Abstract
Sirtuin-1 (SIRT1) is a critical nuclear deacetylase that participates in a wide range of biological processes. We hereby employed quantitative acetyl-proteomics to globally reveal the landscape of SIRT1-dependent acetylation in colorectal cancer (CRC) cells stimulated by specific SIRT1 inhibitor Inauhzin (INZ). We strikingly observed that SIRT1 inhibition enhances protein acetylation levels, with the multisite-acetylated proteins (acetyl sites >4/protein) mainly enriched in mitochondria. INZ treatment increases mitochondrial fission and depolarization in CRC cells. The acetylation of mitochondrial proteins promoted by SIRT1 inhibition prevents the recruitment of ubiquitin and LC3 for mitophagic degradation. We then found that, SIRT1 inhibition increases the acetylation of mitochondrial calcium uniporter (MCU) at residue K332, resulting in mitochondrial Ca2+ overload and depolarization, and ultimately CRC apoptosis. Arginine substitution of the K332 (K332R) dramatically decreases the mitochondrial Ca2+ influx, mitochondrial membrane potential loss and ROS burst induced by INZ. This finding uncovers a non-canonical role of SIRT1 in regulating mitochondrial function and implicates a possible way for anticancer intervention through SIRT1 inhibition.
Keywords: SIRT1, Inauhzin, MCU, Mitochondria, Acetylation, Calcium
Abbreviations: CCCP, carbonyl cyanide m-chlorophenylhydrazone; CO-IP, co-immunoprecipitation; CRC, colorectal cancer; DAPI, 4′,6-diamidino-2-phenylindole; Drp1, dynamin-related protein 1; FIS1, mitochondrial fission 1 protein; INZ, Inauhzin; K–Ac, lysine acetylation; KO, knockout; LC3, microtubule-associated protein 1 light chain 3 beta; LC-MS, liquid chromatography-mass spectrometry; MAPs, multisite-acetylated proteins; MCU, mitochondrial calcium uniporter; MFN1, mitofusin 1; MFN2, mitofusin 2; MIM, mitochondria inner membrane; MMP, mitochondrial membrane potential; MOM, mitochondria outer membrane; PARP, poly (ADP-ribose) polymerase 1; PLGEM, the power law global error model; ROS, reactive oxygen species; SIRT1, Sirtuin-1; TEM, Transmission electron microscope; Tom20, translocase of outer mitochondrial membrane 20 homolog (yeast); Ub, ubiquitin
Graphical abstract
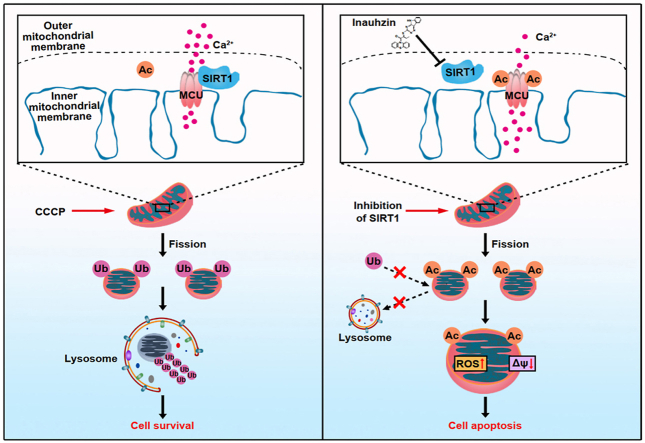
The inhibition of SIRT1 induces acetylation of mitochondrial Ca2+ transporter, MCU, promoting mitochondrial Ca2+ overload, ROS burst and loss of mitochondrial membrane potential. Global acetylation of mitochondrial proteins due to SIRT1 inhibition obstructs their ubiquitin and LC3-labeling, preventing mitophagy-associated clearance, and ultimately causing cell apoptosis.
Highlights
-
•
SIRT1 inhibition induces mitochondrial fission, depolarization and ROS burst.
-
•
SIRT1 inhibition induces the mitochondrial acetylation.
-
•
Mitochondrial acetylation prevents the recruitment of ubiquitin and LC3 for mitophagic degradation.
-
•
SIRT1 inhibition increases the acetylation of MCU at residue K332, resulting in mitochondrial Ca2+ overload.
1. Introduction
Lysine acetylation is an important post-translational modification that regulates a variety of molecular processes in mammalian cells. Nuclear acetylation mainly occurs on histone that is linked to gene transcriptional regulation. However, accumulated studies supported that mitochondria are the acetylation-enriched place in extranucleus [[1], [2], [3]], since acetyl coenzyme A (Acyl-CoA) is highly abundant in mitochondria for participation in tricarboxylic acid (TCA) circle. Proteins in mitochondria appear to be acetylated easily through a non-enzymatic mechanism. Three members of sirtuin family, SIRT3, SIRT4, and SIRT5 are specific deacetylases in mitochondria [2,4,5]. In contrast, SIRT1 is a conservative NAD+-dependent deacetylase that mainly locates in nucleus, playing important functions in energy metabolism, inflammation, oxidative stress and life span prolonging [[6], [7], [8], [9]] by deacetylating many nuclear transcriptional factors, such as p53, E2F1, p65 and FOXO3 [10,11]. Interestingly, many extranuclear targets of SIRT1 were recently identified [9,12], suggesting a critical need to better understand the landscape of SIRT1-mediated acetylome. We herein found that SIRT1 inhibition significantly increases acetylation level of mitochondria, indicating a new horizon of SIRT1 functioning in deacetylation regulation in mitochondria.
Mitochondria are highly dynamic organelles in mammalian cells that play important roles in energy production, biogenesis and calcium maintenance [13]. They are switching between fission and fusion to guarantee requisite number of functional mitochondria to meet the demands of the cell. Upon cellular stress, overload of mitochondrial calcium induces loss of mitochondrial membrane potential (MMP) and reactive oxygen species (ROS) burst. It has been reported that the elevated mitochondrial Ca2+ is able to increase the activity of calcium/calmodulin-dependent protein kinase II (CaMKII), which phosphorylates the dynamin-related protein 1 (Drp1) at Ser616 for mitochondrial fission [14]. Damaged mitochondria can be fragmented and ubiquitinated, recruiting autophagy receptor proteins, like LC3, to deliver them to the autophagosome for autophagic degradation [[15], [16], [17]]. However, it remains unclear whether the elevated acetylation level on mitochondria influences their ubiquitination and autophagic degradation when mitochondria are in injury.
In this study, we observed that SIRT1 inhibition promotes mitochondrial acetylation and damage. Globally acetylated mitochondrial proteins prevent their LC3-labeling and mitophagy-associated clearance, aggravating cell apoptosis. Particularly, SIRT1 inhibition results in the specific acetylation of mitochondrial calcium uniporter (MCU) at residue K332, which increases the mitochondrial Ca2+ influx, leading to the mitochondrial dysfunction of tumor cells.
2. Materials and methods
2.1. Cell culture and chemical reagents
Human colorectal cancer cell lines HCT116 and DLD1 were obtained from ATCC (MD, USA) and cultured in RPMI 1640 medium (Gibco, MD, USA) supplemented with 10% fetal bovine serum (Gibco) at 37 °C in 5% CO2 atmosphere. Inauhzin (INZ), from Targetmol (MA, USA), was dissolved in dimethyl sulfoxide (DMSO, Sigma, MO, USA). Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was purchased from Sigma. The Mitochondria Isolation Kit was obtained from Beyotime (Jiangsu, China).
2.2. Plasmids and transfections
Full-length human MCU and SIRT1 were generated by PCR amplification using HCT116 cDNA cloned into the pLVX-puro vector (TransheepBio, Shanghai, China) with HA-tag or flag-tag, respectively. Human LC3B was generated by PCR amplification using HCT116 cDNA and constructed on the pcDNA3.1 vector (Invitrogen, CA, USA) after fusion with GFP. For the generation of plasmids pLVX-HA-MCU-K332R, the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) was employed using pLVX-HA-MCU as a template. The primer sequences for MCU-K332R were: FOR 5′- GGACCTTAgGAGACTGAGAGACCCATTACAAGTACA-3'; REV 5′-TCAGTCTCcTAAGGTCCATTTCTGCCTGAGCA-3'. The sgRNA sequences of SIRT1 were cloned into lentiCRISPR v2 vector (addgene #52961) to knockout relevant genes. SIRT1 sgRNA#1:5′-GTTGACTGTGAAGCTGTACG-3'; SIRT1 sgRNA#2: 5′-AACAGGTTGCGGGAATCCAA-3'. For cell transfection and infection, the established pLVX vector or lentiCRISPR v2 vector was co-transfected with two helper plasmids pSPAX2 and pMD2.G (addgene #12259 and addgene #12260) into 293T cells (ATCC) for 48 h by using Lipofectamine™3000 reagent (Invitrogen), and the supernatant was collected to infect CRC cells to establish stable cells via puromycin selection [18].
2.3. WST-1 assay
The cell viability was determined by WST-1 (Beyotime). CRC cells were inoculated in 96-well plates (3 × 103 cells/well in 100 μL) and treated with 0, 0.8, 3.2 and 12.8 μM of INZ for 48 and 72 h, respectively. The WST-1 reagent was added to each well and incubated at 37 °C for 1.5 h, then the absorbance was measured at 450 nm using an automated microplate spectrophotometer (BioTek Instruments, Vermont, USA).
2.4. Colony formation assay
Both CRC cells were seeded in 6-well plates (1 × 103 cells/well) and cultured with 0, 0.8, 3.2 and 12.8 μM INZ for 14 days. Cells were immobilized with methanol for 15 min, and stained with 0.5% crystal violet for 10 min. The number of colonies was calculated by ImageJ software (version 1.44I).
2.5. Detection of apoptotic cell death
HCT116 and DLD1 cells were treated with INZ (up to 12.8 μM) for 48 h and detected with Annexin V-FITC/PI staining, followed by detection of apoptosis with Annexin V-FITC/PI Apoptosis Detection Kit (KeyGen, Jiangsu, China). Cells were collected and stained with Annexin V-FITC and PI according to the manufacturer's protocol. The apoptotic cells were assessed and analyzed by flow cytometer (BD Biosciences, CA, USA).
2.6. Measurement of the ROS level and MMP
Intracellular ROS levels of CRC cells were measured by DCFH-DA (Beyotime). MMP was measured by JC-1 assay kit (Beyotime). After treated with INZ (up to 12.8 μM) for 48 h, HCT116 and DLD1 cells were stained with DCFH-DA or JC-1 according to their manufacturer's manual and analyzed by flow cytometer (BD Biosciences), respectively.
2.7. Immunoblotting and immunoprecipitation
For immunoblotting, the cell lysates were extracted by cell lysis buffer (Cell Signaling Technology, MA, USA), dissolved for 30 min on ice and centrifuged at 12,000×g for 30 min at 4 °C. The concentration of protein samples was detected by BCA kit (Thermo Fisher Scientific, MA, USA). Protein samples were subjected to sodium dodecyl sulfate (SDS)-PAGE gels, followed by polyacrylamide gel electrophoresis. Electrotransfer was then performed to transfer the proteins into PVDF membrane (Bio-Rad, CA, USA). After blocking with 5% skim milk for 1.5 h, the membrane was enriched with primary antibody for 2 h at room temperature (RT) and washed 3 times for 10 min each with 1 × Tween Tris Buffered Saline (TBST). The washed membranes were incubated with the corresponding secondary antibody for 1–2 h at RT or overnight at 4 °C. After 3 times of TBST washing, the membranes were imaged by using Clarity Western ECL substrate (Bio-Rad) and the signal was obtained by a Tanon 5200-Multi (Tanon Science & Technology Co.,Ltd, Shanghai, China). If the same samples were used to blot for different proteins with similar molecular weight, the “loading controls” were run on separate gels. Antibodies including PARP, cleaved PARP, caspase-3, cleaved caspase-3, OPA1, Drp1, MFN1 and MFN2 were purchased from Cell Signaling Technology, and Tom20, SIRT1, Tubulin, GAPDH, Actin, HA, Flag, ubiquitin and FIS1 were obtained from Proteintech (Hubei, China). Antibody acetyl-p53-K382 and MCU were purchased from ABclonal (Hubei, China).
For immunoprecipitation, HCT116 were transiently transfected with Flag-SIRT1 and HA-MCU for 48 h. Cells were dissolved in cell lysis buffer for Western and IP (Beyotime) for 30 min on ice, and centrifuged at 12,000×g for 30 min at 4 °C. Anti-HA-tag antibody was added to the supernatant and incubated under low rotation overnight at 4 °C, then the protein A/G Sepharose beads (Santa Cruz, CA, USA) were added to the lysate and incubated for another 4 h at 4 °C. Finally, the mixture was eluted with 2 × SDS/PAGE loading buffer after 5 times of washing with IP-lysis buffer. Finally, the boiled samples were separated by 10% or 12% SDS-PAGE.
2.8. Confocal assay
HCT116 cells transiently transfected with pcDNA3.1-GFP-LC3 vector for 48 h was treated with DMSO or INZ (12.8 μM) for 24 h, the cells were respectively incubated with or without 10 μM CCCP for 2 h. The mitochondria were stained with 100 nM Mito-Tracker Red (Thermo Fisher Scientific) for 20 min and then subjected to confocal laser scanning microscope (LSM 880 with AiryScan, Carl Zeiss) for analysis. For colocalization of mitochondria and lysosome, the mitochondria were stained with Mito-Tracker Green (100 nM, Beyotime) for 15 min; lysosome was stained with Lyso-Tracker Red (50 nM, Beyotime) for 10 min.
For determination of mitochondrial Ca2+, cells were loaded with Rhod-2AM (1 μM) at 37 °C for 30 min. Subsequently, the treated cells were loaded with Mito-Tracker Green (Beyotime) at 37 °C for 15 min. After fixing with 4% paraformaldehyde, the cells were stained with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, 1 μg/mL, Beyotime) for 10 min, and observed with confocal microscope (LSM 880 with AiryScan, Carl Zeiss).
2.9. Transmission electron microscope (TEM)
After treatment with INZ (up to 12.8 μM), DLD1 cells were fixed with TEM fixer in 4 °C for 4 h. Then pre-embed the treated samples in 1% agarose and immobilized in 1% Osmium Tetroxide. After dehydration with ethanol at RT, the cells were embed in Poly/Bed 812 resin and polymerized at 65 °C. Ultrathin sections were stained with 2% uranium acetate saturated alcohol solution and stain with 2.6% lead citrate. Morphological analysis was performed using transmission electron microscope (Tecnai G2 Spirit, Fei Czech Co., Ltd.).
2.10. Quantitative acetyl-proteomics and bioinformatics analysis
For mass spectrometry (MS) analysis, the cells from different treatment were dissolved in cell lysis buffer. After trypsinization using the filter-assisted sample preparation (FASP) method, peptides were vacuum lyophilized. Enrichment of Kac peptides from different fractions was carried out using anti-acetyllysine antibody conjugated agarose beads (PTM Biolabs Inc., Hangzhou, China) according to the manufacturer's manual. The iRT-Standard (Biognosys, MA, USA) was added into each sample and data-dependent acquisition (DDA)/data-independent acquisition (DIA) MS were collected by Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). The DDA raw data were processed and searched with Proteome Discoverer, version 2.1 (Thermo Fisher Scientific). All the data were searched against UniProt Human protein database (http://www.uniprot.org). Carbamidomethylation (C) was specified as fixed modification. Oxidation (M), protein N-terminal acetylation, and lysine acetylation were specified as variable modifications. The FDR thresholds of peptide, protein and modification site identification were specified at 1%. We quantified the protein and peptide expression levels by Spectronaut software (Omicsolution Co., Ltd, Shanghai, China) for DIA raw data. FDR was set to 1% for protein identification. Differentially expressed proteins were filtrated by the power law global error model and analyzed by Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA, USA). The Gene Ontology (GO) analysis was determined using DAVID Bioinformatics Resources 6.8 [19]. Subcellular distribution was annotated by COMPARTMENTS database [20].
2.11. Tumor xenograft experiments and histologic staining
Tumor xenograft experiment was performed according to our previous study [21]. BALB/c nude mice aged 5–6 weeks (Guangdong GemPhamatech Co., Ltd, Guangdong, China) were maintained under standard conditions at the Animal Experiment Center of the College of Medicine (SPF grade) of Jinan University according to the institutional guidelines for animal care. All the animal experiments were approved by the Animal Ethics Committee of Jinan University. The nude mice were subcutaneously inoculated with HCT116 cells (1 × 106 cells/injection). When solid tumor reached about 5 mm in diameter, the mice were divided into three groups (6 mice/group) randomly. The treatment groups received oral gavage of INZ (15 mg/kg) or INZ (30 mg/kg) dissolved in corn oil every 2 days for 14 days, whereas the control group received the vehicle only. The tumor size and body weight were monitored every two days, and the tumor volume was calculated by the following equation: V = (length × width2)/2. At the end of the study, tumors were harvested for TUNEL assay, and several organs (liver, kidney, lung, and heart) were harvested for histologic analyses by Hematoxylin and Eosin (H&E) staining as previously described [22]. Briefly, the paraffin-embedded tissues fixed by paraformaldehyde were stained. After sectioning, the tumors were stained with TUNEL BrightGreen Apoptosis Detection Kit (Vazyme).
2.12. Statistical analysis
All in vitro experiments were performed in 3 independent replicates, and all values are expressed as mean ± standard deviation. GraphPad Prism software (San Diego, CA, USA) was used, and tests were performed using the Student's t-test. All statistical tests were two-sided, and p < 0.05 was considered statistically significant. Significance levels * P < 0.05; **P < 0.01; ***P < 0.001.
3. Results
3.1. Acetyl-proteomics reveals that SIRT1 inhibition promotes mitochondrial acetylation
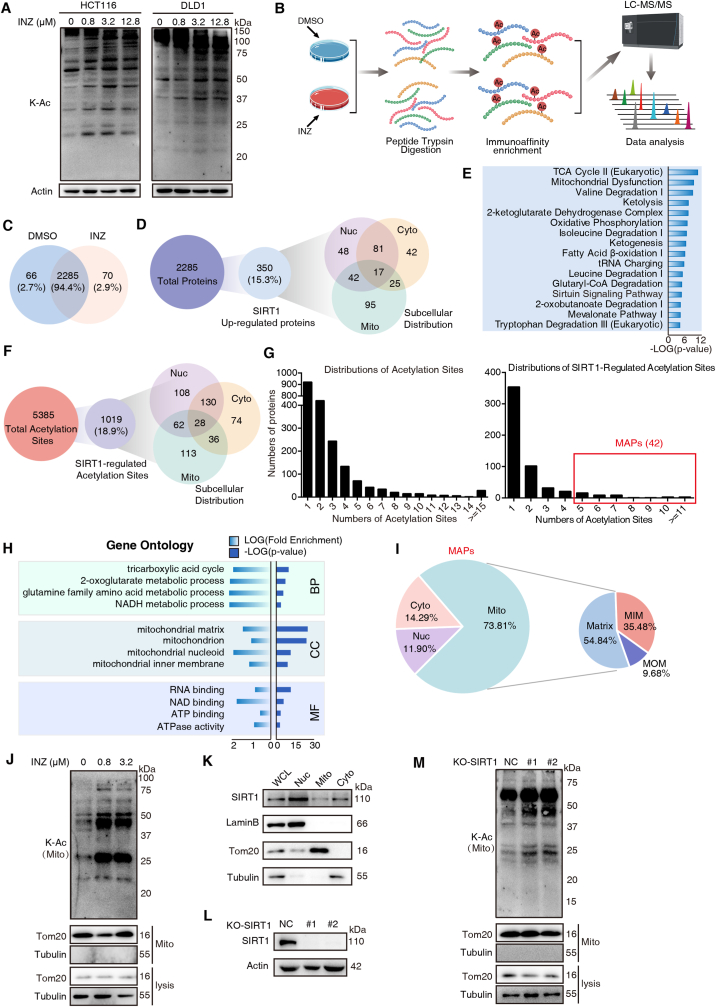
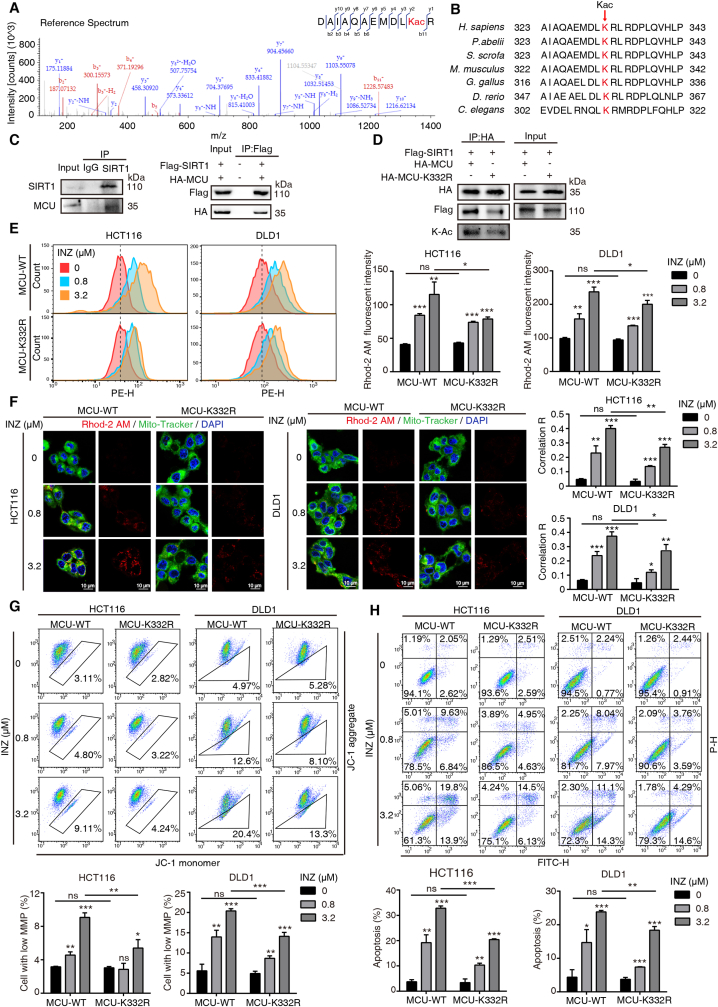
SIRT1 is a nuclear deacetylase [23]; however, increasing extranuclear targets of SIRT1 were recently identified, suggesting that SIRT1 may have extranuclear function in acetylation. To systematically depict SIRT1-associated deacetylation, we employed a specific SIRT1 inhibitor Inauhzin (INZ) [24] to study the alteration of protein acetylation upon INZ treatment. As expected, treatment of INZ in both HCT116 and DLD1 cells globally increased the acetylation level of proteins in a dose-dependent fashion (0–12.8 μM) (Fig. 1A). And the effect was confirmed by the increased acetylation of p53-K382 used as the positive control (Fig. S1A). We then selected 0.8 μM of INZ treatment for acetyl-proteomic analysis based on an affinity-directed mass spectrometry (MS) method (Fig. 1B). Lysates from HCT116 cells with or without INZ treatment (48 h) were subjected to tryptic digestion, Kac peptides enrichment and DIA-based LC-MS/MS analysis (Fig. 1B). With three biological experimental repetitions, a total of 5888 unique Kac sites on 2555 proteins were identified, and a total of 2285 proteins were overlaid in both control and INZ-treated groups (Fig. 1C).
Fig. 1.
SIRT1 inhibition promotes mitochondrial acetylation. (A) Western blot analysis of lysine-acetylation (K–Ac) expressions in whole-cell lysates of HCT116 and DLD1 cells treated with increasing concentrations of INZ (up to 12.8 μM) for 48 h. (B) Schematic representation of experimental workflow for the identification of acetyl-lysine-modified peptides regulated by SIRT1 inhibition. Lysates from HCT116 cells incubated with or without INZ (0.8 μM) were subjected to trypsin digestion and acetylation antibody-based enrichment, and followed by MS analysis. (C) Venn diagram showing the overlap of protein numbers in DMSO-treated and INZ-treated groups. (D) The number of SIRT1 up-regulated proteins and their subcellular distribution as determined by the COMPARTMENTS database. (E) Ingenuity Pathway Analysis (IPA) of the acetylated proteins regulated by SIRT1 (fold change ≥1.5, P < 0.05). The bar graphs showing the top 15 canonical pathways enriched by IPA. (F) Description of the SIRT1-regulated acetylation sites and their subcellular distribution. (G) Histogram showing the distributions of the number of acetylation sites (left) and SIRT1-regulated sites (right) per protein. Multisite-acetylated proteins (MAPs) with more than 4 acetylation sites are indicated in red pane. (H) DAVID Gene Ontology (GO) analysis of hyperacetylated proteins regulated by SIRT1 inhibition. Significantly enriched pathways (FDR <0.01) were categorized by GO different modules for visualization (BP: biological process; CC: cellular component; MF: molecular function). (I) Pie chart showing the proportion of MAPs in mitochondrial, nuclear and cytoplasmic fractions according to cellular component analysis, as well as the distribution of MAPs in mitochondria inner membrane (MIM), mitochondria outer membrane (MOM) and matrix. (J) The acetylation level of mitochondria extracted from HCT116 cells treated with 0.8 and 3.2 μM of INZ. Tom20 and Tubulin were used as markers for mitochondria and cytoplasma, respectively. (K) The expression level of SIRT1 in different cellular components extracted from HCT116 cells, respectively. Nuc, nucleus; mito, mitochondria; cyto, cytosol. (L) Western blot analysis confirming the effect of SIRT1 knockout in HCT116 cells. (M) Western blot analysis of the acetylation level of mitochondria in HCT116 cells after knockout of SIRT1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next performed the power law global error model (PLGEM) to analyze differentially expressed proteins (DEPs), determining the protein abundance with a slope of 0.93 and an adjusted r2 of 0.995 (Pearson r = 0.826; Fig. S1B). The model showed that the residual standard deviations of the data fitted normal distribution and the residuals distributed evenly, independent of the rank of mean abundances (Fig. S1B). As compared to the control group, 350 proteins were up-regulated in the INZ group (P-value ≤ 0.05, fold change ≥1.5) and thus considered as SIRT1 up-regulated proteins (Supplementary Table 1). Cellular component analysis of these proteins showed that mitochondrial proteins occupied a higher proportion than nuclear and cytoplasmic fractions (Fig. 1D). Ingenuity Pathway Analysis (IPA) on the 350 SIRT1-related acetylated proteins showed that TCA cycle, mitochondrial dysfunction and sirtuin signaling pathways were all enriched (Fig. 1E).
We also analyzed the lysine-acetylated peptides (5385 acetylated sites) and found that INZ-regulated acetylation sites (1019 sites, Supplementary Table 2) determined by PLGEM (Fig. S1C) did not show significant subcellular distribution (Fig. 1F). Though the majority of proteins were bearing 1–4 acetylated sites, a number of INZ-regulated proteins (n = 42) had more than 4 acetylated sites (Fig. 1G), and we here name these proteins as “multisite-acetylated proteins (MAPs)”. Upon GO analysis, these SIRT1-regulated MAPs were significantly enriched in mitochondria-associated biological progress (BP) and cellular component (CC) (Fig. 1H). Note that among the 42 MAPs, mitochondrial proteins account for 73.81%, while only 11.9% are nuclear proteins (Fig. 1I). To confirm the acetylation of mitochondria regulated by SIRT1 inhibition, we isolated mitochondria from HCT116 cells with 48 h of INZ treatment (0.8 and 3.2 μM). The immunoblotting assay showed that lysine-acetylation in mitochondria fraction was up-regulated by INZ in a dose-dependent manner (Fig. 1J). We next determined the subcellular distribution of SIRT1, and found that a portion of SIRT1 was detected in mitochondria, though it was predominantly located in nucleus (Fig. 1K). To further ask whether the acetylation was directly regulated by SIRT1, we generated the SIRT1 knockout (KO) cell line of HCT116 (Fig. 1L) by CRISPR/Cas9 and found that the mitochondrial acetylation was globally increased after SIRT1 deletion (Fig. 1M). These results implicate a non-canonical role of SIRT1 in mitochondrial function, by which SIRT1 inhibition induces the promotion of mitochondrial acetylation.
3.2. Inhibition of SIRT1 leads to mitochondrial dysfunction but prevents mitophagy
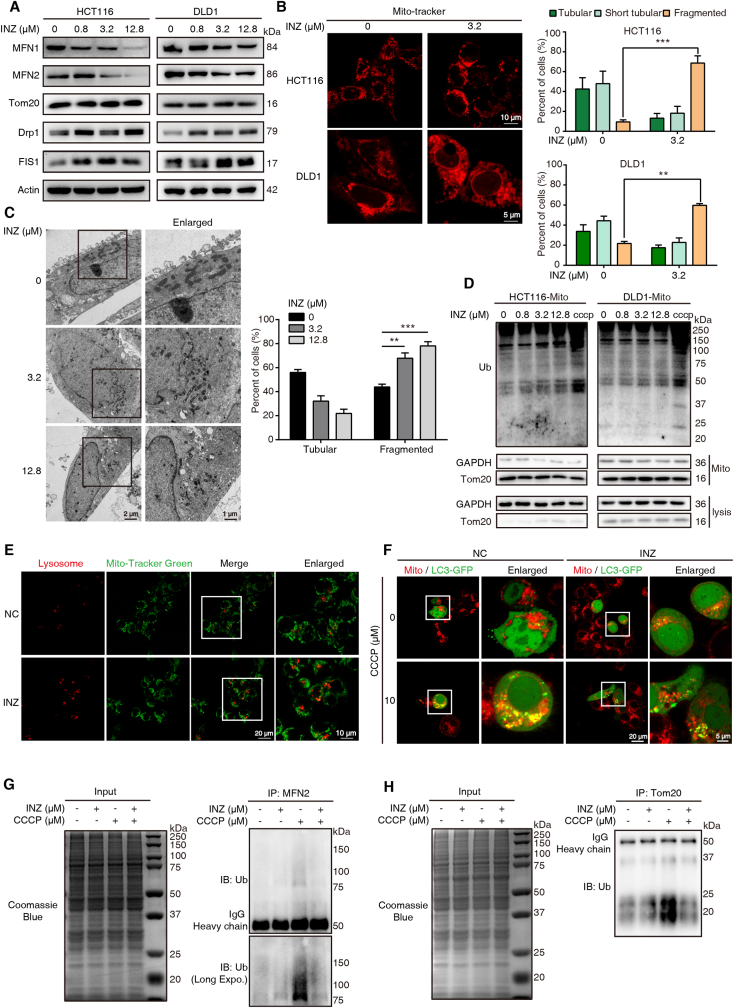
The balance of mitochondrial dynamics is a principal procedure to sustain cellular homeostasis [25]. We determined the effect of SIRT1 inhibition on mitochondrial dynamics. Immunoblotting assay of the markers related to mitochondrial dynamics demonstrated that, the expression level of fusion indicators (MFN1 and MFN2) was remarkably increased, whereas the expression of fission indicators (Drp1 and FIS1) was significantly reduced in HCT116 and DLD1 cells upon INZ treatment (Fig. 2A). Meanwhile, we determined the mitochondria morphological characteristics by laser scanning confocal microscope and electron microscope, and found that the mitochondria were broken from tubular to point-like morphology after INZ stimulation (Fig. 2B). The regulation of INZ on mitochondrial morphology was also consistently detected by transmission electron microscope (TEM), showing that the overall shapes of the mitochondria were fragmented and grossly distorted (Fig. 2C).
Fig. 2.
Inhibition of SIRT1 leads to mitochondrial dysfunction and mitophagy suppression. (A) Western blot analysis of the expression of mitochondrial dynamics-related proteins, MFN1, MFN2, Tom20, Drp1 and FIS1, in HCT116 and DLD1 cells treated with different concentration of INZ (0, 0.8, 3.2 and 12.8 μM) for 48 h. (B) Mitochondrial morphology of HCT116 and DLD1 with or without INZ treatment (3.2 μM, 48 h) was determined by confocal microscope (n = 3). Cells were labeled with Mito-tracker (red) for visualization, mitochondria in tubular, short tubular and fragment were statistically analyzed. Scale bars, 10 μm for HCT116 and 5 μm for DLD1. (C) Representative TEM images of DLD1 treated with INZ (up to 12.8 μM) for 48 h, and the mitochondrial morphology with tubular and fragment were statistically analyzed (n = 3). Scale bars, 1 μm and 2 μm. (D) Western blot determining the ubiquitin level of mitochondria from HCT116 and DLD1cells treated with INZ (for 48 h) or CCCP (for 2 h, positive control). (E) The localization of mitochondria and lysosome in HCT116 cells treated with INZ (12.8 μM, 48 h) was visualized by confocal microscope (n = 3). Green, Mito-tracker; Red, Lyso-Tracker. Scale bars, 10 μm and 20 μm. (F) Confocal microscopy determining the recruitment of LC3 to mitochondria in HCT116 with indicated treatment (n = 3). Green, LC3-GFP; Red, Mito-Tracker. Scale bars, 5 μm and 20 μm. (G, H) Effects of CCCP-induced ubiquitination of MFN2 and Tom20 with or without INZ treatment. HCT116 cells were pretreated with 12.8 μM INZ for 24 h and then treated with CCCP (10 μM) for 6 h, then the ubiquitination of MFN2 and Tom20 was analyzed. Bars, SD; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fission of mitochondria subsequently recruits ubiquitin and LC3 for autophagic degradation and clearance [16,26]. However, our isolated mitochondrial fraction did not show significant ubiquitination after INZ treatment in HCT116 and DLD1, as compared to CCCP treatment (Fig. 2D). Meanwhile, our confocal results showed that fragmented mitochondria after INZ treatment did not co-localize with lysosomes (Fig. 2E), indicating that SIRT1 inhibition-induced enhancement of mitochondrial acetylation prevents mitophagic removement of damaged mitochondria. With the CCCP-mediated mitophagy cell model, we also found that LC3 was not significantly recruited to the mitochondria of INZ-treated HCT116 cell (Fig. 2F), showing the rapid aggregation of LC3 punctate dots on mitochondria in response to CCCP (10 μM) stimuli. This result suggests that SIRT1 inhibition-mediated fission prevents CCCP-induced mitophagy. Mitochondrial outer membrane (MOM) proteins, such as MFN2 and Tom20, are well recognized as the substrates of ubiquitination in response to CCCP [[27], [28], [29], [30]]. Here, we measured the effect of INZ on the ubiquitination level of MFN2 and Tom20 treated with CCCP and observed that INZ pretreatment abolished CCCP-induced ubiquitination in MFN2 and Tom20 (Fig. 2G–H). These data demonstrate a dynamic balance between acetylation and ubiquitination in mitochondria, showing that SIRT1 inhibition results in mitochondrial acetylation and fission but prevents mitochondrial ubiquitination and mitophagic degradation.
3.3. Inhibition of SIRT1 induces mitochondrial calcium overload and ROS burst
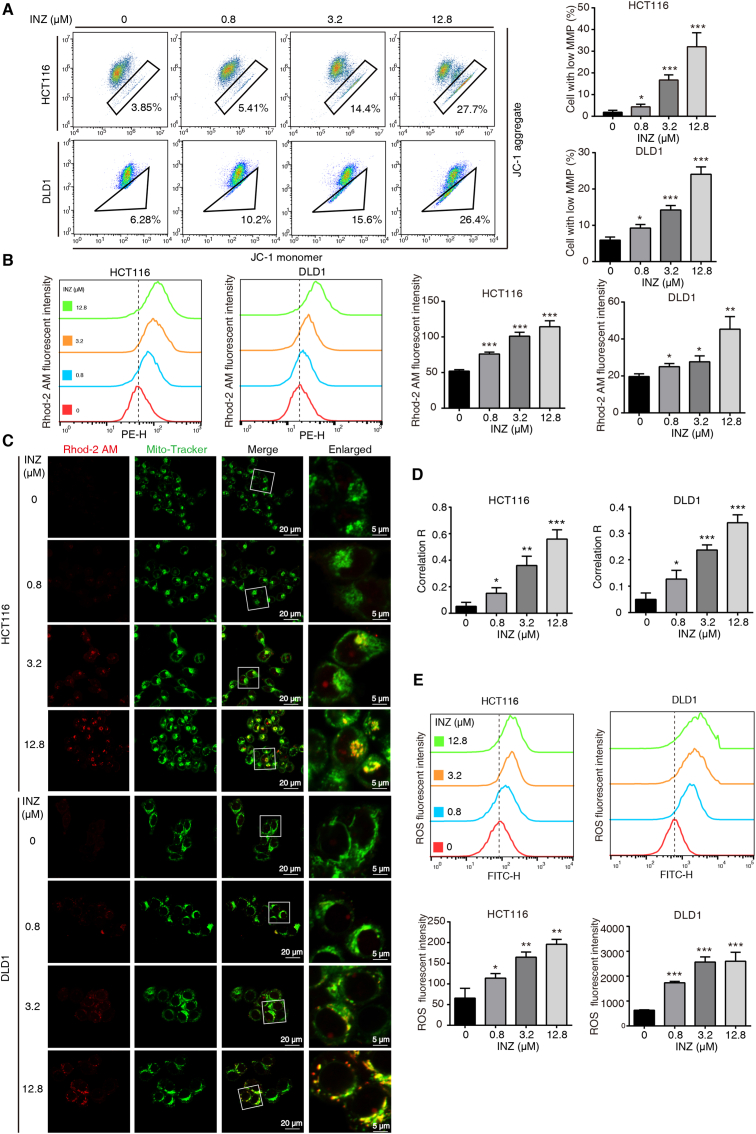
To further explore the effect of SIRT1 inhibition on mitochondria, we determined the mitochondrial membrane permeability (MMP) by using JC-1 assay. Both HCT116 and DLD-1 cells treated with elevating dosages of INZ (up to 12.8 μM) showed significant decreases of MMP in a dose-dependent manner (Fig. 3A). We next explored the mechanism for how SIRT1 inhibition induces loss of MMP. It was reported that mitochondrial calcium overload caused by calcium influx leads to the loss of MMP, oxidative damage and ultimate apoptosis [31]. Therefore, we determined the mitochondrial Ca2+ content and the ROS level in CRC cells upon SIRT1 inhibition. Rhod-2 AM is an effective probe that is widely used to determine the mitochondrial calcium ions [32]. Our confocal result showed that Rhod-2 AM labeled mitochondrial Ca2+ was increased and co-localized with mitochondria after histamine stimuli, while the Rhod-2 AM fluorescence was hardly improved in the presence of MCU inhibitor MCU-i4 [33] (Fig. S2), suggesting Rhod-2 AM is specific to mitochondrial calcium ions in our experimental system. By using the Rhod-2/AM staining and flow cytometry analysis, we found that INZ-treated CRC cells markedly increased mitochondrial Ca2+ in a dose-dependent manner (Fig. 3B). The confocal imaging also confirmed that Ca2+ was increased in the mitochondria of both HCT116 and DLD1 cells after INZ treatment (Fig. 3C–D). In addition, INZ treatment raised the intracellular ROS level in both CRC cells (Fig. 3E). These results demonstrate that SIRT1 inhibition enhances mitochondrial Ca2+, resulting in calcium overload, mitochondrial depolarization and ROS burst.
Fig. 3.
Inhibition of SIRT1 induces mitochondrial calcium influx and ROS burst. (A) JC-1 assay evaluation of the mitochondrial membrane potential level of HCT116 and DLD1 cells treated with the indicated concentrations of INZ (up to 12.8 μM) for 48 h. (B) The alteration of mitochondrial calcium level in HCT116 and DLD1 cells treated with different concentrations of INZ (0, 0.8, 3.2 and 12.8 μM) for 48 h was determined by flow cytometry with Rhod-2AM staining. (C, D) Confocal images of mitochondrial calcium in HCT116 and DLD1 cells treated with INZ (up to 12.8 μM) for 48 h (n = 3). Red, Rhod-2AM; Green, Mito-Tracker. The location overlap between Rhod-2 AM and mitochondria was determined by the correlation R (Pearson's correlation coefficient). Scale bars, 5 μm and 20 μm. (E) Representative flow cytometric analysis of ROS production in HCT116 and DLD1 treated with INZ (up to 12.8 μM) for 48 h. Bars, SD; *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. SIRT1-mediated deacetylation of mitochondrial calcium uniporter (MCU) at K332 is critical for mitochondrial calcium balance
Ca2+ signaling is important for cellular homeostasis and signaling cascade. As a main Ca2+ storage place, mitochondria play a critical role in maintaining Ca2+ levels between the cytosol and endoplasmic reticulum [34]. In our acetyl proteomics, we found that inhibition of SIRT1 significantly up-regulated the acetylation level of MCU at K332 site, a conservative residue (Fig. 4A–B; Supplementary Table 2). As MCU is the chief calcium transporter across the mitochondrial inner membrane to matrix [35], we asked whether SIRT1 is the major deacetylase for MCU and thus affects its function. We firstly verified that MCU directly interacted with SIRT1 endogenously and exogenously (Fig. 4C). To validate that the acetylation of K332 in MCU is regulated by SIRT1, we generated K332R mutation in MCU and found that arginine substitution of K332 remarkably reduced the acetylation of MCU, and decreased the MCU-SIRT1 interaction (Fig. 4D). To investigate the effect of the mutation K332R on Ca2+ uptake, we transfected wild-type (WT) MCU and mutant K332R respectively into HCT116 and DLD1 to compare their effects in response to SIRT1 inhibition. As anticipated, the mitochondrial Ca2+ uptake in WT was significantly increased, but it was weaker in K332R-expressing CRC cells (Fig. 4E). This functional effect was further confirmed by confocal imaging, showing that K332R reduced the co-localization of Ca2+ in mitochondria after INZ stimuli, as compared to WT (Fig. 4F). Moreover, both HCT116 and DLD1 cells bearing K332R mutation showed partial resistance to the loss of MMP (Fig. 4G) and apoptosis (Fig. 4H) induced by SIRT1 inhibition. Taken together, these data indicate that SIRT1 inhibition promotes the acetylation of MCU at K332, contributing to the increase of mitochondrial Ca2+ influx and the loss of MMP.
Fig. 4.
SIRT1 regulates the acetylation of MCU at K332 and influences mitochondrial calcium influx. (A) MS/MS spectra of MCU acetyl-Lys332 peptides derived from HCT116 cells by SIRT1 inhibition. (B) Lys332 in MCU is conserved in various species. (C) CO-IP assay was performed to determine the interaction of SIRT1 and MCU. Expression of MCU was determined in SIRT1 immuonprecipitates by Western blotting in HCT116 cells (left). HA-MCU and Flag-SIRT1 were co-transfected into 293T cells, and co-immunoprecipitated by anti-Flag antibody. (D) The interaction of MCU and SIRT1 was decreased in mutant K332R of MCU. CO-IP assay was performed in HCT116 cells expressed with Flag-SIRT1 and HA-MCU or HA-MCU-K332R plasmid. (E) Representative flow cytometric analysis of mitochondrial Ca2+ signal by Rhod-2AM staining in HCT116 and DLD1, which was transfected with wild-type or K332R MCU plasmids and treated with INZ for 48 h. (F) HCT116 and DLD1 cells transfected with wild-type or K332R MCU plasmids were incubated with INZ (up to 3.2 μM) for 48 h, respectively. Then the cells loaded with 1 μM Rhod-2AM (Red), and 100 nM Mito-Tracker (Green) were observed under confocal microscopy (n = 3). Blue, DAPI. The location overlap between Rhod-2 AM and mitochondria was determined by the correlation R (Pearson's correlation coefficient). Scale bar, 10 μm. (G) JC-1 assay analysis of mitochondrial membrane potential in HCT116 and DLD1 cells with indicated treatment. Cells with low mitochondrial membrane potential were quantified. (H) Annexin V/PI assay analysis of the apoptotic cells with indicated treatment. The apoptotic cells including early apoptosis and late apoptosis were statistically presented. Bars, SD; *P < 0.05; **P < 0.01; ***P < 0.001; ns P > 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. SIRT1 inhibition suppresses tumor progress of CRC
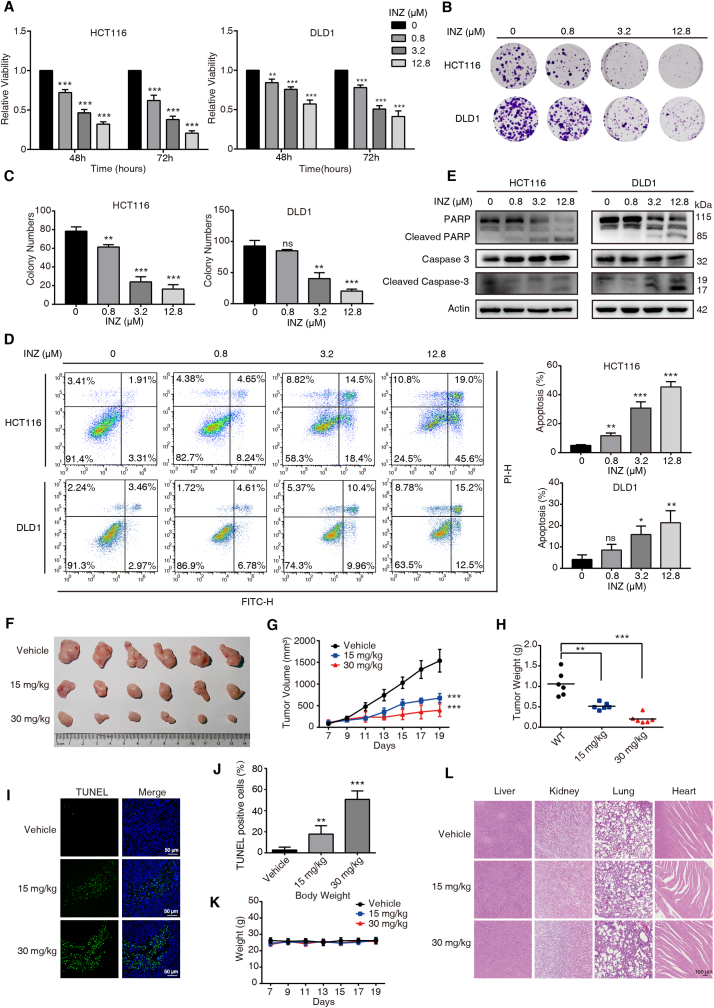
Based on above findings, we further determined the anticancer effect of INZ on CRC. Both HCT116 and DLD-1 cells were incubated with elevating dosages of INZ (up to 12.8 μM) for 48 and 72 h, respectively. WST-1 assay showed that INZ suppressed the growth of CRC cells in dose- and time-dependent manners (Fig. 5A). Colony formation assay confirmed that INZ remarkably decreased the ability of HCT116 and DLD-1 cells to form colonies (Fig. 5B–C). We also studied the effect of INZ on cell apoptosis by using Annexin V/PI staining assay. After 48 h of INZ treatment, apoptotic cells of HCT116 and DLD-1 were elevated in a dose-dependent manner (Fig. 5D), accompanied with the increased expressions of cleaved-PARP and cleaved-caspase-3 (Fig. 5E).
Fig. 5.
SIRT1 inhibitor INZ suppresses tumor growth of CRC in vivo and in vitro. (A) HCT116 and DLD1 cells were incubated with indicated concentrations of INZ for 48 and 72 h, the cell viability was determined by WST-1 assay. (B, C) INZ inhibited the colony formation of HCT116 and DLD1 cells. (D) HCT116 and DLD1 cells were treated with INZ for 48 h, and apoptotic cells were detected by flow cytometry. (E) Western blotting analysis of the expression levels of PARP, cleaved PARP, caspase-3 and cleaved caspase-3 in INZ-treated CRC cells. (F–L) Representative image of tumors with or without INZ treatment (F). HCT116 cells were subcutaneously injected into nude mice to establish tumor xenograft. In the seventh day, the mice were randomized into three groups to receive oral gavage of INZ (15 mg/kg or 30 mg/kg) or vehicle every two days, respectively. (G, H) Tumor curves showing the inhibitory effect of INZ on tumor growth, and the tumor weight was statistically presented. (I, J) Cell apoptosis in tumor tissues was analyzed by TUNEL assay (I), and statistically presented (J). (K) The body weight of nude mice during the experimental period was shown. (L) Hematoxylin and eosin (H&E) staining of the livers, kidneys, lung and heart collected from treatment and control groups. Scale bar, 100 μm. Bars, SD; *P < 0.05; **P < 0.01; ***P < 0.001; ns P > 0.05.
The therapeutic potential of INZ was further investigated in vivo. We employed subcutaneous tumor xenografts using nude mice for orally administration with 15 mg/kg and 30 mg/kg of INZ, and observed the effect of INZ on tumor growth. As shown in Fig. 5F–H, the tumor burden including tumor volume (Fig. 5G) and tumor weight (Fig. 5H) was significantly suppressed by INZ treatment with the decrease of 50% and 75%, respectively. In addition, our TUNEL assay confirmed that INZ significantly induced cell apoptosis in tumor tissues (Fig. 5I–J). Meanwhile the total body weights of each group did not show significant alteration during administration of INZ (Fig. 5K). Also, some vital organs, including the livers, kidneys, lungs and hearts, were evaluated in histological, and no overt change in morphology was found (Fig. 5L), indicating a significantly inhibitory effect of INZ on CRC without toxic effects on animals.
4. Discussion
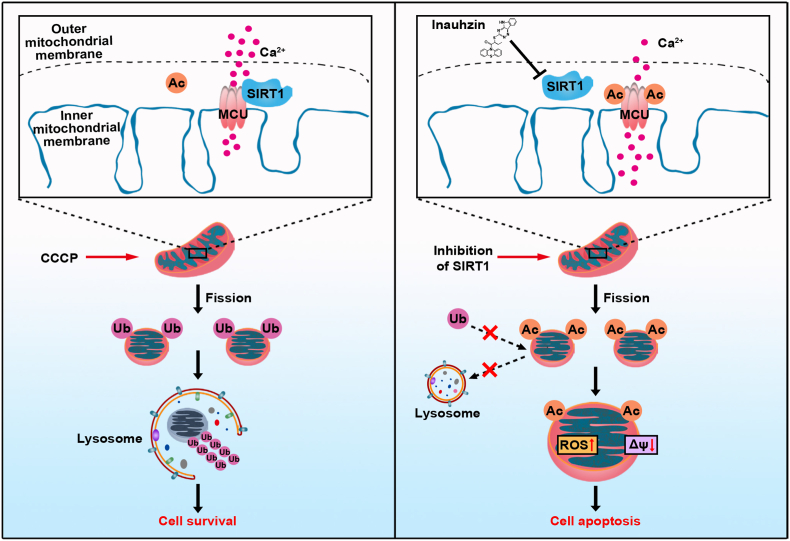
This study uncovered a non-canonical role of SIRT1 in the regulation of deacetylation in mitochondria that is usually mainly mediated by SIRT3, SIRT4 and SIRT5 (Fig. 6). In particular, SIRT1 can directly interact with the major mitochondrial calcium transport protein MCU and deacetylate it at residue K332 to sustain mitochondrial Ca2+ homoeostasis and integrity. Systematically, functional loss of SIRT1 with SIRT1 inhibition triggers the promotion of mitochondrial acetylation, reciprocally reduces mitochondrial ubiquitination for lysosomal degradation, resulting in mitochondrial fragmentation and depolarization. Such a promotion of mitochondrial acetylation prevents LC3 recruitment-mediated mitophagic degradation and clearance, inducing MMP loss and thus aggravating cell apoptosis.
Fig. 6.
Schematic diagram of the action mechanism SIRT1 inhibition on mitochondrial dysfunction. The inhibition of SIRT1 induces acetylation of mitochondrial Ca2+ transporter, MCU, promoting mitochondrial Ca2+ overload, ROS burst and loss of mitochondrial membrane potential. Global acetylation of mitochondrial proteins due to SIRT1 inhibition obstructs their ubiquitin and LC3-labeling, preventing mitophagy-associated clearance, and ultimately causing cell apoptosis.
SIRT1 is widely recognized as a nuclear deacetylase to regulate many critical transcriptional factors [8,36]. However, emerging extranuclear substrates of SIRT1 were recently identified [9,12,36]. For example, SIRT1 induces the deacetylation of Beclin 1 to promote the autophagic degradation of E-cadherin, accelerating the epithelial–mesenchymal transition (EMT) of melanoma [37]. In this connection, we herein using acetyl-proteomics identified a large number of mitochondrial proteins to be highly acetylated upon SIRT1 inhibition (Fig. 1I), suggesting an unexpected role of SIRT1 in mitochondria. This observation was supported by the fact that SIRT1 does exist in mitochondria and its deletion induces mitochondrial acetylation (Fig. 1K, M). It is likely that SIRT1 serves as a guardian to protect mitochondria from oxidative stress-triggered acylation accumulation, such as acetylation and succinylation [38].
Particularly, we identified that SIRT1 can interact with a mitochondrial calcium uniporter MCU, resulting in its deacetylation at K332 to initiate a functional cascade. MCU regulates the entry of ions such as calcium or zinc from the cytosol to mitochondria along with the electrochemical gradient uncouple of ATP hydrolysis [39]. MCU-mediated Ca2+ homeostasis in mitochondria is associated with a variety of cellular responses, including energy metabolism, cell division, cell survival and death [32,40], as well as tumor progression [34,41]. Phosphorylation of serines 57 and 92 on MCU by CaMKII was reported to promote mitochondrial Ca2+ entry [42]. However, acetylation on MCU and its correlation with Ca2+ balance are poorly understood. This study for the first time revealed that K332 on MCU can be deacetylated by SIRT1, along with the fact that CRC cells bearing MCU-K332R mutation display resistant to SIRT1 inhibitor-mediated mitochondrial Ca2+ influx, MMP loss and apoptosis (Fig. 4E–H). Interestingly, modulation of MCU level to induce mitochondrial Ca2+ imbalance, MMP loss and ROS burst has been regarded as a potential strategy for anticancer therapy [43]. Our study further unveiled that acetylation of MCU by SIRT1 inhibition is another way to cause mitochondrial injury leading to cell death. This can be supported by our in vivo and in vitro experiments, showing that SIRT1 inhibition displays significant anticancer effects on CRC, as demonstrated by the decrease of cell viability and colony formation and the increase of apoptosis after INZ treatment (Fig. 5A–E).
As observed in this experiment and other studies [[44], [45], [46]], SIRT1 expression is much higher in tumor cells than non-malignant cells, this may make tumor cells more susceptible to SIRT1 inhibitor INZ-induced cell death, the hypothesis warrants further investigation. Regarding the possibility for INZ-induced Ca2+ overload and mitochondrial depolarization leading to an off-target cell death, it may occur but much be insignificant under the experimental conditions with low concentrations of INZ in this study, as evidenced by the fact that no significant side effect of INZ treatment was observed in our experiments (Fig. 5K and L).
Mitophagy is a cargo-specific autophagic elimination of damaged mitochondria in mammals [47]. Depolarized mitochondria are labeled with ubiquitin at outer membrane by E3 ligases (eg. parkin), activating ubiquitin–autophagy adaptors (eg. FUNDC1, NIX, BNIP3 etc.) to recruit ATG8 family proteins (eg. LC3) for lysosomal degradation. Reversible PTMs serve as regulatory mechanisms to modulate the activity of mitophagy receptors, for example, phosphorylation of BNIP3 at Ser17 and Ser24 is critical for BNIP3-LC3 bindings [48]. However, whether acetylation modulates mitophagy receptors remains unclear. Here, we found that SIRT1 inhibition-mediated mitochondrial acetylation links to mitochondrial fission, an important step to mitophagy, however the acetylated mitochondria are unexpectedly not colocalized with lysosome and LC3 (Fig. 2E–F). We thus looked into mitophagy receptors, MFN2 and Tom20, which are usually ubiquitylated in response to mitochondrial depolarization [[27], [28], [29], [30]]. We interestingly found that the ubiquitination of MFN2 and Tom20 is handicapped by INZ treatment with mitochondrial injury (Fig. 2G–H). This is probably due to a competition between acetylation and ubiquitination, mitochondrial acetylation by SIRT1 inhibition may occupy the ubiquitination sites of the mitophagy proteins. As a result, un-ubiquitinated mitochondria fail to recruit LC3 for mitophagy, leading to the accumulation of damaged mitochondria and ultimately cell death.
In summary, we here discover a novel molecular mechanism of SIRT1 in regulating mitochondria dynamics. SIRT1 inhibition significantly enhances protein acetylation in mitochondria, which hinders protein ubiquitination and inhibits mitophagy by preventing the recruitment of LC3 for mitochondrial homeostatic degradation. SIRT1 inhibition particularly increases the acetylation of MCU at K332, the major Ca2+ transporter, which triggers mitochondrial Ca2+ influx and depolarization, and ultimately apoptosis. This study also highlights the anticancer effect of SIRT1 inhibition through mitochondrial acetylation, implicating a possible alternative for anticancer therapy.
Funding statement
This work was supported by the National Key R & D Program of China (2017YFA0505100, 2020YFE0202200), the National Natural Science Foundation of China (82103208).
Author contributions
Y.W. and Q.Y.H. designed the research, Y.S., Y.Y.M., Y.Y.H. and L.O. performed research, Y.W., Y.S., and Y.Y.M. analyzed data, X.F.Y., Z.H.S. and N.L. performed MS analysis. Y.W., and Q.Y.H. supervised research and wrote the paper.
Ethics approval and consent to participate
The study is approved by the Ethical Review Board of Jinan University. All participants provided written informed consent. All of the animal researches were conducted complying with Guidance for the Care and Use of Laboratory Animals of Jinan University.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102334.
Contributor Information
Qing-Yu He, Email: tqyhe@email.jnu.edu.cn.
Yang Wang, Email: wangyang0507@jnu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1. (A) Western blotting analysis of the acetylation levels of p53-K382 in INZ-treated HCT116 and DLD1 cells. (B) PLGEM analysis of the abundance of INZ-regulated proteins. (C) PLGEM analysis of the abundance of INZ-regulated acetylated peptides.
Fig. S2. Determination on the specificity of Rhod-2 AM in measuring mitochondrial calcium. DLD1 cells loaded with Rhod-2 AM (1 μM) were treated with or without histamine (100 μM) or MCU-i4 (10 μM, 90 min) as indicated. The cells were analyzed by confocal imaging after staining of Mito-Tracker (n = 3). Red, Rhod-2AM; Green, Mito-Tracker. The colocalization between Rhod-2 AM and mitochondria was determined by the correlation R (Pearson’s correlation coefficient). Scale bars, 5 μm and 20 μm.
References
- 1.Baeza J., Smallegan M.J., Denu J.M. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 2016;41:231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J., Xiang H., Liu J., Chen Y., He R.R., Liu B. Mitochondrial Sirtuin 3: new emerging biological function and therapeutic target. Theranostics. 2020;10:8315–8342. doi: 10.7150/thno.45922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrico C., Meyer J.G., He W., Gibson B.W., Verdin E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metabol. 2018;27:497–512. doi: 10.1016/j.cmet.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C., Wang Y., Shen L. Mitochondrial proteins in heart failure: the role of deacetylation by SIRT3. Pharmacol. Res. 2021;172 doi: 10.1016/j.phrs.2021.105802. [DOI] [PubMed] [Google Scholar]
- 6.Imai S., Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herranz D., Serrano M. SIRT1: recent lessons from mouse models. Nat. Rev. Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai H., Sinclair D.A., Ellis J.L., Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogueiras R., Habegger K.M., Chaudhary N., Finan B., Banks A.S., Dietrich M.O., Horvath T.L., Sinclair D.A., Pfluger P.T., Tschöp M.H. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol. Rev. 2012;92:1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W.Y., Wang D.H., Yen R.C., Luo J., Gu W., Baylin S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Kwon H.S., Ott M. The ups and downs of SIRT1. Trends Biochem. Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., Ciriolo M.R. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickles S., Vigié P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018;28:R170–r185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koval O.M., Nguyen E.K., Santhana V., Fidler T.P., Sebag S.C., Rasmussen T.P., Mittauer D.J., Strack S., Goswami P.C., Abel E.D., Grumbach I.M. Loss of MCU prevents mitochondrial fusion in G(1)-S phase and blocks cell cycle progression and proliferation. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aav1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu L., Hu C., Xu M., Yu J., He H., Lin J., Sha H., Lu B., Engelender S., Guan M., Song Z. ATAD3B is a mitophagy receptor mediating clearance of oxidative stress-induced damaged mitochondrial DNA. EMBO J. 2021;40 doi: 10.15252/embj.2020106283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole L.P., Macleod K.F. Mitophagy in tumorigenesis and metastasis. Cell. Mol. Life Sci. 2021;78:3817–3851. doi: 10.1007/s00018-021-03774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Zhou Y., Li N., Liu W.T., Liang J.Z., Sun Y., Zhang W.X., Fang R.D., Huang S.L., Sun Z.H., Wang Y., He Q.Y. Curcumol overcomes TRAIL resistance of non-small cell lung cancer by targeting NRH:quinone oxidoreductase 2 (NQO2) Adv. Sci. 2020;7 doi: 10.1002/advs.202002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O'Donoghue S.I., Schneider R., Jensen L.J. Database; Oxford: 2014. COMPARTMENTS: Unification and Visualization of Protein Subcellular Localization Evidence. (2014) bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Zhang J., Zheng C.C., Huang Z.J., Zhang W.X., Long Y.L., Gao G.B., Sun Y., Xu W.W., Li B., He Q.Y. C20orf24 promotes colorectal cancer progression by recruiting Rin1 to activate Rab5-mediated mitogen-activated protein kinase/extracellular signal-regulated kinase signalling. Clin. Transl. Med. 2022;12:e796. doi: 10.1002/ctm2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Zhang J., Li Y.J., Yu N.N., Liu W.T., Liang J.Z., Xu W.W., Sun Z.H., Li B., He Q.Y. MEST promotes lung cancer invasion and metastasis by interacting with VCP to activate NF-κB signaling. J. Exp. Clin. Cancer Res. 2021;40:301. doi: 10.1186/s13046-021-02107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalkiadaki A., Guarente L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q., Zeng S.X., Zhang Y., Zhang Y., Ding D., Ye Q., Meroueh S.O., Lu H. A small molecule Inauhzin inhibits SIRT1 activity and suppresses tumour growth through activation of p53. EMBO Mol. Med. 2012;4:298–312. doi: 10.1002/emmm.201100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adebayo M., Singh S., Singh A.P., Dasgupta S. Mitochondrial fusion and fission: the fine-tune balance for cellular homeostasis. Faseb. J. 2021;35 doi: 10.1096/fj.202100067R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roca-Portoles A., Tait S.W.G. Mitochondrial quality control: from molecule to organelle. Cell. Mol. Life Sci. 2021;78:3853–3866. doi: 10.1007/s00018-021-03775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J.R., Martinez A., Lane J.D., Mayor U., Clague M.J., Urbé S. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 2015;16:618–627. doi: 10.15252/embr.201439820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLelland G.L., Goiran T., Yi W., Dorval G., Chen C.X., Lauinger N.D., Krahn A.I., Valimehr S., Rakovic A., Rouiller I., Durcan T.M., Trempe J.F., Fon E.A. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. 2018;7 doi: 10.7554/eLife.32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S., Vollmer S., Golombek S., Kahle P.J. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J. Cell Sci. 2014;127:3280–3293. doi: 10.1242/jcs.146035. [DOI] [PubMed] [Google Scholar]
- 31.Aranda-Rivera A.K., Cruz-Gregorio A., Aparicio-Trejo O.E., Pedraza-Chaverri J. Mitochondrial redox signaling and oxidative stress in kidney diseases. Biomolecules. 2021:11. doi: 10.3390/biom11081144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giorgi C., Marchi S., Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 2018;19:713–730. doi: 10.1038/s41580-018-0052-8. [DOI] [PubMed] [Google Scholar]
- 33.Di Marco G., Vallese F., Jourde B., Bergsdorf C., Sturlese M., De Mario A., Techer-Etienne V., Haasen D., Oberhauser B., Schleeger S., Minetti G., Moro S., Rizzuto R., De Stefani D., Fornaro M., Mammucari C. A high-throughput screening identifies MICU1 targeting compounds. Cell Rep. 2020;30:2321–2331. doi: 10.1016/j.celrep.2020.01.081. e2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchi S., Giorgi C., Galluzzi L., Pinton P. Ca(2+) fluxes and cancer. Mol. Cell. 2020;78:1055–1069. doi: 10.1016/j.molcel.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 35.Sancak Y., Markhard A.L., Kitami T., Kovács-Bogdán E., Kamer K.J., Udeshi N.D., Carr S.A., Chaudhuri D., Clapham D.E., Li A.A., Calvo S.E., Goldberger O., Mootha V.K. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Peterson L.M., Li X. Trending topics of SIRT1 in tumorigenicity. Biochim. Biophys. Acta Gen. Subj. 2021;1865 doi: 10.1016/j.bbagen.2021.129952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun T., Jiao L., Wang Y., Yu Y., Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis. 2018;9:136. doi: 10.1038/s41419-017-0167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Han Z.Q., Wang Y., He Q.Y. Alteration of mitochondrial protein succinylation against cellular oxidative stress in cancer. Mil. Med. Res. 2022;9:6. doi: 10.1186/s40779-022-00367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirichok Y., Krapivinsky G., Clapham D.E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 40.Garbincius J.F., Elrod J.W. Mitochondrial calcium exchange in physiology and disease. Physiol. Rev. 2022;102(2):893–992. doi: 10.1152/physrev.00041.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui C., Merritt R., Fu L., Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B. 2017;7:3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joiner M.L., Koval O.M., Li J., He B.J., Allamargot C., Gao Z., Luczak E.D., Hall D.D., Fink B.D., Chen B., Yang J., Moore S.A., Scholz T.D., Strack S., Mohler P.J., Sivitz W.I., Song L.S., Anderson M.E. CaMKII determines mitochondrial stress responses in heart. Nature. 2012;491:269–273. doi: 10.1038/nature11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Yu X., Deng L., Zhou S., Wang Y., Zheng X., Chu Q. Neochlorogenic acid anchors MCU-based calcium overload for cancer therapy. Food Funct. 2021;12:11387–11398. doi: 10.1039/d1fo01393a. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y.H., Song N.Y., Suh J., Kim D.H., Kim W., Ann J., Lee J., Baek J.H., Na H.K., Surh Y.J. Curcumin suppresses oncogenicity of human colon cancer cells by covalently modifying the cysteine 67 residue of SIRT1. Cancer Lett. 2018;431:219–229. doi: 10.1016/j.canlet.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 45.Huffman D.M., Grizzle W.E., Bamman M.M., Kim J.S., Eltoum I.A., Elgavish A., Nagy T.R. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 46.So D., Shin H.W., Kim J., Lee M., Myeong J., Chun Y.S., Park J.W. Cervical cancer is addicted to SIRT1 disarming the AIM2 antiviral defense. Oncogene. 2018;37:5191–5204. doi: 10.1038/s41388-018-0339-4. [DOI] [PubMed] [Google Scholar]
- 47.Huang M.-Y., Zhang J., Ouyang L., Wang Y. ISGylation orchestrates the degradation flux of cellular cargo toward lysosome. Genes Dis. 2021 doi: 10.1016/j.gendis.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., Brady N.R. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Western blotting analysis of the acetylation levels of p53-K382 in INZ-treated HCT116 and DLD1 cells. (B) PLGEM analysis of the abundance of INZ-regulated proteins. (C) PLGEM analysis of the abundance of INZ-regulated acetylated peptides.
Fig. S2. Determination on the specificity of Rhod-2 AM in measuring mitochondrial calcium. DLD1 cells loaded with Rhod-2 AM (1 μM) were treated with or without histamine (100 μM) or MCU-i4 (10 μM, 90 min) as indicated. The cells were analyzed by confocal imaging after staining of Mito-Tracker (n = 3). Red, Rhod-2AM; Green, Mito-Tracker. The colocalization between Rhod-2 AM and mitochondria was determined by the correlation R (Pearson’s correlation coefficient). Scale bars, 5 μm and 20 μm.