Abstract
A novel α-glucosidase with an apparent subunit mass of 59 ± 0.5 kDa was purified from protein extracts of Rhizobium sp. strain USDA 4280, a nodulating strain of black locust (Robinia pseudoacacia L), and characterized. After purification to homogeneity (475-fold; yield, 18%) by ammonium sulfate precipitation, cation-exchange chromatography, hydrophobic chromatography, dye chromatography, and gel filtration, this enzyme had a pI of 4.75 ± 0.05. The enzyme activity was optimal at pH 6.0 to 6.5 and 35°C. The activity increased in the presence of NH4+ and K+ ions but was inhibited by Cu2+, Ag+, Hg+, and Fe2+ ions and by various phenyl, phenol, and flavonoid derivatives. Native enzyme activity was revealed by native gel electrophoresis and isoelectrofocusing-polyacrylamide gel electrophoresis with fluorescence detection in which 4-methylumbelliferyl α-glucoside was the fluorogenic substrate. The enzyme was more active with α-glucosides substituted with aromatic aglycones than with oligosaccharides. This α-glucosidase exhibited Michaelis-Menten kinetics with 4-methylumbelliferyl α-d-glucopyranoside (Km, 0.141 μM; Vmax, 6.79 μmol min−1 mg−1) and with p-nitrophenyl α-d-glucopyranoside (Km, 0.037 μM; Vmax, 2.92 μmol min−1 mg−1). Maltose, trehalose, and sucrose were also hydrolyzed by this enzyme.
Rhizobia are gram-negative soil bacteria belonging to the family Rhizobiaceae (α subgroup of the class Proteobacteria) that are capable of infecting and nodulating the roots of their hosts, leguminous plants (for a review see reference 4). Establishment of an intracellular symbiosis between rhizobia and plants, which leads to nitrogen fixation, requires mutual recognition and signalling, penetration of the host cell, and finally nodulation and survival of bacteria in bacteroids. Glycosidases are probably involved in all of the stages of infection.
Higher plants produce aromatic compounds, such as phenols, flavonoids, and phytohormones in glycosylated forms, that play important roles in bacterial sensing and induction of infection by members of the Rhizobiaceae. Agrobacterium tumefaciens glycosidases have been implicated in the deglycosylation of phenolic derivatives that lead to crown gall tumor formation (9, 27). The gene of a coniferin β-glucosidase from A. tumefaciens B3/73 has also been cloned (6). In addition, the rolC gene of Agrobacterium rhizogenes has been characterized as a cytokinin β-glucosidase gene, and it is suspected that rolA is an auxin β-glucosidase gene (10, 11). In contrast, some particularly interesting bacterial enzymes, such as the α-amylase of Bacillus subtilis X23, can glycosylate various phenolic compounds in vitro (29).
It is also thought that glycosidases play an important role in degradation of the plant cell wall. For a long time, it has been thought that cell wall degradation is caused by bacterial enzymes secreted locally on infection threads (12, 24, 32) or synergistically with plant enzymes (39). Recently, workers have proposed a two-step process involving synthesis of plant enzymes in response to Nod factors, followed by final degradation by rhizobial glycosidases (38). Interestingly, β-glucosidase, α-glucosidase, and β-galactosidase activities have been reported to be associated with 45 strains of rhizobia (15, 34), and some cellulolytic and pectinolytic activities have been detected in Rhizobium leguminosarum (18, 22, 23). Several glycosidases have also been detected in Bradyrhizobium lupini (41). A β-glucosidase that is particularly active with cellobiose has been purified from Agrobacterium faecalis (8, 16). In addition, a β-galactosidase and a β-glucosidase have been purified from the periplasmic space of Rhizobium trifolii (1), and an endoglucanase gene from Azorhizobium caulinodans has been cloned (14). However, the cellulase gene of Erwinia carotovora expressed in Rhizobium fredii has no effect on nodulation of cowpea (20).
It has been shown that a supply of carbohydrate is a major factor in determining each step of nodule formation (2). During soybean nodule development, trehalose, maltose, and sucrose accumulate in the root nodules (35, 37), and trehalose is a major and essential component of nodules (13, 31, 33). Corresponding enzymes for disaccharides have also been detected in nodules (19, 36), and the different compartments of nodules contain numerous glycosidase activities (26). Accumulation of starch also seems to be important during nodulation. Amyloplasts have been found to accumulate in nodules and in dividing inner cortical cells of alfalfa nodulated by Rhizobium meliloti (3). In addition, an R. meliloti nodF nodL mutant which is not able to penetrate the normal host accumulated a substantial amount of starch granules (3). This accumulation was confirmed by the observation that a nonnodulating mutant of Bradyrhizobium japonicum accumulates starch in soybean nodules (28).
It seems clear that glycosidases, particularly glucosidase, maltase, amylase, trehalase, and cellulase, are necessary for the establishment of nodule symbiosis. We are interested in understanding the molecular mechanisms implied in symbiosis and in detecting glycosidases and, therefore, we purified and characterized an α-glucosidase constitutively expressed in Rhizobium sp. strain USDA 4280.
MATERIALS AND METHODS
Microorganism and culture maintenance.
Rhizobium sp. strain USDA 4280 nodulating Robinia pseudoacacia (black locust) was obtained from Peter van Berkum (25). Bacteria were maintained on tryptone yeast (TY) agar (5 g of tryptone per liter, 3 g of yeast per liter, 1 g of CaCl2 per liter, 15 g of agar per liter; pH 7.2) and were cultured aerobically in TY broth at 28°C with agitation. For protein purification, bacteria were grown at 28°C in a 100-liter reactor (Biolafitte, Saint Germain, France) in TY medium with stirring and gentle aeration. Rhizobia were harvested in the late exponential phase and were recovered by centrifugation at 20,000 × g. A 110-g bacterial pellet was obtained from the cultures, divided into 11-g aliquots, and frozen at −80°C.
Protein extraction.
All extraction steps were performed at 4°C. A frozen bacterial pellet (11 g) was resuspended in 60 ml of 50 mM Tris-HCl buffer (pH 8.5) supplemented with 140 mM NaCl and 20 mM benzamidinium chloride with magnetic stirring for 1 h. The sample was subjected to three passages through a French press (American Instrument Company, Silver Spring, Md.) at 1.1 × 107 Pa. The crude extract was stirred in the presence of 1 M NaCl and 1 mM phenylmethylsulfonyl fluoride for 30 min before centrifugation at 25,000 × g for 30 min. The supernatant was then stirred with 0.05% (vol/vol) polyethyleneimine (molecular weight, 50,000; Sigma Chemical Co.) for 30 min. The supernatant obtained after centrifugation at 25,000 × g for 30 min was precipitated with ammonium sulfate at 70% saturation for 16 h with gentle stirring. The precipitated proteins were recovered by centrifugation at 10,000 × g for 1 h and were dialyzed against 20 mM sodium phosphate buffer (pH 7.2) supplemented with 1 mM EDTA and 1 mM dithiothreitol (DTT). The residual insoluble debris was removed after dialysis by centrifugation at 10,000 × g for 1 h.
Purification of α-glucosidase from crude protein extract.
α-Glucosidase was purified from crude protein extract by a four-step chromatographic procedure. The α-glucosidase activity was monitored by using 4-methylumbelliferyl (4-MUF) α-glucoside as the fluorogenic substrate. All steps were performed at 4°C in a cold room.
(i) Ion-exchange chromatography on DEAE-TrisAcryl M.
Approximately 50 ml of clear crude extract was loaded onto a DEAE-TrisAcryl M anion-exchange column (BioSepra; 15 by 1.5 cm; 1 ml/min) that had been equilibrated previously in 20 mM sodium phosphate buffer (pH 7.2) supplemented with 1 mM EDTA and 1 mM DTT. The α-glucosidase was eluted with a linear 20 to 100 mM sodium phosphate buffer (pH 7.2) gradient. Fractions (5 ml) were collected and tested for α-glucosidase activity. Active fractions were pooled, dialyzed, and equilibrated with 20 mM sodium phosphate buffer (pH 7.2) supplemented with 1 mM EDTA and 1 mM DTT.
(ii) Hydrophobic chromatography on phenyl-Sepharose.
NaCl (0.5 M) and 1 mM phenylmethylsulfonyl fluoride were added to α-glucosidase fractions before they were loaded onto a phenyl-Sepharose column (13 by 1 cm; 1 ml/min; Sigma). The column was extensively washed first with 20 mM sodium phosphate buffer supplemented with 1 mM EDTA, 1 mM DTT, and 0.5 M NaCl and then with sodium phosphate buffer without NaCl. The α-glucosidase was eluted with distilled water and then with 4 M urea. Active fractions were pooled and concentrated with an Ultrafree-15 membrane (Amicon) to a final volume of approximately 5 ml.
(iii) Dye chromatography on Reactive Blue 2.
α-Glucosidase fractions were loaded onto a Reactive Blue 2 column (0.5 by 5 cm; 1 ml/min; Sigma) in 20 mM sodium phosphate buffer (pH 7.2). α-Glucosidase passed through the column, and fractions were collected and concentrated with an Ultrafree-15 membrane before equilibration with 20 mM sodium phosphate buffer (pH 7.2).
(iv) Gel filtration with an Ultrogel AcA 44 column.
Fractions containing α-Glucosidase were loaded onto an Ultrogel AcA 44 column (110 by 2 cm; 10 ml/h; BioSepra) in phosphate-buffered saline (PBS)–1 mM EDTA–1 mM DTT. Active fractions were pooled, concentrated with an Ultrafree-15 membrane, and equilibrated with 20 mM sodium phosphate buffer (pH 7.2).
Assay for glycosidase activities.
Glycosidase activities were routinely assayed by using 4-MUF glycosides (Sigma) as substrates. Previous kinetic experiments showed that enzymatic activity was linear after more than 45 min of incubation with the substrate concentration used. Thus, 50 μl of a 10 mM 4-MUF glycoside solution and 20 μl of enzyme extract were added to 1 ml of PBS. The mixture was incubated at 37°C for 30 min in the dark, and the reaction was stopped by adding 500 μl of 1 M sodium carbonate. The fluorescence intensity was measured with a spectrofluorometer (Fluoroskan II; Titertek) by using an excitation wavelength of 365 nm and an emission wavelength of 446 nm. The following 4-MUF glycosides were used: α-d-glucopyranoside, α-l-fucopyranoside, β-l-fucopyranoside, β-d-glucopyranoside, α-l-arabinopyranoside, β-d-acetamido-2-deoxyglucopyranoside, α-d-acetamido-2-deoxyglucopyranoside, α-d-galactopyranoside, β-d-galactopyranoside, α-d-mannopyranoside, β-d-mannopyranoside, β-d-xylopyranoside, β-d-acetamido-2-deoxygalactopyranoside, and β-d-glucuronic acid. 4-MUF was used as the standard. The substrate specificity was determined by measuring the level of free glucose with a model 510-A glucose oxidase kit as described by the manufacturer (Sigma); glucose was used as the standard. The assay for p-nitrophenyl (pNP) α-glucopyranoside hydrolysis was performed in 1 ml of PBS containing 10 mM pNP glucopyranoside and 20 μl of enzyme extract. The mixture was incubated at 37°C for 60 min in the dark, and hydrolysis was stopped by adding 500 μl of 1 M sodium carbonate. The absorbance of the p-nitrophenol released from the pNP substrates was measured at 450 nm. One unit of α-glucosidase activity was defined as the amount of enzyme that produced 1 μmol of glucose or 4-MUF or pNP per min per mg of protein; 1 kat was defined as 1 mol per s per g of protein. Kinetic constants were calculated from double-reciprocal Lineweaver-Burk plots.
Analytical methods.
Protein contents were determined by the Bradford method (5) by using the Bio-Rad protein assay reagent and bovine serum albumin (BSA) as the standard. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was carried out on 12.5% polyacrylamide gels in the presence of β-mercaptoethanol with a Bio-Rad apparatus by using the method of Laemmli (21), and the gels were stained with Coomassie blue R250. The low-molecular-weight protein markers, isoelectrofocusing (IEF) markers, and prestained markers used for blotting were purchased from Bio-Rad. α-Glucosidase activity was routinely detected by native PAGE on 7.5% polyacrylamide gels without SDS and β-mercaptoethanol as described by Davies (7). The gels were soaked in PBS at 37°C for 2 min, and enzyme activity was revealed by incubating the gels with 10 mM 4-MUF α-glucopyranoside in PBS for 10 min at 37°C. Fluorescence was visualized under UV light. Mini IEF-PAGE under nondenaturating conditions was performed with a Bio-Rad apparatus as recommended by the manufacturer by using Bio-Rad IEF markers and Bio-lyte carrier ampholytes in the pH range from 3.0 to 10.0.
Carbohydrate analysis.
Protein glycosylation was analyzed by a modified periodic acid-Schiff procedure. Following SDS-PAGE on 12.5% acrylamide gels, the enzyme was fixed with 5% phosphotungstic acid in 2 N HCl for 1 h. SDS was eliminated from the gels by two 1-h washes with ethanol-acetic acid-H2O (14:7:79). Proteins were oxidized for 1 h in a solution containing 1% (wt/vol) periodic acid and 7% (wt/vol) trichloracetic acid. After a 1-h rinse with 0.5% (wt/vol) sodium metabisulfite in 0.1 N HCl, the Schiff reagent was added, and the gels were soaked overnight at 4°C. Enhanced visualization was accomplished by incubating the gels in 40% (vol/vol) ethanol–5% (vol/vol) acetic acid–0.5% (wt/vol) sodium metabisulfite for 90 min at 55°C. After 24 h of decoloration with 40% (vol/vol) ethanol–5% (vol/vol) acetic acid, glycosylation was observed. A second procedure based on fixation of biotinylated concanavalin A (ConA) was also tested. After SDS-PAGE, the proteins were blotted for 50 min onto a Hybond membrane. The membrane was satured with Tris-buffered saline supplemented with 1% BSA for 1 h at 37°C. The proteins were incubated with biotinylated ConA (4 μg/ml in Tris-buffered saline supplemented with 0.2% BSA and 0.01 mM MnCl2) for 1 h at 37°C. Biotinylated ConA binding was revealed by the following two methods: the avidin peroxidase-chloronaphthol procedure and the avidin-phosphatase (alkaline)-nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) procedure.
NH2-terminal amino acid sequencing.
Sequencing was carried out by using the Edman procedure and an Applied Biosystems model Procise 492 protein sequencer. SDS-PAGE gels were stained for 15 min at 20°C with 0.2% (wt/vol) Coomassie blue R250–20% (vol/vol) methanol and were destained with 30% (vol/vol) methanol. After four washes (30 min each) in bidistilled water, α-glucosidase bands were excised and eluted overnight at 37°C with 200 μl of 0.1 M sodium acetate (pH 8.5) containing 0.1% (wt/vol) SDS. The proteins were transferred onto a ProSorb polyvinylidene difluoride cartridge membrane (Applied Biosystems). The membrane was used directly for protein sequencing.
RESULTS AND DISCUSSION
Glycosidase activities in Rhizobium sp. strain USDA 4280.
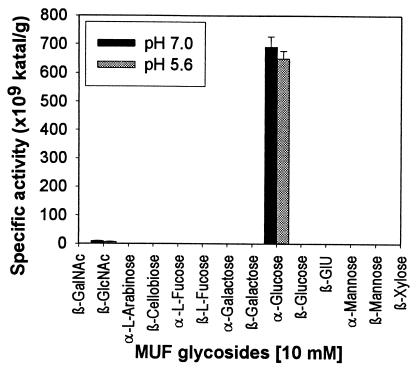
Protein extracts from Rhizobium sp. strain USDA 4280 were assayed to determine their abilities to hydrolyze various kinds of 4-MUF glycosides at pH 5.6 and 7.0 (Fig. 1). High levels of α-glycosidase activity were detected at both pHs. Low levels of activity were recovered from the filtered culture supernatants of bacterial cell cultures, indicating that the enzyme was not secreted. No other 4-MUF glycosides with high levels of hydrolysis activity were detected, although a low level of N-acetyl-glucosaminidase activity was detected.
FIG. 1.
Hydrolysis of various 4-MUF glycosides in 0.1 M sodium acetate buffer (pH 5.6) and in 0.1 M sodium phosphate buffer (pH 7.0) by crude protein extract from Rhizobium sp. strain USDA 4280. GlU, glucuronic acid.
Physical properties of the α-glucosidase.
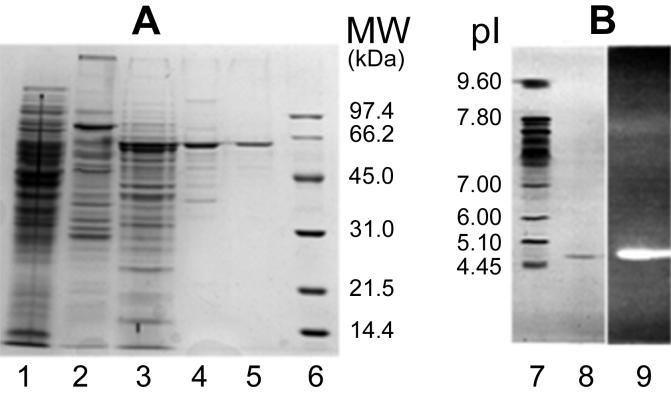
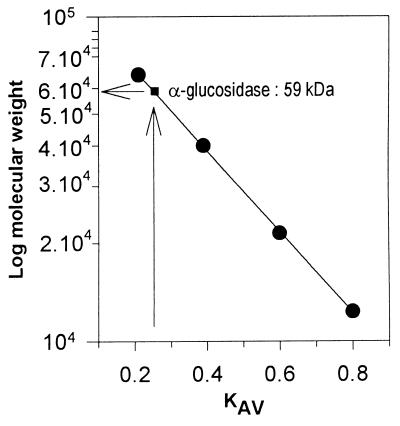
The enzyme was purified by a four-step chromatographic procedure. The final gel filtration step yielded approximately 90 μg of electrophoretically pure α-glucosidase (Table 1). The enzyme was purified 475-fold; the specific activity was 5.71 U/mg, and the yield was 18%. The relative molecular weight (Mr) of the α-glucosidase was estimated by several methods, and 12.5% SDS–PAGE in the presence of β-mercaptoethanol clearly showed that the polypeptide chains did not contain any interchain disulfide bridges (Fig. 2A). The purified α-glucosidase appeared to be a monomer since electrophoresis under both reducing and nonreducing conditions revealed a single band at an Mr of 59,000 ± 500. More extreme reducing conditions, such as 3% SDS, 1.5 mM β-mercaptoethanol, and boiling for 20 min or treatment with 6 M guanidinium chloride, did not release peptides. During Ultrogel AcA 44 gel filtration chromatography, the native enzyme produced a single symmetrical peak corresponding to an Mr of ∼59,000 after calibration with standard proteins (Fig. 3). The purity of the enzyme permitted N-terminal sequencing; however, unfortunately, the N-terminal end of the α-glucosidase was blocked. The pI of the α-glucosidase was determined by analytical electrofocusing. IEF-PAGE of the purified native enzyme from the Ultrogel AcA 44 column resulted in a single polypeptide band at a pI of 4.75 ± 0.5 after coloration with Coomassie blue (Fig. 2B, lane 8). Furthermore, the same polypeptide band was detected in IEF-PAGE gels by the strong fluorescence of 4-MUF resulting from hydrolysis of 4-MUF α-glucoside by the α-glucosidase (Fig. 2B, lane 9). The two different methods used did not permit us to detect glycosylation of the enzyme. First, the enzyme did not bind to the ConA lectin, and second, the enzyme was not stained by the periodic acid-Shiff method.
TABLE 1.
Purification of α-glucosidase activity from Rhizobium sp. strain USDA 4280
| Purification stepa | Vol (ml) | Protein concn (mg/ml) | Total amt of protein (mg) | Activity (μmol/min)b | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|---|
| Dialyzed protein extract | 50 | 4.75 | 238 | 2.9 | 0.012 | 100 | |
| DEAE-TrisAcryl M | 60 | 0.346 | 21 | 2.4 | 0.110 | 9.2 | 82 |
| Phenyl-Sepharose | 5.4 | 1.180 | 6.4 | 0.87 | 0.73 | 60.8 | 30 |
| Reactive Blue 2 | 5.4 | 0.202 | 1.09 | 0.82 | 0.75 | 62.5 | 28 |
| Ultrogel AcA 44 | 12.2 | 0.007 | 0.091 | 0.52 | 5.71 | 475 | 18 |
An 11-g bacterial pellet was used for purification.
α-Glucosidase activity was assayed with 10 mM 4-MUF α-glucoside in PBS at 37°C.
FIG. 2.
(A) 12.5% Polyacrylamide SDS-PAGE of α-glucosidase from Rhizobium sp. strain USDA 4280 in the presence of β-mercaptoethanol. Lane 1, crude protein extract; lane 2, DEAE-TrisAcryl M fractions; lane 3, phenyl-Sepharose fractions; lane 4, Reactive Blue 2 fractions; lane 5, purified α-glucosidase after gel filtration on an Ultrogel AcA 44 column; lane 6, low-molecular-weight Bio-Rad standard markers. (B) IEF-PAGE performed under nonreducing conditions. Lane 7, pI standard markers; lane 8, purified α-glucosidase after gel filtration on an Ultrogel AcA 44 column; lane 9, enzymatic activity of purified α-glucosidase revealed by fluorescence under a UV lamp when 4-MUF α-glucoside (10 mmol liter−1) was used as the substrate. MW, molecular weight.
FIG. 3.
Determination of the molecular weight of the α-glucosidase from Rhizobium sp. strain USDA 4280 by Ultrogel AcA 44 gel filtration in PBS supplemented with 1 mM EDTA and 1 mM DTT. KAV was defined as (Ve − V0)/(Vt − V0), where Ve is the elution volume for the protein, V0 is the void volume (115 ml, as determined with blue dextran), and Vt is the total bed volume (345 ml). The Mr standards (Sigma) used were BSA (molecular weight, 66,200), orosomucoid (40,000), soybean trypsin inhibitor (21,500), and cytochrome c (12,327).
Enzymatic properties of α-glucosidase.
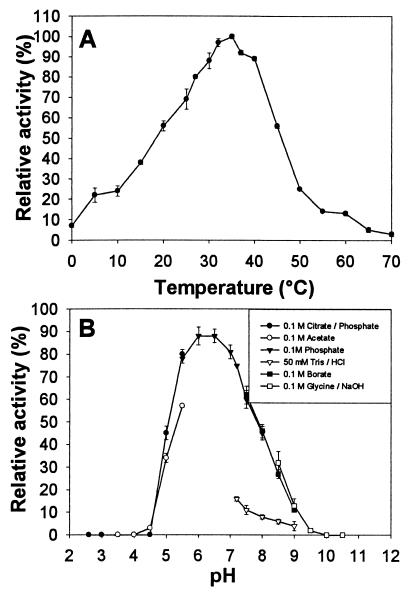
α-Glucosidase appears to be stable for months when it is stored at −20°C and for several weeks when it is stored at 4°C. Freeze-drying does not affect the stability of the enzyme. Optimal activity was observed at pH 6 to 6.5 and at 35°C (Fig. 4). However, the buffer used was important, and the highest levels of activity were obtained with PBS at pH 7.4, suggesting that different ions had cooperative effects. Enzyme activity was optimal with sodium phosphate buffers, in contrast to Tris buffers, which had an inhibitory effect on activity. The effects of various cations at a concentration of 10 mM on the activity of the enzyme were assessed (Table 2). NH4+ and K+ ions were strong enhancers of α-glucosidase activity, while Ag+, Cu2+, Hg2+, and Fe3+ were absolute inhibitors. A similar enhancement of the α-glucosidase activities of members of the Rhizobiaceae by NH4+ and K+ monovalent cations has been described previously (17), which suggests that enzymes could play a role in nodules in which NH4+ is produced. Other agents, such 0.1% SDS, 4 M urea, 50 mM DTT, and phenols (such as acetosyringone or flavonoids) at a concentration of 10 mM, inactivated the enzyme. It has also been reported that phenols and flavonoids, such as p-N-coumaroyltyramine and quercetin isolated from Tochu-cha and Welsh onion, could be potent α-glucosidase inhibitors (30, 40).
FIG. 4.
Physical characterization of the purified α-glucosidase from Rhizobium sp. strain USDA 4280. (A) Optimum temperature for 4-MUF α-glucoside hydrolysis in PBS. The assay results obtained at 35°C were defined as 100% relative activity. (B) Optimum pH for 4-MUF α-glucoside hydrolysis at 37°C with PBS. The experiments were repeated at least four times, and 100% relative activity was defined as 5.71 μmol of 4-MUF liberated/min/mg of protein.
TABLE 2.
Effects of various cations (10 mmol liter−1) on purified Rhizobium sp. strain USDA 4280 α-glucosidase activity
| Cationa | Relative activity (%)b |
|---|---|
| NH4+ | 207.0 ± 7.0 |
| K+ | 141.0 ± 10.0 |
| Mn2+ | 108.5 ± 13.5 |
| Zn2+ | 85.0 ± 9.0 |
| Li2+ | 84.0 ± 10.0 |
| Ca2+ | 81.5 ± 9.5 |
| Mg2+ | 80.5 ± 10.5 |
| Co2+ | 77.5 ± 8.5 |
| Tris+ | 63.5 ± 1.5 |
| Fe2+ | 16.3 ± 4.3 |
Ag+, Cu2+, Hg2+, and Fe3+ completely inhibited α-glucosidase activity.
The buffer used was 100 mM sodium phosphate (pH 7.2), and 100% relative activity was equivalent to 5.12 μmol of 4-MUF liberated/min/mg of protein. PBS, which was used for detection of α-glucosidase, gave a relative activity of 135% ± 4.5%. The experiments were repeated at least four times.
Substrate specificity of α-glucosidase.
The glucosyl hydrolase activity of the enzyme with oligosaccharides was estimated by measuring the levels of free glucose released when a glucose oxidase kit was used. The α-glucosidase exhibited some specificity for aromatic aglycones, such as 4-MUF and pNP α-glucosides (Table 3), and exhibited simple Michaelis-Menten kinetics for 4-MUF α-glucoside (Km, 0.141 μM; Vmax, 6.79 μmol min−1 mg−1) and pNP α-glucoside (Km, 0.037 μM; Vmax, 2.92 μmol min−1 mg−1). Maltose, trehalose, and sucrose were also hydrolyzed by the enzyme, but at a lower rate (Table 3), which revealed that the enzyme had a preference for such dissaccharides. No release of glucose was observed with α-linked oligosaccharides and polymers and β-linked glucosides. This result supports identification of the enzyme as an α-glucosidase (EC 3.2.1.20) with a broad range of activity. However, such an enzyme could specifically interact with aromatic aglycones from plants, such as phenyl, flavonoid, or hormone glycosides.
TABLE 3.
Substrate specificity of the purified α-glucosidase from Rhizobium sp. strain USDA 4280
| Substratea | Glycosidic bond | Relative activity (%)b |
|---|---|---|
| 4-MUF α-glucoside | Glc-α-1-MUF | 100 ± 12 |
| pNP α-glucoside | Glc-α-1-pNP | 100 ± 9 |
| Phenyl α-glucoside | Glc-α-1-phenyl | 80 ± 17 |
| Maltose | Glc-α-1,4-Glc | 51 ± 6 |
| Sucrose | Glc-α-1,2-β-Fru | 28 ± 9 |
| Trehalose | Glc-α-1,1-α-Glc | 14 ± 9 |
Substrates were used at a concentration of 10 mM or 2 mg/ml and were dissolved in PBS. α-Glucosidase did not liberate glucose from methyl α-glucoside, maltotriose, maltotetraose, maltopentaose, maltohexaose, maltoheptaose, maltose 1-phosphate, turanose, maltitol, maltotriitol, isomaltose, isomaltitol, panose, palatinose, palatinitol, starch, amylopectin, β-cyclodextrin, dextran, pullulan, cellobiose, cellulose, lactose, and gentiobiose.
The experiments were repeated five times, and 100% relative activity was equivalent to 6.42 μmol of glucose liberated/min/mg of protein.
ACKNOWLEDGMENTS
We thank P. van Berkum for providing Rhizobium sp. strain USDA 4280 and F. Schoentgen for sequencing the protein. We also thank F. Piller and V. Piller and M. Monsigny for many valuable discussions. S. Hawkins is acknowledged for critically reading the manuscript.
K. Berthelot received a fellowship from MENESRIP (Ministère de l’Éducation Nationale, de l’Enseignement Supérieur et de la Recherche, et de l’Insertion Professionnelle).
REFERENCES
- 1.Abe M, Higashi S. β-Glucosidase and β-galactosidase from the periplasmic space of Rhizobium trifolii cells. J Gen Appl Microbiol. 1982;28:551–562. [Google Scholar]
- 2.Allison F E. Carbohydrate supply as a primary factor in legume symbiosis. Soil Sci. 1935;39:123–143. [Google Scholar]
- 3.Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé J-C, Dénarié J, Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron C, Zambryski P C. Notes from the underground: highlights from plant-microbe interactions. Trends Biotechnol. 1995;13:356–362. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Castle L A, Smith K D, Morris R O. Cloning and sequencing of an Agrobacterium tumefaciens β-glucosidase gene involved in modifying a vir-inducing plant signal molecule. J Bacteriol. 1992;174:1478–1486. doi: 10.1128/jb.174.5.1478-1486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis B J. Clinical applications. Disc electrophoresis. II. Method and application to human serum proteins. Ann NY Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 8.Day A G, Withers S G. The purification and characterization of a β-glucosidase from Alcaligenes faecalis. Biochem Cell Biol. 1986;64:914–922. doi: 10.1139/o86-122. [DOI] [PubMed] [Google Scholar]
- 9.Delay D, Dyé F, Wisniewski J-P, Delmotte F. Synthesis and Agrobacterium vir-inducing activities of coniferyl alcohol β-glycosides. Phytochemistry. 1994;36:289–298. [Google Scholar]
- 10.Estruch J J, Chriqui D, Grossmann K, Schell J, Spena A. The plant oncogene rolC is responsible for the release of cytokinins from glucoside conjugates. EMBO J. 1991;10:2889–2895. doi: 10.1002/j.1460-2075.1991.tb07838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estruch J J, Schell J, Spena A. The protein encoded by the rolB plant oncogene hydrolyses indole glucosides. EMBO J. 1991;10:3125–3128. doi: 10.1002/j.1460-2075.1991.tb04873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fåhraeus G, Ljunggren H. The possible significance of pectic enzymes in root hair infection by nodule bacteria. Physiol Plant. 1959;12:145–154. [Google Scholar]
- 13.Farías-Rodríguez R, Mellor R B, Arias C, Peña-Cabriales J J. The accumulation of trehalose in nodules of several cultivars of common bean (Phaseolus vulgaris) and its correlation with resistance to drought stress. Physiol Plant. 1998;102:353–359. [Google Scholar]
- 14.Geleen D, Van Montagu M, Holsters M. Cloning of an Azorhizobium caulinodans endoglucanase gene and analysis of its role in symbiosis. Appl Environ Microbiol. 1995;61:3304–3310. doi: 10.1128/aem.61.9.3304-3310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glenn A R, Dilworth M J. The uptake and hydrolysis of disaccharides by fast- and slow-growing species of Rhizobium. Arch Microbiol. 1981;129:233–239. [Google Scholar]
- 16.Han Y W, Srinivasan V R. Purification and characterization of β-glucosidase of Alcaligenes faecalis. J Bacteriol. 1969;100:1355–1363. doi: 10.1128/jb.100.3.1355-1363.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoelzle I, Streeter J G. Stimulation of α-glucosidases from fast-growing rhizobia and Agrobacterium tumefaciens by K+, NH4+ and Rb+ Can J Microbiol. 1990;36:223–227. [Google Scholar]
- 18.Hubbell D H, Morales V M, Umali-Garcia M. Pectolytic enzymes in Rhizobium. Appl Environ Microbiol. 1978;35:210–213. doi: 10.1128/aem.35.1.210-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinnback A, Werner D. Glucosidases (α, β) and trehalase (α) in the peribacteroid space and the bacteroid periplasm of Glycine max root nodules. Plant Sci. 1991;77:47–55. [Google Scholar]
- 20.Krishnan H B, Pueppke S G. A cloned cellulase gene from Erwinia carotovora subsp. carotovora is expressed in Rhizobium freddii but does not influence nodulation of cowpea. FEMS Microbiol Lett. 1994;119:289–294. [Google Scholar]
- 21.Laëmmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Molina E, Morales V M, Hubbell D H. Hydrolytic enzyme production by Rhizobium. Appl Environ Microbiol. 1979;38:1186–1188. doi: 10.1128/aem.38.6.1186-1188.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateos P F, Jimenez-Zurdo J I, Chen J, Squartini A S, Haack S K, Martinez-Molina E, Hubbell D H, Dazzo F B. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1992;58:1816–1822. doi: 10.1128/aem.58.6.1816-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy E. Infection by Bact. Radicicola in relation to the microchemistry of the host’s cell walls. Proc R Soc London B Biol Sci. 1932;110:514–533. [Google Scholar]
- 25.McCray-Batzli J, Graves W R, van Berkum P. Diversity among rhizobia effective with Robinia pseudoacacia L. Appl Environ Microbiol. 1992;58:2137–2143. doi: 10.1128/aem.58.7.2137-2143.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellor R B. Bacteroids in the Rhizobium-legume symbiosis inhabit a plant internal lytic compartment: implications for other microbial endosymbioses. J Exp Bot. 1989;40:831–839. [Google Scholar]
- 27.Morris J W, Morris R O. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc Natl Acad Sci USA. 1990;87:3614–3618. doi: 10.1073/pnas.87.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller J, Staehelin C, Boller T, Wiemken A. Carbohydrate pools in nodules of “nonnodulating” and “supernodulating” soybean (Glycine max L. Merr. cv. Bragg) mutants. J Plant Physiol. 1995;145:759–762. [Google Scholar]
- 29.Nishimura T, Kometani T, Takki H, Terada Y, Okada S. Purification and some properties of α-amylase from Bacillus subtilis X-23 that glucosylates phenolic compounds such as hydroquinone. J Ferment Bioeng. 1994;78:31–36. [Google Scholar]
- 30.Nishioka T, Watanabe J, Kawabata J, Niki R. Isolation and activity of N-p-coumaroyltyramine, an α-glucosidase inhibitor in Welsh onion (Allium fistulosum) Biosci Biotechnol Biochem. 1997;61:1138–1141. [Google Scholar]
- 31.Reibach P H, Streeter J G. Metabolism of 14C-labeled photosynthate and distribution of enzymes of glucose metabolism in soybean nodules. Plant Physiol. 1983;72:634–640. doi: 10.1104/pp.72.3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridge R W, Rolfe B G. Rhizobium sp. degradation of legume root hair cell wall at the site of infection thread origin. Appl Environ Microbiol. 1985;50:717–720. doi: 10.1128/aem.50.3.717-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salminen S O, Streeter J G. Enzymes of α,α-trehalose metabolism in soybean nodules. Plant Physiol. 1986;81:538–541. doi: 10.1104/pp.81.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A P, Singh J B. Differences in α- and β-glucosidase and β-galactosidase activity among fast- and slow-growing species of Rhizobium and Agrobacterium tumefaciens. Microbios. 1985;43:169–176. [Google Scholar]
- 35.Streeter J G. Carbohydrate in soybean nodules. II. Distribution of compounds in seedlings during the onset of nitrogen fixation. Plant Physiol. 1980;66:471–476. doi: 10.1104/pp.66.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streeter J G. Enzymes of sucrose, maltose, and α,α-trehalose catabolism in soybean root nodules. Planta. 1982;155:112–115. doi: 10.1007/BF00392540. [DOI] [PubMed] [Google Scholar]
- 37.Streeter J G. Accumulation of α,α-trehalose by Rhizobium bacteria and bacteroids. J Bacteriol. 1985;164:78–84. doi: 10.1128/jb.164.1.78-84.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Spronsen P C, Bakhuisen R, van Brussel A A N, Kijne J W. Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. Eur J Cell Biol. 1994;64:88–94. [PubMed] [Google Scholar]
- 39.Verma D P S, Zogbi V. A cooperative action of plant and Rhizobium to dissolve the host cell wall during development of root nodule symbiosis. Plant Sci Lett. 1978;13:137–142. [Google Scholar]
- 40.Watanabe J, Kawabata J, Kurihara H, Niki R. Isolation and identification of α-glucosidase inhibitors from Tochu-cha (Eucommia ulmoides) Biosci Biotechnol Biochem. 1997;61:177–178. doi: 10.1271/bbb.61.177. [DOI] [PubMed] [Google Scholar]
- 41.Wisniewski J-P, Monsigny M, Delmotte F M. Purification of an α-l-fucoside-binding protein from Rhizobium lupini. Biochimie. 1994;76:121–128. doi: 10.1016/0300-9084(94)90003-5. [DOI] [PubMed] [Google Scholar]