Abstract
The M1 strain, able to grow on β-myrcene as the sole carbon and energy source, was isolated by an enrichment culture and identified as a Pseudomonas sp. One β-myrcene-negative mutant, called N22, obtained by transposon mutagenesis, accumulated (E)-2-methyl-6-methylen-2,7-octadien-1-ol (or myrcen-8-ol) as a unique β-myrcene biotransformation product. This compound was identified by gas chromatography-mass spectrometry. We cloned and sequenced the DNA regions flanking the transposon and used these fragments to identify the M1 genomic library clones containing the wild-type copy of the interrupted gene. One of the selected cosmids, containing a 22-kb genomic insert, was able to complement the N22 mutant for growth on β-myrcene. A 5,370-bp-long sequence spanning the region interrupted by the transposon in the mutant was determined. We identified four open reading frames, named myrA, myrB, myrC, and myrD, which can potentially code for an aldehyde dehydrogenase, an alcohol dehydrogenase, an acyl-coenzyme A (CoA) synthetase, and an enoyl-CoA hydratase, respectively. myrA, myrB, and myrC are likely organized in an operon, since they are separated by only 19 and 36 nucleotides (nt), respectively, and no promoter-like sequences have been found in these regions. The myrD gene starts 224 nt upstream of myrA and is divergently transcribed. The myrB sequence was found to be completely identical to the one flanking the transposon in the mutant. Therefore, we could ascertain that the transposon had been inserted inside the myrB gene, in complete agreement with the accumulation of (E)-2-methyl-6-methylen-2,7-octadien-1-ol by the mutant. Based on sequence and biotransformation data, we propose a pathway for β-myrcene catabolism in Pseudomonas sp. strain M1.
Terpenes are important flavor and fragrance compounds widely distributed in nature. They are isolated from plants or chemically synthesized, but interest in processes based on microbial transformation has greatly increased during the past decade. In particular, attention has been focused on the production of oxygenated derivatives of terpenes, commonly called terpenoids, which have a stronger odor.
Several reviews concerning the opportunities for microbial transformation of terpenes have been published recently (32, 35, 48, 52–54, 57).
Many microorganisms are able to partly oxidize terpenes by cometabolism, often giving rise to the accumulation of different metabolites, which makes the purification of a single product very difficult (1, 3, 12, 15, 49). Other microorganisms, mainly belonging to the genus Pseudomonas, are able to completely mineralize terpenes, using these substrates as the only source of carbon and energy. This opens the possibility of producing single metabolites by the use of specific mutants or by cloning genes for single enzymatic activities. However, information on the genetics of terpene degradation is lacking, and only few genes have been cloned (8, 10, 34, 44, 46, 56).
β-Myrcene is an acyclic monoterpene found in the essential oils of several useful plants, such as lemongrass, hops, bay, and verbena. While this monoterpene has been extensively studied in relation to its effects on human health (19, 21, 23), its microbial metabolism has been little investigated. Basidiomycetes cometabolize β-myrcene, leading to the accumulation of several oxidated derivatives (16). The only β-myrcene-utilizing bacterium described so far is Pseudomonas putida S4-2, for which a catabolic pathway has been proposed, based on the identification of β-myrcene metabolites from the culture broth (40).
In this paper we report the characterization of a new Pseudomonas sp. strain, called M1, able to grow on β-myrcene as the sole carbon and energy source and of a β-myrcene-negative mutant. The β-myrcene catabolism genes were also identified and sequenced.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. coli | ||
| S17.1 | recA pro thi hsdR RP4-2-Tc::Mu-Km::Tn7 Tra+ Tpr Smr | 50 |
| HB101 | recA13 proA leuB6 hsdR rpsL20 (Smr) | 13 |
| CC118λpir | Δ(ara-leu) araDE ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE (Am) recA1 λpir; phage lysogen | 29 |
| DH5α | supE44 thi-1 recA1 hsdR17 endA1 gyrA (Nalr) Δ(lacIZYA-argF)U169 deoR [φ80 dlacΔ(lacZ)M15] | 27 |
| Pseudomonas sp. strain M1 | Myr+ | This study |
| P. putida NCIMB 8248 | NCIMB wild-type strainb | |
| Pseudomonas sp. strain N22 | Myr− mutant obtained by transposon mutagenesis | This study |
| pRK2013 | RK2-Tra+ RK2-Mob+ Kmrori ColE1 | 22 |
| pUT mini-Tn5 Km | Apr Kmr; ori R6K RP4 oriT delivery plasmid for mini-Tn5 Km | 20 |
| pBluescript KS(+) | Apr α-lac/MCS | Stratagene |
| pLAFR3 | Broad-host-range cosmid vector IncP1 RK2 replicon; λcos+lacZα Mob+ Tra− Tcr | 9 |
| pC7 | Tcr; pLAFR3 with a 22-kb chromosomal fragment from Pseudomonas sp. strain M1 | This study |
| pB22Sc | Apr Kmr; pBluescript KS(+) with the 7.7-kb SacI fragment containing mini-Tn5 | This study |
| pB2.3 BgSc | Apr; pBluescript KS(+) with the 2.3-kb BglII-SacI fragment from pB22Sc | This study |
| pB5.3 Sc | Apr; pBluescript KS(+) with the 5.3-kb SacI fragment from pC7 | This study |
Myr, myrcene-degradative phenotype; Apr, ampicillin resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance; MCS, multiple cloning site.
NCIMB, National Collections of Industrial and Marine Bacteria, Aberdeen, Scotland, United Kingdom.
The strains were grown at 37 (Escherichia coli) or 30°C (Pseudomonas spp.) in Luria-Bertani LB broth, M9 medium (39), or Pseudomonas standard mineral base medium (PSMBM) (18) supplemented with glucose (0.2%), sodium lactate (0.2%), or sodium succinate (0.2%). Thiamine (1 mM) and proline (40 μg/ml) were added when required. Antibiotics were routinely used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; tetracycline, 15 μg/ml. The rich medium used to store cosmid clones in 96-well microtiter plates was prepared according to the method of Gergen et al. (24). Growth of Pseudomonas sp. strain M1 on β-myrcene (Fluka) was performed under a saturated atmosphere of this compound on both solid and liquid media. Biotransformation experiments were performed in 1× M9 salts solution (50 mM Na2HPO4 · 7H2O, 20 mM KH2PO4, 8.5 mM NaCl, 20 mM NH4Cl) (37).
Isolation of Pseudomonas sp. strain M1.
Pseudomonas sp. strain M1 was isolated from an enrichment culture containing a sediment sample (10 g) from the Rhine River, Wageningen, the Netherlands. The sediment sample was diluted in 30 ml of PSMBM containing 1 mM myrcene as the sole source of carbon and energy in a serum flask with a butyl rubber stopper (130 ml). After incubating this culture for 2 weeks on a shaker at 30°C with two successive transfers (3% inoculum) into fresh medium, samples of the enrichments were plated onto PSMBM agar plates. These plates were incubated in a desiccator in which β-myrcene was supplied via the gas phase. The colonies that developed were isolated and checked for purity by plating them on yeast extract-glucose plates.
Construction of Pseudomonas sp. strain M1 genomic library.
The M1 wild-type genomic library was obtained as previously described (38) with the following modifications: chromosomal DNA was partially digested with 0.005 U of Sau3AI/μg for 1 h at 37°C, and the selected fractions were ligated in a 1:3 ratio to BamHI-digested pLAFR3. A total of 4,000 cosmid clones were stored at −80°C in 96-well microtiter plates containing rich medium supplemented with tetracycline (25 μg/ml).
Transposon mutagenesis.
Conjugal transfer of the mini-Tn5 Km transposon was performed by triparental mating. The donor strain, E. coli CC118λpir(pUT mini-Tn5 Km), was grown overnight with shaking at 37°C in LB medium containing 100 μg of ampicillin/ml and 50 μg of kanamycin/ml; E. coli HB101(pRK2013) was grown overnight in LB medium containing 50 μg of kanamycin/ml. The recipient strain, Pseudomonas sp. strain M1, was grown overnight at 30°C in LB medium. A 1.5-ml volume of each culture was harvested, and the cells were washed twice with 0.9% NaCl and resuspended in the same solution. The suspensions were combined, and the mating mix was transferred onto a 0.45-μm-pore-size cellulose filter membrane placed on the surface of an LB medium plate. After overnight incubation at 30°C, the bacteria were resuspended in 10 ml of 0.9% NaCl and plated (100 μl/plate) on appropriate selective medium. Kanamycin-resistant transconjugants were screened for the loss of the ability to grow on β-myrcene (Myr− phenotype) by replica plating.
DNA manipulation.
Plasmid preparation and restrictions and ligations were performed by standard procedures (37). Restriction endonucleases and T4 DNA ligase were from Boehringer Mannheim. DNA fragments were purified from agarose with the Qiaquick gel extraction kit (Qiagen). Purified DNA fragments were labeled with digoxigenin (DIG)-11-dUTP with the DIG DNA labeling kit (Boehringer). For Southern experiments, DNA was transferred onto positively charged nylon membrane (Boehringer) and the DIG labels were visualized with a chemiluminescence detection kit (Boehringer) according to the supplier’s instructions.
The nucleotide sequence was determined with an Applied Biosystems automated sequencer (model 373 Stretch) with a DyeDeoxy terminator cycle sequencing kit (Perkin-Elmer). Both commercially available and synthetic primers were used for sequencing reactions. The oligonucleotides corresponding to the I and O ends of mini-Tn5 were 5′-TGTCCACTACGTGAAAGGC-3′ and 5′-CGAACTTGTGTATAAGAGTCAG-3′, respectively. DNA sequences were identified by similarity searches with the BLAST 2.0 (6) and FASTA 3 (42) programs. The deduced amino acid sequences were analyzed by the pattern and profile programs at the ExPASy World Wide Web Molecular Biology Server, Swiss Institute of Bioinformatics, Geneva, Switzerland (7).
Culture conditions for metabolite analysis.
A 25-ml culture of Pseudomonas sp. strain N22 was grown overnight at 30°C with shaking in PSMBM supplemented with 0.2% sodium lactate as the carbon source. Experimental cultures were obtained by inoculating 5% (vol/vol) of the overnight culture into two flasks containing 50 ml of 0.2% lactate PSMBM, to one of which β-myrcene was supplied via the gas phase. When the cultures reached an optical density at 600 nm of 1.5, the cells were harvested by centrifugation at 4,000 rpm and washed twice with 1× M9 salts solution. Finally, the pellets were resuspended in an appropriate volume of the same salts solution to an optical density at 600 nm of 10. β-Myrcene was directly added (1 μl/ml) to one of the two cell suspensions, and both flasks were incubated at 30°C for 1 h. As a control, a flask containing only 1× M9 salts solution and 1 μl of β-myrcene/ml was incubated under the same conditions.
Identification of the metabolites.
Cells were separated by centrifugation at 8,000 rpm for 15 min. The cell-free solution was saturated with NaCl and extracted three times with one-third volume of a hexane-ether mixture (1:1), and the combined extracts were dried over anhydrous sodium sulfate and concentrated to a volume of 1 ml with a rotative evaporator (20 mm of Hg; bath temperature, 25°C).
Analysis of the extracts was performed with a gas chromatograph (GC) (Fisons GC 8000) coupled with a mass spectrometer (MS) (Fisons MD 800). A methyl-phenyl silicon (SE 52 MS) capillary column ([inside diameter]; 30 m by 0.25 mm; MEGA) was used, with a temperature program of 50°C for the first 4 min to 250°C at a heating rate of 10°C/min and a flow rate of the carrier gas, helium, of 1 ml per min; 1 μl was injected at 200°C in the splitless mode. Mass spectra were recorded in electron impact ionization mode at 70 eV by scanning the mass range of 30 to 450 m/z.
Nucleotide sequence accession number.
The nucleotide sequence of 5,370 bp has been deposited in GenBank under accession no. AF112883.
RESULTS
Characterization of strain M1.
The M1 strain was one of the five microorganisms isolated from an enrichment culture, as described in Materials and Methods. It is an oxidase-positive, gram-negative, motile, rod-shaped bacterium identified as Pseudomonas sp. by the API 20NE system.
The maximum growth rate on β-myrcene was obtained by using β-myrcene-saturated PSMBM inoculated with 5% (vol/vol) of an overnight culture grown on the same medium containing 0.2% lactate. Under these conditions, the lag phase lasted approximately 2 h and the duplication time during the exponential growth phase was about 1 h 35 min. Since the M1 strain was newly isolated, we investigated the characteristics that are important to carry out genetic and physiological studies.
The stability of the β-myrcene-positive phenotype was studied by replica plating after growth on lactate or glucose for 20 generations, and we found that it was 100% stable. This is an important result in view of the possibility of isolating mutants unable to grow on β-myrcene.
The antibiotic sensitivity test showed that the M1 strain is sensitive to kanamycin (20 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (25 μg/ml), whose resistance genes are the genetic markers of the majority of the Pseudomonas vectors. The M1 strain was also tested as the recipient strain in conjugation experiments by using E. coli S17.1 containing the pLAFR3 cosmid as the donor strain. The resulting transconjugants contained a plasmid whose restriction pattern was identical to the original one. As a whole, the results obtained indicated that this newly isolated strain could be manipulated by the classical genetic and molecular techniques.
Isolation and characterization of β-myrcene-negative mutants.
In order to characterize the genes of the β-myrcene metabolic pathway, transposon mutagenesis was used to generate mutants unable to grow on β-myrcene.
Conjugal transfer of the mini-Tn5 Km transposon from E. coli CC118λpir into Pseudomonas sp. strain M1 was performed by triparental mating with the helper plasmid pRK2013, as described in Materials and Methods. Transconjugants were selected on glucose PSMBM in the presence of kanamycin, the selective marker of the transposon. By a screening of 6,000 transconjugants for growth on β-myrcene, we have isolated two mutants, named N22 and N59, unable to grow on β-myrcene and three leaky mutants whose growth on this substrate was very poor.
The N22 mutant was further analyzed for the accumulation of intermediates of β-myrcene catabolism by biotransformation experiments.
β-Myrcene biotransformation by N22 mutant.
N22 resting cells, prepared as described in Materials and Methods, were incubated in the presence and in the absence of β-myrcene for 1 h at 30°C. Control experiments were performed by incubating β-myrcene-containing 1× M9 salts solution without cells. Extracts were then analyzed by GC-MS. While β-myrcene did not undergo spontaneous transformation in the absence of cells (data not shown), a comparison of the chromatograms of the N22 extracts showed a single and abundant difference peak at a retention time of 14.24 min, present only in the extracts from the sample containing β-myrcene (Fig. 1A and B).
FIG. 1.
GC-MS chromatograms of the N22 biotransformation extracts performed in the presence (A) and in the absence (B) of β-myrcene. (C) Mass spectrum of the (E)-2-methyl-6-methylen-2,7-octadien-1-ol metabolite (the boxed peak in panel A). rt, retention time.
The mass spectrum of the peak at 14.24 min (Fig. 1C) showed a weak parent peak (152 m/z; 0.24%) and the first significant fragment, derived by the loss of water (134 m/z; 17%), consistent with the presence of an alcoholic function that, due to the existence of the M-31 fragment (121 m/z; 9%), should be primary. The residual fragmentation pattern was almost superimposable on that of β-myrcene itself. The base peak at 93 m/z (100%) and the peak at 79 m/z (45%) are in agreement with the formation of carbocationic fragments derived from the loss of 3- and 4-carbon alcoholic subunits from the isopropyl moiety of the molecule.
These data suggested that the likely structure could be (Z)- or (E)-2-methyl-6-methylen-2,7-octadien-1-ol (or myrcen-8-ol), derived from β-myrcene by oxidation with oxygen insertion in the C-8–H position.
Definitive structure and (E) stereochemistry were assigned to the metabolite by comparison of its GC retention time and MS spectrum with that of an authentic sample of (E)-2-methyl-6-methylen-2,7-octadien-1-ol obtained by oxidation of myrcene with SeO2 (14). The mass spectra and retention times of the two alcohols were identical within the limits of experimental error (not shown).
Cloning of the N22 chromosomal DNA regions flanking the mini-Tn5 insertion.
The insertion of the mini-Tn5 transposon into the N22 chromosome was verified by Southern blot analysis of the chromosomal DNA digested with SacI, XhoI, and ClaI, restriction enzymes which do not cut inside the transposon. The probe used was almost all of mini-Tn5, as a 2,260-bp HindIII fragment from pUT mini-Tn5 Km. Only one hybridization band was present in each digestion, indicating that a single insertion event had occurred (data not shown).
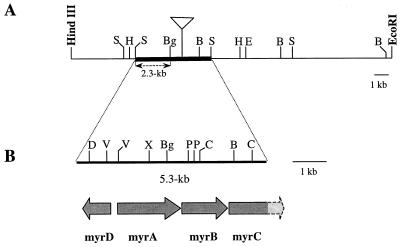
To clone the DNA fragment containing the transposon and its flanking regions, we constructed a minilibrary of N22 chromosomal DNA digested with SacI, using the pBluescript KS(+) plasmid as the cloning vector and E. coli DH5α as the host strain. Transformants containing the transposon were selected on kanamycin and ampicillin plates. All the transformants analyzed contained the same SacI fragment of approximately 7.7 kb. The restriction map of this fragment showed that it contains a chromosomal region of 3.1 kb upstream and one of 2.2 kb downstream of the transposon. The 2.3-kb SacI-BglII fragment from the upstream region (shown in Fig. 2A) was used as a probe in Southern blot analysis of the SacI-digested chromosomal DNAs from the M1 and N22 strains. The results obtained are in complete agreement with what was expected. In fact, only one hybridization band was present in M1, whose size (5.3 kb) corresponds to the sum of the sizes of the two chromosomal regions flanking the transposon in the 7.7-kb SacI fragment. In N22, the hybridization band was approximately 2.4 kb longer, consistent with the insertion of the mini-Tn5 transposon (data not shown).
FIG. 2.
(A) Restriction map of the 22-kb genomic insert of the pC7 gene bank clone. The boldface EcoRI and HindIII sites are in the polylinker of the pLAFR3 cosmid vector. The insertion site of the mini-Tn5 transposon in the N22 mutant chromosome is indicated by the open triangle. The 2.3-kb SacI-BglII fragment used as a probe in the hybridization experiments is also indicated. (B) Restriction map of the sequenced 5.3-kb SacI fragment. The arrows indicate the transcription direction of the four open reading frames identified. The light-grey dashed end of myrC indicates the unsequenced 3′ end of this gene. B, BamHI; Bg, BglII; C, ClaI; D, DraI; E, EcoRI; V, EcoRV; H, HindIII; P, PstI; S, SacI; X, XhoI.
When the mini-Tn5 probe was used, in N22 the hybridization band had the same size as that obtained with the 2.3-kb SacI-BglII probe, while no signal was detected in M1 (data not shown).
These data confirm that the cloned 7.7-kb fragment corresponds to the wild-type chromosomal region interrupted by the transposon in the mutant.
Identification of the gene interrupted by the transposon in the N22 mutant.
The transposon-containing 7.7-kb SacI fragment was used to identify the disrupted gene of the mutant. We sequenced the mini-Tn5 flanking regions with two oligonucleotides corresponding to the I and O ends of the transposon, respectively (see Materials and Methods). We obtained two sequences of approximately 300 nt each, and a comparison analysis of the deduced amino acid sequences against protein databases revealed a high level of similarity to those of alcohol dehydrogenases (see below). This result is in complete agreement with the accumulation of (E)-2-methyl-6-methylen-2,7-octadien-1-ol by the N22 mutant.
Construction of an M1 genomic library and identification of gene bank clones containing the β-myrcene degradative genes.
In order to obtain the wild-type copy of the gene mutagenized by the mini-Tn5 transposon in the N22 mutant, we constructed a genomic library of strain M1 as described in Materials and Methods. We obtained approximately 4,000 clones containing genomic inserts with an average size of 23 kb, which means that the M1 chromosome is represented about three times in the genomic library. Colony hybridization experiments were performed on the gene bank clones, using the 2.3-kb SacI-BglII fragment as a probe (Fig. 2A). We identified four positive clones containing the same 5.3-kb SacI fragment, in agreement with the size of the hybridization band found in M1. The restriction map of the recombinant cosmid (called pC7) of one of these clones is shown in Fig. 2A.
The conjugal transfer of this recombinant clone into the Myr− mutant N22 restored the β-myrcene-positive phenotype. Moreover, pC7 cosmid, transferred by conjugation into P. putida NCIMB 8248, which is unable to grow on β-myrcene, conferred the ability to grow on this substrate on the transconjugants.
Sequencing of the M1 5.3-kb SacI fragment.
The 5.3-kb SacI fragment was subcloned and sequenced. The entire fragment was found to be 5,370 bp long. Sequence analysis showed the presence of three open reading frames, named myrA, myrB, and myrC, localized on one DNA strand and one open reading frame, named myrD, on the opposite strand (Fig. 2B). The start codon of each open reading frame is preceded by a potential ribosomal binding site. The first open reading frame (myrA) shows a GTG start codon at nt 1338 and ends at nt 2798. It could encode a protein of 486 amino acids (aa) (with a theoretical molecular mass of 54.3 kDa). A database search of proteins related to MyrA revealed a significant similarity to prokaryotic and eukaryotic aldehyde dehydrogenases (39 to 48% identity in the first 10 best scores). The highly conserved glutamic acid and cysteine residues implicated in the catalytic activity (4) are located at positions 232 and 266, respectively, in MyrA. The second open reading frame (myrB), extending from nt 2818 to 3921, could encode a polypeptide of 367 aa with an estimated molecular mass of 38 kDa. MyrB showed homology with several alcohol dehydrogenases. The best scores (31 to 46% identity) were with zinc-containing long-chain alcohol dehydrogenases (43, 51), and a protein ScanProsite search (7) showed the presence of a zinc-binding signature at positions 60 to 74. XylB and TerpD alcohol dehydrogenases, involved in the degradation of xylenes (28) and α-terpineol (44), respectively, showed 45% identity with MyrB. The myrB sequence was found to be completely identical to the one flanking the transposon in the N22 mutant. Therefore, we could ascertain that the transposon had been inserted between nt 3142 and 3143, inside the myrB gene, which, according to the homologies found, could code for an alcohol dehydrogenase. The third open reading frame, myrC, starts at position 3958 and extends to the end of the clone without encountering a stop codon, giving a truncated protein of 471 amino acid residues. Comparison of MyrC with protein databases showed similarity (26 to 31% amino acid identity) with long-chain fatty-acid CoA ligases (EC 6.2.1.3) and 4-coumarate CoA ligases (EC 6.2.1.12). Actually, the best score (47% identity) was with Mycobacterium tuberculosis fadD4 protein (AC Z92669), which in turn shares 30% identity with 4-coumarate CoA ligase (P41636). All these enzymes contain the putative AMP-binding domain signature (36, 55), which extends from position 164 to 175 in the MyrC sequence. On the complementary strand, we found an open reading frame which starts at nt 1113 and stops at nt 277 with two stop codons. The resulting polypeptide of 277 aa, MyrD (calculated molecular mass, 30.4 kDa), belongs to the enoyl-CoA hydratase (ECH) family, including an ECHH (for enoyl-CoA hydratase homolog) from Rhodobacter capsulatus (31.8% identity; AC P24162), possibly involved in fatty acid oxidation (11), and the paaG product (33.8% identity; AC P77467) and PhaB (30.7% identity; AC AF029714) from E. coli K-12 and P. putida U (41), respectively, both involved in the phenylacetic acid catabolic pathway. Furthermore, a protein ProfileScan search (7) showed the presence of the ECH family signature pattern: the conserved region rich in glycine and hydrophobic residues extending from positions 116 to 202 in the MyrD sequence. On the same strand, downstream of myrD, we identified the 5′ end of a possible open reading frame (nt 117 to 1). In fact, the amino acid sequence deduced from this truncated open reading frame revealed a high homology with the N terminus of positive regulators belonging to the LysR family (47). The best score (97% identity) was with the putative regulatory protein BphR (AC D38633 [unpublished]) from the soil bacterium Pseudomonas sp. strain KKS102, which has biphenyl- and polychlorinated biphenyl-degrading activities (33).
DISCUSSION
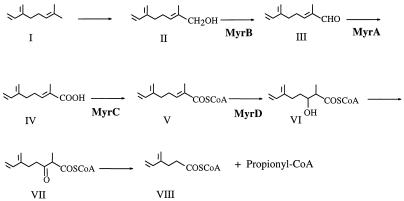
This study provides new information on β-myrcene catabolism genes which had not been previously described. We have identified four open reading frames which potentially code for an alcohol dehydrogenase, an aldehyde dehydrogenase, an acyl-CoA synthetase, and an enoyl-CoA hydratase. The relationship between these genes and β-myrcene catabolism is strongly suggested by the following facts: (i) the pC7 M1 gene bank cosmid, containing the corresponding genes, restored the wild-type Myr+ phenotype when transferred into the Pseudomonas sp. strain N22 mutant, (ii) the same gene bank clone conferred the ability to grow on β-myrcene on a P. putida strain unable to utilize this substrate, and (iii) the disruption of the alcohol dehydrogenase gene in the mutant N22 led to the accumulation of the β-myrcene oxidized derivative (E)-2-methyl-6-methylen-2,7-octadien-1-ol. This compound was identified by the comparison of its MS spectrum with that of the authentic chemically synthesized compound. Although (E)- and/or (Z)-myrcen-8-ols have already been found in insect pheromones and essential oils and recognized by their mass spectra (5, 26, 30, 31), we failed to find any reported mass spectral data in the literature. To our knowledge, this is the first report of the mass spectrum of 2-methyl-6-methylen-2,7-octadien-1-ol. Based on this evidence, we propose the metabolic pathway of β-myrcene shown in Fig. 3. The β-myrcene (Fig. 3, I) hydroxylation to 2-methyl-6-methylen-2,7-octadien-1-ol (Fig. 3, II) is demonstrated by the accumulation of this compound by the mutant, while the subsequent proposed degradation to 2-methyl-3-hydroxy-6-methylen-7-octenoyl-CoA (Fig. 3, VI) via β-oxidation is based on sequencing data. Experimental evidence for 2-methyl-3-keto-6-methylen-7-octenoyl-CoA (Fig. 3, VII) and 4-methylen-5-hexenoic acid (Fig. 3, VIII) formation is lacking. However, the last compound and 2-methyl-6-methylen-2,7-octadienoic acid (Fig. 3, IV) have been identified as β-myrcene metabolites in the β-myrcene-degrading P. putida S4-2 strain (40). Studies of biotransformation of β-myrcene with fungi showed the formation of a series of oxidized compounds derived from hydration or epoxydation at different double bonds, and no hydroxylation of the terminal carbon could be observed (16). Formation of mixtures of (E)- and (Z)-myrcen-8-ols, together with other minor components, have been described in Nocardia and Streptomyces and in fungi only when the dienyl moiety of β-myrcene was protected by a dienophile, such as sulfur dioxide (2). Up to now, the β-myrcene catabolic pathway proposed here has been found exclusively in M1 and S4-2, the only two Pseudomonas strains in which β-myrcene catabolism has been studied.
FIG. 3.
Proposed catabolic pathway of β-myrcene. I, β-myrcene (7-methyl-3-methylen-1,6-octadien); II, 2-methyl-6-methylen-2,7-octadien-1-ol; III, 2-methyl-6-methylen-2,7-octadien-1-al; IV, 2-methyl-6-methylen-2,7-octadienoic acid; V, 2-methyl-6-methylen-2,7-octadienoyl-CoA; VI, 2-methyl-3-hydroxy-6-methylen-7-octenoyl-CoA; VII, 2-methyl-3-keto-6-methylen-7-octenoyl-CoA; VIII, 4-methylen-5-hexenoyl-CoA. MyrB, alcohol dehydrogenase; MyrA, aldehyde dehydrogenase; MyrC, CoA ligase; MyrD, enoyl-CoA hydratase.
As far as the organization of the catabolic genes is concerned, myrA, myrB, and myrC should be organized in an operon, since they are separated by only 19 and 36 nt, respectively, and no promoter-like sequences have been found in these regions. The myrD gene starts 224 nt upstream of myrA and is divergently transcribed. In the DNA region between these two genes we have found two stretches of sequence (nt 1181 to 1200 and 1271 to 1290) with an imperfect dyad symmetry, containing the proposed LysR motif (5′-T-N11-A-3′), involved in specific binding to LysR proteins (25). Interestingly, a LysR-like regulator is probably present downstream of myrD, and it could be involved in the expression of the catabolic genes. In the sequenced fragment we did not find the monooxygenase gene responsible for the conversion of β-myrcene into (E)-myrcen-8-ol. However, this gene could be contained in the pC7 gene bank cosmid, which confers the ability to grow on β-myrcene on P. putida NCIMB 8248, even if the expression of a related activity encoded by the host strain cannot be ruled out.
The N22 mutant is able to accumulate (E)-myrcen-8-ol. This compound has been used as a synthon for the production of zoapatanol or its derivatives (17), chemicals with antifertility activity obtained from plants belonging to the genus Montanoa (45). Although terpenoids containing terminal allylic alcohols [such as (E)-myrcen-8-ol] can be obtained by chemical synthesis, this oxidation step requires very toxic chemicals, difficult to separate from the product. In contrast, biotransformations have the advantage of proceeding under milder and safer conditions. Moreover, both the N22 mutant and the genes isolated in this study could represent an important opportunity for the production of fine chemicals, due to the low substrate specificity of many Pseudomonas spp. catabolic enzymes.
ACKNOWLEDGMENTS
This work was supported by a grant from the European Community (BIO4-CT95-0049).
We thank D. Leak (Imperial College of Science Technology and Medicine, London, England) for providing P. putida NCIMB 8248.
REFERENCES
- 1.Abraham W R, Hoffmann H M R, Kieslich K, Reng G, Stumpf B. Microbial transformations of some monoterpenoids and sesquiterpenoids. Ciba Found Symp. 1985;111:146–160. doi: 10.1002/9780470720929.ch11. [DOI] [PubMed] [Google Scholar]
- 2.Abraham W R, Arfmaan H A. Microbial hydroxylation of activated acyclic monoterpene hydrocarbons. Tetrahedron. 1992;48:6681–6686. [Google Scholar]
- 3.Abraham W R. Phylogenetic influences in microbial hydroxylation of terpenoids. World J Microbiol Biotechnol. 1994;10:88–92. doi: 10.1007/BF00357570. [DOI] [PubMed] [Google Scholar]
- 4.Abriola D P, Fields R, Stein S, MacKarrel A D, Jr, Pietruszko R. Active site of human liver dehydrogenase. Biochemistry. 1987;26:5679–5684. doi: 10.1021/bi00392a015. [DOI] [PubMed] [Google Scholar]
- 5.Adzet T, Canigueral S, Gabalda N, Ibanez C, Tomas X, Vila R. Composition and variability of the essential oil of Thymus willkomii. Phytochemistry. 1991;30:2289–2293. [Google Scholar]
- 6.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 8.Aramaki H, Koga H, Sagara Y, Hosoi M, Horiuchi T. Complete nucleotide sequence of the 5-exo-hydroxycamphor dehydrogenase gene on the CAM plasmid of Pseudomonas putida (ATCC 17453) Biochim Biophys Acta. 1993;1174:91–94. doi: 10.1016/0167-4781(93)90098-x. [DOI] [PubMed] [Google Scholar]
- 9.Bally M, Wretlind B, Lazdunski A. Protein secretion in Pseudomonas aeruginosa: molecular cloning and characterization of the xcp-1 gene. J Bacteriol. 1989;171:4342–4348. doi: 10.1128/jb.171.8.4342-4348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbirato F, Verdoes J C, de Bont J A, van der Werf M J. The Rhodococcus erythropolis DCL14 limonene-1,2-epoxide hydrogenase gene encodes an enzyme belonging to a novel class of epoxide hydrolases. FEBS Lett. 1998;438:293–296. doi: 10.1016/s0014-5793(98)01322-2. [DOI] [PubMed] [Google Scholar]
- 11.Beckman D L, Kranz R G. A bacterial homolog to the mitochondrial enoyl-CoA hydratase. Gene. 1991;107:171–172. doi: 10.1016/0378-1119(91)90313-z. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya P K, Prema B R, Kulkarni B D, Pradham S K. Microbial transformation of terpens: hydroxylation of α-pinene. Nature. 1960;187:689–690. doi: 10.1038/187689b0. [DOI] [PubMed] [Google Scholar]
- 13.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 14.Büchi G, Wüest H. Eine Synthese des Beta-Sinensals. Helv Chim Acta. 1967;50:2440–2445. [Google Scholar]
- 15.Busmann D, Berger R G. Oxyfunctionalization of α- and β-pinene by selected basidiomycetes. Z Naturforsch. 1994;49:545–552. [Google Scholar]
- 16.Busmann D, Berger R G. Conversion of myrcene by submerged cultures of basidiomycetes. J Biotechnol. 1994;37:39–43. [Google Scholar]
- 17.Chen, R. June 1978. U.S. patent 4177194.
- 18.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetics studies of pigment synthesis by non-sulphur purple bacteria. J Cell Comp Physiol. 1957;44:25–28. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 19.Delgado I F, Nogueira A C, Souza C A, Costa A M, Figueiredo L H, Mattos A P, Chahoud I, Paumgartten F J. Peri- and postnatal developmental toxicity of beta-myrcene in the rat. Food Chem Toxicol. 1993;31:623–628. doi: 10.1016/0278-6915(93)90044-y. [DOI] [PubMed] [Google Scholar]
- 20.De Lorenzo V, Herrero M, Jakubziz U, Timmis K. Mini-Tn5 transposon derivatives for insertion mutagenesis promoter probing and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Oliveira A C, Ribeiro-Pinto L F, Paumgartten J R. In vitro inhibition of CYP2B1 monooxygenase by beta-myrcene and other monoterpenoid compounds. Toxicol Lett. 1997;92:39–46. doi: 10.1016/s0378-4274(97)00034-9. [DOI] [PubMed] [Google Scholar]
- 22.Figurski D H, Helinski D R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freitas J C, Presgrave O A, Fingola F F, Menezes M A, Paumgartten F J. Effect of beta-myrcene on pentobarbital sleeping time. Braz J Med Biol Res. 1993;26:519–523. [PubMed] [Google Scholar]
- 24.Gergen P J, Stern R H, Wensink P C. Filter replicas and permanent collection of recombinant DNA plasmids. Nucleic Acids Res. 1979;7:2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goethals K, van Montagu M, Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurudutt K N, Naik J P, Srinivas P, Ravindranath B. Volatile constituents of large Cardamon (Amomum subulatum Roxb) Flavour Fragr J. 1996;11:7–9. [Google Scholar]
- 27.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 28.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWWO from Pseudomonas putida and identification of gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrero M, De Lorenzo V, Timmis K M. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt D W A, Borden J H. Terpene alcohol pheromone production by Dendroctonus ponderosae and Ips paraconfusus (Coleoptera: Scolytidae) in the absence of readily culturable microorganism. J Chem Ecol. 1989;15:1433–1463. doi: 10.1007/BF01012375. [DOI] [PubMed] [Google Scholar]
- 31.Ivarsson P, Birgersson G. Regulation and biosynthesis of pheromone components in the double spined bark beetle Ips duplicatus (Coleoptera: Scolytidae) J Insect Physiol. 1995;41:843–849. [Google Scholar]
- 32.Janssens L, De Pooter H L, Schamp N M, Vandamme E J. Production of flavours by microorganism. Proc Biochem. 1992;27:195–198. [Google Scholar]
- 33.Kikuchi Y, Yasukochi Y, Nagata Y, Fukuda M, Takagi M. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J Bacteriol. 1994;176:4269–4276. doi: 10.1128/jb.176.14.4269-4276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koga H, Yamaguchi E, Matsunaga K, Aramaki H, Horiuchi T. Cloning and nucleotide sequences of NADH-putidaredoxin reductase gene (camA) and putidaredoxin gene (camB) involved in cytochrome P-450cam hydroxylase of Pseudomonas putida. J Biochem. 1989;106:831–836. doi: 10.1093/oxfordjournals.jbchem.a122939. [DOI] [PubMed] [Google Scholar]
- 35.Krings U, Berger R G. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol. 1998;49:1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- 36.Mallonee D H, Adams J L, Hylemon P B. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 38.Marconi A M, Beltrametti F, Bestetti G, Solinas F, Ruzzi M, Galli E, Zennaro E. Cloning and characterization of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1996;62:121–127. doi: 10.1128/aem.62.1.121-127.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Narushima H, Omori T, Minoda Y. Microbial oxidation of beta-myrcene. In: Verina C, Singh K, editors. Advances in biotechnology. Vol. 3. Oxford, England: Pergamon Press; 1982. pp. 525–531. [Google Scholar]
- 41.Olivera E R, Miñambres B, García B, Muñiz C, Moreno M A, Ferrández A, Diaz E, García J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson N R. Rapid and sensitive sequence comparison with FASTAP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 43.Persson B, Hallborn J, Walfridsson M, Hahn-Hagerdal B, Keranen S, Penttila M, Jornvall H. Dual relationships of xylitol and alcohol dehydrogenases in families of two protein types. FEBS Lett. 1993;324:9–14. doi: 10.1016/0014-5793(93)81522-2. [DOI] [PubMed] [Google Scholar]
- 44.Peterson J A, Lu J Y, Geisselsoder J, Graham-Lorence S, Carmona C, Witney F, Lorence M C. Cytochrome P-450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J Biol Chem. 1992;267:14193–14203. [PubMed] [Google Scholar]
- 45.Ponce-Monter H, Campos-Lara G, Pedron N, De la Torre L, Villaneuva T, Gallegos A, Romo de Vivar A, Azpeitia E, Perez A. The zoapatle. XV. Activity of 16 alpha-hydroxy-ent-kauran-19-oic acid isolated from Montanoa hibiscifolia, and its methyl ester on rat and guinea pig uterus. J Ethnopharmacol. 1988;24:127–134. doi: 10.1016/0378-8741(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 46.Ropp J D, Gunsalus I C, Sligar S G. Cloning and expression of a member of a new cytochrome P-450 family: cytochrome P-450lin (CYP111) from Pseudomonas incognita. J Bacteriol. 1993;175:6028–6037. doi: 10.1128/jb.175.18.6028-6037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 48.Seitz E W. Fermentation production of pyrazines and terpenoids for flavors and fragrances. In: Gabelman A, editor. Bioprocess production of flavor, fragrance, and color ingredients. New York, N.Y: John Wiley & Sons; 1994. pp. 95–136. [Google Scholar]
- 49.Shukla O P, Bhattacharyya P K. Microbiological transformation of terpenes. Part XI. Pathways of degradation of α- and β-pinenes in a soil pseudomonad (PL strain) Indian J Biochem. 1968;5:92–101. [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 51.Sun H W, Plapp B V. Progressive sequence alignment and molecular evolution of Zn-containing alcohol-dehydrogenase family. J Mol Evol. 1992;34:522–535. doi: 10.1007/BF00160465. [DOI] [PubMed] [Google Scholar]
- 52.Trudgill P W. Terpenoid metabolism by Pseudomonas. In: Sokatch J R, editor. The bacteria, a treatise on structure and function. X. New York, N.Y: Academic Press; 1986. pp. 483–525. [Google Scholar]
- 53.Trudgill P W. Microbial metabolism of monoterpenes—recent developments. Biodegradation. 1990;1:93–105. doi: 10.1007/BF00058829. [DOI] [PubMed] [Google Scholar]
- 54.Trudgill P W. Microbial metabolism and transformation of selected monoterpenes. In: Ratledge C, editor. Biochemistry of microbial degradation. Dordrecht, The Netherlands: Kluwer; 1994. pp. 33–61. [Google Scholar]
- 55.Turgay K, Krause M, Marahiel M A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 56.Unger B P, Gunsalus I C, Sligar S G. Nucleotide sequence of the Pseudomonas putida cytocrome P-450cam gene and its expression in Escherichia coli. J Biol Chem. 1986;261:1158–1163. [PubMed] [Google Scholar]
- 57.Van der Werf M J, De Bont J A M, Leak D J. Opportunities in microbial biotransformation of monoterpenes. Adv Biochem Eng Biotechnol. 1997;55:147–177. [Google Scholar]