Highlights
-
•
It was first time to combined transcriptomic and proteome analyses to investigate the ultrasound pretreated peanut sprouts.
-
•
A total of 1104 DEGs and 399 DEPs between ultrasound pretreated and nontreated peanut sprouts.
-
•
Ultrasound upregulated three key genes that could have increased the content of resveratrol via phenylpropanoid biosynthesis.
-
•
The genes and proteins related to phenylpropanoid biosynthesis, flavonoids biosynthesis, and lipid metabolism.
Keywords: Peanut sprout, Transcriptome, Proteome, Phenylpropanoids, Flavonoids
Abstract
Combined transcriptomic and proteome analyses were carried out to investigate the influence of ultrasound pretreatment on peanut sprouts. In total, 1104 differentially expressed genes (upregulated:538, downregulated:521) and 399 differentially accumulated proteins (upregulated: 197, downregulated: 202) were identified between ultrasound pretreated and nontreated peanut sprouts. These genes and proteins were related to a series of crucial biomolecular processes, including the metabolism of carbohydrates, terpenoids, and polyketides. The most enriched pathways were further analyzed in each category. Importantly, ultrasound upregulated three key genes namely the arahy. Tifrunner. gnm1.ann1.DXZI51, arahy.Tifrunner.gnm1.ann1.VGN2GE, and arahy.Tifrunner.gnm1.ann1.Y23DM6 that could have increased the content of resveratrol via phenylpropanoid biosynthesis. Furthermore, this study shows that B3, MYB transcription factor-like families play a significant role in response to ultrasound treatment. Overall, this study provides useful transcriptomics and proteomics information highlighting the molecular mechanisms that influence nutritional differences in peanut sprouts.
1. Introduction
Peanut, one of the most popular oil and food resources, is rich in essential fatty acids, protein, fat-soluble vitamins, polyphenols, mineral elements, and other beneficial constituents. Peanut seeds consist of 22–30% protein and 44–56% oil (Camargo et al., 2017). Peanuts contain about four times more unsaturated fatty acids than saturated fatty acids. The main fatty acids in peanuts are oleic acid and linoleic acid, accounting for ∼ 80%. In addition, peanuts contain various trace elements, such as VB, VC, calcium, iron, magnesium, phosphorus, manganese, etc. (Xiao, Liu, & Li, 2021). Peanuts are also rich in physiologically active substances such as phytosterols, saponins, resveratrol, proanthocyanins, and flavonoids (Adhikari et al., 2018).
Peanuts can be consumed as a sprout that has high protein but lower fat contents (Adhikari et al., 2018; Yuan, et al.). Compared to other vegetable sprouts, peanut sprouts have a crisp peanut-specific rich flavor and rich in resveratrol, which makes them ideal as a functional food ingredient (Miao et al., 2016). Studies suggest that the germination process significantly changes the protein content in peanut seeds; the nitrogen content in the protein hydrolysate decreases, while the peptide and amino acid content increases (Yu et al., 2021). The change in resveratrol content during germination is also an important research area. Limmongkon et al., 2017, Limmongkon et al., 2019) found that resveratrol content increases significantly during peanut germination. Wang et al. (2017) reported that the resveratrol content varies with peanut parts; a higher polyphenol content and antioxidant activity were reported in the germinated peanut extract, which exerts neuroprotective effects.
Ultrasound treatment for seed germination is a new technology to enrich biologically active substances. YANG et al.(Lo Porto et al., 2018, Perera and Alzahrani, 2021) studied the effect of ultrasonic treatment on soybean seeds and the nutritional quality of soybean sprouts; they found that ultrasonic treatment improved the germination rate, germination length, gamma-aminobutyric acid content, and nutritional quality of soybeans. Previously, we explored the effect of ultrasonic treatment on the accumulation of resveratrol during the germination of three varieties of peanuts. We reported that ultrasonic treatment significantly improved the seed germination rate and resveratrol content (Yu, Liu, Shi, Liu, & Wang, 2016). Furthermore, to explore the molecular mechanism for such changes in peanut sprouts, we identified some macroscopic changes due to ultrasonic action. However, the detailed molecular mechanism remained elusive, which required modern methods. Transcriptome sequencing technology is a new type of high-throughput sequencing technology, which has the advantages of large amount of information, less data redundancy, and accurate analysis (Huang et al., 2019). Tandem mass spectrometry tagging (TMT), as a new quantitative research technology of proteomics, has good quantitative effect and high reproducibility, and is widely used in food science, botany, microbiology and medicine and other fields (Luo et al., 2019). These two technologies have been extensively applied in many small crops bind the gene level to the protein level, but surprisingly, study on peanut sprouts has been rarely reported. A combination of transcriptome, proteome, and metabolome analysis can effectively identify metabolic-related functional genes. Since most studies majorly focused on the change in nutrient contents, the detailed molecular mechanism remained unclear in the lack of functional analysis.
Accordingly, this study performed the multi-omics analysis of transcriptome and proteome to study the functional classification, metabolic pathways, and biological processes that may regulate peanut sprout under ultrasonic induction. This study reports key genes and proteins that are associated with the metabolism and accumulation of nutrients and highlights the molecular mechanism of changes in peanut sprouts under ultrasonic pretreatment.
2. Materials and methods
2.1. Materials and treatments
Seeds of the peanut cultivar Fuhua 23 were supplied by the Liaoning Academy of Agricultural Sciences in mid-October 2020.
The methods of ultrasonic pretreatment and peanut germination were reported previously (Miao et al., 2016, Yu et al., 2016). Samples were sonicated with an ultrasonic cleaner bath (KQ-300VDE, Kunshan Ultrasonic Instrument Co., Ltd, Kunshan City, China) for 30 min at 35 °C, 85 kHz, and 240 W. The ultrasound pretreated peanut seeds (CS; n = 600, three parallel groups of 200 each) were germinated in a constant temperature (27 °C) and relative humidity (90%) incubator (160HL, Jinyi Instrument, Jiangsu, China); another batch of 600 seeds (three parallel groups of 200 each) without ultrasound was used as a control group (KB). At the grain germination period and 72 h after germination, peanut sprouts were frozen in liquid nitrogen to store at −80 °C and then sent to Shanghai Meiji Biological Co., Ltd for further analysis.
2.2. RNA extraction and Illumina sequencing
Total RNA was extracted from peanut sprouts using the CTAB method (Cao, Xu, Chen, & Ma, 2016). RNA quantity and quality were determined by NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA), respectively. RNA integrity was determined by 1% agarose gel electrophoresis and concentration was adjusted for uniformity. Three biological replicates and three technical replicates were assessed.
The mRNA was isolated from total RNA using the oligo (dT) magnetic beads. The cDNA was synthesized using a cDNA Synthesis Kit (TaKaRa) and the sequencing adapters were attached to both ends (Chai et al., 2014). The prepared libraries were sequenced on an Illumina HiSeq TM2500 platform. Sequence data with base-pair qualities Q ≥ 20 were extracted. The filtered reads were mapped to Arachis_hypogaea reference genome (https://download.maizegdb.org/B73_RefGen_v3/). The DEGs had a criterion of log2 (fold change) ≥ 1 and corrected P ≤ 0.005. All DEGs were subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses using GOseq and KOBAS software, respectively.
2.3. Protein extraction, protein digestion, and TMT (Tandem Mass Tags) labeling
Protein was extracted from respective peanut sprouts using trichloroacetic acid/acetone. The protein extraction and digestion were performed as described previously (Zhang et al., 2016). Briefly, the total protein (100 μg) extracted from each sample was mixed with 100 μL of lysis buffer. This was added with Tris(2-chloroethyl) phosphate (10 mM) and stored at 37 °C. After 60 min, iodoacetamide (40 nM) was added to the mixture, which was stored at room temperature in the dark for 40 min. Then, six volumes of cold acetone were added to precipitate protein at −20 °C for 4 h. The mixture was centrifuged at 10,000g for 20 min at 4 °C, and the precipitate was resuspended in 100 µL of 50 mM TEAB buffer. Trypsin was added to protein at a 1:50 ratio and the mixture was incubated overnight at 37 °C. 1 unit of Tandem Mass Tags reagent was thawed and reconstituted in 50 µL of acetonitrile at room temperature for 2 h, and hydroxylamine was added for reaction at room temperature for 15 min. Finally, all samples were mixed, desalted, and vacuum dried. Three biological replicates and three technical replicates were assessed.
2.4. LC–MS/MS analysis
The 9RKFSG2_NCS-3500R system (Thermo, USA) connected to the Q_Exactive HF-X system (Thermo, USA) via a nanoelectrospray ion source was used for the study. The labeled peptides were analyzed by online nanoflow liquid chromatography-tandem mass spectrometry. Briefly, a c18 reversed-phase column (75 μm × 25 cm, Thermo, USA) was equilibrated with solvent A (2% formic acid and 0.1% formic acid) and solvent B (80% acetonitrile and 0.1% formic acid). The elution conditions were as follows: 0–2 min, 0–3% B gradient elution; 2–92 min, 5–25% B; 92–102 min, 25–45% B; 102–105 min, 45–100% B; 105–120 min, 100–0% B; flow rate, 300 nL/min. Q_Exactive HF-X was operated in data-dependent acquisition mode (DDA) to automatically switch between full-scan MS and MS/MS acquisition. In Orbitrap, the full-scan mass spectrometry in the range of m/z 350–1500 was obtained with a resolution of 70,000. The automatic gain control (AGC) target was 3e6, and the maximum filling time was 20 ms. The first 20 precursor ions were selected for entry into the collision unit for high-energy collision dissociation (HCD) fragmentation. The MS/MS resolution was set to 35,000 (m/z 100); the automatic gain control (AGC) target was set to 1e5; the maximum fill time was 50 MS, and the dynamic rejection time was 30 s. Three biological replicates and three technical replicates were assessed.
2.5. Protein identification and data analysis
RAW data was analyzed with Proteome Discoverer (Thermo Scientific, Version 2.2). MS/MS search conditions were as follows: mass tolerance 20 ppm Da 0.02 MS and MS/MS Tolerance; trypsinase 2 missed cleavage were allowed. Ureidocysteine methylation and TMT N-terminus and lysine side chain peptide were used as a fixed modification, and methionine oxidation was used as a dynamic modification, respectively. The false discovery rate of peptide recognition was set to FDR ≤ 0.01. At least one unique peptide identification was used to support protein identification. GO (https://geneontology.org/) and KEGG pathway (https://www.genome.jp/kegg/) annotations were performed for all identified proteins. String v10.5 was used for protein–protein interaction analysis.
3. Results and discussion
3.1. Transcriptome difference between the CS and KB peanut sprouts
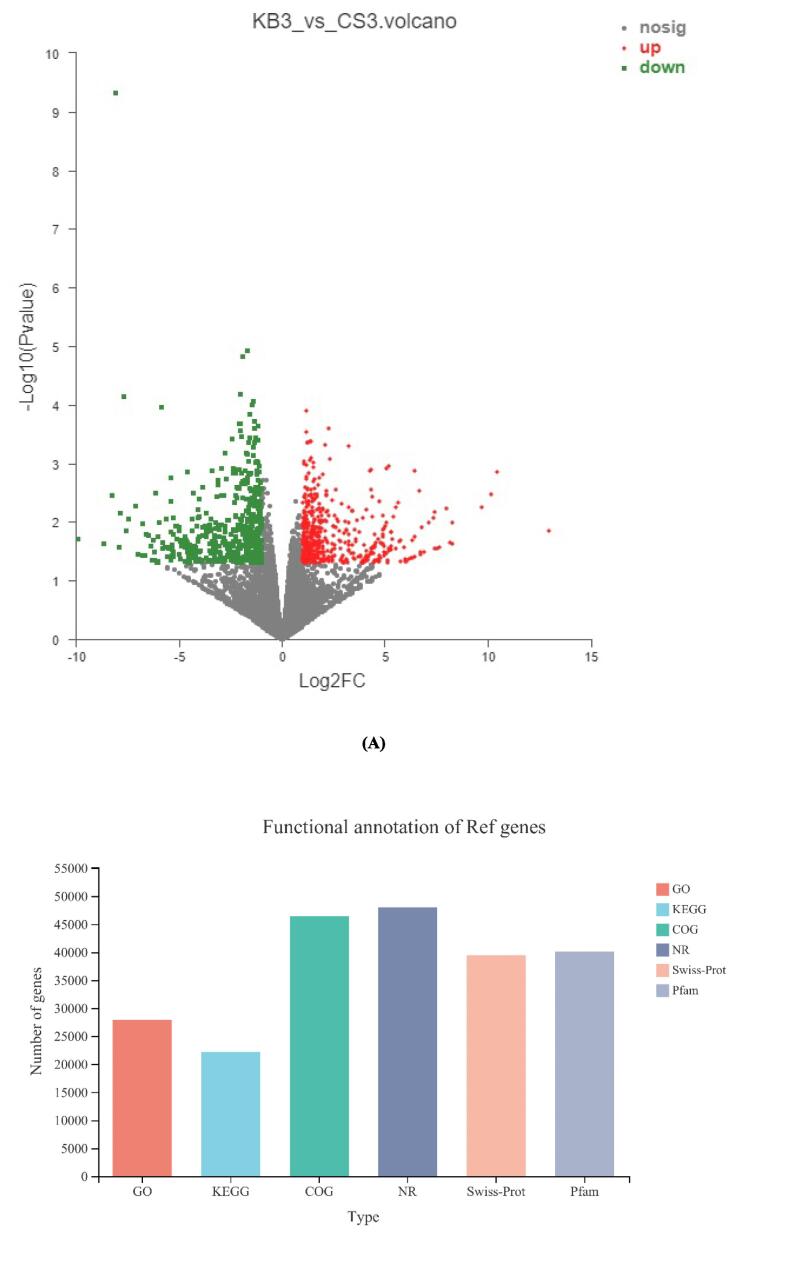
RNA-Seq produced 46,208,369 and 49,367,041 clean reads from CS and KB libraries, respectively. Clean data were from 6 libraries, with 3 replicates for each group. The total number of bases was > 6 GB with Q30 > 94% (sequences with a sequencing error rate < 0.1%). The average GC content was 45%. Overall, the data was of high quality and could be used for further analysis. Comparing the two groups, there were 1104 DEGs, including 583 upregulated and 521 downregulated genes between the CS and KB groups (Fig. 1A).
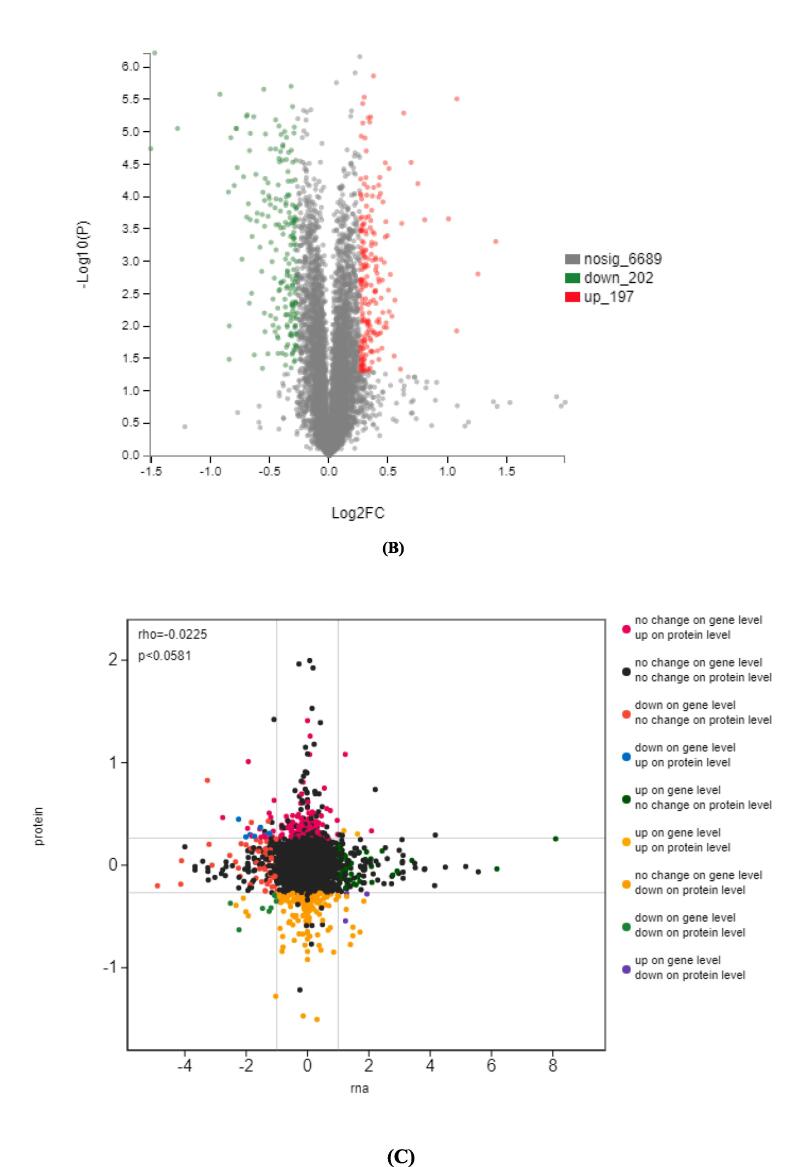
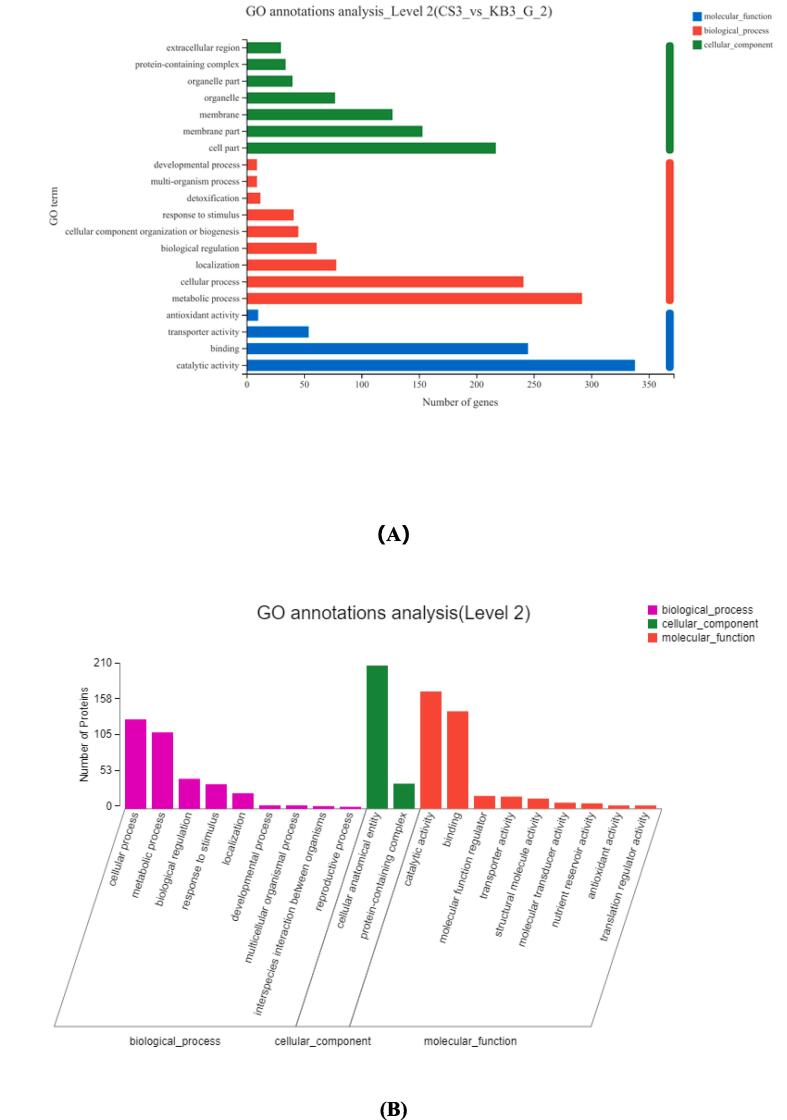
Fig. 1.
Volcano map of the differential genes under ultrasound induction (A). Volcano map of differential proteins under ultrasound induction (B). Scatter diagram of the correlation between the proteins and transcripts in the CS and KB groups (Pearson correlation:0.2204, p-value:0 (C).
All unigenes were annotated using BLASTX searches against the NCBI Nr protein sequences, Swiss-Prot, Pfam, EggNOG, GO, and KEGG databases to obtain 48,660 annotated unigenes (Table.1). The maximum annotated unigenes were from the Nr database (98.64%). 27,880 (57.30%) and 46,411 (95.38%) unigenes were from GO and COG databases, respectively. Only 22,200 (45.62%) genes were from the KEGG database.
Table 1.
Functional annotation of ref genes.
| Sample | Raw reads | Raw bases | Clean reads | GC (%) |
|---|---|---|---|---|
| GO | 27880(0.573) | 32507(0.5851) | 33775(0.5032) | 43951(0.5188) |
| KEGG | 22200(0.4562) | 26104(0.4699) | 28853(0.4298) | 37824(0.4465) |
| COG | 46411(0.9538) | 53312(0.9596) | 60883(0.907) | 77079(0.9099) |
| NR | 47998(0.9864) | 54935(0.9888) | 65247(0.972) | 82113(0.9693) |
| Swiss-Prot | 39446(0.8106) | 45725(0.8231) | 49409(0.7361) | 63545(0.7501) |
| Pfam | 40083(0.8237) | 45639(0.8215) | 50644(0.7545) | 63691(0.7518) |
| Total_anno | 48056(0.9876) | 54980(0.9896) | 65374(0.9739) | 82254(0.971) |
| Total | 48660(1.0) | 55555(1.0) | 67124(1) | 84714(1) |
3.2. Proteomics characterization by TMT
The total proteins, extracted from the CS and KB groups at the filling stage, were subjected to TMT and 2D LC–MS/MS analysis to complement the transcriptome analysis. A total of 374,782 spectra, 56,802 Identified spectra, and 38,423 peptides were obtained from proteomic analysis; in total, 7088 proteins were identified. A total of 399 proteins were identified as DEPs (differentially expressed proteins) based on criteria fold change (>1.2 or < 0.83) and P-value < 0.05 (Fig. 1B). These included 197 upregulated and 202 downregulated proteins between the CS and KB groups.
3.3. Analysis of DEGs and DEPs
To examine the congruence between transcriptome and proteome, we conducted a global correlation analysis between the protein and mRNA data. In total, 6562 proteins matched to transcripts (|log2 (ratio of transcript) | < 1, |log2 (ratio of protein) | < 0.25). Also, the expression of 21 DEPs, including 10 upregulated and 11 downregulated proteins, was consistent with the transcriptome. Overall, the results showed a poor correlation (r Pearson correlation = 0.2204) between tests, which could be due to inconsistent space and time between transcription and protein expression data. The time-dependent delay or regulatory processes from transcript-to-protein may play important roles in protein production. The low correlation between transcription and protein expression demonstrates that the abundances of transcripts are not perfect proxies for protein abundances causing a reasonable difference between the mono- and two-omics data analyses.
As shown in Fig. 1C, the proteins of the 3rd and 7th quadrants exhibited a similar trend between transcripts and proteins levels, while the proteins of the 1st and 9th quadrants showed an opposite trend between the two. Concerning the 1st and 9th quadrants, the SGS domain and lytic trans-glycosylase are involved in carbohydrate transport and metabolism showing an increase in protein and transcript levels. In the 3rd and 7th quadrants, alternative oxidase, sugar transport protein, O-methyltransferase, and xyloglucan endotransglucosylase are involved in carbohydrate transport and metabolism. Proteins were down-regulated in the 3rd and 7th quadrants. These proteins are associated with the nutritional quality of peanut sprouts and should be further analyzed.
3.4. GO functional classification
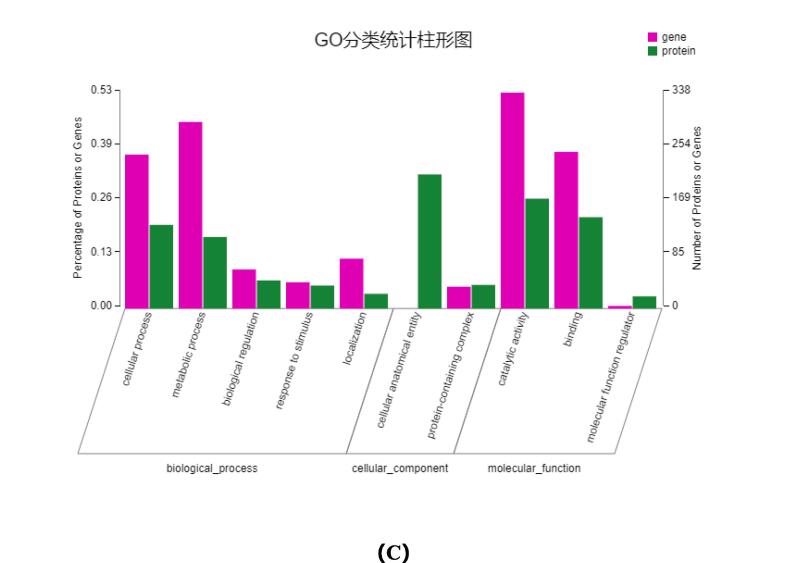
Next, the DEGs and DEPs in the two groups were subjected to GO functional cluster analysis. Gene ontology (GO) terms were assigned to the annotated DEGs. A total of 808, 692, and 668 genes were classified into three GO categories: ‘biological process’, ‘cellular component’, and ‘molecular function’ in the comparison group. Within the biological process category, the greatest abundance was of ‘metabolic process’, ‘biological regulation’, ‘localization’, and ‘cellular process’. Within the cellular component category, the most enriched terms were ‘cell part’, ‘cell’, ‘membrane part’, and ‘membrane’ and ‘organelle’. Within the molecular function category, the most highly represented terms were ‘catalytic activity’ and ‘binding’.
To obtain a global picture of the proteomic changes, 399 DEPs were annotated with GO analysis; 172 proteins were annotated to 20 terms. The functional classifications of DEPs were consistent with DEGs at the transcription level (Fig. 2). Among them, most DEPs were linked to response to stimulus (24), response to stress (23), molecular function regulator (16), enzyme activity inhibitor (16), and enzyme activity regulator (16).To predict the molecular mechanisms that regulate the physiological state of peanut sprouts, we compared the transcriptome and proteome data. Comparative analysis of GO terms for DEGs and DEPs showed that 15, 12, and 12 GO terms overlapped in the category of biological process, molecular function, and cellular component, respectively. GO terms with significant enrichment mainly included cellular process (21, 131), metabolic process (22, 112), biological regulation (5, 44), response to stimulus (4, 36), localization (5, 23), cellular anatomical entity (0, 210), protein-containing complex (2, 172), catalytic activity (26, 143), binding (22, 19), and molecular function regulator (19, 4). The bold and regular numbers in parenthesis show the numbers of enriched DEGs and DEPs in respective GO terms.
Fig. 2.
GO function analysis. (a) Annotations analysis of DEGs. (b) Annotations analysis of DEGs. (C) The GO categories assigned to DEGs and DEPs in CS and KB.
3.5. KEGG pathway enrichment analysis
The DEGs were subjected to KEGG pathways analysis to examine their potential involvement in specific metabolic pathways. A comparison of DEGs between CS and KB identified 97 KEGG pathways. The maximum DEGs mapped to phenylpropanoid biosynthesis, followed by plant hormone signal transduction, endocytosis, plant-pathogen interaction, and flavonoid biosynthesis.
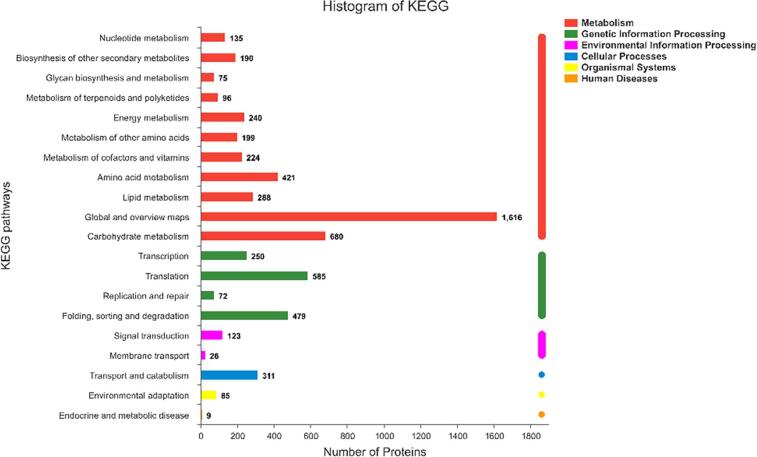
KEGG analysis assigned the DEPs to 20 pathways; of these, 11 (55.0 %) are the metabolism pathways (Fig. 3). In addition, the metabolism pathways contained eleven secondary classifications including “Nucleotide metabolism”, “Biosynthesis of other secondary metabolites”, “Glycan biosynthesis and metabolism”, “Metabolism of terpenoids and polyketides”, “Energy metabolism”, “Metabolism of other amino acids”, “Metabolism of cofactors and vitamins”, “Amino acid metabolism”, “Lipid metabolism”, “Global and overview maps” and “Carbohydrate metabolism”. These results indicate that many DEPs are linked to metabolic pathways in peanut sprouts.
Fig. 3.
Bubble chart of 20 metabolic pathways in KEGG analysis of DEPs according to the size of p-value between the CS and KB peanut sprouts.
3.6. Phenylpropanoid biosynthesis pathway
Notably, the phenylpropanoid biosynthesis metabolic pathway showed the maximum DEGs enrichment. A total of 24 DEGs were enriched, including 10 peroxidases (E1.11.1.7) (3 downregulated and 7 upregulated), 1 β-glucosidase (EC3.2.1.21; upregulated), 4 Hydrocannabinic acid synthase (all 4 upregulated), 1 mannitol dehydrogenase 2 (upregulated), 1 reticulase-like protein (upregulated), 2 4-coumarate-CoA ligase (both up-regulated), 1 spermidine hydroxylcinnamoacyltransferase (upregulated), 2 cyano β-glucosidase (1 upregulated and 1 downregulated), 2 caffeoyl-CoA O-methyltransferase (both upregulated), and 2 cinnamoyl-CoA reductase (both upregulated) (Supplementary S1).
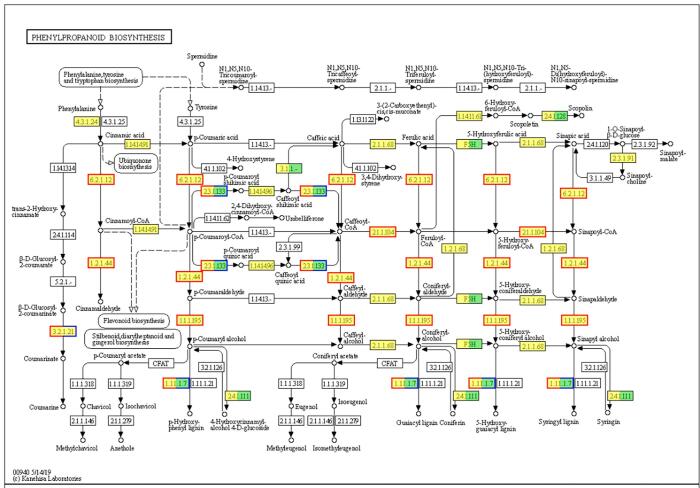
Previous studies reported (Bru, Belchi-Navarro, Pedreno, Almagro, & Martínez-Márquez, 2015) that plants produce resveratrol through the phenylpropanoid pathway using phenylpropanoid as a substrate. The transcripts identified in this study are related to the phenylpropane synthesis pathway (Map00940) (Fig. 4). The results showed that the three genes, named arahy.Tifrunner.gnm1.ann1.DXZI51, arahy.Tifrunner.gnm1.ann1.VGN2GE, and arahy.Tifrunner.gnm1.ann1.Y23DM6, were significantly up-regulated in peanut sprouts and related to the resveratrol synthesis pathway. arahy.Tifrunner.gnm1.ann1.DXZI51 is a cinnamoyl-CoA reductase, which participates in the synthesis of cinnamic acid. PHE (Phenylalanine) undergoes deamination by PAL (phenylalanine ammonia-lyase) and is then converted to cinnamic acid, which then passes through C4H (cinnamate-4-hydroxylase) mediated hydroxylation of phospholipids to produce p-coumaric acid (Cinnamoyl-COA). In this pathway, arahy.Tifrunner.gnm1.ann1.VGN2GE, and arahy.Tifrunner.gnm1.ann1.Y23DM6 are 4-coumarate--CoA ligase, which participates in the biosynthesis of coenzyme A. A previous report [5] showed that 4-coumaroyl-COA stimulated by external factors initiates the pathway of resveratrol production, which involves 1 molecule of coumarin-CoA and 3 molecules of Malonyl-COA. The reaction is catalyzed by stilbene synthase (STS) to produce 1 molecule of trans-resveratrol. It could be that ultrasound induction promotes the enrichment of resveratrol in peanut sprouts by regulating the arahy.Tifrunner.gnm1.ann1.DXZI51, arahy.Tifrunner.gnm1.ann1.VGN2GE, and arahy.Tifrunner.gnm1.ann1.Y23DM6 genes.
Fig. 4.
KEGG map of phenylpropanoid biosynthesis pathway.
The KEGG pathway database annotation of 310 proteins revealed that these are mainly involved in metabolic pathways. DEGs and DEPs involved in flavonoid biosynthesis are significant factors. Flavonoids, a group of polyphenols secondary metabolites, are widely distributed in plants and are of great interest to scientists. The evolution of flavonoids began when plants started to grow on land. They play important role in protecting plants from UV damage, reactive oxygen species, etc. To date, >8000 different flavonoid compounds have been identified (Yuan, et al.). Based on basic molecular structure, flavonoids can be divided into six categories, namely flavanones, flavones, flavonols, isoflavones, anthocyanins, and flavanols (Camargo et al., 2017, Ju, 2019). Many ancient herbal preparations contain flavonoids as the principal physiologically active constituents that are used to treat human diseases, such as inflammation, heart disease, cancer, and so on (Camargo et al., 2017, Exploration of the Effects of Different Blue LED, 2020).
As shown in Supplementary S2, five proteins were found to be involved in flavonoid metabolic process including three chalcone-flavanone isomerase (arahy.Tifrunner.gnm1.ann1.425ZXH.1, arahy.Tifrunner.gnm1.ann1.6JHV2K.1, and arahy.Tifrunner.gnm1.ann1.2GDU51.1), putative stilbene synthase (arahy.Tifrunner.gnm1.ann1.95I963.1) and stilbene synthase (arahy.Tifrunner.gnm1.ann1.5YH4B6.1). All of these were upregulated. Overall, these results suggest that ultrasound pretreatment greatly increases the functional properties of peanut sprouts.
Concerning the secondary metabolic pathways of plants, most phenolic substances such as flavonoids, lignin, and resveratrol are synthesized through the phenylpropanoid metabolic pathway (Chen et al. (2016)). The key enzymes involved in these pathways respond to external biological signals to regulate secondary metabolism during abiotic adversity. For example, the increase in PAL activity enhances the synthesis of lignin in soybean roots; the increase in licorice flavonoids is inseparable from the increased activity of PAL and C4H (Zahra, Khatereh, Maryam, & Reza, 2019). The above results indicate that ultrasound induction upregulates the genes of the phenylpropane synthesis pathway, in turn, altering the downstream products in germinating peanuts. Early experiments found that ultrasound induction significantly increases the resveratrol content of germinated peanuts. Cho, Hong, Chun, Sang, and Min (2006) used ultrasound for resveratrol enrichment in grapes involving the upregulated activity of resveratrol synthase. Likewise, Sharma et al. (Sales and Resurreccion, 2010, Sharma et al., 2022) showed that ultrasound combined with phenylpropanoid induction has a synergistic effect on the enrichment of resveratrol in peanut buds which involved upregulated activity of cinnamic acid-4-hydroxylase, coumaric acid-CoA ligase, and flavonoid 3-O-glucosyltransferase. Ling, Jia, Shao, Li, Jin, and Zheng (2018) found that ultrasonic treatment (400 W, 6 min) and 0.4% peracetic acid at 20 °C reduced the total flavonoid content in loquat fruit. The highest value was observed on the 3rd day compared to the control group. This may be because ultrasound combined with peracetic acid treatment increased the activities of polyphenol oxidase and peroxidase in loquat fruits. These two enzymes are involved in the phenylpropane pathway and oxidation process signifying a variety of structure and defense functions of phenolic compounds. In addition, Shazini, Patimah, and Asmah (2014) showed that ultrasonic treatment effectively promoted the germination of Tartary buckwheat seeds and thereby increased the enrichment of flavonoids in buckwheat sprouts. These effects can be attributed to the fact that ultrasonic treatment effectively activates the activities of various enzymes during the germination of plant seeds (Arefi-Oskoui et al., 2019, Chen et al., 2020), significantly increasing the seed germination rate, and some biologically active components (Sangronis & Machado, 2007). These changes are consistent with this study, indicating that ultrasound induction regulates genes related to phenylpropane biosynthetic pathways in sprouting peanuts.
3.7. Analysis of ultrasound-induced transcription factors in peanut sprouts
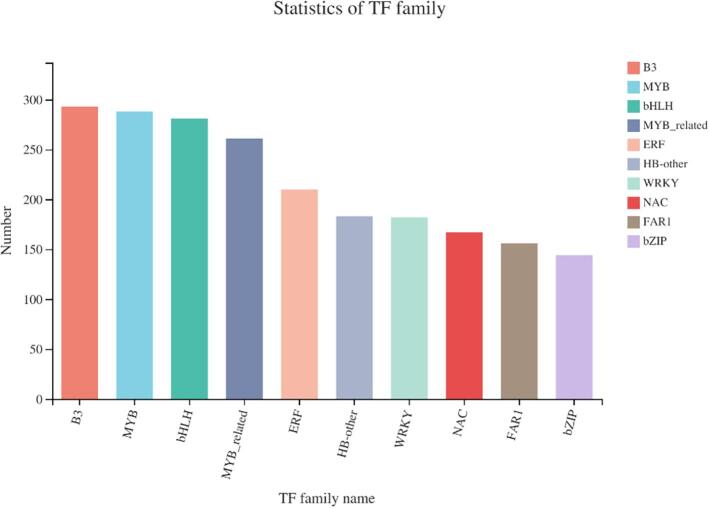
Transcription factors play an important role in regulating plant resistance to abiotic stress (Liu, Osbourn, & Ma, 2015). This study analyzed the ultrasound-induced DEGs in peanut sprouts, which belong to 48 transcription factor families (Fig. 5). Among them, the B3 family contains 293 genes, followed by the MYB family with 288, bHLH family with 281, MYB_related family with 261, and ERF family with 210 genes. This indicates that ultrasound-induced the expression of transcription factors in these families in germinating peanuts. Notably, the B3 transcription factor family showed the highest changes. Studies showed that changes in the expression of MYB, B3, WRKY, bHLH, and bZIP transcription factors can improve plants' resistance to adversity (Fan, Wang, Wen, & Guijie, 2020). The B3 transcription factors have a highly conserved structural domain that specifically binds to DNA, while the other conserved domains include AP2, auxin response factor, auxin/indole-3-acetic acid, and so on (King, Chanson, Mccallum, Ohme-Takagi, Byriel, & Hill, 2013). The bHLH transcription factor family is one of the largest gene families in plants, which is widely involved in plant metabolism, development, and stress response. The MYB transcription factor family is also one of the largest protein groups in plants (Murakami, Kakutani, Kuroyanagi, Iwai, Hori, Shimojima, & Ohta, 2020). It is widely involved in plant physiological and biochemical processes, including plant epidermal tissue cell differentiation, response to external environmental factors, and hormone response. Studies showed that the MYB transcription factors have diverse functions, which are essential for the growth and development of plants (Kim et al., 2013, Millard et al., 2019, Physiology, 2006). For example, MYB transcription factors can regulate the cell cycle by controlling the cell division period, and the synthesis of secondary metabolites such as anthocyanins and flavonoids by regulating the expression of related genes. In addition, MYB transcription factors can also regulate the response to plant hormones. Our study shows that B3, MYB, and other transcription factor-like families play a significant role in response to ultrasound treatment.
Fig. 5.
Statistics of transcription factors.
4. Conclusion
To examine the effect of ultrasonic external field induction in germinating peanuts, this study screened the prominent regulatory genes. This study selected the suitable concentration of resveratrol peanut varieties as the test material to examine the transcriptomic differences between ultrasound-induced sprouted peanut (CS) and normal sprouted peanut (KB). The results revealed that ultrasound induced the biosynthesis of phenylpropanoids in sprouted peanuts. A total of 1104 DEGs and 399 DEPs were screened from the two groups of samples. The functional classification, metabolic pathways, and biological processes for the involved genes were analyzed. The DEGs were mainly enriched in genetic information, metabolic pathways, and protein conversion. These results provide a certain reference for the regulation of the phenylpropanoid biosynthesis pathway, which can be used to regulate the nutritional and functional components of sprouted peanuts for human health benefits.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors acknowledge technical assistance and guidance from the food nutrition and quality safety team of LAAS for allowing usage of their facility. This work is supported by the Doctoral Scientific Research Foundation of Liaoning Province (2020-BS-038);2021 Liaoning Province “Revealing the List and Taking Command” Science and Technology Research Project: Nutritional Analysis and Quality Control of Peanut Processing (2020DD123502).
The authors would like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for their linguistic assistance during the preparation of this manuscript.
Contributions
Mengxi Xie designed the study, collected data, and performed analysis. Miao Yu contributed to data collection and analysis. Liangchen Zhang contributed to data collection. Taiyuan Shi contributed to study design and manuscript revisions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2022.100102.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adhikari B., Dhungana S.K., Ali M.W., Adhikari A., Kim I.D., Shin D.H. Resveratrol, total phenolic and flavonoid contents, and antioxidant potential of seeds and sprouts of Korean peanuts. Food Science and Biotechnology. 2018;27(5):1275–1284. doi: 10.1007/s10068-018-0364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefi-Oskoui, S., Khataee, A., Safarpour, M., Orooji, Y., & Vatanpour, V. J. U. S. (2019). A review on the applications of ultrasonic technology in membrane bioreactors. 58, 104633. [DOI] [PubMed]
- Bru R., Belchi-Navarro S., Pedreno M.A., Almagro L., Martínez-Márquez A. Enhanced extracellular production of trans-resveratrol in Vitis vinifera suspension cultured cells by using cyclodextrins and coronatine. Plant Physiology and Biochemistry. 2015;97:361–367. doi: 10.1016/j.plaphy.2015.10.025. [DOI] [PubMed] [Google Scholar]
- Camargo A., Regitano-D'Arce M.A.B., Rasera G.B., Canniatti-Brazaca S.G., Prado-Silva L.D., Alvarenga Verônica.O., et al. Phenolic acids and flavonoids of peanut by-products: Antioxidant capacity and antimicrobial effects. Food Chemistry. 2017;237:538. doi: 10.1016/j.foodchem.2017.05.046. [DOI] [PubMed] [Google Scholar]
- Cao L., Xu X., Chen S., Ma H. Cloning and expression analysis of Ficus carica anthocyanidin synthase 1 gene. Scientia Horticulturae. 2016 [Google Scholar]
- Chai L., Li Y., Chen S., Perl A., Zhao F., Ma H. RNA sequencing reveals high resolution expression change of major plant hormone pathway genes after young seedless grape berries treated with gibberellin. Plant Science. 2014;229:215–224. doi: 10.1016/j.plantsci.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Chen F.Y., Zhang M., Yang C.H. Application of ultrasound technology in processing of ready-to-eat fresh food: A review. Ultrasonics Sonochemistry. 2020;63 doi: 10.1016/j.ultsonch.2019.104953. [DOI] [PubMed] [Google Scholar]
- Fan F., Wang Q., Wen X., Guijie D. Transcriptome-wide identification and expression profiling of Pinus massoniana MYB transcription factors responding to phosphorus deficiency. Food Chemistry. 2020;31(03):214–224. [Google Scholar]
- Chen Y.-Y., Zhang Z.-H., Zhong C.-Y., Song X.-M., Lin Q.-H., Huang C.-M., Huang R.-H., Chen W. Functional analysis of differentially expressed proteins in Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruits during ripening. Food Chemistry. 2016;190:763–770. doi: 10.1016/j.foodchem.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Cho Y.J., Hong J.Y., Chun H.S., Sang K.L., Min H.Y. Ultrasonication-assisted extraction of resveratrol from grapes. Journal of Food Engineering. 2006;77(3):725–730. [Google Scholar]
- Exploration of the Effects of Different Blue LED Light Intensities on Flavonoid and Lipid Metabolism in Tea Plants via Transcriptomics and Metabolomics %J International Journal of Molecular Sciences. (2020). 21(13). [DOI] [PMC free article] [PubMed]
- Huang Q., Xu M., Zhang H., He D., Kong Y., Chen L., et al. Transcriptome and proteome analyses of the molecular mechanisms associated with coix seed nutritional quality in the process of breeding. Food chemistry. 2019;272:549–558. doi: 10.1016/j.foodchem.2018.07.116. [DOI] [PubMed] [Google Scholar]
- Ju J.S. Using a high-performance bioreactor system to cultivate peanut kernels for the induction of stilbenoids. Journal of the Chinese Institute of Engineers, Transactions of the Chinese Institute of Engineers, Series A. 2019;42(6):498–506. [Google Scholar]
- Kim, Y. B., Sun, J. K., Park, W. T., Kwon, D. Y., Yun, J. P., Kim, H. H., et al. (2013). Functional characterization of R2R3-MYB transcription factors in glucosinolate biosynthesis and abiotic stresses in Chinese cabbage (Brassica rapa ssp.pekinensis). 66(2), 102-116.
- King, G. J., Chanson, A. H., Mccallum, E. J., Ohme-Takagi, M., Byriel, K., Hill, J. et al. (2013). The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface. 288(5), 3198-3207. [DOI] [PMC free article] [PubMed]
- Limmongkon, A., Janhom, P., Amthong, A., Kawpanuk, M., Nopprang, P., Poohadsuan, J., et al. (2017). Antioxidant activity,total phenolic,and resveratrol content in five cultivars of peanut sprouts. (4).
- Limmongkon A., Pankam J., Somboon T., Wongshaya P., Nopprang P. Evaluation of the DNA damage protective activity of the germinated peanut (Arachis hypogaea) in relation to antioxidant and anti-inflammatory activity. LWT. 2019;101(C):259–268. [Google Scholar]
- Ling, C., Jia, X., Shao, S., Li, W., Jin, P., & Zheng, Y. J. J. o. F. Q. (2018). Effect of Ultrasonic Treatment Combined with Peracetic Acid Treatment Reduces Decay and Maintains Quality in Loquat Fruit. 2018, 1-8.
- Liu, J., Osbourn, A., & Ma, P. J. M. P. (2015). MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. 8(5), 689-708. [DOI] [PubMed]
- Lo Porto C., Ziuzina D., Los A., Boehm D., Palumbo F., Favia P., et al. Plasma activated water and airborne ultrasound treatments for enhanced germination and growth of soybean. Innovative Food Science and Emerging Technologies. 2018;49:13–19. [Google Scholar]
- Luo G., Zhao L., Xiaojin X., Qin Y., Huang L., Yongquan S., et al. Integrated dual RNA-seq and dual iTRAQ of infected tissue reveals the functions of a diguanylate cyclase gene of Pseudomonas plecoglossicida in host-pathogen interactions with Epinephelus coioides. Fish and Shellfish Immunology. 2019;95:481–490. doi: 10.1016/j.fsi.2019.11.008. [DOI] [PubMed] [Google Scholar]
- Miao Y., Hong-zhi L., Ying Y., Ai-min S., Li L., Hui H., et al. Optimising germinated conditions to enhance yield of resveratrol content in peanut sprout using response surface methodology. International Journal of Food Science and Technology. 2016;51(8):1754–1761. [Google Scholar]
- Millard, P. S., Kragelund, B., & Burow, M. J. T. i. P. S. (2019). R2R3 MYB Transcription Factors – Functions outside the DNA-Binding Domain. 24(10). [DOI] [PubMed]
- Murakami, H., Kakutani, N., Kuroyanagi, Y., Iwai, M., Hori, K., Shimojima, M., & Ohta, H. J. F. L. (2020). MYB‐like transcription factor NoPSR1 is crucial for membrane lipid remodeling under phosphate starvation in the oleaginous microalga Nannochloropsis oceanica. [DOI] [PubMed]
- Perera, C. O., Alzahrani, M. J. L.-F. S., & Technology. (2021). Ultrasound as a Pre-treatment for Extraction of Bioactive Compounds and Food Safety: A review. 142(11), 111114.
- L., & Physiology, D. J. P. (2006). Characterization of a Grapevine R2R3-MYB Transcription Factor That Regulates the Phenylpropanoid Pathway. [DOI] [PMC free article] [PubMed]
- Sales, J. M., & Resurreccion, A. J. F. C. (2010). Phenolic profile, antioxidants, and sensory acceptance of bioactive-enhanced peanuts using ultrasound and UV. 122(3), 795-803.
- Sangronis, E., Machado, C. J. J. L.-F. S., & Technology. (2007). Influence of germination on the nutritional quality of Phaseolus vulgaris and Cajanus cajan. 40(1), 116-120.
- Sharma, A., Goel, H. C., Sinha, A. K., Joshi, B. P., & Prasad, J. Ultrasound-assisted conversion of toxic beta-asarone into nontoxic bioactive phenylpropanoid: isoacoramone, a metabolite of Piper marginatum and Acorus tararinowii. [DOI] [PubMed]
- Shazini, R. N., Patimah, I., & Asmah, R. (2014). Influence of conventional and ultrasonic-assisted extraction on phenolic contents, betacyanin contents, and antioxidant capacity of red dragon fruit (Hylocereus polyrhizus). 2014, 964731. [DOI] [PMC free article] [PubMed]
- Wang G., Lei Z., Zhong Q., Wu W., Zhang H., Min T., et al. Enrichment of caffeic acid in peanut sprouts and evaluation of its in vitro effectiveness against oxidative stress-induced erythrocyte hemolysis. Food Chemistry. 2017;217:332–341. doi: 10.1016/j.foodchem.2016.07.126. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Liu H., Li H., et al. Widely targeted metabolomics characterizes the dynamic changes of chemical profile in postharvest peanut sprouts grown under the dark and light conditions. Lwt. 2021;152 [Google Scholar]
- Yu M., Liu H., Shi A., Liu L., Wang Q. Preparation of resveratrol-enriched and poor allergic protein peanut sprout from ultrasound treated peanut seeds. Ultrasonics Sonochemistry. 2016;334 doi: 10.1016/j.ultsonch.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Yu M., Liu H., Yang Y., Shi A., Liu L., Hui H., et al. Optimisation for resveratrol accumulation during peanut germination with phenylalanine feeding & ultrasound-treatment using response surface methodology. International Journal of Food Science and Technology. 2016;51(4):938–945. [Google Scholar]
- Yu M., Zhou Y., Wang X., Xie M., Zhang B., Yu H., et al. Effect of ultrasonic pre-treatment on Ara h 1 in peanut sprouts. Ultrasonics Sonochemistry. 2021;75 doi: 10.1016/j.ultsonch.2021.105607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahra K., Khatereh K., Maryam M., Reza H. Low-level laser irradiation potentiates anticancer activity of p-coumaric acid against human malignant melanoma cells. Melanoma Research. 2019 doi: 10.1097/CMR.0000000000000603. [DOI] [PubMed] [Google Scholar]
- Zhang C.W., Wei Y.P., Xiao D., Gao L.W., Lyu S.W., Hou X.L., et al. Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. Journal of Proteomics. 2016;144:1–10. doi: 10.1016/j.jprot.2016.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.