPC-transplanted mice (Figure S2). The absence of tumor formation in brains of ASPA-CD NPC-transplanted mice was further verified by immunostaining for Ki67, a proliferative marker. We detected a low mitotic index, as revealed by the low percentage of hNu-positive and Ki67-positive cells in ASPA-CD NPC-grafted brains (Figures 5C and 5D). These results together confirm preliminary safety of ASPA-CD NPCs.
CR-ASPA-CD NPCs derived from CRISPR/Cas9-edited CD iPSCs can rescue motor function deficits in CD mice
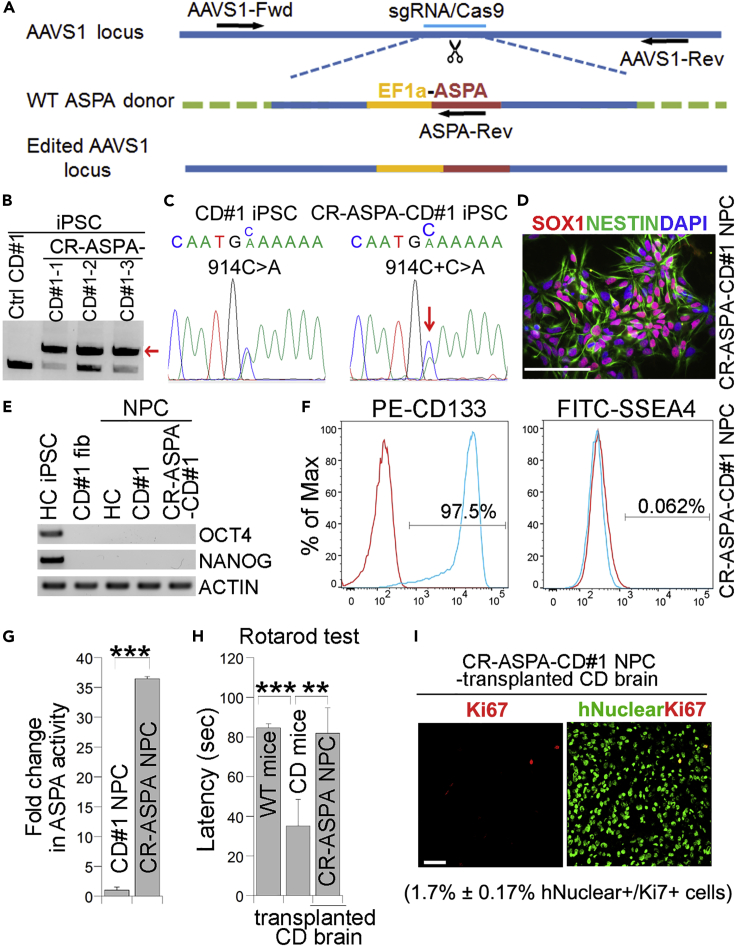
In addition to introducing the WT ASPA gene into CD iPSCs through lentiviral transduction, we introduced the WT ASPA gene into the AAVS1 safe harbor site in CD iPSCs by CRISPR/Cas9-based gene editing (Figure 6A) (Cong et al., 2013; Blair et al., 2016; Oceguera-Yanez et al., 2016). Three clones were selected for further analysis based on the genotyping result showing successful introduction of the WT ASPA gene (Figure 6B). The presence of the WT ASPA gene in edited clones was confirmed by genomic DNA sequencing (Figure 6C). The edited iPSCs were called CRISPR (CR)-ASPA-CD iPSCs. One of the clones (CR-ASPA-CD#1-1 iPSCs) was selected for further characterization and was termed CR-ASPA-CD#1 iPSCs or CR-ASPA-CD iPSCs in short. Immunostaining revealed that the edited clone expressed the pluripotency factors OCT4, SOX2, and NANOG, and the human ESC surface markers SSEA4, TRA-1-60, and TRA-1-81 (Figure S3A). The CR-ASPA-CD iPSCs maintained their developmental potential as revealed by teratoma formation assay (Figure S3B), exhibited normal karyotype (Figure S3C) and lacked off-target editing (Table S1).
Figure 6.
CR-ASPA-CD NPCs derived from CRISPR/Cas9-edited iPSCs can rescue the motor function deficits in CD mice
(A) Schematic for introducing the WT ASPA gene into the AAVS1 locus by CRISPR/Cas9 gene editing.
(B) PCR-based genotyping of CR-ASPA-CD iPSC clones. The PCR product of the edited allele is indicated by arrow.
(C) Genomic DNA sequencing confirms the presence of the WT ASPA sequence in CR-ASPA-CD iPSCs.
(D) Expression of the NPC markers NESTIN and SOX1 in CR-ASPA-CD NPCs as revealed by immunostaining.
(E) Lack of expression of the pluripotency factors in HC (healthy control), CD, and CR-ASPA-CD NPCs as revealed by RT-PCR.
(F) CR-ASPA-CD NPCs are positive for the neural progenitor marker CD133 but negative for the human ESC marker SSEA4 as revealed by FACS analysis.
(G) CR-ASPA-CD NPCs displayed potent ASPA enzymatic activity, compared to the control CD NPCs.
(H) Transplantation with CR-ASPA-CD NPCs rescued the motor function deficits in CD mice as revealed by rotarod test 3 months post grafting (n = 5 WT, 4 CD, and 7 CR-ASPA-CD NPC-transplanted mice).
(I) CR-ASPA-CD NPC-transplanted CD mouse brains were immunostained for the human nuclear antigen and Ki67 (n = 3 mice analyzed). Scale bar: 50 μm for panels (D and I).
Error bars are SE of the mean for panels (G and H). ∗∗p < 0.01 and ∗∗∗p < 0.001 by Student’s t test for panel (G) and one-way ANOVA for panel (H). See also Figure S3.
We then differentiated CR-ASPA-CD iPSCs into NPCs. The derived CR-ASPA-CD NPCs expressed typical NPC markers, including SOX1, SOX2, and NESTIN (Figures 6D and S3D). In contrast, no expression of the pluripotency factors OCT4 and NANOG was detected in the NPCs (Figure 6E). FACS analysis revealed that most of the CR-ASPA-CD NPCs were CD133-positive, with minimal contamination of undifferentiated iPSCs as revealed by the negligible percentage of SSEA4-positive cells (Figure 6F). Moreover, the CR-ASPA-CD NPCs displayed potent ASPA activity when compared to control CD NPCs (Figure 6G). Together, these results demonstrate the identity, purity, and activity of the CR-ASPA-CD NPCs.
CR-ASPA-CD NPCs were then transplanted into CD mice for efficacy study as described above. Staining for human nuclear antigen revealed that transplanted CR-ASPA-CD NPCs were able to survive in brains of CD mice (Figure S3E). Three months after transplantation, CR-ASPA-CD NPC-transplanted CD mice were tested on accelerating rotarod. CR-ASPA-CD NPCs improved rotarod performance in transplanted CD mice substantially, compared to CD mice without transplantation (Figure 6H). These results indicate that transplantation with CR-ASPA-CD NPCs can improve motor function in CD mice.
We observed no sign of tumor formation or other adverse effect during three months after transplantation with CR-ASPA-CD NPCs. Ki67 staining showed low percentage of human antigen and Ki67 double-positive cells (1.7% ± 0.17%) in CR-ASPA-CD NPC-grafted brains (Figure 6I). These results indicate that CR-ASPA-CD NPCs exhibit preliminary safety.
Discussion
In this study, we introduced a WT ASPA gene into CD patient iPSCs and differentiated the resultant ASPA-CD iPSCs into ASPA-CD NPCs. We further established preclinical efficacy for ASPA-CD NPCs, in order to develop this cellular product as a candidate cell therapy for CD, a devastating neurological disease that has neither a cure nor a standard treatment. Because CD is caused by genetic mutations in the ASPA gene, we have genetically engineered CD patient iPSCs by introducing the WT ASPA gene through lentiviral transduction. We further differentiated the resultant ASPA-CD iPSCs into NPCs. The ASPA-CD NPCs exhibited potent ASPA activity. We transplanted the ASPA-CD NPCs into a CD mouse model. ASPA-CD NPCs were able to survive after transplantation. Moreover, the transplanted cells were able to increase the ASPA activity, reduce the elevated NAA level, and rescue the sponge degeneration in brains of transplanted CD mice. Transplantation with ASPA-CD NPCs also rescued motor function deficiency in CD mice. Importantly, no tumorigenesis or other adverse effect was observed in transplanted mice. Furthermore, we demonstrated that NAA level in the CSF of ASPA-CD NPC-transplanted CD mice was reduced in a manner that correlates with the reduction of NAA level in transplanted brains and the rescue of other CD pathological phenotypes in the transplanted mice, indicating that the CSF NAA level could be an effective biomarker for monitoring the efficacy of ASPA-CD NPCs in vivo. These results together indicate that ASPA-CD NPCs have the potential to serve as a cell therapy candidate for CD and that the CSF NAA level can be used to monitor the outcome of the treatment for CD.
In addition to introducing the WT ASPA gene into CD iPSCs by lentiviral transduction, we knocked in the WT ASPA gene into the AAVS1 safe harbor site in CD iPSCs by CRISPR/Cas9-mediated gene editing. The CRISPR/Cas9-edited ASPA iPSCs were then differentiated into NPCs (CR-ASPA-CD NPCs). After being transplanted into CD mouse brains, these cells rescued the behavioral phenotype of CD mice to a level that is comparable to that by ASPA-CD NPCs in which the WT ASPA gene was introduced lentivirally. Notably, we detected no tumorigenesis or other adverse effects in mice transplanted with either ASPA-CD NPCs or CR-ASPA-CD NPCs. These results indicate that both ASPA-CD NPCs and CR-ASPA-CD NPCs are promising cell therapy candidates for CD.
Huge efforts have been devoted into therapeutic development for CD. Some of the approaches such as direct gene therapy to introduce a functional ASPA gene have resulted in substantial functional recovery in preclinical studies, although clinical benefits remain to be demonstrated (Leone et al., 2000; Janson et al., 2002; Matalon et al., 2003; McPhee et al., 2005; McPhee et al., 2006; Leone et al., 2012; Ahmed et al., 2013; Ahmed et al., 2016; Francis et al., 2016; Gessler et al., 2017; von Jonquieres et al., 2018). Likewise, reducing NAA level by knocking down the NAA-synthesizing enzyme Nat8l also resulted in rescue of disease phenotypes in preclinical studies (Bannerman et al., 2018; Hull et al., 2020), suggesting that lowering NAA level via inhibition of Nat8l expression or function could also be a potential approach for the treatment of CD, although the clinical benefit of this approach remains to be determined. Until now, there remains no cure for patients with CD and treatment is symptomatic only.
The application of cell therapy is gaining great momentum because it could have broad therapeutic impact. Compared to direct gene therapy, the combined cell and gene therapy approach offers an opportunity for extensive evaluation of the safety and efficacy of the genetically modified cells in vitro and selection of the best candidate for advancing into in vivo study. Moreover, the cellular product could provide a reservoir for sustained ASPA activity and have regenerative potential (Shi et al., 2017; Feng et al., 2020). In a previous study (Feng et al., 2020), we demonstrated the preclinical efficacy of ASPA iNPCs, a cellular product in which a functional ASPA gene with a point mutation (ASPA R132G) was introduced into CD patient iPSC-derived NPCs by lentiviral transduction. In the current study, the WT ASPA gene was introduced into CD iPSCs via lentiviral transduction or CRISPR/Cas9 editing. A CD iPSC clone carrying the WT ASPA gene was picked and characterized extensively. Well-characterized ASPA-CD iPSCs were then differentiated into ASPA-CD NPCs. One advantage of the current approach is that less virus will be needed to make the cellular product because viral transduction will be conducted at the initial iPSC stage instead of at the later NPC stage when cells have been expanded. Moreover, all of the resulted ASPA-CD NPCs should carry the ASPA transgene because these NPCs should all be generated from one iPSC clone that is originated from one single iPSC cell.
A major obstacle for moving cell therapy to humans is to have enough cells for transplantation into patients. iPSCs could provide an unlimited source of cells that are otherwise not possible to obtain for cell replacement therapy due to their easy accessibility and extensive expandability. Moreover, human iPSC-derived NPCs are also largely expandable, allowing us to reach human doses without much technical challenges. In this study, both ASPA-CD NPCs and CR-ASPA-CD NPCs are readily expandable. Furthermore, patient-specific iPSCs could provide a source of autologous cells that may avoid immunogenicity associated with allogeneic cell transplantation, therefore offering an exciting opportunity for the treatment of human diseases using cell replacement therapy, especially for orphan disease such as Canavan disease, for which a master cell bank may not be necessary (Shi et al., 2017).
Although there is a safety concern when iPSC-based products are developed for therapeutic applications because undifferentiated iPSCs could generate teratomas, it is worth noting that the differentiated product of iPSCs has not been shown to form teratomas. To address the safety concern associated with potential development of teratoma from iPSC products, it is important to make sure the final iPSC-derived products do not include undifferentiated cells. We have used a protocol that can differentiate human iPSCs into NPCs at very high efficiency (Liu et al., 2012). FACS analysis of our ASPA-CD NPCs revealed minimal residual undifferentiated cells. More importantly, close observation of the ASPA-CD NPC-transplanted mice for 3 months revealed no sign of tumorigenesis.
Monitoring treatment outcome effectively using biomarkers is of significant clinical importance. High level of NAA has been found in the CSF of patient with CD (Hamaguchi et al., 1993). In this study, we demonstrated that the level of NAA in the CSF of ASPA-CD NPC-transplanted CD mice was reduced substantially compared to control CD mice, correlating with reduced brain NAA level and improved cellular and behavioral phenotypes in ASPA-CD NPC-transplanted mice, indicating that the CSF NAA level can be an effective biomarker for tracking the treatment outcome in patients with CD. This biomarker can be used for monitoring the therapeutic outcome of not only our cellular therapies but also any NAA-lowering therapies including direct ASPA gene therapy and Nat8l-targeting therapies.
In summary, this study provides critical preclinical data on efficacy and preliminary safety for developing a human iPSC-based cell therapy candidate for CD, a neurological disease that has no cure or disease-modifying treatments. We also presented data to demonstrate that the CSF NAA level can be an effective biomarker for monitoring the outcome for CD treatments. The combination of gene therapy with cell therapy as described in this study provides tremendous hope for a variety of genetic disorders that have no treatment options.
Limitations of the study
This study provides clear evidence that neural progenitor cells derived from CD patient iPSCs that were introduced with a WT ASPA gene by either lentiviral transduction or CRISPR/Cas9 editing could provide robust disease-rescuing effect. However, there are several limitations that remain to be addressed in the future. Firstly, the number of patient iPSC line is limited. Because we have shown that NPC lines that carry different ASPA gene mutations and were transduced with a functional (but mutant) ASPA gene exhibit comparable disease-modifying effects, it is plausible to speculate that the outcome of ASPA-CD NPCs derived from different CD patient lines would be comparable. Nevertheless, confirming the effects of ASPA-CD NPCs generated from different CD patient lines is part of our future plan. Secondly, we were not able to determine the engraftment efficiency properly because the engrafted ASPA-CD NPCs were localized together, thus it is difficult to count individual engrafted cells accurately. Thirdly, while we have provided evidence of behavioral rescue by CR-ASPA-CD NPCs in which the WT ASPA gene was introduced by CRISPR/Cas9 editing, we did not have direct experimental data to confirm improved myelination and reduced vacuolation. Future studies are needed to assess the extent of rescue in terms of myelination and spongy degeneration by CR-ASPA-CD NPCs. Another limitation is that we did not keep the transplanted mice long enough to check the lifespan of these mice. Longer follow-up will also be needed to evaluate long-term survival and integration of transplanted cells in the engrafted brains and for future tumorigenicity study. Despite these limitations, this study provides robust disease-rescuing effect of ASPA-CD NPCs that were derived from CD patient iPSCs and introduced with the WT ASPA gene. This cellular product represents a promising cell therapy candidate for CD, a disease that has neither a cure nor a standard treatment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-NANOG | Cell Signaling | Cat # 4903; RRID:AB_10559205 |
| Mouse monoclonal anti-OCT4 | Santa Cruz | Cat # sc-5279; RRID:AB_628051 |
| Goat polyclonal anti-SOX2 | Santa Cruz | Cat # sc-17320; RRID:AB_2286684 |
| Mouse monoclonal anti-SSEA4 | Santa Cruz | Cat # sc-21704; RRID:AB_628289 |
| Mouse monoclonal IgM anti-Tra-1-60 | Santa Cruz | Cat # sc-21705; RRID:AB_628385 |
| Mouse monoclonal IgM anti-Tra-1-81 | Santa Cruz | Cat # sc-21706; RRID:AB_628386 |
| Mouse monoclonal anti-NESTIN | BD Biosciences | Cat # 611659; RRID:AB_399177 |
| Goat polyclonal anti-SOX1 | R&D | Cat # AF3369; RRID:AB_2239879 |
| FITC-conjugated anti-SSEA4 | Miltenyi Biotec | Cat # 130-098-371; RRID:AB_2653517 |
| PE-conjugated anti-CD133/1 | Miltenyi Biotec | Cat # 130-113-108; RRID:AB_2725937 |
| Mouse monoclonal anti-human nuclear antigen antibody [235-1], hNuclear | Abcam | Cat # Ab191181; RRID:AB_2885016 |
| Rabbit polyclonal anti-PAX6 | Biolegend | Cat # 901301; RRID:AB_2565003 |
| Rabbit polyclonal anti-OLIG2 | Millipore | Cat # AB9610; RRID:AB_570666 |
| Goat polyclonal anti-SOX9 | R&D | Cat # AF3075; RRID:AB_2194160 |
| Rabbit polyclonal anti-NEUN | Genetex | Cat # GTX16208 |
| Rabbit monoclonal anti-Ki67 | ThermoFisher Scientific | Cat # RM-9106-S0; RRID:AB_2341197 |
| Chemicals, peptides, and recombinant proteins | ||
| Essential 8TM basal medium | Gibco | A15169 |
| Essential 8TM supplement | Gibco | A15171 |
| Matrigel | Corning | 354230 |
| DMEM/F12 basal medium | Gibco | 11330-032 |
| N-2 supplement | Gibco | 17502-048 |
| B-27TM supplement | Gibco | 17504-044 |
| GlutaMAXTM supplement | Gibco | 35050-061 |
| MEM Non-Essential Amino Acids Solution (NEAA) | Gibco | 11140-050 |
| Y27632 | Cellagen Technology | C9127 |
| CHIR99021 | Cellagen Technology | C2447 |
| SB431542 | Cellagen Technology | C7243 |
| Dorsomorphin | Sigma | P5499 |
| Retinoic acid | Sigma | R2625 |
| Animal-free recombinant human EGF | Peprotech | AF-100-15 |
| Animal-free recombinant human FGF-basic | Peprotech | AF-100-18B |
| Experimental models: Cell lines | ||
| CD fibroblast | Coriell | GM00059 |
| ASPA-CD iPSC | This paper | N/A |
| ASPA-CD NPC | This paper | N/A |
| CR-ASPA-CD iPSC | This paper | N/A |
| CR-ASPA-CD NPC | This paper | N/A |
| Experimental models: Organisms/strains | ||
| ASPAnur7/J mice | Jackson Laboratory | 008607 |
| B6(Cg)-Rag2tm1.1Cgn/J mice | Jackson Laboratory | 008449 |
| ASPAnur7/nur7/Rag2−/− | This paper | N/A |
| NSG mice | Jackson Laboratory | 005557 |
| Oligonucleotides | ||
| Primers for ASPA exon sequencing, RT-PCR and qRT-PCR, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pLV- EF1α -hASPA | This paper | N/A |
| AAVS1-CAG-hrGFP vector | Addgene | 52344 |
| pAAVS1-EF1α-ASPA-T2A-CD19t | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 9.0 | GraphPad Prism Software, Inc. | https://www.graphpad.com/ |
| Image-Pro Premier 9.3 | Media Cybernetics, Inc. | https://www.mediacy.com/ |
| ImageJ | Public resource | https://imagej.nih.gov/ij/ |
| NIS-Elements AR 5.20.02 | Nikon Instruments Inc. | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Adobe Photoshop 2022 | Adobe | https://www.adobe.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yanhong Shi (yshi@coh.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
ASPA lentiviral preparation and generation of ASPA-CD iPSC
To make ASPA-expressing lentivirus, we PCR-amplified the human ASPA coding sequence using the human ASPA cDNA (ATCC, MGC-34517) as the template and cloned the PCR product into the lentiviral vector pLV-EF1α downstream of the human EF1α promoter. To package the ASPA-expressing virus, 15 μg of pLV- EF1α -hASPA, 5 μg of VSV-G, 5 μg of REV and 15 μg of MDL were transfected into HEK 293T cells using calcium phosphate transfection method as we described previously (Cui et al., 2016, 2017, 2021; Feng et al., 2020). Forty-eight to seventy-two hr after transfection, virus-containing medium was harvested and filtered through 0.45 μm filter. For viral transduction, 625 μL ASPA-expressing virus-containing medium was added to 2.5 × 104 CD iPSCs that were dissociated into single cells. The CD iPSCs was derived from CD patient fibroblast CD59 (Coriell, GM00059) as described (Feng et al., 2020) in our laboratory under the approved IRB and SCRO protocols. Twenty-four hr after viral transduction, cells were fed with fresh iPSC culture media and subjected to daily medium change for 5 days. Then ASPA-CD iPSC clones were dissociated into single cells and re-seeded onto a 48-well plate to get single clones. The selected ASPA-CD iPSC clones were expanded and characterized.
Differentiation of human iPSCs into NPCs
NPCs were derived from human iPSCs following an established protocol (Liu et al., 2012; Feng et al., 2020). To start neural induction, human iPSCs were dissociated into single cells with Accutase (Sigma, A6964) and passaged onto Matrigel-coated 6-well plates at 1 × 105/well in E8 medium with 10 μM ROCK inhibitor. After 24 hr, cells were switched to Neural Induction Medium 1 (NIM-1) containing Advanced DMEM/F12 (Gibco, 11330-032), N2 (Gibco, 17502-048), B27 (Gibco, 17504-044), GlutaMAX (Gibco, 35050-061), NEAA (Gibco, 11140-050), 4 μM CHIR99021 (Cellagen Technology, C2447), 3 μM SB431542 (Cellagen Technology, C7243), 2 μM Dorsomorphin (Sigma, P5499), 0.1 μM Retinoic acid (Sigma, R2625), 10 ng/mL EGF (Peprotech) and 10 ng/mL FGF (Peprotech). Cells were maintained in NIM-I for 2 days, then changed to Neural Induction Medium II (NIM-II) that has the same components of NIM-1 medium but without Dorsomorphine for another 5 days. Cells were then dissociated with Accutase and maintained in Neural Progenitor Cell Maintenance Medium (NPMM) containing DMEM/F12, N2, B27, GlutaMAX, NEAA, 3 μM CHIR99021, 2 μM SB431542, 0.1 μM Retinoic acid, 10 ng/mL EGF and 10 ng/mL FGF.
Generation of CR-ASPA-CD iPSCs
CR-ASPA-CD iPSCs were generated by CRISPR/Cas9 gene editing of CD iPSCs. We used the published sgRNA sequence for CRISPR/Cas9-mediated targeting of the AAVS1 locus: 5'-GGGGCCACTAGGGACAGGATTGG-3′ (Blair et al., 2016; Oceguera-Yanez et al., 2016). The donor vector was prepared by introducing the EF1α-ASPA-T2A-CD19t cassette into the AAVS1-CAG-hrGFP vector (Addgene, #52344) between the two arms of AAVS1. The CRISPR/Cas9/sgRNA and the donor vector were electroporated into CD59 iPSCs. The gene-edited iPSCs were subjected to fluorescence-activated cell sorting (FACS) using a CD19 antibody. The CD19-positive cells were collected and plated as single cells. The resultant iPSC colonies were screened by genomic DNA PCR. Three primers, AAVS1-Fwd 5'-CTCTAACGCTGCCGTCTCTC-3′; AAVS1-Rev 5'-GCTTCTCCTCTTGGGAAGTG-3′ and ASPA-Rev 5'-AGCTCATTCCCATGGGTTCC-3′, were designed to genotype the clones.
Generation and maintenance of immunodeficient CD mice
All animals were maintained in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the Institutional Animal Care and Use Committee of City of Hope. The immunodeficient CD mice were generated by breeding ASPAnur7/+ (ASPAnur7/J, 008607, Jackson Laboratory) and Rag2−/− mice (B6(Cg)-Rag2tm1.1Cgn/J, 008449, Jackson Laboratory). The resultant ASPAnur7/+ mice were backcrossed with Rag2−/− mice for four generations. Homozygous ASPAnur7/nur7 and Rag2−/− mice were selected by genotyping and was called CD mice. The WT, Het, CD mice and the ASPA-CD NPC-transplanted CD mice were monitored for three months. About three-month-old mice were harvested for efficacy and preliminary safety study. Both male and female mice were used in all assays.
Method details
Analysis of potential off-target effect induced by CRISPR/Cas9 editing
We selected potential off-target sites as described (Cameron et al., 2017) and combined them with the off-target sites predicted by CHOPCHOP (Montague et al., 2014; Labun et al., 2016) (Table S1). Primers were designed according to the potential off-target sites. Genomic DNA of CD iPSCs and CR-ASPA-CD iPSCs were extracted and amplified by PCR. The sequences of the PCR products were compared between the parental CD iPSCs and the gene-edited CR-ASPA-CD iPSCs.
Teratoma formation
iPSCs were dissociated with Accutase at 1 to 2 dilution in PBS and re-suspended in ice cold mixture of E8 medium and Matrigel (1:1) at the density of 1 × 107 cells/mL. 100 μL of cell suspension (1 × 106 cells) were injected subcutaneously into the dorsal flank of immunodeficient Nod Scid Gamma (NSG) mice. Eight to twelve weeks after injection, teratoma was dissected and fixed in formalin. Fixed tissues were embedded in paraffin, sectioned and stained with hemotoxylin and eosin (H&E).
Immunostaining
Immunostaining was performed as we describe previously (Feng et al., 2020) using antibodies listed in STAR Methods.
ASPA enzymatic activity assay in cell
The ASPA enzymatic assay was performed as described (Madhavarao et al., 2002; Feng et al., 2020). After reactions, the absorbance was measured at 340 nm using a plate reader. The ASPA activity of cell is defined by the amount of aspartate (nmol)/mg protein lysate in 1 hr at 37°C.
Stereotaxic transplantation
NPCs were transplanted bilaterally into brains of P1-4 pups at 600,000 cells in 1.5 μL per site and six sites per pup brain by following a published method as we described (Feng et al., 2020) We used the coordinates of (+0.5, ±1.0, −2.5) for the subcortical region, (+3.0, ±1.6, −1.3) for the corpus callosum region, and (−1.6, ±0.8, −3.0) for the brain stem region. The coordinates were adjusted a little for pups weighted more than 2 g or with bigger head size as the following: (0.5, ±1.0, −2.5) for the subcortical region, (+3.5, ±1.7, −1.4) for the corpus callosum, and (−1.6, ±1.0, −3.1) for the brain stem. The coordinates are (A, L, V) relative to Lambda. A: anteroposterior from midline; L: lateral from midline; V: ventral from the surface of brain.
Immunohistochemistry
Immunohistochemistry was performed following the published protocol we described (Feng et al., 2020). Cell fate and proliferation status were assessed by double immunostaining for the human nuclear antigen along with PAX6, NeuN, SOX9, OLIG2, or Ki67. For quantification, slides in every sixth section from each mouse brain were selected, and 9 images of the engrafted cells in each targeting region were taken. The tiling image of the sagittal section was taken by Nikon Ti-2 microscope.
Vacuolation analysis
The hematoxylin and eosin (H&E) staining was performed in every sixth slides from each mouse brain as described (Feng et al., 2020). The subcortical, cerebellum, and brain stem regions were selected as regions of interest (ROI) for vacuolation measurement using Image-Pro Premier 9.3. The vacuolated region was distinguished from the intact region by the color of the image, with the white region corresponding to the vacuolated region. Because the value of the image histogram could affect the color of the image, we set the value of the image histogram to 155 for all H&E images. Under this condition, the region with color (>1 pixel2) was classified as the intact brain region, whereas the region without color (white region) was classified as the vacuolated region. The area of the intact brain region was measured by the software automatically by sampling the region with color and the percentage of the intact area was calculated by the software accordingly. The % vacuolated region was calculated using the following formular: % vacuolated are = 100% - % intact area.
Electron microscopy (EM)
The detailed heavy metal staining process were developed by Dr. Mark Ellisman’s group (West et al., 2010) and modified following the published protocol (Feng et al., 2020). For quantification, twelve to fifteen images with one axon per image were randomly taken at 67,000x magnification for each sample in the brain stem region. The layer number of myelin sheaths per axon was quantified. Twelve axons were sampled for WT mice, 12 for CD mice, and 15 for ASPA-CD transplanted mice. The intact and compact myelin sheath layer were counted manually. The compact myelin structure was judged by morphology. Those without open and loose myelin sheaths were considered the intact and compact myelin sheaths. The number of compact myelin structure was counted from 8 images randomly taken from the brain stem region. The denominator for the percentage of the compact myelin sheaths is the number of compact myelin sheaths from the brain stem region of WT mice.
Rotarod test
The motor performance of mice was tested by rotarod treadmill (Columbus Instruments) as described (Traka et al., 2008; Feng et al., 2020). Three-month-old Het, CD, and ASPA-CD NPC- transplanted CD mice were assessed. Briefly, the mice were put on the rotating rod at the accelerating speed from 2 rpm to 65 rpm in 2 min. Each mouse was monitored for the latency 4 times per test. At least 6 mice were tested for each group.
Grip strength test
The forelimb strength of the control mice and transplanted mice was measured using a grip strength meter (BIO-GS3, Bioseb) to evaluate the neuromuscular function. Three-month-old Het mice, CD mice, or CD mice transplanted with ASPA-CD NPCs were assessed by following the published protocol (Feng et al., 2020). Each mouse was tested with four sequential measurements. At least 6 mice were tested for each group.
Quantification and statistical analysis
Data are shown as means ± se as specified in the figure legends. The number of mice analyzed per treatment group is indicated as “n” in the corresponding figure legends. No exclusion criteria were applied. Animals were assigned randomly to treatment groups. One-Way ANOVA followed by Tukey’s or Dunnett’s multiple comparison tests were used for statistical analysis as reported in each figure legend. p <0.05 was considered statistically significant.
Acknowledgments
We thank Dr. Lucy Ghoda for suggestions. J.C. was a Herbert Horvitz Fellow. This work was supported by the Louise and Herbert Horvitz Charitable Foundation, California Institute for Regenerative Medicine TRAN1-08525, and NIH NINDS U01NS122101 to Y.S. Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572 and included work performed in the Animal Resources Center, Flow Cytometry Core, and Electron Microscopy Core supported by the National Cancer Institute of the National Institutes of Health (award number P30CA33572). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, Y.S.; Experimental Design, J.C., L.F., and Y.S.; Methodology, J.C., L.F., X.C., and P.Y.; Investigation, J.C., L.F., P.Y., X.C., Q.C., G.S., T.Z., E.T., W.L., and W.H.; Writing - Original Draft, J.C.; Writing – Review & Editing, Y.S. and J.C.; Supervision, Y.S., A.D.R., and R.M.; Funding Acquisition, Y.S.
Declaration of interests
A patent application related to this work has been filed. The authors declare no other competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104391.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- Ahmed S.S., Li H., Cao C., Sikoglu E.M., Denninger A.R., Su Q., Eaton S., Liso Navarro A.A., Xie J., Szucs S., et al. A single intravenous rAAV injection as late as P20 achieves efficacious and sustained CNS Gene therapy in Canavan mice. Mol. Ther. 2013;21:2136–2147. doi: 10.1038/mt.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.S., Schattgen S.A., Frakes A.E., Sikoglu E.M., Su Q., Li J., Hampton T.G., Denninger A.R., Kirschner D.A., Kaspar B., et al. rAAV gene therapy in a canavan's disease mouse model reveals immune impairments and an extended pathology beyond the central nervous system. Mol. Ther. 2016;24:1030–1041. doi: 10.1038/mt.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun P., Madhavarao C.N., Moffett J.R., Hamilton K., Grunberg N.E., Ariyannur P.S., Gahl W.A., Anikster Y., Mog S., Hallows W.C., et al. Metabolic acetate therapy improves phenotype in the tremor rat model of Canavan disease. J. Inherit. Metab. Dis. 2010;33:195–210. doi: 10.1007/s10545-010-9100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerman P., Guo F., Chechneva O., Burns T., Zhu X., Wang Y., Kim B., Singhal N.K., McDonough J.A., Pleasure D. Brain Nat8l Knockdown suppresses Spongiform leukodystrophy in an aspartoacylase-deficient canavan disease mouse model. Mol. Ther. 2018;26:793–800. doi: 10.1016/j.ymthe.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslow M.H. N-acetylaspartate in the vertebrate brain: metabolism and function. Neurochem. Res. 2003;28:941–953. doi: 10.1023/a:1023250721185. [DOI] [PubMed] [Google Scholar]
- Blair J.D., Bateup H.S., Hockemeyer D.F. Establishment of genome-edited human pluripotent stem cell lines: from targeting to isolation. J. Vis. Exp. 2016:e53583. doi: 10.3791/53583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A.M., Ream M.A. Recent advancements in the diagnosis and treatment of leukodystrophies. Semin. Pediatr. Neurol. 2021;37:100876. doi: 10.1016/j.spen.2021.100876. [DOI] [PubMed] [Google Scholar]
- Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M., Gradia S., Vidal B., Garner E., Slorach E.M., et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods. 2017;14:600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- Canavan M. Schilder's encephalitis perioxalis diffusa. Neurology. 1931;25:299–308. doi: 10.1001/archneurpsyc.1931.02230020085005. [DOI] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G., et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Yang S., Ye P., Tian E., Sun G., Zhou J., Sun G., Liu X., Chen C., Murai K., et al. Downregulation of TLX induces TET3 expression and inhibits glioblastoma stem cell self-renewal and tumorigenesis. Nat. Commun. 2016;7:10637. doi: 10.1038/ncomms10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Yin K., Zhang X., Ye P., Chen X., Chao J., Meng H., Wei J., Roeth D., Li L., et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat. Cancer. 2021;2:932–949. doi: 10.1038/s43018-021-00238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Chao J., Tian E., Li L., Ye P., Zhang M., Chen X., Cui Q., Sun G., Zhou T., et al. Stem cell therapy: cell-based therapy for canavan disease using human iPSC-derived NPCs and OPCs (adv. Sci. 23/2020) Adv. Sci. (Weinh) 2020;7:2070131. doi: 10.1002/advs.202070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J.S., Markov V., Leone P. Dietary triheptanoin rescues oligodendrocyte loss, dysmyelination and motor function in the nur7 mouse model of Canavan disease. J. Inherit. Metab. Dis. 2014;37:369–381. doi: 10.1007/s10545-013-9663-6. [DOI] [PubMed] [Google Scholar]
- Francis J.S., Markov V., Wojtas I.D., Gray S., McCown T., Samulski R.J., Figueroa M., Leone P. Preclinical biodistribution, tropism, and efficacy of oligotropic AAV/Olig001 in a mouse model of congenital white matter disease. Mol. Ther. Methods Clin. Dev. 2021;20:520–534. doi: 10.1016/j.omtm.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J.S., Wojtas I., Markov V., Gray S.J., McCown T.J., Samulski R.J., Bilaniuk L.T., Wang D.J., De Vivo D.C., Janson C.G., et al. N-acetylaspartate supports the energetic demands of developmental myelination via oligodendroglial aspartoacylase. Neurobiol. Dis. 2016;96:323–334. doi: 10.1016/j.nbd.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler D.J., Li D., Xu H., Su Q., Sanmiguel J., Tuncer S., Moore C., King J., Matalon R., Gao G. Redirecting N-acetylaspartate metabolism in the central nervous system normalizes myelination and rescues Canavan disease. JCI Insight. 2017;2:e90807. doi: 10.1172/jci.insight.90807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Bannerman P., Mills Ko E., Miers L., Xu J., Burns T., Li S., Freeman E., McDonough J.A., Pleasure D. Ablating N-acetylaspartate prevents leukodystrophy in a Canavan disease model. Ann. Neurol. 2015;77:884–888. doi: 10.1002/ana.24392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi H., Nihei K., Nakamoto N., Ezoe T., Naito H., Hara M., Yokota K., Inoue Y., Matsumoto I. A case of Canavan disease: the first biochemically proven case in a Japanese girl. Brain Dev. 1993;15:367–371. doi: 10.1016/0387-7604(93)90123-p. [DOI] [PubMed] [Google Scholar]
- Hoshino H., Kubota M. Canavan disease: clinical features and recent advances in research. Pediatr. Int. 2014;56:477–483. doi: 10.1111/ped.12422. [DOI] [PubMed] [Google Scholar]
- Hull V., Wang Y., Burns T., Zhang S., Sternbach S., McDonough J., Guo F., Pleasure D. Antisense oligonucleotide reverses leukodystrophy in canavan disease mice. Ann. Neurol. 2020;87:480–485. doi: 10.1002/ana.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson C., McPhee S., Bilaniuk L., Haselgrove J., Testaiuti M., Freese A., Wang D.J., Shera D., Hurh P., Rupin J., et al. Gene therapy of canavan disease: AAV-2 vector for neurosurgical delivery of aspartoacylase gene (ASPA) to the human brain. Hum. Gene Ther. 2002;13:1391–1412. doi: 10.1089/104303402760128612. [DOI] [PubMed] [Google Scholar]
- Kaul R., Gao G.P., Aloya M., Balamurugan K., Petrosky A., Michals K., Matalon R. Canavan disease: mutations among Jewish and non-Jewish patients. Am. J. Hum. Genet. 1994;55:34–41. [PMC free article] [PubMed] [Google Scholar]
- Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone P., Janson C.G., Bilianuk L., Wang Z., Sorgi F., Huang L., Matalon R., Kaul R., Zeng Z., Freese A., et al. Aspartoacylase gene transfer to the mammalian central nervous system with therapeutic implications for Canavan disease. Ann. Neurol. 2000;48:27–38. doi: 10.1002/1531-8249(200007)48:1<27::aid-ana6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Leone P., Shera D., McPhee S.W., Francis J.S., Kolodny E.H., Bilaniuk L.T., Wang D.J., Assadi M., Goldfarb O., Goldman H.W., et al. Long-term follow-up after gene therapy for canavan disease. Sci. Transl. Med. 2012;4:165ra163. doi: 10.1126/scitranslmed.3003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H., Qu J., Suzuki K., Nivet E., Li M., Montserrat N., Yi F., Xu X., Ruiz S., Zhang W., et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotun A., Gessler D.J., Gao G. Canavan disease as a model for gene therapy-mediated myelin repair. Front. Cell Neurosci. 2021;15:661928. doi: 10.3389/fncel.2021.661928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavarao C.N., Arun P., Anikster Y., Mog S.R., Staretz-Chacham O., Moffett J.R., Grunberg N.E., Gahl W.A., Namboodiri A.M.A. Glyceryl triacetate for Canavan disease: a low-dose trial in infants and evaluation of a higher dose for toxicity in the tremor rat model. J. Inherit. Metab. Dis. 2009;32:640. doi: 10.1007/s10545-009-1155-3. [DOI] [PubMed] [Google Scholar]
- Madhavarao C.N., Hammer J.A., Quarles R.H., Namboodiri M.A. A radiometric assay for aspartoacylase activity in cultured oligodendrocytes. Anal. Biochem. 2002;308:314–319. doi: 10.1016/s0003-2697(02)00225-7. [DOI] [PubMed] [Google Scholar]
- Matalon R., Michals K., Kaul R. Canavan disease: from spongy degeneration to molecular analysis. J. Pediatr. 1995;127:511–517. doi: 10.1016/s0022-3476(95)70105-2. [DOI] [PubMed] [Google Scholar]
- Matalon R., Michals K., Sebesta D., Deanching M., Gashkoff P., Casanova J., Optiz J.M., Reynolds J.F. Aspartoacylase deficiency and N-acetylaspartic aciduria in patients with Canavan disease. Am. J. Med. Genet. 1988;29:463–471. doi: 10.1002/ajmg.1320290234. [DOI] [PubMed] [Google Scholar]
- Matalon R., Rady P.L., Platt K.A., Skinner H.B., Quast M.J., Campbell G.A., Matalon K., Ceci J.D., Tyring S.K., Nehls M., et al. Knock-out mouse for Canavan disease: a model for gene transfer to the central nervous system. J. Gene Med. 2000;2:165–175. doi: 10.1002/(sici)1521-2254(200005/06)2:3<165::aid-jgm107>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Matalon R., Surendran S., Rady P.L., Quast M.J., Campbell G.A., Matalon K.M., Tyring S.K., Wei J., Peden C.S., Ezell E.L., et al. Adeno-associated virus-mediated aspartoacylase gene transfer to the brain of knockout mouse for canavan disease. Mol. Ther. 2003;7:580–587. doi: 10.1016/s1525-0016(03)00066-2. [DOI] [PubMed] [Google Scholar]
- Matalon R.M., Michals-Matalon K. Spongy degeneration of the brain, Canavan disease: biochemical and molecular findings. Front. Biosci. 2000;5:D307–D311. doi: 10.2741/matalon. [DOI] [PubMed] [Google Scholar]
- McPhee S.W., Francis J., Janson C.G., Serikawa T., Hyland K., Ong E.O., Raghavan S.S., Freese A., Leone P. Effects of AAV-2-mediated aspartoacylase gene transfer in the tremor rat model of Canavan disease. Brain Res. 2005;135:112–121. doi: 10.1016/j.molbrainres.2004.12.007. [DOI] [PubMed] [Google Scholar]
- McPhee S.W., Janson C.G., Li C., Samulski R.J., Camp A.S., Francis J., Shera D., Lioutermann L., Feely M., Freese A., et al. Immune responses to AAV in a phase I study for Canavan disease. J. Gene Med. 2006;8:577–588. doi: 10.1002/jgm.885. [DOI] [PubMed] [Google Scholar]
- Mendell J.R., Al-Zaidy S.A., Rodino-Klapac L.R., Goodspeed K., Gray S.J., Kay C.N., Boye S.L., Boye S.E., George L.A., Salabarria S., et al. Current clinical applications of in vivo gene therapy with AAVs. Mol. Ther. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann N., Tkachev D., Jelinek R., Roth P.T., Mobius W., Ruhwedel T., Ruhle S., Weber-Fahr W., Sartorius A., Klugmann M. Aspartoacylase-lacZ knockin mice: an engineered model of Canavan disease. PLoS One. 2011;6:e20336. doi: 10.1371/journal.pone.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesuta O., Thomas A.G., Alt J., Hin N., Neuzilova A., Long S., Tsukamoto T., Rojas C., Wei H., Slusher B.S. High throughput screening cascade to identify human aspartate N-acetyltransferase (ANAT) inhibitors for canavan disease. ACS Chem. Neurosci. 2021;12:3445–3455. doi: 10.1021/acschemneuro.1c00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oceguera-Yanez F., Kim S.I., Matsumoto T., Tan G.W., Xiang L., Hatani T., Kondo T., Ikeya M., Yoshida Y., Inoue H., et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43–55. doi: 10.1016/j.ymeth.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pleasure D., Guo F., Chechneva O., Bannerman P., McDonough J., Burns T., Wang Y., Hull V. Pathophysiology and treatment of canavan disease. Neurochem. Res. 2020;45:561–565. doi: 10.1007/s11064-018-2693-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel R., Anikster Y., Zevin S., Steinberg A., Gahl W.A., Fisher D., Staretz-Chacham O., Zimran A., Altarescu G. A safety trial of high dose glyceryl triacetate for Canavan disease. Mol. Genet. Metab. 2011;103:203–206. doi: 10.1016/j.ymgme.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J., Bannerman P., Guo F., Burns T., Miers L., Croteau C., Singhal N.K., McDonough J.A., Pleasure D. Suppressing N-Acetyl-l-Aspartate synthesis prevents loss of neurons in a murine model of canavan leukodystrophy. J. Neurosci. 2017;37:413–421. doi: 10.1523/jneurosci.2013-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Traka M., Wollmann R.L., Cerda S.R., Dugas J., Barres B.A., Popko B. Nur7 is a nonsense mutation in the mouse aspartoacylase gene that causes spongy degeneration of the CNS. J. Neurosci. 2008;28:11537–11549. doi: 10.1523/jneurosci.1490-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bogaert L., Bertrand I. Sur une idiotie familiale avec degerescence sponglieuse de neuraxe (note preliminaire) Acta Neurol. 1949;49:572–587. [Google Scholar]
- von Jonquieres G., Spencer Z.H.T., Rowlands B.D., Klugmann C.B., Bongers A., Harasta A.E., Parley K.E., Cederholm J., Teahan O., Pickford R., et al. Uncoupling N-acetylaspartate from brain pathology: implications for Canavan disease gene therapy. Acta Neuropathol. 2018;135:95–113. doi: 10.1007/s00401-017-1784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.B., Fu Z., Deerinck T.J., Mackey M.R., Obayashi J.T., Ellisman M.H. Structure-function studies of blood and air capillaries in chicken lung using 3D electron microscopy. Respir. Physiol. Neurobiol. 2010;170:202–209. doi: 10.1016/j.resp.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.