Abstract
MicroRNAs (miRNAs) are evolutionarily conserved small non-coding RNAs, that are involved in the multistep process of carcinogenesis, contributing to all established hallmarks of cancer. In this review, implications of miRNAs in hematological malignancies and their clinical utilization fields are discussed. As components of the complex regulatory network of gene expression, influenced by the tissue microenvironment and epigenetic modifiers, miRNAs are “micromanagers” of all physiological processes including the regulation of hematopoiesis and metabolic pathways. Dysregulated miRNA expression levels contribute to both the initiation and progression of acute leukemias, the metabolic reprogramming of malignantly transformed hematopoietic precursors, and to the development of chemoresistance. Since they are highly stable and can be easily quantified in body fluids and tissue specimens, miRNAs are promising biomarkers for the early detection of hematological malignancies. Besides novel opportunities for differential diagnosis, miRNAs can contribute to advanced chemoresistance prediction and prognostic stratification of acute leukemias. Synthetic oligonucleotides and delivery vehicles aim the therapeutic modulation of miRNA expression levels. However, major challenges such as efficient delivery to specific locations, differences of miRNA expression patterns between pediatric and adult hematological malignancies, and potential side effects of miRNA-based therapies should be considered.
Keywords: microRNA, acute leukemia, carcinogenesis, differential diagnosis, chemoresistance, prognostic stratification, biomarker, precision oncology, system biology approach
1. Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, single-stranded non-coding RNAs that are 18 to 25 nucleotides in length [1,2]. The first identified miRNA is transcribed from the Caenorhabditis elegans lin-4 locus. The first mammalian miRNA let-7, was discovered seven years later in 2000 [3]. The biogenesis of miRNAs is a multistep process, including the transcription of primary double-stranded transcripts, nuclear processing by the RNase III endonuclease enzyme Drosha, nucleocytoplasmic export by Exportin-5, cytoplasmic cleavage by Dicer, and formation of the RNA-induced silencing complex (RISC) with Argonaute (Ago) proteins [4]. miRNAs are posttranscriptional regulators of gene expression, that function primarily by binding to the 3′ untranslated regions (3′UTR) of their target messenger RNAs (mRNAs), resulting in the inhibition of translation [5,6]. Depending on the microenvironment, miRNAs can target multiple mRNAs, and a single mRNA can also be targeted by several different miRNAs [7]. miRNAs are involved in a wide range of biological processes such as hematopoiesis [8], while dysregulation of miRNA expression levels plays pivotal roles in numerous diseases [6]. The finding that approximately 50 percent of human miRNAs are located at cancer-associated genomic regions (CAGR) suggests that miRNAs are important puzzle-pieces in the pathogenesis of cancer [9].
In this review, causes and consequences of dysregulated miRNA expression patterns are discussed in hematological malignant diseases, including implication of miRNAs in the well-established hallmarks of cancer and widespread interactions of miRNAs with epigenetic modifiers. Special emphasis is placed on the applications of miRNAs in the differential diagnosis and prognostic stratification of acute leukemias. miRNAs are also novel biomarkers for the prediction of chemoresistance, while modulation of miRNA expression levels provides novel therapeutic opportunities and challenges in hematological malignancies.
2. Implication of miRNAs in Carcinogenesis and Leukemogenesis
2.1. Dysregulation of miRNA Expression Levels in Cancer
The first evidence of miRNA involvement in cancer was published by Calin et al. in 2002, in a study of chronic lymphocytic leukemia (CLL), describing miR-15 and miR-16 as frequently deleted anti-oncomiRs encoded in chromosomal region 13q14 [10]. During the previous twenty years, growing amount of evidence confirmed that expression signatures of miRNAs are associated with types and grades of tumors [11]. Moreover, evaluation of cancer-specific miRNA patterns is supposed to be a more accurate method of classifying cancer subtypes compared to mRNA signatures [12], especially in case of poorly differentiated tumors [13].
Dysregulation of miRNA expression levels is a hallmark of hematological malignancies [12], that is associated with distinct subtypes of acute leukemia, featured by characteristic cytogenetic and molecular genetic alterations. In pediatric acute lymphoblastic leukemia (ALL), seven different subtypes were identified based on miRNA expression profiles, such as down-regulation of miR-127 and miR-143 in precursor B-cell ALL [14]. In patients with acute myeloid leukemia (AML), up-regulation of miR-126 was associated with inv(16)(p13q22) [15], while high expression level of miR-10 was characteristic to the subgroup of NPM1-mutated patients [16]. Altered miRNA expression levels are involved in the pathogenesis of malignant hematological diseases, such as miR-128a and miR-130 were identified in 2021 as downstream targets of MLL-AF4 fusion oncoprotein, that drive the transition from a pre-leukemic stage to acute leukemia in a murine model [17,18]. MiR-192 was confirmed to play a key role in the pathogenesis of CLL through the regulation of Bcl-2, p21, and p53 [19].
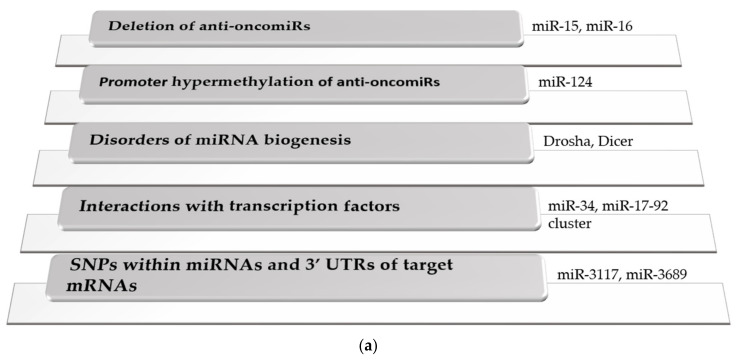
According to recent studies, changes in miRNA expression levels are early events during the multistep process of carcinogenesis [20], that are mediated by several different mechanisms (Figure 1a).
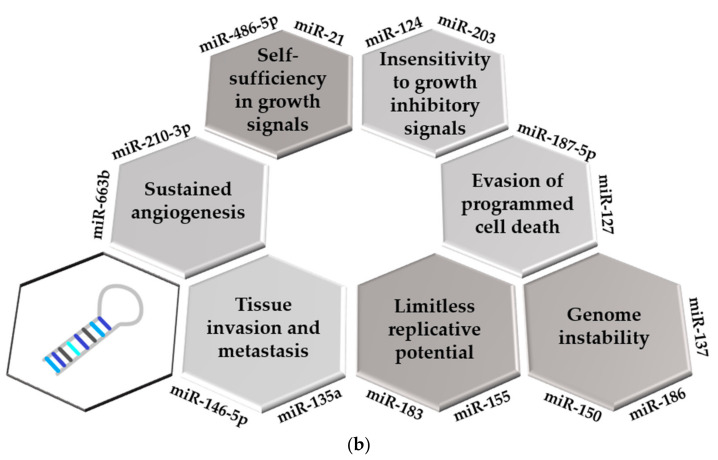
Figure 1.
(a) Mechanisms of dysregulated miRNA expression in hematological malignancies. Abbreviations: SNP: single nucleotide polymorphism, UTR: untranslated region. (b) Implication of miRNAs in the development of the hallmarks of cancer.
Widespread interactions have been confirmed between miRNAs and transcription factors, among which the most well-known is the tumor suppressor p53, controlling the expression of miR-34 family [21,22], while the proto-oncogenic c-Myc transcription factor is implicated in the regulation of miR-17-92 cluster [23]. Disorders of miRNA biogenesis include aberrant promoter methylation of Drosha and Dicer enzymes, that can be a novel biomarker for the early detection of lung cancer [24]. Expression of Drosha was shown to be down-regulated in many types of cancer [25], and the E518K mutation of DGCR8, an RNA-binding protein associated with Drosha, was found in Wilms tumor, resulting in selectively reduced expression of numerous canonical miRNAs [26]. Global inhibition of miRNA biogenesis by depletion of Dicer promoted cell growth in human cancer cell lines [27]. Phosphorylation of Exportin-5 correlated with the global down-regulation of miRNAs and unfavorable prognosis of patients with hepatocellular carcinoma (HCC), while Ago2 protein was found to be up-regulated in various types of cancer [28].
Single-nucleotide polymorphisms (SNPs) are involved in aberrant miRNA biogenesis and disruption of miRNA targets. Drosha rs6877842 and DGCR8 rs417309 SNPs were confirmed to play pivotal roles in carcinogenesis [29]. SNPs disrupting miRNA targets differentiated patients at high risk versus low risk for long-term genitourinary toxicity after prostate cancer radiation therapy [30]. In the let-7 binding site of interleukin 23 receptor (IL23R) mRNA, rs10880677 A > C polymorphism was described as a potential screening biomarker for colorectal cancer (CRC) [31]. rs6426881 at pre-B cell leukemia transcription factor 1 (PBX1) 3′UTR was associated with poor prognosis in breast and gastric cancer by altering the affinity of miR-522-3p to PBX1 [32]. Numerous SNPs within miRNAs have been associated with increased risk of hematological malignancies and with chemotherapy-induced toxicity. SNPs rs12402181 in miR-3117, rs62571442 in miR-3689 and rs10406069 in miR-5196 were associated with increased risk of pediatric ALL [33,34], while rs2114358 in miR-1206 indicated increased risk of chronic myeloid leukemia (CML) [35]. Previous studies suggested that rs2648841 in miR-1208 and rs2114358 in miR-1206 contribute to increased risk of hepatotoxicity and methotrexate-induced oral mucositis in ALL, respectively [36,37].
Epigenetic regulation of miRNAs results in significant alterations of their expression levels and function [38]. Promoter hypermethylation of tumor suppressor miRNAs is a frequent mechanism of gene inactivation in cancer [39]. Silencing of miR-124 by promoter methylation was detected in a wide variety of hematological malignancies including multiple myeloma (MM), ALL, AML, CLL, and non-Hodgkin lymphomas (NHL) [40]. In classic Hodgkin lymphoma, mir-148a was identified as a tumor suppressor miRNA, inactivated by DNA hypermethylation [41], while homozygous methylation of miR-203 was detected in MM cell lines [42].
2.2. Implication of miRNAs in the Development of the Hallmarks of Cancer
Altered expression levels of miRNAs are involved in both the initiation and progression of malignant diseases, [13], contributing to all established hallmarks of cancer [43] (Figure 1b). miRNAs play important role in the development of self-sufficiency in growth signals and insensitivity to growth inhibitory signals. They can act as both oncogenes or tumor suppressors (oncomiRs and anti-oncomiRs, respectively), depending on the tissue where they are expressed [13]. MiR-146a is a dual nature miRNA showing both oncogenic and tumor suppressor properties in different types of leukemias [44]. The first tumor suppressor miRNAs to be established were miR-16-1 and miR-15a, located in the most frequently deleted genomic region (13q14) in CLL [45,46]. Dysregulation of the anti-oncomiR let-7a was confirmed to be essential for sustaining the survival of MLL-rearranged leukemic cells [47]. MiR-20a-5p, miR-203, and miR-939 function as potent tumor suppressors by targeting protein phosphatase 6 catalytic subunit (PPP6C) in AML, modulating JUNB transcription factor in pediatric anaplastic large cell lymphoma (ALCL), and inhibiting cellular proliferation by targeting CREB1 in MM, respectively [42,48,49]. MiR-124 is generally considered as an anti-oncomiR in hematological malignancies [40], while miR-155 acts as a powerful oncomiR in lymphomas and several types of solid tumors by the down-regulation of SHIP1 protein phosphatase [3]. MiR-155 and miR-183 promote cell proliferation by targeting the transcriptional repressor ZNF238 protein in pediatric ALL and regulating programmed cell death 6 (PCD6) protein in pediatric AML, respectively [50,51]. MiR-486-5p is an erythroid oncomiR that cooperates with GATA1 transcription factor in supporting the growth and survival of leukemic cells, contributing to aberrant erythroid phenotype of the megakaryocytic leukemias in Down syndrome [52]. Amplification of the oncogenic miR-21 was registered in breast, hepatocellular, lung, ovarian and prostate cancers [3]. Besides limitless replicative potential, cancer cells are also featured by the evasion of programmed cell death. In B-cell precursor ALL, up-regulated miR-187-5p induces Bcl-2 protein, resulting in the apoptosis resistance of cells [53], while silencing of miR-127 by promoter hypermethylation contributes to increased expression level of Bcl-6 in bladder cancer [54]. The oncogenic miR-155 is implicated in sustained angiogenesis of tumors by down-regulation of von Hippel–Lindau (VHL) tumor suppressor protein [55]. Accumulating evidence indicates that tumor-derived exosomal miRNAs promote angiogenesis, such as miR-210-3p and miR-663b in oral squamous cell carcinoma (OSCC) and cervical cancer, respectively [56,57].
Epithelial-to-mesenchymal transition (EMT) is linked to a variety of cancer-related activities, including tissue invasion and metastasis [58]. MiR-338-3p was confirmed to suppress EMT pathway in neuroblastoma cells by targeting matrix metalloproteinase-2 (MMP-2) [59]. Reduced expression of miR-146b-5p in T-cell precursor ALL resulted in the up-regulation of IL17A, which promotes cell migration and invasion [60]. In non-small-cell lung carcinoma (NSCLC) and osteosarcoma cells, the central role for miR-124-3p in carcinoma cell invasion and metastasis has been established [61], while miR-135a enhances metastasis formation in breast cancer by targeting homeobox A10 (HOXA10) protein [62]. Recent evidence suggests the role of miRNAs in genomic instability of cancer cells [63]. Low expression level of miR-137 was accompanied by increased frequency of IgH translocations in MM [63], up-regulation of miR-22 and miR-150 increased genomic instability in CML patients by decreasing the activity of an alternative form of nonhomologous end-joining (NHEJ) [64], and increased expression level of miR-186 resulted in chromosomal instability of arsenic-exposed human keratinocytes [65].
miRNAs can be delivered from cancer cells to the tumor microenvironment (TME) and vice versa, thereby affecting growth and evolution of transformed cells [66]. Moreover, increased miR-155 and decreased miR-320 expression levels mediate the transformation of normal fibroblasts to cancer-associated fibroblasts (CAFs), providing a stromal framework for cancer cells [3,67]. In step with progression of MM, up-regulation of miR-27b-3p and miR-214-3p promotes apoptosis resistance of bone marrow fibroblasts [68]. Antitumor effect of miR-340 in pancreatic ductal adenocarcinoma is exerted by promoting macrophages to acquire M1-like phenotype [69], while exosomal delivery of oncomiRs such as miR-126, miR-144 and miR-155 from breast cancer cells to TME adipocytes results in their conversion into cancer-associated adipocytes [66]. In T-cell precursor ALL, increased level of miR-29b was confirmed to decrease cytotoxicity by natural killer (NK) cells, thereby contributing to the escape of leukemic cells from immune surveillance [70].
2.3. miRNAs and Metabolic Reprogramming in Leukemia
Aberrant nature of cancer metabolism was first recognized by Otto Warburg in the early 1920s when he postulated that tumor tissues have an increased rate of glucose uptake [71]. Besides elevated rate of glycolysis, cancer cells undergo complex metabolic reprogramming, including increased glutamine consumption and enhanced anabolism. TME cells are also featured by active metabolism, resulting in the suppression of antitumor immune response [72]. Cancer metabolic networks control not only energy balance and growth of tumor cells but are also important modulators of epigenetic mechanisms [71]. Leukemic progenitors activate different metabolic programs in association with genetic alterations and the levels of their differentiation blockade [73]. Metabolic alterations of leukemic cells play a role in leukemogenesis and have an impact on prognosis [73]. In serum samples of patients with AML, significant metabolomic changes have been detected [74]. Glutamine level was elevated among patients with French-American-British (FAB) subgroups M4 and M5, the level of alanine was higher in FLT3-ITD positive patients, and significant decrement of glutamate and phosphocoline levels were registered in the NPM1-mutated subgroup of patients [73]. Comparing FLT3-ITD positive and FLT3 wild-type pediatric AML patients, differential abundance of 21 metabolites in plasma and 33 metabolites in leukemic cells have been detected [75]. In the cytogenetically normal subgroup of adult AML patients (CN-AML), a panel of six metabolites (lactate, 2-oxoglutarate, pyruvate, 2-hydoxyglutarate, glycerol-3-phosphate, citrate) was able to independently assess prognostic potential [76].
In recent years, implication of miRNAs in the metabolic reprogramming of tumor cells has been confirmed, including the regulation of glycolysis, glutaminolysis and anabolic pathways, such as miR-210, miR-23a, and miR-33a, respectively [77,78]. According to the latest data, impaired expression of miR-652-5p leads to decreased levels of ATP, lactate, and pyruvate in T-ALL cell lines, thereby providing a potential drug target [79]. A special subgroup of miRNAs, so-called mitomiRs, are localized in the mitochondria and play an essential role in the control of cancer cell metabolism by targeting key transporters, metabolic enzymes and several oncogenic pathways [80]. Expression levels of metabolism-regulating miRNAs in leukemic stem and progenitor cells have to be further evaluated.
2.4. Interactions between miRNAs and Epigenetic Regulatory Circuits
miRNAs are involved in the regulation of those epigenetic mechanisms that play an important role in early stages of leukemogenesis [81,82]. According to a prominent body of evidence, these so-called epi-miRNAs are implicated in the regulation of both DNA-methylation and histone modification [82,83].
In AML, miR-29b induces global DNA hypomethylation and re-expression of tumor suppressor genes by targeting DNA methyltransferase (DNMT) enzymes [84]. In estrogen receptor (ER)-positive breast cancer, miR-29c-5p was found to trigger early abnormal DNA-methylation through direct targeting of DNMT3A [85]. DNMT1 enzyme was identified as a target of miR-152 [86], and miR-137 inhibits the invasion and metastasis of nasopharyngeal cancer cells by regulating the histone lysine demethylase enzyme LSD1 [58]. In breast cancer, miR-22 was identified as an oncomiR that targets the methylcytosine dioxygenase TET2 enzyme and promotes cell migration [87]. The potent tumor suppressor role of miR-377 in osteosarcoma is associated with the inactivation of histone acetyltransferase 1 (HAT1)-mediated Wnt signaling pathway [88]. In Waldenström macroglobulinaemia, the HAT enzyme KAT6A is down-regulated by miR-206-3p, while histone deacetylase (HDAC) enzymes HDAC4 and HDAC5 are targeted by miR-9-3p [89]. Cancer cell proliferation and metastasis in HCC were found to be suppressed by miR-20a through the regulation of the histone methyltransferase enzyme EZH1 [90].
3. Regulation of Hematopoiesis by miRNAs
Lineage- and maturation-specific expression of miRNAs in hematopoietic cells was first reported by Chen and colleagues in 2004 [91]. Since then, numerous miRNAs have been confirmed to interact with transcription factors crucial for normal development of hematopoietic progenitor cells [38]. Early stages of hematopoiesis, stem cell maintenance, lineage selection, and terminal differentiation of hematopoietic cells are all affected by miRNAs that function as downstream effectors of transcription factors [38,92].
In hematopoietic stem cells (HSCs), high expression of miR-125b was described [81], while enforced expression of this miRNA resulted in the blockade of granulocytic differentiation in murine 32D cells [93]. MiR-223 is a fine-tuner of granulocytic differentiation [94], regulated by NFIA and C/EBPα transcription factors [95]. MiR-150 and miR-486-3p were identified as critical factors in the lineage selection of megakaryocyte-erythrocyte progenitors, by targeting MYB and c-MAF transcription factors, respectively [92,94]. While miR-150 was preferentially expressed in megakaryocytic lineage [94], enforced expression of miR-486-3p increased erythropoiesis and blocked megakaryocytopoiesis in hematopoietic stem and progenitor cells [92]. MiR-17-5p, miR-20a, miR-106a, and miR-129 inhibit monocytopoiesis by repressing RUNX1 transcription factor [92]. FOXP1, a transcription factor that is required for early B-cell development, was identified as a key target of miR-34a [96]. Besides miR-34 family, the miR-17-92 cluster, miR-21-5p, miR-29 family, miR-125b-5p, miR-150-5p, miR-155-5p, and miR-181 also appeared as important regulators of B-cell development [38]. MiR-17-92 promotes early differentiation of T cells [97], and miR-150 is implicated in T lymphopoiesis through the regulation of NOTCH3 transcription factor [98]. Development of NK cells is regulated by a wide range of miRNAs including miR-155 and miR-181a [99].
4. Clinical Applications of miRNAs Focusing on Hematological Malignancies
4.1. Differential Diagnosis and Early Detection of Hematological Malignancies
Alterations of miRNA expression levels can be applied as non-invasive biomarkers to make advances in both differential diagnosis and early detection of hematological malignancies (Table 1). Based on the expression signature of four miRNAs (miR-128a, miR-128b, let-7b, miR-223), ALL and AML can be distinguished with an accuracy rate of >95 percent [100]. MiR-200c and miR-326 could serve as diagnostic biomarkers for pediatric ALL [101], while a panel of 16 miRNAs may differentiate between pediatric T-ALL and B-ALL, among which miR-29c-5p was identified as the best discriminator [102]. According to recently published data, the expression ratio of miR-92a/miR-638 in blood plasma could be applied as a novel biomarker for AML [103]. In pediatric AML, levels of miR-25, miR-155, miR-196b, and miR-370 are potential diagnostic biomarkers [104,105], among which miR-196b is specific for FAB subgroups M4 and M5 [104].
Table 1.
Clinical applications of miRNAs in hematological malignancies. See references in the text.
| Differential Diagnosis | Prognostic Stratification | Chemoresistance | |
|---|---|---|---|
| ALL | miR-128a, miR-128b, let-7b, miR-223 | miR-146a, miR-429, miR-124 hypermethylation, miR-99a, miR-100, miR-16 | miR-206, miR-34a, miR-142-3p, miR-17-92 cluster |
|
Pediatric
ALL |
miR-29c-5p, miR-326, miR-200c | miR-10a, miR-134, miR-214, miR-708, miR-99a, miR-151-5p, miR-451, miR-1290, miR-155, miR-181a, miR-143, miR-182, miR-152 promoter methylation, miR-101-3p, miR-4774-5p, miR-1324, miR-631, miR-4699-5p, miR-922 | miR-125b, miR-99a, miR-100, miR-324-3p, miR-508-5p, miR-18a, miR-532, miR-218, miR-625, miR-193a, miR-638, miR-550, miR-633, miR-21, miR-326, miR-221 |
| AML | miR-128a, miR-128b, let-7b, miR-223, miR-92a/miR-638 ratio | miR-181 family, miR-504-3p, miR-191, miR-199a, miR-195, miR-363 | miR-874-3p, miR-15a-5p, miR-21-5p, miR-9, miR-217, miR-143, miR-204 |
|
Pediatric
AML |
miR-196b, miR-155, miR-25, miR-370 | miR-193b-3p, miR-370, miR-29a, miR-509, miR-542, miR-146a, miR-3667, miR-199a | miR-29a, miR-100, miR-125b |
| CLL | miR-192, miR-32-5p, miR-98-5p, miR-374b-5p, miR-145-5p, miR-185-5p | miR-181b, miR-650, miR-708, miR-29b, miR-29c, miR-18a, miR-19b-1, miR-92a-1, miR-17 | miR-148a, miR-222, miR-21, miR-181a, miR-155 |
| CML | miR-451, miR-222, miR-126, miR-155 | miR-486-5p, miR-320a, miR-150 | miR-217, miR-199b, miR-221, miR-577, miR-451, miR-146a, miR-9, miR-142-5p, miR-365a-3p |
| Lymphomas | let-7f, miR-9, miR-27a, miR-142-3p, miR-155, miR-203, miR-30c | miR-130a, miR-199a, miR-497, miR-34a-5p, miR-22, miR-129-5p, miR-27b, miR-7, miR-223 | miR-125b-5p, miR-155, miR-1244, miR-193b-5p, miR-223-3p |
| MM | miR-34a, let-7e, miR-4254 | miR-223-3p, miR-744, let-7e, miR-720, miR-1246 | miR-145-3p, miR-155, miR-221, miR-222 |
While miR-32-5p, miR-98-5p, miR-145-5p, miR-185-5p, miR-192, and miR-374b-5p are potential biomarkers for the early detection of CLL [19,106,107], a miRNA expression signature composed of 13 genes can differentiate between subgroups of CLL patients, based on the level of ZAP-70 expression and immunoglobulin heavy chain variable region gene (IgHV) mutational status [5]. MiR-451 was found to be up-regulated in plasma samples of CML patients in the chronic phase [108], while miR-126, miR-155, and miR-222 were up-regulated in samples obtained during blast crisis [109].
The expression of let-7f, miR-9, and miR-27a can serve as biomarkers to distinguish classic Hodgkin lymphoma from other B-cell lymphomas [110]. Activated B-cell and germinal center-like diffuse large B-cell lymphoma (DLBCL) cell lines can be differentiated based on a panel of nine miRNAs [111]. In lymph gland samples, a set of 20 miRNAs were identified as judgmental indicators to differentiate between reactive lymphoid hyperplasia (RLH) and peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS) [112]. The expression levels of miR-142-3p, miR-155, and miR-203 might be helpful biomarkers for the differential diagnosis between chronic gastritis and gastric MALT lymphomas [113]. The level of miR-30c in the cerebrospinal fluid (CSF) can be applied to differentiate between patients with primary lymphomas of the central nervous system (CNS) and secondary spread of systemic lymphoma to the CNS [114].
In MM, miR-4254 was found to be the most promising diagnostic biomarker [115]. In addition, a combination of up-regulated miR-34a and down-regulated let-7e could distinguish MM from both control and monoclonal gammopathy of undetermined significance (MGUS) with high sensitivity and specificity [116].
4.2. Advances in Prognostic Stratification of Hematological Malignancies
Growing number of miRNAs has been identified as novel biomarkers for prognostic stratification in hematological malignancies, contributing to optimization of therapy in numerous subgroups of patients (Table 1).
While the overall survival (OS) rate of ALL patients was better with low expression of miR-429 [117] and high expression level of miR-16 was also found to be a good prognostic factor [118], up-regulation of miR-146a, promoter hypermethylation of miR-124 and low expression levels of miR-99a and miR-100 correlated with poor prognosis in adult ALL patients [44,118,119].
In pediatric ALL, miR-10a, miR-134, and miR-214 were linked with a favorable outcome arising from their tumor suppressor activity [14]. Increased expression of miR-155 and down-regulation of miR-99a and miR-708 were associated with poor prognosis in childhood ALL [50,120], and the high degree of miR-152 promoter methylation indicated poor clinical outcome among t(4;11)-positive infant ALL patients [86]. Up-regulation of miR-155 and miR-181a was described in pediatric ALL patients with high levels of minimal residual disease (MRD) [121]. In pediatric precursor B-cell ALL, down-regulation of miR-151-5p and miR-451, up-regulation of miR-1290, or a combination of all three predicted inferior relapse-free survival (RFS) [122]. Increased expression of miR-143 and/or miR-182 at the end of induction treatment was associated with significantly higher risk for short-term relapse and death [123], while a set of six miRNAs (miR-101-3p, miR-4774-5p, miR-1324, miR-631, miR-4699-5p, and miR-922) was confirmed to be up-regulated in early relapse in pediatric B-ALL [124].
In AML patients, higher expression of the miR-181 family was reported to positively correlate with favorable prognosis [19,125]. Up-regulation of miR-195 was associated with favorable outcome in the CN-AML group [126]. Down-regulation of miR-504-3p and high expression levels of miR-363, miR-191 and miR-199a were related to poor prognosis, shorter event-free survival (EFS), and worse OS data in AML patients [81,127,128].
Low expression of miR-193b-3p and increased levels of miR-509 and miR-542 were found to be poor prognostic factors, whereas up-regulation of miR-146a and miR-3667 were linked with favorable outcome in pediatric and adolescent AML [129,130]. Pediatric AML patients with reduced levels of miR-29a and miR-370 had shorter RFS and OS [19,131], while down-regulation of miR-199a correlated with shorter EFS [132].
In CLL, down-regulation of miR-29b, miR-29c, and miR-181b were associated with unfavorable prognosis [8,133]. In Egyptian CLL patients, increased expression levels of miR-18a, miR-19b-1, and miR-92a-1 also had an adverse prognostic value [134]. Up-regulation of miR-650 and miR-708 was associated with favorable prognosis in CLL [135].
In CML, decreased expression of miR-320a was associated with worse OS and RFS rates [136], patients with early treatment response (ETR) had significantly higher levels of miR-150 [137], while up-regulation of miR-486-5p was associated with favorable prognosis [138].
In DLBCL, low expression levels of miR-27b and miR-129-5p were linked with poor clinical outcome [139,140]. Up-regulation of miR-130a was associated with adverse prognosis of primary gastrointestinal DLBCL [141], high expression of miR-22 correlated with a worse progression-free survival (PFS) [142], whereas increased expression levels of miR-199a and miR-497 indicated favorable outcome in DLBCL patients [143]. MiR-34a-5p was identified as a novel biomarker for the transformation of gastric MALT lymphoma to gastric DLBCL [144]. Down-regulation of miR-7 and miR-223 was associated with inferior OS data in NHL and mantle cell lymphoma, respectively [145,146].
Low levels of let-7e, miR-223-3p and miR-744 were associated with shorter remission and OS in MM patients [147,148], while up-regulation of miR-720 and miR-1246 correlated with shorter PFS data [149].
4.3. Implication of miRNAs in Chemosensitivity, Prediction of Chemoresistance in Hematological Malignancies
Drug resistance is one of the topmost challenges of cancer chemotherapy, caused by many mechanisms including decreased intake or increased release of drugs, degradation or deactivation, target modification, enhanced DNA damage repair activity, and alterations of cell cycle checkpoints [21,150]. Altered miRNA expression levels have been associated with chemoresistance in hematological malignancies (Table 1).
Up-regulation of miR-142-3p and the miR-17-92 cluster in B-ALL was associated with acquired resistance to dexamethasone [151], while transfection of Jurkat cells with miR-34a resulted in increased sensitivity to doxorubicin [152]. MiR-206 is implicated in chemoresistance of B-lymphoblasts by targeting the guanine nucleotide exchange factor NET1 [153]. In pediatric ALL, resistance to daunorubicin and vincristine was characterized by the up-regulation of miR-99a, miR-100, and miR-125b [14]. According to recently published results, good and poor prednisone response can be distinguished by the evaluation of a miRNA signature composed of 8 genes (miR-18a, miR-193a, miR-218, miR-532, miR-550, miR-625, miR-633, and miR-638) [154]. Increased expression of miR-221 and miR-21 correlated with poor response to induction therapy in T-ALL and B-ALL, respectively [155,156]. Down-regulation of miR-326 was introduced as a biomarker for drug resistance in childhood ALL, showing inverse correlation with the expression level of ABCA2 transporter [101]. A negative correlation was also identified between the expression of two miRNAs (miR-324-3p and miR-508-5p) and ABCA3 transporter, that is associated directly with chemoresistance [157].
Decreased expression of miR-874-3p and up-regulation of miR-15a-5p and miR-21-5p contribute to the development of chemoresistance to cytosine arabinoside in AML and to doxorubicin in CN-AML, respectively [158,159]. MiR-143 sensitizes AML cells to cytosine arabinoside via targeting autophagy-related proteins ATG2B and ATG7 [160]. MiR-15a-5p induces resistance, whereas miR-9 enhances sensitivity of AML cells to daunorubicin through the abrogation of autophagy and by targeting the EIF5A2/MCL-1 axis, respectively [161,162]. MiR-217 enhances chemosensitivity of AML cells to doxorubicin by targeting KRAS [163], while miR-204 potentiates the sensitivity of acute promyelocytic leukemia (APL) cells to arsenic trioxide (ATO) [164]. MiR-29a and miR-100 were identified as significant predictors of chemotherapy response in pediatric AML [165]. Up-regulation of miR-125b indicated increased drug resistance in pediatric APL [166].
In CLL, fludarabine resistance was associated with increased expression levels of miR-21, miR-148a, miR-155, and miR-222 [8,167]. On the other hand, increased expression of miR-181a was found to sensitize CLL cells to fludarabine [168]. MiR-9 was confirmed to play a critical role in the development of multidrug resistance in CML by targeting ABCB1 transporter [169]. Down-regulation of miR-142-5p, miR-199b, miR-217, miR-221, and miR-365a-3p correlated with resistance to tyrosine kinase inhibitors (TKIs) in CML [8,170,171]. Measurement of circulating miR-146a and miR-451 expression can also predict treatment response to TKIs [172,173]. MiR-577 promotes sensitivity of CML cells to imatinib by targeting the NUP160 subcomplex of the nuclear pore [174].
In DLBCL, high expression of miR-155 and down-regulation of miR-193b-5p and miR-1244 were associated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) treatment failure [175,176]. Overexpression of miR-125b-5p sensitized cutaneous T-cell lymphoma cells to bortezomib via modulation of MAD4 protein [21], while miR-223-3p is implicated in ibrutinib resistance through regulation of the conserved helix-loop-helix ubiquitous kinase (CHUK)-NFκB signaling pathway in mantle cell lymphoma [177].
Down-regulation of miR-155 and miR-145-3p was associated with bortezomib resistance [178,179], while up-regulation of miR-221 and 222 was found to inhibit autophagy, thereby promoting dexamethasone resistance in MM cells [180].
4.4. Changes of miRNA Expression Levels during Chemotherapy
The first report to show that chemotherapy influences miRNA expression in human cancer cells was published in 2006, discussing the changes of miRNA profiles in cholangiocarcinoma cells during gemcitabine treatment [181]. Growing number of evidence suggests that chemotherapy causes significant alterations of miRNA expression profiles in solid tumors and hematological malignancies. In ALL cell lines, increased levels of miR-15a and miR-16-1 were registered after prednisolone treatment [182]. In ALL patients, decreased expression level of miR-146a and up-regulation of let-7e was detected after 14 days of treatment [44,183]. Obinutuzumab-induced CD16 stimulation and bortezomib led to an up-regulation of miR-155-5p and miR-29b in follicular lymphoma and in murine models of cutaneous T-cell lymphoma, respectively [184,185]. In CML, dasatinib affected the expression of miR-let-7d, miR-let-7e, miR-15a, miR-16, miR-21, miR-130a and miR-142-3p, while imitanib was shown to modulate miR-15a, miR-21, miR-122, miR-126, and miR-130a levels [186,187,188].
4.5. miRNA-Based Anticancer Therapeutic Approaches
The Nobel Prize in Medicine was awarded to Andrew Z. Fire and Craig C. Mello in 2006 for the discovery of RNA interference, gene silencing by double-stranded RNA molecules [189]. The first miRNA-targeted drug was miravirsen (SPC3649), a locked nucleic acid (LNA)-modified antisense oligonucleotide targeting miR-122, that is currently in phase II clinical trial for the treatment of hepatitis C virus infection [190,191]. MRX34 (miRNA Therapeutics), containing a double-stranded miR-34a mimic encapsulated in ionizable liposomes, was the first miRNA mimic to enter a phase I clinical trial in 2013 (NCT01829971) in several solid tumors and hematological malignancies [3,192]; however, the trial was closed due to severe immune-related adverse events [6].
Modulation of miRNA expression levels is possible with both mimic and inhibitor molecules. Increased stability, longer half-lives, and higher efficiency are aimed by the optimization of delivery and chemical modifications of oligonucleotide molecules [3].
In AML cell lines THP-1 and U937, overexpression of miR-20a-5p suppressed cell proliferation, induced cell cycle arrest and apoptosis, representing a novel therapeutic target for AML [48]. Overexpression of miR-150 in NK/T-cell lymphoma cells resulted in substantially enhanced sensitivity to ionizing radiation treatment [193], while miR-28-5p was found to suppress the growth of DLBCL cells by inhibiting the expression of YWHAZ protein [194]. In animal models, targeted delivery of miR-200c with nanoparticles suppressed proliferation of triple-negative breast cancer cells [195]. Up to recently published results, miR-15 and miR-16 mimics could be used for the therapy of Bcl-2-overexpressing tumors [45]. Delivery of miR-16 using an EnGeneIC Delivery Vehicle (EDV) nanocell system targeting epidermal growth factor receptor (EGFR) in NSCLC and malignant pleural mesothelioma resulted in significant tumor reduction [196]. In NSCLC and breast cancer cells, overexpression of miR-20b-5p combined with pembrolizumab potentiated sensitivity of tumor cells to radiotherapy by repressing PD-L1/PD1 [197].
miRNA sponges are synthetic RNA molecules containing multiple high affinity miRNA binding sites to reduce the abundance of oncogenic miRNAs within the cell [62]. Overexpression of the long non-coding RNA molecule, MIR17HG promoted homoharringtonine (HHT)-induced apoptosis of AML cells by sponging miR-21, resulting in the up-regulation of the tumor suppressor PTEN protein phosphatase [198]. MiR-10b sponge was confirmed to inhibit metastasis formation in breast cancer by the up-regulation of HOXD-10 [199]. Silencing of miR-146a, miR-155, miR-181a, and let-7e enhanced the effect of prednisolone treatment in pediatric ALL [200], while blockade of let-7a-5p may be a novel therapeutic strategy in APL through the activation of caspase-3 [201]. Inhibition of miR-182 by the transfection of LNA-anti-miR-182 resulted in decreased proliferation of APL cells through the modulation of CASP9 expression [202].
Besides the modulation of their expression levels by mimics and inhibitors, miRNAs can be applied in anticancer treatment by further approaches including miRNA-level based therapeutic decisions and epigenetic drugs restoring altered miRNA expression levels. In AML, patients with high expression levels of miR-363 may be highly recommended for early allo-HSCT regimen [128]. Demethylation treatment contributes to the restoration of tumor suppressor miRNAs. Tumor-specific promoter hypermethylation and silencing of miR-124 can be reversed by the hypomethylating agent 5-aza-2-deoxycytidine, that is associated with consequent down-regulation of CDK6 enzyme [40]. In the t(4;11)-positive B-ALL cell line SEMK2, expression levels of seven miRNAs (miR-10a, miR-152, miR-200a, miR200b, miR429, miR-432, and miR-503) increased following demethylation by the cytidine analog Zebularine [86]. In MM cell lines, 5-aza-2-deoxycytidine led to promoter demethylation and re-expression of miR-203 [42]. HDAC inhibitors (HDACi) have also been confirmed to modify miRNA expression levels. Sapacitabine induced the expression of miR-182 in primary AML blasts, thereby sensitizing AML cells to DNA-damaging agents [203]. In synergism with imatinib mesylate, vorinostat was found to elicit marked inhibition of CML stem cells by the up-regulation of miR-196a [204]. Pan-HDACIs vorinostat and panobinostat inhibit metastasis formation in advanced cutaneous T-cell lymphoma by restoring the tumor suppressor miR-150 [205]. In gastric cancer, panobinostat increased the expression of the tumor suppressor miR-874, thereby inhibiting proliferation of tumor cells [206].
4.6. Clinical Applications of Exosomal miRNAs
Exosomes are highly stable endosomal sorting pathway products with a diameter of 30–100 nm. Accumulating evidence highlights their roles in the pathogenesis of cancer, and numerous exosomal miRNAs in serum have been identified as novel biomarkers for early detection and prognosis prediction of malignant diseases [207]. Furthermore, exosomal miRNAs derived from cancer cells modulate chemosensitivity and TME, providing promising therapeutic targets [11].
In 2015, Hornick et al. developed a panel of three exosomal miRNAs (miR-150, miR-155, and miR-1246) as a minimally invasive early biomarker of MRD in AML [208]. Exosomal miR-7-5p and miR-425-5p, derived from bone marrow mesenchymal stem cells (BM-MSCs), were confirmed to inhibit proliferation, and promote apoptosis of AML cells by targeting PI3K/AKT/mTOR signaling pathway and Wilms tumor 1-associated protein (WTAP), respectively [209,210]. Exosomal miR-125-5p reduced sensitivity of DLBCL cells to rituximab treatment [211], while increased levels of exosomal miR-99a-5p and miR-125b-5p in sera samples of DLBCL patients were associated with shorter PFS data [212]. In MM patients, down-regulation of four exosomal miRNAs (miR-5a-5p, miR-16-5p, miR-17-5p and miR-20a-5p) was registered in the bortezomib resistance group [207], and high levels of exosomal miR-1305 indicated poor OS [213]. MiR-221 and miR-222 secreted by CRC cells are implicated in the formation of a hospitable metastatic environment in the liver [214].
5. Concluding Remarks and Future Perspectives
Resulting from advances in molecular diagnostic evaluation, chemotherapeutic protocols and supportive treatment of patients, survival outcomes in hematological malignancies have considerably improved during the previous decades. The 5-year OS rate for pediatric acute leukemia is now more than 90% in ALL and 60–70% in AML [12]. Chemoresistance and toxicity are still among the major causes of treatment failure and mortality [215]. According to recent evidence, identification of new biomarkers can be utilized in early identification, differential diagnosis, non-invasive MRD monitoring, and prognostic stratification of acute leukemias [81].
miRNAs are a class of highly stable, non-coding small RNAs that can be easily quantified in body fluids and paraffin-embedded tissues [38]. Besides the high number of currently ongoing studies to assess their potential as MRD biomarkers [206], miRNAs are novel candidates for therapeutic intervention [216]. However, modulation of miRNA levels holds great opportunities, major challenges must be overcome to provide specificity and avoid side effects including activation of innate immune system and undesirable toxicities [217]. Resulting from the great numbers of their targets and the complexity of the regulatory network that determines their levels, the biological impact of a certain miRNA expression pattern is highly dependent on the tissue microenvironment. Since the biological behavior of pediatric malignancies is markedly different from their adult counterparts [218], it is not surprising that major differences between miRNA expression signatures of pediatric and adult acute leukemias have been described [19].
Based on the complex pathogenesis of acute leukemias and recent innovation in molecular genetic diagnostics and bioinformatics, major focuses of onco-hematology include precision oncology and system biology approach, aiming the clinical translation of novel biomarkers and personalized treatment opportunities. Application of miRNAs in the early detection, prognostic stratification and therapy of hematological malignancies can contribute to more efficient and less toxic antileukemic treatment, thereby improving survival and life quality of patients.
Abbreviations
3′UTR: 3′ untranslated region, Ago: Argonaute, ALL: acute lymphoblastic leukemia, ALCL: anaplastic large cell lymphoma, AML: acute myeloid leukemia, ANK1: ankyrin 1, APL: acute promyelocytic leukemia, ATO: arsenic trioxide, Bcl-2: B-cell lymphoma 2, BM-MSC: bone marrow mesenchymal stem cell, CAF: cancer-associated fibroblast, CAGR: cancer-associated genomic regions, C/EBPα: CCAAT-enhancer-binding protein α, CHUK: conserved helix-loop-helix ubiquitous kinase, CDK: cyclin-dependent kinase, CLL: chronic lymphocytic leukemia, CML: chronic myeloid leukemia, CN: cytogenetically normal, CNS: central nervous system, CRC: colorectal cancer, CREB1: cAMP-responsive element binding protein 1, CSF: cerebrospinal fluid, DLBCL: diffuse large B-cell lymphoma, DNMT: DNA methyltransferase, EDV: EnGeneIC Delivery Vehicle, EFS: event-free survival, EGFR: epidermal growth factor receptor, EMT: epithelial-to-mesenchymal transition, ER: estrogen receptor, EZH1: enhancer of zeste 1 polycomb repressive complex 2 subunit, FAB: French-American-British classification, FLT3: fms-like tyrosine kinase-3, FOXP1: forkhead box protein 1, GATA1: GATA-binding factor 1, HAT1: histone acetyltransferase 1, HCC: hepatocellular carcinoma, HDAC: histone deacetylase enzyme, HDACi: HDAC inhibitor, HHT: homoharringtonine, HOXA10: homeobox A10, HSC: hematopoietic stem cell, HSCT: hematopoietic stem cell transplantation, IgHV: immunoglobulin heavy chain variable region, IL17A: interleukin-17A, IL23R: interleukin 23 receptor, ITD: internal tandem duplication, LNA: locked nucleic acid, LSD1: lysine-specific demethylase 1, MAD4: Max-interacting transcriptional repressor, MAF: musculoaponeurotic fibrosarcoma, MALT: mucosa-associated lymphoid tissue, MGUS: monoclonal gammopathy of undetermined significance, miRNA: microRNA, MLL: mixed lineage leukemia, MM: multiple myeloma, MMP-2: matrix metalloproteinase-2, MRD: minimal residual disease, mRNA: messenger RNA, mTOR: mammalian target of rapamycin, NET1: neuroepithelial cell transforming 1, NFIA: nuclear factor 1 A-type protein, NHEJ: nonhomologous end-joining, NHL: non-Hodgkin lymphoma, NK: natural killer, NPM1: nucleophosmin 1, NSCLC: non-small-cell lung carcinoma, NUP160: nucleoporin 160, OS: overall survival, OSCC: oral squamous cell carcinoma, PBX1: pre-B cell leukemia transcription factor 1, PCD6: programmed cell death 6, PD-L1: programmed death-ligand 1, PFS: progression-free survival, PI3K: phosphoinositide 3-kinase, PPP6C: protein phosphatase 6 catalytic subunit, PTCL-NOS: peripheral T-cell lymphoma not otherwise specified, PTEN: phosphatase and tensin homolog, RFS: relapse-free survival, RISC: RNA-induced silencing complex, RLH: reactive lymphoid hyperplasia, RUNX1: runt-related transcription factor, SHIP1: SH-2 containing inositol 5′ polyphosphatase 1, SNP: single nucleotide polymorphism, TET2: TET methylcytosine dioxygenase 2, TKI: tyrosine kinase inhibitor, TME: tumor microenvironment, VHL: von Hippel–Lindau, WTAP: Wilms tumor 1-associated protein, YWHAZ: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activating protein zeta.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The author declares no conflict of interest.
Funding Statement
This research was funded by Foundation for Children with Leukemia and University of Debrecen University and National Library.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tafrihi M., Hasheminasab E. miRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. Microrna. 2019;8:4–27. doi: 10.2174/2211536607666180827111633. [DOI] [PubMed] [Google Scholar]
- 2.Lai P., Wang Y. Epigenetics of cutaneous T-cell lymphoma: Biomarkers and therapeutic potentials. Cancer Biol. Med. 2021;18:34–51. doi: 10.20892/j.issn.2095-3941.2020.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rupaimoole R., Slack F. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 4.Matsuyama H., Suzuki H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019;21:132. doi: 10.3390/ijms21010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z., Xu Y., Wan M., Zeng X., Wu J. miR-340: A multifunctional role in human malignant diseases. Int. J. Biol. Sci. 2021;17:236–246. doi: 10.7150/ijbs.51123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artemaki P.I., Letsos P.A., Zoupa I.C., Katsaraki K., Karousi P., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. The Multifaceted Role and Utility of MicroRNAs in Indolent B-Cell Non-Hodgkin Lymphomas. Biomedicines. 2021;9:333. doi: 10.3390/biomedicines9040333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh C.H., Moles R., Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol. Cancer. 2016;15:37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Y., Croce C. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szczepanek J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J. Clin. Oncol. 2020;11:348–369. doi: 10.5306/wjco.v11.i6.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Schotte D., De Menezes R.X., Akbari Moqadam F., Khankahdani L.M., Lange-Turenhout E., Chen C., Pieters R., Den Boer M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703–711. doi: 10.3324/haematol.2010.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Nguyen L.X.T., Chen Y.C., Wu D., Cook G.J., Hoang D.H., Brewer C.J., He X., Dong H., Li S., et al. Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance. Nat. Commun. 2021;12:6154. doi: 10.1038/s41467-021-26420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havelange V., Ranganathan P., Geyer S., Nicolet D., Huang X., Yu X., Volinia S., Kornblau S.M., Andreeff M., Croce C.M., et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood. 2014;123:2412–2415. doi: 10.1182/blood-2013-10-532374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malouf C., Antunes E.T.B., O’Dwyer M., Jakobczyk H., Sahm F., Landua S.L., Anderson R.A., Soufi A., Halsey C., Ottersbach K. miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute leukemia. Blood. 2021;138:2066–2092. doi: 10.1182/blood.2020006610. [DOI] [PubMed] [Google Scholar]
- 18.Schwaller J. MLL-AF4+ infant leukemia: A microRNA affair. Blood. 2021;138:2014–2015. doi: 10.1182/blood.2021012818. [DOI] [PubMed] [Google Scholar]
- 19.Mardani R., Jafari Najaf Abadi M.H., Motieian M., Taghizadeh-Boroujeni S., Bayat A., Farsinezhad A., Hayat S.M.G., Motieian M., Pourghadamyari H. MicroRNA in leukemia: Tumor suppressors and oncogenes with prognostic potential. J. Cell. Physiol. 2019;234:8465–8486. doi: 10.1002/jcp.27776. [DOI] [PubMed] [Google Scholar]
- 20.Ryu J.K., Hong S.M., Karikari C.A., Hruban R.H., Goggins M.G., Maitra A. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10:66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manfe V., Biskup E., Willumsgaard A., Skov A.G., Palmieri D., Gasparini P., Lagana A., Woetmann A., Odum N., Croce C.M., et al. cMyc/miR-125b-5p signalling determines sensitivity to bortezomib in preclinical model of cutaneous T-cell lymphomas. PLoS ONE. 2013;8:e59390. doi: 10.1371/journal.pone.0059390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermeking H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 23.O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 24.Szczyrek M., Grenda A., Kuźnar-Kamińska B., Krawczyk P., Sawicki M., Batura-Gabryel H., Mlak R., Szudy-Szczyrek A., Krajka T., Krajka A., et al. Methylation of DROSHA and DICER as a Biomarker for the Detection of Lung Cancer. Cancers. 2021;13:6139. doi: 10.3390/cancers13236139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S.S., Min H., Ha J.Y., Kim B.H., Choi M.S., Kim S. Dysregulation of the miRNA biogenesis components DICER1, DROSHA, DGCR8 and AGO2 in clear cell renal cell carcinoma in both a Korean cohort and the cancer genome atlas kidney clear cell carcinoma cohort. Oncol. Lett. 2019;18:4337–4345. doi: 10.3892/ol.2019.10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardapour R., Kehl T., Kneitz S., Ludwig N., Meese E., Lenhof H.P., Gessler M. The DGCR8 E518K mutation found in Wilms tumors leads to a partial miRNA processing defect that alters gene expression patterns and biological processes. Carcinogenesis. 2021;43:82–93. doi: 10.1093/carcin/bgab110. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez-Moya J., Wert-Lamas L., Riesco-Eizaguirre G., Santisteban P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene. 2019;38:5486–5499. doi: 10.1038/s41388-019-0804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali Syeda Z., Langden S.S.S., Munkhzul C., Lee M., Song S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020;21:1723. doi: 10.3390/ijms21051723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen J., Lv Z., Ding H., Fang X., Sun M. Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: A systematic review and meta-analysis. Biosci. Rep. 2018;38:BSR20180072. doi: 10.1042/BSR20180072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishan A.U., Marco N., Schulz-Jaavall M.B., Steinberg M.L., Tran P.T., Juarez J.E., Dang A., Telesca D., Lilleby W.A., Weidhaas J.B. Germline variants disrupting microRNAs predict long-term genitourinary toxicity after prostate cancer radiation. Radiother. Oncol. 2022;167:226–232. doi: 10.1016/j.radonc.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosallaei M., Simonian M., Esmaeilzadeh E., Bagheri H., Miraghajani M., Salehi A.R., Mehrzad V., Salehi R. Single nucleotide polymorphism rs10889677 in miRNAs Let-7e and Let-7f binding site of IL23R gene is a strong colorectal cancer determinant: Report and meta-analysis. Cancer Genet. 2019;239:46–53. doi: 10.1016/j.cancergen.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Mohammadi M., Salehzadeh A., Talesh Sasani S., Tarang A. rs6426881 in the 3′-UTR of PBX1 is involved in breast and gastric cancers via altering the binding potential of miR-522-3p. Mol. Biol. Rep. 2021;48:7405–7414. doi: 10.1007/s11033-021-06756-5. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez-Camino A., Richer C., St-Onge P., Lopez-Lopez E., Bañeres A.C., de Andoin N.G., Sastre A., Astigarraaga I., Martin-Guerrero I., Sinnett D., et al. Role of rs10406069 in miR-5196 in hyperdiploid childhood acute lymphoblastic leukemia. Epigenomics. 2020;12:1949–1955. doi: 10.2217/epi-2020-0152. [DOI] [PubMed] [Google Scholar]
- 34.Gutierrez-Camino A., Martin-Guerrero I., Dolzan V., Jazbec J., Carbone-Bañeres A., Garcia de Andoin N., Sastre A., Astigarraga I., Navajas A., Garcia-Orad A. Involvement of SNPs in miR-3117 and miR-3689d2 in childhood acute lymphoblastic leukemia risk. Oncotarget. 2018;9:22907–22914. doi: 10.18632/oncotarget.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan F.M. The Association of rs2114358 in the miR-1206 Polymorphism to Chronic Myeloid Leukemia. Microrna. 2019;8:248–252. doi: 10.2174/2211536608666190102143439. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez-Camino A., Umerez M., Santos B., Martin-Guerrero I., García de Andoin N., Sastre A., Navajas A., Astigarraga I., Garcia-Orad A. Pharmacoepigenetics in childhood acute lymphoblastic leukemia: Involvement of miRNA polymorphisms in hepatotoxicity. Epigenomics. 2018;10:409–417. doi: 10.2217/epi-2017-0138. [DOI] [PubMed] [Google Scholar]
- 37.Gutierrez-Camino A., Oosterom N., den Hoed M.A.H., Lopez-Lopez E., Martin-Guerrero I., Plujim S.M.F., Pieters R., de Jonge R., Tissing W.J.E., Heil S.G., et al. The miR1206 microRNA variant is associated with methotrexate-induced oral mucositis in pediatric acute lymphoblastic leukemia. Pharmacogenet. Genom. 2017;27:303–306. doi: 10.1097/FPC.0000000000000291. [DOI] [PubMed] [Google Scholar]
- 38.Katsaraki K., Karousi P., Artemaki P.I., Scorilas A., Pappa V., Kontos C.K., Papageorgiou S.G. MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia. Cancers. 2021;13:593. doi: 10.3390/cancers13040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chim C.S., Wan T.S., Wong K.Y., Fung T.K., Drexler H.G., Wong K.F. Methylation of miR-34a, miR-34b/c, miR-124-1 and miR-203 in Ph-negative myeloproliferative neoplasms. J. Transl. Med. 2011;9:197. doi: 10.1186/1479-5876-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong K.Y., So C.C., Loong F., Chung L.P., Lam W.W., Liang R. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE. 2011;6:e19027. doi: 10.1371/journal.pone.0019027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paczkowska J., Janiszewska J., Bein J., Schneider M., Bednarek K., Ustaszewski A., Hartmann S., Hansmann M.L., Giefing M. The Tumor Suppressive mir-148a Is Epigenetically Inactivated in Classical Hodgkin Lymphoma. Cells. 2020;9:2292. doi: 10.3390/cells9102292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong K.Y., Liang R., So C.C., Jin D.Y., Costello J.F., Chim C.S. Epigenetic silencing of MIR203 in multiple myeloma. Br. J. Haematol. 2011;154:569–578. doi: 10.1111/j.1365-2141.2011.08782.x. [DOI] [PubMed] [Google Scholar]
- 43.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 44.Shahid S., Shahid W., Shaheen J., Akhtar M.W., Sadaf S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021;11:22783. doi: 10.1038/s41598-021-02257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calin G.A., Croce C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 47.Zhu S., Cheng X., Wang R., Tan Y., Ge M., Li D., Xu Q., Sun Y., Zhao C., Chen S., et al. Restoration of microRNA function impairs MYC-dependent maintenance of MLL leukemia. Leukemia. 2020;34:2484–2488. doi: 10.1038/s41375-020-0768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao F., Zhang L., Pei X., Lian C., Liu Y., Tan H., Lei P. MiR-20a-5p functions as a potent tumor suppressor by targeting PPP6C in acute myeloid leukemia. PLoS ONE. 2021;16:e0256995. doi: 10.1371/journal.pone.0256995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garbin A., Lovisa F., Holmes A.B., Damanti C.C., Gallingani I., Carraro E., Accordi B., Veltri G., Pizzi M., d’Amore E.S.G., et al. miR-939 acts as tumor suppressor by modulating JUNB transcriptional activity in pediatric anaplastic large cell lymphoma. Haematologica. 2021;106:610–613. doi: 10.3324/haematol.2019.241307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang C., Li Y., Wang L.N., Zhang L., Luo J.S., Peng C.J., Tang W.Y., Huang L.B., Tang Y.L., Luo X.Q. Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology. 2021;26:16–25. doi: 10.1080/16078454.2020.1860187. [DOI] [PubMed] [Google Scholar]
- 51.Wang X., Zuo D., Yuan Y., Yang X., Hong Z., Zhang R. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 2017;143:169–180. doi: 10.1007/s00432-016-2277-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaham L., Vendramini E., Ge Y., Goren Y., Birger Y., Tijssen M.R., McNulty M., Geron I., Schwartzman O., Goldberg L., et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood. 2015;125:1292–1301. doi: 10.1182/blood-2014-06-581892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tapeh B.E.G., Alivand M.R., Solali S. The role of microRNAs in acute lymphoblastic leukaemia: From biology to applications. Cell Biochem. Funct. 2020;38:334–346. doi: 10.1002/cbf.3466. [DOI] [PubMed] [Google Scholar]
- 54.Saito Y., Liang G., Egger G., Friedman J.M., Chuang J.C., Coetzee G.A., Jones P.A. Specific activation of microRNA-127 with downregulation of the proto-Oncogene BCL6 by chromatin-Modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 55.Kong W., He L., Richards E.J., Challa S., Xu C.X., Permuth-Wey J., Lancaster J.M., Coppola D., Sellers T.A., Djeu J.Y., et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.You X., Sun W., Wang Y., Liu X., Wang A., Liu L., Han S., Sun Y., Zhang J., Guo L., et al. Cervical cancer-derived exosomal miR-663b promotes angiogenesis by inhibiting vinculin expression in vascular endothelial cells. Cancer Cell Int. 2021;21:684. doi: 10.1186/s12935-021-02379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., Wang L., Zhou X., Luo X., Liu K., Jiang E., Chen Y., Shao Z., Shang Z. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. BioMed Res. Int. 2020;2020:2125656. doi: 10.1155/2020/2125656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu H.Q., Wang R.K., Wang H.R., Zhou G.Q., Zhang Y.S. miR-137 Inhibition of the Invasion, Metastasis, and Epithelial-Mesenchymal Transition of Nasopharyngeal Cancer by Regulating KDM1A. J. Oncol. 2021;2021:6060762. doi: 10.1155/2021/6060762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan H., Liu F., Ma T., Zeng Z., Zhang N. miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2. Open Life Sci. 2021;16:198–209. doi: 10.1515/biol-2021-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu Z., Xiong J., Xiao R., Shao L., Yang X., Zhou L., Yuan W., Wang M., Yin Q., Wu Y., et al. Loss of miR-146b-5p promotes T cell acute lymphoblastic leukemia migration and invasion via the IL-17A pathway. J. Cell. Biochem. 2019;120:5936–5948. doi: 10.1002/jcb.27882. [DOI] [PubMed] [Google Scholar]
- 61.Deng L., Petrek H., Tu M.J., Batra N., Yu A.X., Yu A.M. Bioengineered miR-124-3p prodrug selectively alters the proteome of human carcinoma cells to control multiple cellular components and lung metastasis in vivo. Acta Pharm. Sin. B. 2021;11:3950–3965. doi: 10.1016/j.apsb.2021.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahman M.M., Brane A.C., Tollefsbol T.O. MicroRNAs and Epigenetics Strategies to Reverse Breast Cancer. Cells. 2019;8:1214. doi: 10.3390/cells8101214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen D., Yang X., Liu M., Zhang Z., Xing E. Roles of miRNA dysregulation in the pathogenesis of multiple myeloma. Cancer Gene Ther. 2021;28:1256–1268. doi: 10.1038/s41417-020-00291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muvarak N., Kelley S., Robert C., Baer M.R., Perrotti D., Gambacorti-Passerini C., Civin C., Scheibner K., Rassool F.V. c-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias. Mol. Cancer Res. 2015;13:699–712. doi: 10.1158/1541-7786.MCR-14-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu J., Ferragut Cardoso A.P., States V.A.R., Al-Eryani L., Doll M., Wise S.S., Rai S.N., States C. Overexpression of hsa-miR-186 induces chromosomal instability in arsenic-exposed human keratinocytes. Toxicol. Appl. Pharmacol. 2019;378:114614. doi: 10.1016/j.taap.2019.114614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cosentino G., Plantamura I., Tagliabue E., Iorio M.V., Cataldo A. Breast Cancer Drug Resistance: Overcoming the Challenge by Capitalizing on MicroRNA and Tumor Microenvironment Interplay. Cancers. 2021;13:3691. doi: 10.3390/cancers13153691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitra A.K., Zillhardt M., Hua Y., Tiwari P., Murmann A.E., Peter M.E., Lengyel E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–1108. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frassanito M.A., Desantis V., Di Marzo L., Craparotta I., Beltrame L., Marchini S., Annese T., Visino F., Arciuli M., Saltarella I., et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J. Pathol. 2019;247:241–253. doi: 10.1002/path.5187. [DOI] [PubMed] [Google Scholar]
- 69.Xi Q., Zhang J., Yang G., Zhang L., Chen Y., Wang C., Zhang Z., Guo X., Zhao J., Xue Z., et al. Restoration of miR-340 controls pancreatic cancer cell CD47 expression to promote macrophage phagocytosis and enhance antitumor immunity. J. Immunother. Cancer. 2020;8:e000253. doi: 10.1136/jitc-2019-000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Del Gaizo M., Sergio I., Lazzari S., Cialfi S., Pelullo M., Screpanti I., Felli M.P. MicroRNAs as Modulators of the Immune Response in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022;23:829. doi: 10.3390/ijms23020829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rashkovan M., Ferrando A. Metabolic dependencies and vulnerabilities in leukemia. Genes Dev. 2019;33:1460–1474. doi: 10.1101/gad.326470.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bao L., Xu T., Lu X., Huang P., Pan Z., Ge M. Metabolic Reprogramming of Thyroid Cancer Cells and Crosstalk in Their Microenvironment. Front. Oncol. 2021;11:773028. doi: 10.3389/fonc.2021.773028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo Presti C., Fauvelle F., Jacob M.C., Mondet J., Mossuz P. The metabolic reprogramming in acute myeloid leukemia patients depends on their genotype and is a prognostic marker. Blood Adv. 2021;5:156–166. doi: 10.1182/bloodadvances.2020002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wojcicki A.V., Kasowski M.M., Sakamoto K.M., Lacayo N. Metabolomics in acute myeloid leukemia. Mol. Genet. Metab. 2020;130:230–238. doi: 10.1016/j.ymgme.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Stockard B., Garrett T., Guingab-Cagmat J., Meshinchi S., Lamba J. Distinct Metabolic features differentiating FLT3-ITD AML from FLT3-WT childhood Acute Myeloid Leukemia. Sci. Rep. 2018;8:5534. doi: 10.1038/s41598-018-23863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen W.L., Wang J.H., Zhao A.H., Xu X., Wang Y.H., Chen T.L., Li J.M., Mi J.Q., Zhu Y.M., Liu Y.F., et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–1654. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao P., Sun L., He X., Cao Y., Zhang H. MicroRNAs and the Warburg Effect: New players in an old arena. Curr. Gene Ther. 2012;12:285–291. doi: 10.2174/156652312802083620. [DOI] [PubMed] [Google Scholar]
- 78.Ozfiliz-Kilbas P., Sonmez O., Obakan-Yerlikaya P., Coker-Gurkan A., Palavan-Ünsal N., Uysal-Onganer P., Arisan E.D. In Vitro Investigations of miR-33a Expression in Estrogen Receptor-Targeting Therapies in Breast Cancer Cells. Cancers. 2021;13:5322. doi: 10.3390/cancers13215322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu S., Wang H., Guo W., Zhou X., Shu Y., Liu H., Yang L., Tang S., Su H., Liu Z., et al. MiR-652-5p elevated glycolysis level by targeting TIGAR in T-cell acute lymphoblastic leukemia. Cell Death Dis. 2022;13:148. doi: 10.1038/s41419-022-04600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rencelj A., Gvozdenovic N., Cemazar M. MitomiRs: Their roles in mitochondria and importance in cancer cell metabolism. Radiol. Oncol. 2021;55:379–392. doi: 10.2478/raon-2021-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prada-Arismendy J., Arroyave J.C., Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76. doi: 10.1016/j.blre.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 82.Mortazavi D., Sohrabi B., Mosallaei M., Nariman-Saleh-Fam Z., Bastami M., Mansoori Y., Daraei A., Vahed S.Z., Navid S., Saadatian Z., et al. Epi-miRNAs: Regulators of the Histone Modification Machinery in Human Cancer. J. Oncol. 2022;2022:4889807. doi: 10.1155/2022/4889807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karimzadeh M.R., Pourdavoud P., Ehtesham N., Qadbeigi M., Asl M.M., Alani B., Mosallaei M., Pakzad B. Regulation of DNA methylation machinery by epi-miRNAs in human cancer: Emerging new targets in cancer therapy. Cancer Gene Ther. 2021;28:157–174. doi: 10.1038/s41417-020-00210-7. [DOI] [PubMed] [Google Scholar]
- 84.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aure M.R., Fleischer T., Bjørklund S., Ankill J., Castro-Mondragon J.A., Børresen-Dale A.L., Tost J., Sahlberg K.K., Mathelier A., Tekpli X., et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021;13:72. doi: 10.1186/s13073-021-00880-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stumpel D., Schotte D., Lange-Turenhout E., Schneider P., Seslija L., de Menezes R.X., Marquez V.E., Pieters R., den Boer M.L., Stam R.W. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: Major matters at a micro scale. Leukemia. 2011;25:429–439. doi: 10.1038/leu.2010.282. [DOI] [PubMed] [Google Scholar]
- 87.Humphries B., Wang Z., Yang C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers. 2019;11:897. doi: 10.3390/cancers11070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xia P., Gu R., Zhang W., Shao L., Li F., Wu C., Sun Y. MicroRNA-377 exerts a potent suppressive role in osteosarcoma through the involvement of the histone acetyltransferase 1-mediated Wnt axis. J. Cell. Physiol. 2019;234:22787–22798. doi: 10.1002/jcp.28843. [DOI] [PubMed] [Google Scholar]
- 89.Roccaro A.M., Sacco A., Jia X., Azab A.K., Maiso P., Ngo H.T., Azab F., Runnels J., Quang P., Ghobrial I.M. microRNA dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Q., Deng X., Tang X., You Y., Mei M., Liu D., Gui L., Cai Y., Xin X., He X., et al. MicroRNA-20a Suppresses Tumor Proliferation and Metastasis in Hepatocellular Carcinoma by Directly Targeting EZH1. Front. Oncol. 2021;11:737986. doi: 10.3389/fonc.2021.737986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 92.Kim M., Civin C.I., Kingsbury T.J. MicroRNAs as regulators and effectors of hematopoietic transcription factors. Wiley Interdiscip. Rev. RNA. 2019;10:e1537. doi: 10.1002/wrna.1537. [DOI] [PubMed] [Google Scholar]
- 93.Surdziel E., Cabanski M., Dallmann I., Lyszkiewicz M., Krueger A., Ganser A., Scherr M., Eder M. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011;117:4338–4348. doi: 10.1182/blood-2010-06-289058. [DOI] [PubMed] [Google Scholar]
- 94.Raghuwanshi S., Dahariya S., Musvi S.S., Gutti U., Kandi R., Undi R.B., Sahu I., Gautam D.K., Paddibhatla I., Gutti R.K. MicroRNA function in megakaryocytes. Platelets. 2019;30:809–816. doi: 10.1080/09537104.2018.1528343. [DOI] [PubMed] [Google Scholar]
- 95.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 96.Rao D.S., O’Connell R.M., Chaudhuri A.A., Garcia-Flores Y., Geiger T.L., Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koenecke C., Krueger A. MicroRNA in T-Cell Development and T-Cell Mediated Acute Graft-Versus-Host Disease. Front. Immunol. 2018;9:992. doi: 10.3389/fimmu.2018.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghisi M., Corradin A., Basso K., Frasson C., Serafin V., Mukherjee S., Mussolin L., Ruggero K., Bonanno L., Guffanti A., et al. Modulation of microRNA expression in human T-cell development: Targeting of NOTCH3 by miR-150. Blood. 2011;117:7053–7062. doi: 10.1182/blood-2010-12-326629. [DOI] [PubMed] [Google Scholar]
- 99.Saultz J.N., Freud A.G., Mundy-Bosse B.L. MicroRNA regulation of natural killer cell development and function in leukemia. Mol. Immunol. 2019;115:12–20. doi: 10.1016/j.molimm.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 100.Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M.B., Wang Y., Qian Z., Jin J., Zhang Y., et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghodousi E.S., Rahgozar S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2018;119:6024–6032. doi: 10.1002/jcb.26800. [DOI] [PubMed] [Google Scholar]
- 102.Almeida R.S., Costa e Silva M., Coutinho L.L., Garcia Gomes R., Pedrosa F., Massaro J.D., Donadi E.A., Lucena-Silva N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol. Oncol. 2019;37:103–112. doi: 10.1002/hon.2567. [DOI] [PubMed] [Google Scholar]
- 103.Tanaka M., Oikawa K., Takanashi M., Kudo M., Ohyashiki J., Ohyashiki K., Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan W., Xu L., Sun Z., Lin Y., Zhang W., Chen J., Hu S., Shen B. MicroRNA biomarker identification for pediatric acute myeloid leukemia based on a novel bioinformatics model. Oncotarget. 2015;6:26424–26436. doi: 10.18632/oncotarget.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin X., Wang Z., Wang Y., Feng W. Serum MicroRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 2015;8:14658–14666. [PMC free article] [PubMed] [Google Scholar]
- 106.Rahimi Z., Ghorbani Z., Motamed H., Jalilian N. Aberrant expression profile of miR-32, miR-98 and miR-374 in chronic lymphocytic leukemia. Leuk. Res. 2021;111:106691. doi: 10.1016/j.leukres.2021.106691. [DOI] [PubMed] [Google Scholar]
- 107.Bagheri M., Khansarinejad B., Mosayebi G., Moradabadi A., Mondanizadeh M. Diagnostic Value of Plasma miR-145 and miR-185 as Targeting of the APRIL Oncogene in the B-cell Chronic Lymphocytic Leukemia. Asian Pac. J. Cancer Prev. 2021;22:111–117. doi: 10.31557/APJCP.2021.22.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keramati F., Jafarian A., Soltani A., Javandoost E., Mollaei M., Fallah P. Circulating miRNAs can serve as potential diagnostic biomarkers in chronic myelogenous leukemia patients. Leuk. Res. Rep. 2021;16:100257. doi: 10.1016/j.lrr.2021.100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poláková K.M., Lopotová T., Klamová H., Burda P., Trněný M., Stopka T., Moravcová J. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. Cancer. 2011;10:41. doi: 10.1186/1476-4598-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paczkowska J., Giefing M. MicroRNA signature in classical Hodgkin lymphoma. J. Appl. Genet. 2021;62:281–288. doi: 10.1007/s13353-021-00614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Culpin R.E., Proctor S.J., Angus B., Crosier S., Anderson J.J., Mainou-Fowler T. A 9 series microRNA signature differentiates between germinal centre and activated B-cell-like diffuse large B-cell lymphoma cell lines. Int. J. Oncol. 2010;37:367–376. doi: 10.1016/S1359-6349(10)71567-6. [DOI] [PubMed] [Google Scholar]
- 112.Denaro M., Navari E., Ugolini C., Seccia V., Donati V., Casani A.P., Basolo F. A microRNA signature for the differential diagnosis of salivary gland tumors. PLoS ONE. 2019;14:e0210968. doi: 10.1371/journal.pone.0210968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fernández C., Bellosillo B., Ferraro M., Seoane A., Sánchez-González B., Pairet S., Pons A., Barranco L., Vela M.C., Gimeno E., et al. MicroRNAs 142-3p, miR-155 and miR-203 Are Deregulated in Gastric MALT Lymphomas Compared to Chronic Gastritis. Cancer Genom. Proteom. 2017;14:75–82. doi: 10.21873/cgp.20020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baraniskin A., Chomiak M., Ahle G., Gress T., Buchholz M., Turewicz M., Eisenacher M., Margold M., Schlegel U., Schmiegel W., et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J. Neurooncol. 2018;137:463–468. doi: 10.1007/s11060-018-2749-0. [DOI] [PubMed] [Google Scholar]
- 115.Xiang Y., Zhang L., Xiang P., Zhang J. Circulating miRNAs as Auxiliary Diagnostic Biomarkers for Multiple Myeloma: A Systematic Review, Meta-Analysis, and Recommendations. Front. Oncol. 2021;11:698197. doi: 10.3389/fonc.2021.698197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kubiczkova L., Kryukov F., Slaby O., Dementyeva E., Jarkovsky J., Nekvindova J., Radova L., Greslikova H., Kuglik P., Vetesnikova E., et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99:511–518. doi: 10.3324/haematol.2013.093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang L.C., Zhong C., Ma Y.J., Liu M., Nong W.X. Expression of MiR-429 in Patients with Acute Lymphoblastic Leukemia and Its Prognostic Value. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28:119–124. doi: 10.19746/j.cnki.issn.1009-2137.2020.01.020. [DOI] [PubMed] [Google Scholar]
- 118.Gębarowska K., Mroczek A., Kowalczyk J.R., Lejman M. MicroRNA as a Prognostic and Diagnostic Marker in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021;22:5317. doi: 10.3390/ijms22105317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agirre X., Vilas-Zornoza A., Jiménez-Velasco A., Martin-Subero J.I., Cordeu L., Gárate L., San José-Eneriz E., Abizanda G., Rogríguez-Otero P., Fortes P., et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 120.Gutierrez-Camino A., Garcia-Obregon S., Lopez-Lopez E., Astigarraga I., Garcia-Orad A. miRNA deregulation in childhood acute lymphoblastic leukemia: A systematic review. Epigenomics. 2020;12:69–80. doi: 10.2217/epi-2019-0154. [DOI] [PubMed] [Google Scholar]
- 121.El-Khazragy N., Noshi M.A., Abdel-Malak C., Zahran R.F., Swellam M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2019;120:6315–6321. doi: 10.1002/jcb.27918. [DOI] [PubMed] [Google Scholar]
- 122.Avigad S., Verly I.R., Lebel A., Kordi O., Shichrur K., Ohali A., Hameiri-Grossman M., Kaspers G.J.L., Cloos J., Fronkova E., et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2016;55:328–339. doi: 10.1002/gcc.22334. [DOI] [PubMed] [Google Scholar]
- 123.Piatopoulou D., Avgeris M., Drakaki I., Marmarinos A., Xagorari M., Baka M., Pourtsidis A., Kossiva L., Gourgiotis D., Scorilas A. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann. Hematol. 2018;97:1169–1182. doi: 10.1007/s00277-018-3292-y. [DOI] [PubMed] [Google Scholar]
- 124.Amankwah E.K., Devidas M., Teachey D.T., Rabin K.R., Brown P.A. Six Candidate miRNAs Associated with Early Relapse in Pediatric B-Cell Acute Lymphoblastic Leukemia. Anticancer Res. 2020;40:3147–3153. doi: 10.21873/anticanres.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weng H., Lal K., Yang F.F., Chen J. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancer Genet. 2015;208:225–229. doi: 10.1016/j.cancergen.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cui L., Zeng T., Zhang L., Liu Y., Wu G., Fu L. High expression of miR-195 is related to favorable prognosis in cytogenetically normal acute myeloid leukemia. Int. J. Clin. Oncol. 2021;26:1986–1993. doi: 10.1007/s10147-021-01999-5. [DOI] [PubMed] [Google Scholar]
- 127.Li S.M., Zhao Y.Q., Hao Y.L., Liang Y.Y. Upregulation of miR-504-3p is associated with favorable prognosis of acute myeloid leukemia and may serve as a tumor suppressor by targeting MTHFD2. Eur. Rev. Med. Pharmacol. Sci. 2019;23:1203–1213. doi: 10.26355/eurrev_201902_17013. [DOI] [PubMed] [Google Scholar]
- 128.Zhang H., Zhang N., Wang R., Shao T., Feng Y., Yao Y., Wu Q., Zhu S., Cao J., Zhang H., et al. High expression of miR-363 predicts poor prognosis and guides treatment selection in acute myeloid leukemia. J. Transl. Med. 2019;17:106. doi: 10.1186/s12967-019-1858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bhayadia R., Krowiorz K., Haetscher N., Jammal R., Emmrich S., Obulkasim A., Fiedler J., Schwarzer A., Rouhi A., Heuser M., et al. Endogenous Tumor Suppressor microRNA-193b: Therapeutic and Prognostic Value in Acute Myeloid Leukemia. J. Clin. Oncol. 2018;36:1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 130.Zhu R., Lin W., Zhao W., Fan F., Tang L., Hu Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J. Cell. Biochem. 2019;120:3958–3968. doi: 10.1002/jcb.27679. [DOI] [PubMed] [Google Scholar]
- 131.Zhu C., Wang Y., Kuai W., Sun X., Chen H., Hong Z. Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin. Biochem. 2013;46:49–53. doi: 10.1016/j.clinbiochem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 132.Qi X., Zhang Y. MicroRNA-199a deficiency relates to higher bone marrow blasts, poor risk stratification and worse prognostication in pediatric acute myeloid leukemia patients. Pediatr. Hematol. Oncol. 2022;19:1–8. doi: 10.1080/08880018.2021.2022045. [DOI] [PubMed] [Google Scholar]
- 133.Marcucci G., Mrózek K., Radmacher M.D., Bloomfield C.D., Croce C.M. MicroRNA expression profiling in acute myeloid and chronic lymphocytic leukaemias. Best Pract. Res. Clin. Haematol. 2009;22:239–248. doi: 10.1016/j.beha.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 134.Khalifa M.M., Zaki N.E., Nazier A.A., Moussa M.A., Haleem R.A., Rabie M.A., Mansour A.R. Prognostic significance of microRNA 17–92 cluster expression in Egyptian chronic lymphocytic leukemia patients. J. Egypt. Natl. Cancer Inst. 2021;33:37. doi: 10.1186/s43046-021-00097-x. [DOI] [PubMed] [Google Scholar]
- 135.Baer C., Oakes C.C., Ruppert A.S., Claus R., Kim-Wanner S.Z., Mertens D., Zenz T., Stilgenbauer S., Byrd J.C., Plass C. Epigenetic silencing of miR-708 enhances NF-κB signaling in chronic lymphocytic leukemia. Int. J. Cancer. 2015;137:1352–1361. doi: 10.1002/ijc.29491. [DOI] [PubMed] [Google Scholar]
- 136.Litwińska Z., Machaliński B. miRNAs in chronic myeloid leukemia: Small molecules, essential function. Leuk. Lymphoma. 2017;58:1297–1305. doi: 10.1080/10428194.2016.1243676. [DOI] [PubMed] [Google Scholar]
- 137.Habib E.M., Nosiar N.A., Eid M.A., Taha A.M., Sherief D.E., Hassan A.E., Abdel Ghafar M.T. MiR-150 Expression in Chronic Myeloid Leukemia: Relation to Imatinib Response. Lab Med. 2022;53:58–64. doi: 10.1093/labmed/lmab040. [DOI] [PubMed] [Google Scholar]
- 138.Ninawe A., Guru S.A., Yadav P., Masroor M., Samadhiya A., Bhutani N., Gupta N., Gupta R., Saxena A. miR-486-5p: A Prognostic Biomarker for Chronic Myeloid Leukemia. ACS Omega. 2021;6:7711–7718. doi: 10.1021/acsomega.1c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jia Y.J., Liu Z.B., Wang W.G., Sun C.B., Wei P., Yang Y.L., You M.J., Yu B.H., Li X.Q., Zhou X.Y. HDAC6 regulates microRNA-27b that suppresses proliferation, promotes apoptosis and target MET in diffuse large B-cell lymphoma. Leukemia. 2018;32:703–711. doi: 10.1038/leu.2017.299. [DOI] [PubMed] [Google Scholar]
- 140.Hedström G., Thunberg U., Berglund M., Simonsson M., Amini R.M., Enblad G. Low expression of microRNA-129-5p predicts poor clinical outcome in diffuse large B cell lymphoma (DLBCL) Int. J. Hematol. 2013;97:465–471. doi: 10.1007/s12185-013-1303-2. [DOI] [PubMed] [Google Scholar]
- 141.Li L., Qiu L., Zhang H., Qian Z., Zhao H. Overexpression of microRNA-130a predicts adverse prognosis of primary gastrointestinal diffuse large B-cell lymphoma. Oncol. Lett. 2020;20:93. doi: 10.3892/ol.2020.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marchesi F., Regazzo G., Palombi F., Terrenato I., Sacconi A., Spagnuolo M., Donzelli S., Marino M., Ercolani C., Di Benedetto A., et al. Serum miR-22 as potential non-invasive predictor of poor clinical outcome in newly diagnosed, uniformly treated patients with diffuse large B-cell lymphoma: An explorative pilot study. J. Exp. Clin. Cancer Res. 2018;37:95. doi: 10.1186/s13046-018-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Troppan K., Wenzl K., Pichler M., Pursche B., Schwarzenbacher D., Feichtinger J., Thallinger G.G., Beham-Schmid C., Neumeister P., Deutsch A. miR-199a and miR-497 Are Associated with Better Overall Survival due to Increased Chemosensitivity in Diffuse Large B-Cell Lymphoma Patients. Int. J. Mol. Sci. 2015;16:18077–18095. doi: 10.3390/ijms160818077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He M., Gao L., Zhang S., Tao L., Wang J., Yang J., Zhu M. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric Cancer. 2014;17:431–441. doi: 10.1007/s10120-013-0313-3. [DOI] [PubMed] [Google Scholar]
- 145.Morales-Martinez M., Vega G.G., Neri N., Nambo M.J., Alvarado I., Cuadra I., Duran-Padilla M.A., Huerta-Yepez S., Vega M.I. MicroRNA-7 Regulates Migration and Chemoresistance in Non-Hodgkin Lymphoma Cells Through Regulation of KLF4 and YY1. Front. Oncol. 2020;10:588893. doi: 10.3389/fonc.2020.588893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou K., Feng X., Wang Y., Liu Y., Tian L., Zuo W., Yi S., Wei X., Song Y., Qiu L. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp. Hematol. 2018;58:27–34.e1. doi: 10.1016/j.exphem.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 147.Papanota A.M., Karousi P., Kontos C.K., Artemaki P.I., Liacos C.I., Papadimitriou M.A., Bagratuni T., Eleutherakis-Papaiakovou E., Malandrakis P., Ntanasis-Stathopoulos I., et al. A Cancer-Related microRNA Signature Shows Biomarker Utility in Multiple Myeloma. Int. J. Mol. Sci. 2021;22:13144. doi: 10.3390/ijms222313144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Desantis V., Solimando A.G., Saltarella I., Sacco A., Giustini V., Bento M., Lamanuzzi A., Melaccio A., Frassanito M.A., Paradiso A., et al. MicroRNAs as a Potential New Preventive Approach in the Transition from Asymptomatic to Symptomatic Multiple Myeloma Disease. Cancers. 2021;13:3650. doi: 10.3390/cancers13153650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ren Y., Li X., Wang W., He W., Wang J., Wang Y. Expression of Peripheral Blood miRNA-720 and miRNA-1246 Can Be Used as a Predictor for Outcome in Multiple Myeloma Patients. Clin. Lymphoma Myeloma Leuk. 2017;17:415–423. doi: 10.1016/j.clml.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 150.Mondal P., Meeran S.M. microRNAs in cancer chemoresistance: The sword and the shield. Noncoding RNA Res. 2021;6:200–210. doi: 10.1016/j.ncrna.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sakurai N., Komada Y., Hanaki R., Morimoto M., Ito T., Nakato D., Hirayama M. Role of microRNAs in glucocorticoid-resistant B-cell precursor acute lymphoblastic leukemia. Oncol. Rep. 2019;42:708–716. doi: 10.3892/or.2019.7191. [DOI] [PubMed] [Google Scholar]
- 152.Najjary S., Mohammadzadeh R., Mansoori B., Vahidian F., Mohammadi A., Doustvandi M.A., Khaze V., Hajiasgharzadeh K., Baradaran B. Combination therapy with miR-34a and doxorubicin synergistically induced apoptosis in T-cell acute lymphoblastic leukemia cell line. Med. Oncol. 2021;38:142. doi: 10.1007/s12032-021-01578-8. [DOI] [PubMed] [Google Scholar]
- 153.Sun H., Zhang Z., Luo W., Liu J., Lou Y., Xia S. NET1 Enhances Proliferation and Chemoresistance in Acute Lymphoblastic Leukemia Cells. Oncol. Res. 2019;27:935–944. doi: 10.3727/096504019X15555388198071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang H., Luo X.Q., Zhang P., Huang L.B., Zheng Y.S., Wu J., Zhou H., Qu L.H., Xu L., Chen Y.Q. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li S.W., Li H., Zhang Z.P., Zhuo F., Li Z.X. Expression and Clinical Significance of MiR-146a and MiR-221 in Childhood Acute T Lymphoblastic Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28:436–441. doi: 10.19746/j.cnki.issn.1009-2137.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 156.Labib H.A., Elantouny N.G., Ibrahim N.F., Alnagar A.A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22:392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 157.Zamani A., Fattahi Dolatabadi N., Houshmand M., Nabavizadeh N. miR-324-3p and miR-508-5p expression levels could serve as potential diagnostic and multidrug-resistant biomarkers in childhood acute lymphoblastic leukemia. Leuk. Res. 2021;109:106643. doi: 10.1016/j.leukres.2021.106643. [DOI] [PubMed] [Google Scholar]
- 158.Zhang H., Liu L., Chen L., Liu H., Ren S., Tao Y. Long noncoding RNA DANCR confers cytarabine resistance in acute myeloid leukemia by activating autophagy via the miR-874-3P/ATG16L1 axis. Mol. Oncol. 2021;15:1203–1216. doi: 10.1002/1878-0261.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vandewalle V., Essaghir A., Bollaert E., Lenglez S., Graux C., Schoemans H., Saussoy P., Michaux L., Valk P.J.M., Demoulin J.B., et al. miR-15a-5p and miR-21-5p contribute to chemoresistance in cytogenetically normal acute myeloid leukaemia by targeting PDCD4, ARL2 and BTG2. J. Cell. Mol. Med. 2021;25:575–585. doi: 10.1111/jcmm.16110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang H., Kang J., Liu L., Chen L., Ren S., Tao Y. MicroRNA-143 sensitizes acute myeloid leukemia cells to cytarabine via targeting ATG7- and ATG2B-dependent autophagy. Aging. 2020;12:20111–20126. doi: 10.18632/aging.103614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bollaert E., Claus M., Vandewalle V., Lenglez S., Essaghir A., Demoulin J.B., Havelange V. MiR-15a-5p Confers Chemoresistance in Acute Myeloid Leukemia by Inhibiting Autophagy Induced by Daunorubicin. Int. J. Mol. Sci. 2021;22:5153. doi: 10.3390/ijms22105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Y., Lei P., Qiao H., Sun K., Lu X., Bao F., Yu R., Lian C., Li Y., Chen W., et al. miR-9 Enhances the Chemosensitivity of AML Cells to Daunorubicin by Targeting the EIF5A2/MCL-1 Axis. Int. J. Biol. Sci. 2019;15:579–586. doi: 10.7150/ijbs.29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiao Y., Deng T., Su C., Shang Z. MicroRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting KRAS. Oncol. Lett. 2017;13:4986–4994. doi: 10.3892/ol.2017.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Z., Fang Z., Lu R., Zhao H., Gong T., Liu D., Hong L., Ma J., Zhang M. MicroRNA-204 Potentiates the Sensitivity of Acute Myeloid Leukemia Cells to Arsenic Trioxide. Oncol. Res. 2019;27:1035–1042. doi: 10.3727/096504019X15528367532612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Said F., Tantawy M., Sayed A., Ahmed S. Clinical Significance of MicroRNA-29a and MicroRNA-100 Gene Expression in Pediatric Acute Myeloid Leukemia. J. Pediatr. Hematol. Oncol. 2022;44:e391–e395. doi: 10.1097/MPH.0000000000002168. [DOI] [PubMed] [Google Scholar]
- 166.Zhang H., Luo X.Q., Feng D.D., Zhang X.J., Wu J., Zheng Y.S., Chen X., Xu L., Chen Y.Q. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. Cancer. 2011;10:108. doi: 10.1186/1476-4598-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fonte E., Apollonio B., Scarfò L., Ranghetti P., Fazi C., Ghia P., Caligaris-Cappio F., Muzio M. In vitro sensitivity of CLL cells to fludarabine may be modulated by the stimulation of Toll-like receptors. Clin. Cancer Res. 2013;19:367–379. doi: 10.1158/1078-0432.CCR-12-1922. [DOI] [PubMed] [Google Scholar]
- 168.Zhu D.X., Zhu W., Fang C., Fan L., Zou Z.J., Wang Y.H., Liu P., Hong M., Miao K.R., Liu P., et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 169.Li Y., Zhao L., Li N., Miao Y., Zhou H., Jia L. miR-9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. Oncol. Rep. 2017;37:2193–2200. doi: 10.3892/or.2017.5464. [DOI] [PubMed] [Google Scholar]
- 170.Jiang X., Cheng Y., Hu C., Zhang A., Ren Y., Xu X. MicroRNA-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Leuk. Lymphoma. 2019;60:1709–1720. doi: 10.1080/10428194.2018.1543875. [DOI] [PubMed] [Google Scholar]
- 171.Klümper T., Bruckmueller H., Diewock T., Kaehler M., Haenisch S., Pott C., Bruhn O., Cascorbi I. Expression differences of miR-142-5p between treatment-naïve chronic myeloid leukemia patients responding and non-responding to imatinib therapy suggest a link to oncogenic ABL2, SRI, cKIT and MCL1 signaling pathways critical for development of therapy resistance. Exp. Hematol. Oncol. 2020;9:26. doi: 10.1186/s40164-020-00183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alves R., Gonçalves A.C., Jorge J., Marques G., Luís D., Ribeiro A.B., Freitas-Tavares P., Oliveiros B., Almeida A.M., Sarmento-Ribeiro A.B. MicroRNA signature refine response prediction in CML. Sci. Rep. 2019;9:9666. doi: 10.1038/s41598-019-46132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Habib E.M., Nosiar N.A., Eid M.A., Taha A.M., Sherief D.E., Hassan A.E., Ghafar M.T.A. Circulating miR-146a expression predicts early treatment response to imatinib in adult chronic myeloid leukemia. J. Investig. Med. 2021;69:333–337. doi: 10.1136/jim-2020-001563. [DOI] [PubMed] [Google Scholar]
- 174.Zhang X.T., Dong S.H., Zhang J.Y., Shan B. MicroRNA-577 promotes the sensitivity of chronic myeloid leukemia cells to imatinib by targeting NUP160. Eur. Rev. Med. Pharmacol. Sci. 2019;23:7008–7015. doi: 10.26355/eurrev_201908_18741. [DOI] [PubMed] [Google Scholar]
- 175.Iqbal J., Shen Y., Huang X., Liu Y., Wake L., Liu C., Deffenbacher K., Lachel C.M., Wang C., Rohr J., et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood. 2015;125:1137–1145. doi: 10.1182/blood-2014-04-566778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bento L., Vögler O., Sas-Barbeito A., Muncunill J., Ros T., Martínez J., Quintero-Duarte A., Ramos R., Asensio V.J., Fernández-Rodríguez C., et al. Screening for Prognostic microRNAs Associated with Treatment Failure in Diffuse Large B Cell Lymphoma. Cancers. 2022;14:1065. doi: 10.3390/cancers14041065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yuan J., Zhang Q., Wu S., Yan S., Zhao R., Sun Y., Tian X., Zhou K. miRNA-223-3p modulates ibrutinib resistance through regulation of the CHUK/Nf-κb signaling pathway in mantle cell lymphoma. Exp. Hematol. 2021;103:52–59.e2. doi: 10.1016/j.exphem.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 178.Wu H., Liu C., Yang Q., Xin C., Du J., Sun F., Zhou L. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy. 2020;16:683–697. doi: 10.1080/15548627.2019.1635380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Amodio N., Gallo Cantafio M.E., Botta C., Agosti V., Federico C., Caracciolo D., Ronchetti D., Rossi M., Driessen C., Neri A., et al. Replacement of miR-155 elicits tumor suppressive activity and antagonizes bortezomib resistance in multiple myeloma. Cancers. 2019;11:236. doi: 10.3390/cancers11020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu J., Su Y., Xu A., Fan F., Mu S., Chen L., Chu Z., Zhang B., Huang H., Zhang J., et al. miR-221/222-mediated inhibition of autophagy promotes dexamethasone resistance in multiple myeloma. Molecules. 2019;27:559–570. doi: 10.1016/j.ymthe.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J.T., Jiang J., Schmittgen T.D., Patel T. Involvement of human microRNAs in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 182.Azimi A., Hagh M.F., Yousefi B., Rahnama M.A., Khorrami A., Heydarabad M.Z., Najafpour M., Hallajzadeh J., Ghahremani A. The Effect of Prednisolone on miR 15a and miR16-1 Expression Levels and Apoptosis in Acute Lymphoblastic Leukemia Cell Line: CCRF-CEM. Drug Res. 2016;66:432–435. doi: 10.1055/s-0042-108640. [DOI] [PubMed] [Google Scholar]
- 183.De Oliveira J.C., Scrideli C.A., Brassesco M.S., Morales A.G., Pezuk J.A., de Queiroz P., Yunes J.A., Brandalise S.R., Tone L.G. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk. Res. 2012;36:293–298. doi: 10.1016/j.leukres.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 184.Capuano C., Pighi C., Maggio R., Battella S., Morrone S., Palmieri G., Santoni A., Klein C., Galandrini R. CD16 pre-ligation by defucosylated tumor-targeting mAb sensitizes human NK cells to gammac cytokine stimulation via PI3K/mTOR axis. Cancer Immunol. Immunother. 2020;69:501–512. doi: 10.1007/s00262-020-02482-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kohnken R., Wen J., Mundy-Bosse B., McConnell K., Keiter A., Grinshpun L., Hartlage A., Yano M., McNeil B., Chakravarti N., et al. Diminished microRNA-29b level is associated with BRD4-mediated activation of oncogenes in cutaneous T-cell lymphoma. Blood. 2018;131:771–781. doi: 10.1182/blood-2017-09-805663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ferreira A.F., Moura L.G., Tojal I., Ambrósio L., Pinto-Simões B., Hamerschlak N., Calin G.A., Ivan C., Covas D.T., Kashima S., et al. ApoptomiRs expression modulated by BCR-ABL is linked to CML progression and imatinib resistance. Blood Cells Mol. Dis. 2014;53:47–55. doi: 10.1016/j.bcmd.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 187.Ali Beg M.M., Guru S.A., Abdullah S.M., Ahmad I., Rizvi A., Akhter J., Goyal J., Verma A.K. Regulation of miR-126 and miR-122 Expression and Response of Imatinib Treatment on Its Expression in Chronic Myeloid Leukemia Patients. Oncol. Res. Treat. 2021;44:530–537. doi: 10.1159/000518722. [DOI] [PubMed] [Google Scholar]
- 188.Mirza M.A.B., Guru S.A., Abdullah S.M., Rizvi A., Saxena A. microRNA-21 Expression as Prognostic and Therapeutic Response Marker in Chronic Myeloid Leukaemia Patients. Asian Pac. J. Cancer Prev. 2019;20:2379–2383. doi: 10.31557/APJCP.2019.20.8.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brüggenwirth I.M.A., Martins P.N. RNA interference therapeutics in organ transplantation: The dawn of a new era. Am. J. Transplant. 2020;20:931–941. doi: 10.1111/ajt.15689. [DOI] [PubMed] [Google Scholar]
- 190.Chakraborty C., Sharma A.R., Sharma G., Lee S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2020;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Van der Ree M.H., van der Meer A.J., van Nuenen A.C., de Bruijne J., Ottosen S., Janssen H.L., Kootstra N.A., Reesink H.W. Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment. Pharmacol. Ther. 2016;43:102–113. doi: 10.1111/apt.13432. [DOI] [PubMed] [Google Scholar]
- 192.Van Roosbroeck K., Calin G.A. MicroRNAs in chronic lymphocytic leukemia: miRacle or miRage for prognosis and targeted therapies? Semin. Oncol. 2016;43:209–214. doi: 10.1053/j.seminoncol.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu S.J., Chen J., Wu B., Wang Y.J., Guo K.Y. MicroRNA-150 enhances radiosensitivity by inhibiting the AKT pathway in NK/T cell lymphoma. J. Exp. Clin. Cancer Res. 2018;37:18. doi: 10.1186/s13046-017-0639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yan S., Chen Y., Yang M., Zhang Q., Ma J., Liu B., Yang L., Li X. hsa-MicroRNA-28-5p Inhibits Diffuse Large B-Cell Lymphoma Cell Proliferation by Downregulating 14-3-3ζ Expression. Evid.-Based Complement. Altern. Med. 2022;2022:4605329. doi: 10.1155/2022/4605329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schilb A.L., Ayat N.R., Vaidya A.M., Hertz L.M., Hall R.C., Scheidt J.H., Sun D., Sun Z., Gopalakrishnan R., Lu Z.R. Efficacy of Targeted ECO/miR-200c Nanoparticles for Modulating Tumor Microenvironment and Treating Triple Negative Breast Cancer as Non-invasively Monitored by MR Molecular Imaging. Pharm. Res. 2021;38:1405–1418. doi: 10.1007/s11095-021-03083-z. [DOI] [PubMed] [Google Scholar]
- 196.Reid G., Williams M., Kirschner M.B., Mugridge N., Weiss J., Brahmbhatt H., Macdiarmid J., Molloy M., Lin R., Van Zandwijk N. Abstract 3976: Targeted delivery of a synthetic microRNA-based mimic as an approach to cancer therapy. Cancer Res. 2015;75((Suppl. 15)) doi: 10.1158/1538-7445.AM2015-3976. [DOI] [Google Scholar]
- 197.Jiang K., Zou H. microRNA-20b-5p overexpression combing Pembrolizumab potentiates cancer cells to radiation therapy via repressing programmed death-ligand 1. Bioengineered. 2022;13:917–929. doi: 10.1080/21655979.2021.2014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yan J., Yao L., Li P., Wu G., Lv X. Long non-coding RNA MIR17HG sponges microRNA-21 to upregulate PTEN and regulate homoharringtonine-based chemoresistance of acute myeloid leukemia cells. Oncol. Lett. 2022;23:24. doi: 10.3892/ol.2021.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liang A.L., Zhang T.T., Zhou N., Wu C.Y., Lin M.H., Liu Y.J. miRNA-10b sponge: An anti-breast cancer study in vitro. Oncol. Rep. 2016;35:1950–1958. doi: 10.3892/or.2016.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Durmaz B., Bagca B.G., Cogulu O., Susluer S.Y., Alpay A., Aksoylar S., Gunduz C. Antileukemic Effects of Anti-miR-146a, Anti-miR-155, Anti-miR-181a, and Prednisolone on Childhood Acute Lymphoblastic Leukemia. BioMed Res. Int. 2021;2021:3207328. doi: 10.1155/2021/3207328. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 201.Chen H., Wang J., Wang H., Liang J., Dong J., Bai H., Jiang G. Advances in the application of Let-7 microRNAs in the diagnosis, treatment and prognosis of leukemia. Oncol. Lett. 2022;23:1–8. doi: 10.3892/ol.2021.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fasihi-Ramandi M., Moridnia A., Najafi A., Sharifi M. Inducing cell proliferative prevention in human acute promyelocytic leukemia by miR-182 inhibition through modulation of CASP9 expression. Biomed. Pharmacother. 2017;89:1152–1158. doi: 10.1016/j.biopha.2017.02.100. [DOI] [PubMed] [Google Scholar]
- 203.Lai T.H., Ewald B., Zecevic A., Liu C., Sulda M., Papaioannou D., Garzon R., Blachly J.S., Plunkett W., Sampath D. HDAC Inhibition Induces MicroRNA-182, which Targets RAD51 and Impairs HR Repair to Sensitize Cells to Sapacitabine in Acute Myelogenous Leukemia. Clin. Cancer Res. 2016;22:3537–3549. doi: 10.1158/1078-0432.CCR-15-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bamodu O.A., Kuo K.T., Yuan L.P., Cheng W.H., Lee W.H., Ho Y.S., Chao T.Y., Yeh C.T. HDAC inhibitor suppresses proliferation and tumorigenicity of drug-resistant chronic myeloid leukemia stem cells through regulation of hsa-miR-196a targeting BCR/ABL1. Exp. Cell Res. 2018;370:519–530. doi: 10.1016/j.yexcr.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 205.Abe F., Kitadate A., Ikeda S., Yamashita J., Nakanishi H., Takahashi N., Asaka C., Teshima K., Miyagaki T., Sugaya M., et al. Histone deacetylase inhibitors inhibit metastasis by restoring a tumor suppressive microRNA-150 in advanced cutaneous T-cell lymphoma. Oncotarget. 2017;8:7572–7585. doi: 10.18632/oncotarget.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang X., Tang J., Zhi X., Xie K., Wang W., Li Z., Zhu Y., Yang L., Xu H., Xu Z. miR-874 functions as a tumor suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway in gastric cancer. Oncotarget. 2015;6:1605–1617. doi: 10.18632/oncotarget.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Moloudizargari M., Hekmatirad S., Mofarahe Z.S., Asghari M.H. Exosomal microRNA panels as biomarkers for hematological malignancies. Curr. Probl. Cancer. 2021;45:100726. doi: 10.1016/j.currproblcancer.2021.100726. [DOI] [PubMed] [Google Scholar]
- 208.Hornick N.I., Huan J., Doron B., Goloviznina N.A., Lapidus J., Chang B.H., Kurre P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015;5:11295. doi: 10.1038/srep11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jiang D., Wu X., Sun X., Tan W., Dai X., Xie Y., Du A., Zhao Q. Bone mesenchymal stem cell-derived exosomal microRNA-7-5p inhibits progression of acute myeloid leukemia by targeting OSBPL11. J. Nanobiotechnol. 2022;20:29. doi: 10.1186/s12951-021-01206-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 210.Zhang L., Khadka B., Wu J., Feng Y., Long B., Xiao R., Liu J. Bone Marrow Mesenchymal Stem Cells-Derived Exosomal miR-425-5p Inhibits Acute Myeloid Leukemia Cell Proliferation, Apoptosis, Invasion and Migration by Targeting WTAP. OncoTargets Ther. 2021;14:4901–4914. doi: 10.2147/OTT.S286326. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 211.Zhang L., Zhou S., Zhou T., Li X., Tang J. Potential of the tumor-derived extracellular vesicles carrying the miR-125b-5p target TNFAIP3 in reducing the sensitivity of diffuse large B cell lymphoma to rituximab. Int. J. Oncol. 2021;58:31. doi: 10.3892/ijo.2021.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Feng Y., Zhong M., Zeng S., Wang L., Liu P., Xiao X., Liu Y. Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics. 2019;11:35–51. doi: 10.2217/epi-2018-0123. [DOI] [PubMed] [Google Scholar]
- 213.Lee J.Y., Ryu D., Lim S.W., Ryu K.J., Choi M.E., Yoon S.E., Kim K., Park C., Kim S.J. Exosomal miR-1305 in the oncogenic activity of hypoxic multiple myeloma cells: A biomarker for predicting prognosis. J. Cancer. 2021;12:2825–2834. doi: 10.7150/jca.55553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tian F., Wang P., Lin D., Dai J., Liu Q., Guan Y., Zhan Y., Yang Y., Wang W., Wang J., et al. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021;112:3744–3755. doi: 10.1111/cas.15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Si W., Shen J., Zheng H., Weimin F. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019;11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ye J., Zou M., Li P., Liu H. MicroRNA Regulation of Energy Metabolism to Induce Chemoresistance in Cancers. Technol. Cancer Res. Treat. 2018;17:1533033818805997. doi: 10.1177/1533033818805997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mollaei H., Safaralizadeh R., Rostami Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 218.De Oliveira J.C., Molinari Roberto G., Baroni M., Bezerra Salomão K., Alejandra Pezuk J., Sol Brassesco M. miRNA Dysregulation in Childhood Hematological Cancer. Int. J. Mol. Sci. 2018;19:2688. doi: 10.3390/ijms19092688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.