Abstract
Inositol-requiring enzyme 1 (IRE1) is an evolutionarily conserved sensor of endoplasmic reticulum (ER) stress and mediates a key branch of the unfolded protein response in eukaryotic cells. It is an ER-resident transmembrane protein that possesses Ser/Thr protein kinase and endoribonuclease (RNase) activities in its cytoplasmic region. IRE1 is activated through dimerization/oligomerization and autophosphorylation at multiple sites, acting through its RNase activity to restore the functional capacity of the ER. However, it remains poorly defined in vivo how the autophosphorylation events of endogenous IRE1 govern its dynamic activation and functional output. Here, we generated a mouse model harboring a S724A knock-in mutation (Ern1S724A/S724A) and investigated the importance of phosphorylation at Ser724 within the kinase activation loop of murine IRE1α. We found that in mouse embryonic fibroblast cells and in primary hepatocytes, S724A mutation resulted in markedly reduced IRE1α autophosphorylation in parallel with blunted activation of its RNase activity to catalyze X-box binding protein 1 (Xbp1) mRNA splicing. Furthermore, ablation of IRE1α phosphorylation at Ser724 exacerbated ER stress–induced hepatic steatosis in tunicamycin-treated Ern1S724A/S724A mice. This was accompanied by significantly decreased hepatic production of spliced XBP1 protein but increased CCAAT-enhancer–binding protein homologous protein (CHOP) level, along with suppressed expression of key metabolic regulators of fatty acid β-oxidation and lipid secretion. These results demonstrate a critical role of phosphorylation at Ser724 of IRE1α in dynamically controlling its kinase activity, and thus its autophosphorylation state, which is coupled to activation of its RNase activity in counteracting hepatic steatosis under ER stress conditions.
Keywords: ER stress, IRE1α, autophosphorylation, Xbp1, RIDD, hepatic steatosis
Abbreviations: ACC1, acetyl-CoA carboxylase 1; Bip, binding immunoglobulin protein; CHOP, CCAAT-enhancer binding protein homologous protein; ER, endoplasmic reticulum; ES, embryonic stem; FLP, flippase; hsSRS, hyperspectral stimulated Raman scattering; IRE1, inositol-requiring enzyme 1; MEF, mouse embryonic fibroblast; PDI, protein disulfide isomerase; PERK, PKR-like endoplasmic reticulum kinase; Pparγ, peroxisome proliferator–activated receptor gamma; RIDD, regulated IRE1-dependent decay; RNase, ribonuclease; Srebp1, sterol regulatory element–binding transcription factor 1; Tg, thapsigargin; TG, triacylglyceride; Tm, tunicamycin; UPR, unfolded protein response; XBP-1, X-box binding protein 1; XBP1s, spliced XBP1
In eukaryotic cells, the endoplasmic reticulum (ER) is a continuous membrane network that performs a variety of crucial functions, including the synthesis, folding, and processing of nearly one-third of the cellular proteome. Excess accumulation of unfolded or misfolded proteins within the ER lumen results in ER stress, triggering the activation of the adaptive cellular response referred to as the unfolded protein response (UPR) (1, 2, 3). Three ER-resident transmembrane signal transducers, PKR-like endoplasmic reticulum kinase (PERK), activating transcription factor 6, and inositol-requiring enzyme 1 (IRE1), constitute the three canonical UPR signaling branches to manage ER stress and restore cellular homeostasis (1, 2). A great number of studies have demonstrated the pivotal roles of the UPR pathways in cell fate decisions (4) as well as their implications in many pathological conditions including cancer and metabolic disorders (5, 6, 7, 8).
IRE1 is an ancient UPR sensor that is highly conserved from yeast to mammals, including humans (9, 10, 11). It possesses the ER lumenal domain sensing unfolded/misfolded proteins and the cytoplasmic region with dual Ser/Thr protein kinase and endoribonuclease (RNase) activities mediating its functional outputs (12, 13, 14). Upon ER stress, IRE1 RNase is activated through dimerization/oligomerization and transautophosphorylation (15, 16, 17, 18), catalyzing the unconventional splicing of the mRNA encoding X-box binding protein 1 (XBP1) to generate a spliced transcriptionally active transcription factor XBP1s (19, 20, 21). The IRE1–XBP1 pathway initiates a key UPR gene expression program that enhances the protein folding capacity of the ER and the ER-associated degradation of unfolded/misfolded proteins (4, 14, 22, 23, 24, 25). Another effector output of the RNase activity of IRE1 is the degradation of select sets of mRNAs or pre-miRNAs in a process termed “regulated IRE1-dependent decay” (RIDD) (26, 27, 28, 29, 30). Emerging lines of evidence have also demonstrated that in mammals, IRE1α not only has a critical prosurvival role in coping with ER stress but also acts as a multifunctional regulator in response to nutrient stress and metabolic cues (3, 31, 32). A multitude of hormonal, metabolic, and immune signals have been shown to instigate the activation of IRE1α signaling in various metabolic tissues (32), but the precise intracellular mechanisms as well as the molecular signatures with regard to its activation mode and functional output have yet to be fully understood.
Autophosphorylation is a key feature of IRE1 activation in response to ER stress, and its RNase activity is thought to depend upon its kinase activity (33, 34, 35). Biophysical structure studies of the cytoplasmic kinase/RNase portion of yeast IRE1 have shown that dimerization (16) or oligomerization assembly (17) may position the kinase domain for transautophosphorylation at multiple sites, for example, up to 17 residues based on a mass spectrometry assessment (17). Transautophosphorylation is proposed to promote the nucleotide binding and further facilitate its dimerization or oligomer assembly, leading to intrinsic conformational changes for activating its C-terminal RNase domain for RNA substrate processing (16, 17). Structural characterization of the cytoplasmic domain of dephosphorylated human IRE1α bound to ADP has revealed a phosphoryl-transfer–competent dimeric face-to-face complex, with the tip of its kinase activation segment between residues 720 and 730 found to be poorly ordered (18). Moreover, phosphorylation of three residues, Ser724, Ser726, and Ser729, within this conserved kinase activation segment in mammalian IRE1α, has been directly identified from overexpressed cytoplasmic portion of human IRE1α protein (36), and phosphorylation of these sites can affect its RNase activity for Xbp1 mRNA splicing as well as RIDD of certain mRNA substrates (36, 37). However, mutational analyses using overexpressed IRE1α proteins also suggested that phosphorylation of the activation loop is not required for Xbp1 mRNA splicing/cleavage, and phosphorylation at Ser724 and Ser726, but not Ser729, can exert an enhancing effect upon its RNase activity (36). Interestingly, a recent study showed that phosphorylation at Ser729 of endogenous IRE1α in B cells or mouse multiple myeloma cells can be robustly and selectively stimulated by lipopolysaccharides and the bacterial subtilase cytotoxin but not by typical ER stressors such as tunicamycin (Tm) (38). In addition, Ser729 phosphorylation was shown to be dispensable for Xbp1 mRNA splicing activity of IRE1α in lipopolysaccharide-stimulated B cells, and it might exert a prominent enhancing effect upon its RIDD activity in vivo. However, it is unclear whether S729A knock-in mutation can alter the kinase activity of IRE1α for phosphorylating other sites in response to stress stimuli in B cells (38). Therefore, phosphorylation of individual sites, particularly the three residues within the kinase activation loop of IRE1α, can respond to distinct stress signals, suggesting that the phosphorylation events serve to govern the activation mode of its RNase for exerting context-specific effector functions.
Despite the recent progress in our understanding of the molecular details in vitro with regard to the phosphorylation–activation features of IRE1α, it remains largely obscure whether phosphorylation within its kinase activation loop is critically implicated in the dynamic regulation of endogenous IRE1α activation during ER stress in vivo. Importantly, phosphorylation at Ser724 has long been widely utilized for monitoring IRE1α activation in response to a variety of stress signals under physiological or pathological conditions. Hence, we generated a mouse model (Ern1S724A/S724A) possessing the S724A knock-in mutation to abolish phosphorylation at this site and investigated the importance of Ser724 phosphorylation–dependent control of the dynamic activation of IRE1α protein during typical ER stress and determined its functional impact in vivo in the context of ER stress–induced hepatic steatosis.
Results
Generation of the Ern1S724A/S724A knock-in mouse model
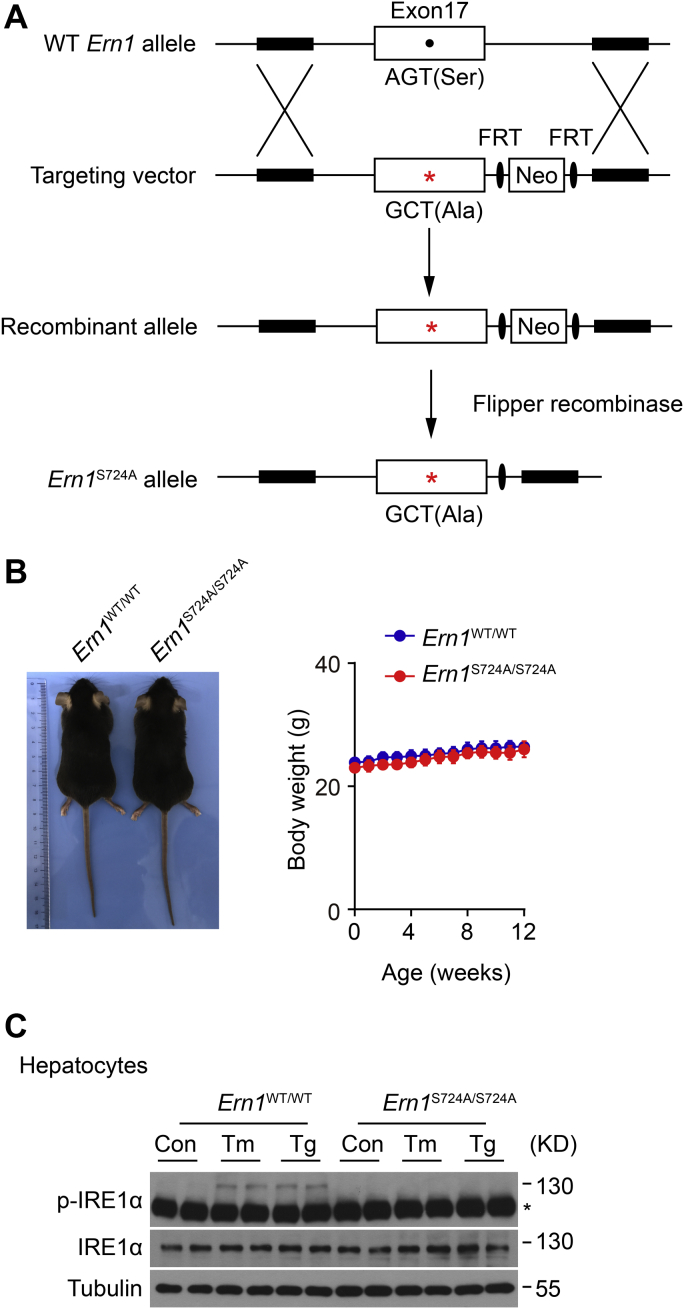
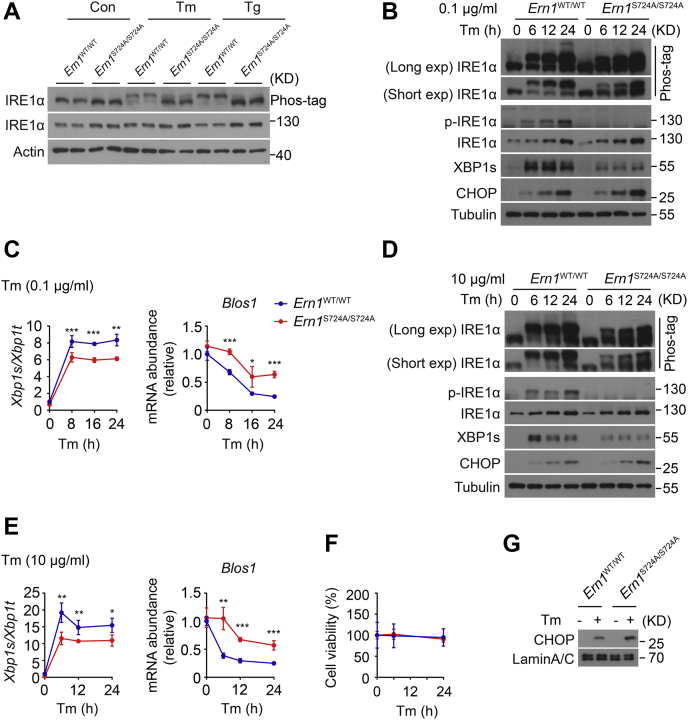
To determine the functional impact of phosphorylation at Ser724 upon the activation of endogenous IRE1α in vivo, we created a knock-in mutant mouse model, denoted Ern1S724A/S724A, in which the Ser724 residue (AGT) was replaced by Ala724 (GCT) within the Ern1 locus (Fig. 1A). After backcrossing into the genetic background of C57BL/6J mice, Ern1S724A/S724A animals were grossly normal, born at normal Mendelian ratios with no overt phenotypic changes in comparison to their Ern1WT/WT littermates such as their body weight (Fig. 1B). Using the commercial antibody directed against phospho-IRE1α at Ser724, we detected robust IRE1α phosphorylation at this site from primary hepatocytes isolated from WT Ern1WT/WT control animals but observed no detectable signals from their Ern1S724A/S724A counterparts upon ER stress induced by Tm or thapsigargin (Tg), the two chemical ER stressors (Fig. 1C). This indicates the successful abrogation of IRE1α phosphorylation at Ser724 within its kinase activation loop in Ern1S724A/S724A knock-in mice.
Figure 1.
Creation of Ern1S724Aknock-in mutant mouse model.A, schematic illustration of the gene targeting strategy for creating Ern1S724A knock-in mutant line. The targeting vector contains exon 17 in which the Ser (S, AGT) residue at 724 was substituted with Ala (A, GCT) in the mouse Ern1 gene, and the FRT-flanked Neomycin (Neo) cassette was subsequently removed by Flipper recombinase. B, photograph of Ern1S724A/S724A mice and Ern1WT/WT littermates and their body weight when maintained on a normal chow diet at the indicated ages (n = 4 per group). C, immunoblot analysis of IRE1α phosphorylation at Ser724 in primary hepatocytes from Ern1WT/WT control and Ern1S724A/S724A mice. Hepatocytes were treated with the chemical ER stressors tunicamycin (Tm, 10 μg/ml) for 4 h or thapsigargin (Tg, 1 μM) for 2 h. The asterisk indicates a nonspecific band detected by anti–phospho-IRE1α antibody. Tubulin was used as the loading control. ER, endoplasmic reticulum; IRE1α, inositol-requiring enzyme 1α.
S724A mutation results in lower autophosphorylation of IRE1α with decreased RNase activation
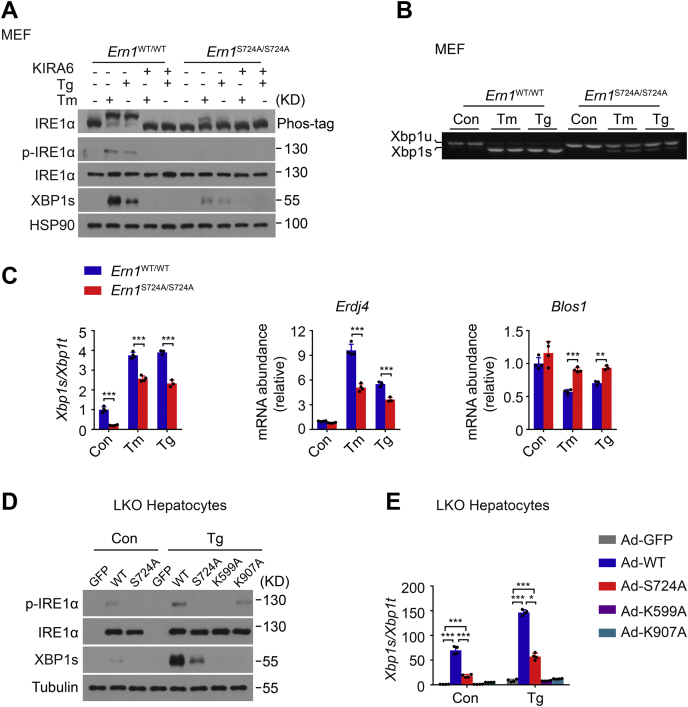
We then generated immortalized mouse embryonic fibroblast (MEF) cell lines from Ern1S724A/S724A mice and their Ern1WT/WT littermates to evaluate the effect of phosphorylation at Ser724 upon the kinase activity of IRE1α for autophosphorylation at other sites and consequently upon the activation of its RNase activity as well. Utilizing Phos-tag gel-based immunoblot analyses (39, 40), we measured the overall phosphorylation states of IRE1α protein in MEF cells in response to chemical ER stress. In Ern1WT/WT control MEF cells treated with 10 μg/ml Tm or 1 μM Tg, we observed apparently slow-migrating IRE1α protein from the Phos-tag gel because of its robust autophosphorylation (Fig. 2A), which was in parallel with phosphorylation at Ser724 as detected by the phospho-IRE1α antibody and could be efficiently blocked by KIRA6, an IRE1α kinase inhibitor (41) (Fig. 2A). By contrast, abrogation of Ser724 phosphorylation markedly reduced autophosphorylation, conceivably at other sites, of IRE1α–S724A protein in Ern1S724A/S724A MEF cells upon ER stress (Fig. 2A). Corresponding to its lower autophosphorylation states, S724A mutation of IRE1α resulted in significant decreases in Xbp1 mRNA splicing and mRNA abundance of Erdj4, an XBP1s target gene, along with significantly increased mRNA abundance of Blos1, a typical RIDD target that affects the cellular behavior and function of lysosomes (42), in Ern1S724A/S724A MEF cells relative to WT Ern1WT/WT control cells (Fig. 2, B and C). To further affirm the effect of S724A mutation upon the RNase activation of IRE1α, we overexpressed by adenovirus infection IRE1α–S724A protein in primary hepatocytes isolated from liver-specific IRE1α knockout (LKO) mice (43) (Fig. 2D). Whereas overexpression of the WT IRE1α protein led to robust Xbp1 mRNA splicing and detectable XBP1s production that could be further increased upon treatment with 1 μM Tg, S724A mutation significantly reduced them under both conditions (Fig. 2E). However, IRE1α–S724A protein possessed significant Xbp1 mRNA splicing activity in comparison to its kinase-dead IRE1α–K599A or RNase-dead K907A mutant protein (44) (Fig. 2E). These results indicate that loss of Ser724 phosphorylation reduces the kinase activity of IRE1α for autophosphorylation at other sites, leading to blunted RNase activity and Xbp1 mRNA splicing during ER stress.
Figure 2.
S724A mutation results in lower IRE1α autophosphorylation with reduced RNase activity upon ER stress.A, MEF cells derived from Ern1WT/WT and Ern1S724A/S724A mice were treated with DMSO (−), 10 μg/ml Tm, for 4 h or 1 μM Tg for 2 h after preincubation for 30 min with DMSO (−) or 10 μM KIRA6, the IRE1α kinase inhibitor. Immunoblot analysis of IRE1α autophosphorylation by Phos-tag gel and its phosphorylation at Ser724 using anti–phospho-IRE1α antibody. HSP90 was used as the loading control. B and C, MEF cells of the indicated genotypes were likewise treated with DMSO (Con), Tm, or Tg. B, agarose gel analysis of Xbp1 mRNA splicing by RT–PCR. Shown are PCR products corresponding to the unspliced Xbp1u and spliced Xbp1s mRNA. C, quantitative RT–PCR analysis of Xbp1 mRNA splicing, shown as the ratio of spliced (Xbp1s) to total (Xbp1t) Xbp1 mRNA, along with the abundance of Erdj4 and Blos1 mRNA. D and E, primary hepatocytes from male liver–specific IRE1α knockout (LKO) mice were infected for 48 h with adenoviruses expressing GFP, WT, or the indicated mutant human IRE1α proteins and subsequently treated for 2 h with DMSO (Con) or 1 μM Tg. D, immunoblot analysis of IRE1α protein and its phosphorylation at Ser724, along with the production of XBP1s protein. Tubulin was used as the loading control. E, quantitative RT–PCR analysis of Xbp1 mRNA splicing. All data are presented as the mean ± SD (n = 2 or 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by two-tailed unpaired Student's t test. DMSO, dimethyl sulfoxide; ER, endoplasmic reticulum; HSP90, heat shock protein 90; IRE1α, inositol-requiring enzyme 1α; MEF, mouse embryonic fibroblast; RNase, ribonuclease; Tg, thapsigargin; Tm, tunicamycin; Xbp1, X-box binding protein 1.
Phosphorylation at Ser724 affects the dynamic activation of IRE1α
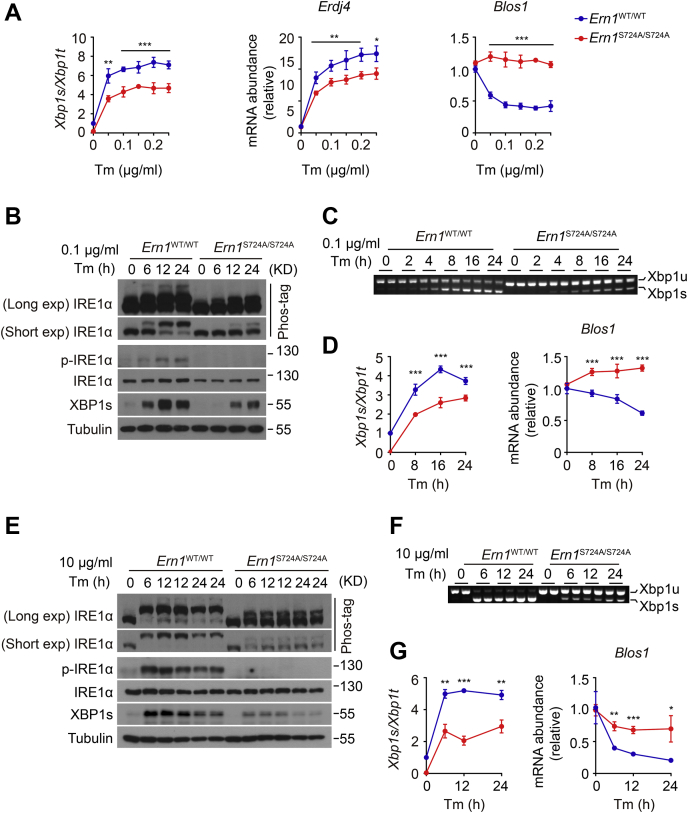
To test whether inactivation of Ser724 phosphorylation influences the dynamic features of IRE1α autophosphorylation in association with its functional outputs of RNase activity, we first measured the dose-responsive activation of IRE1α RNase activity in MEF cells treated for 24 h with Tm at a range of 0.05 to 0.25 μg/ml. Quantitative RT–PCR analysis revealed rapid elevations in Xbp1 mRNA splicing and Erdj4 mRNA levels, as well as a swift decrease in Blos1 mRNA abundance, upon treatment with 0.05 μg/ml Tm, reaching a maximum at 0.15 μg/ml Tm for Xbp1 mRNA splicing and at 0.1 μg/ml Tm for RIDD of Blos1 mRNA in Ern1WT/WT control MEF cells (Fig. 3A). By contrast, regardless of the Tm doses tested, abrogation of Ser724 phosphorylation resulted in a steady ∼40% reduction in the activity of IRE1α for Xbp1 mRNA splicing, and a complete loss of its activity for RIDD of Blos1 mRNA, in Ern1S724A/S724A MEF cells (Fig. 3A). Next, we examined by Phos-tag gel analysis, the phosphorylation states of IRE1α at different time intervals during relatively mild ER stress induced by 0.1 μg/ml Tm, and we detected a majority of robustly phosphorylated IRE1α protein that increased in a time-dependent fashion after Tm treatment in Ern1WT/WT cells (Fig. 3B). Interestingly, we also observed some very minor super-shifted species, likely representing IRE1α protein molecules that possessed more residues undergoing autophosphorylation. In comparison, S724A mutation resulted in dramatic decreases in the amount of autophosphorylated IRE1α–S724A protein in Ern1S724A/S724A cells even after 24 h of Tm treatment, which also exhibited an appreciably lower shift relative to its WT counterpart in Ern1WT/WT cells (Fig. 3B). In parallel with the phosphorylation levels of IRE1α protein, production of XBP1s protein (Fig. 3B) as well as its mRNA splicing (Fig. 3, C and D) reached to their highest levels at 12 and 16 h, respectively, after Tm treatment in Ern1WT/WT MEF cells; whereas significant reductions in XBP1s protein production (Fig. 3B) and its mRNA splicing (Fig. 3, C and D) were observed in Ern1S724A/S724A cells. Consistently, no decrease in Blos1 mRNA abundance was detected in Ern1S724A/S724A MEF cells (Fig. 3D). Then, we treated MEF cells with higher concentration of Tm at 10 μg/ml and asked if S724A mutation could impact the kinase/RNase activity of IRE1α under a more severe ER stress condition. Phos-tag gel analysis showed marked phosphorylation of IRE1α protein at 6 h following high Tm treatment, which was sustained at similar levels thereafter in Ern1WT/WT cells (Fig. 3E). This was accompanied by highly robust Xbp1 mRNA splicing maintained at similar levels throughout the period of high Tm treatment from 6 to 24 h (Fig. 3, F and G), along with rapid and gradual decreases in Blos1 mRNA abundance (Fig. 3G). In Ern1S724A/S724A MEF cells, by contrast, stimulation with high Tm resulted in similarly rapid occurrence of phosphorylated IRE1α–S724A protein that was obviously less abundant and shifted lower in comparison to their WT counterparts and did not increase in a time-dependent manner (Fig. 3E). In correspondence to the altered phosphorylation states of IRE1α–S724A protein, ∼40 to 60% reductions were observed in its activities for Xbp1 mRNA splicing and Blos1 mRNA degradation (Fig. 3, F and G). Unlike that under mild ER stress state, IRE1α–S724A protein exhibited a considerable RIDD activity for Blos1 mRNA upon severe ER stress induced by high Tm (Fig. 3G). Interestingly, we observed a high amount of XBP1s protein induced at 6 h that decreased in Ern1WT/WT cells at 24 h following high Tm treatment, while detecting an apparently lower amount of XBP1s protein that similarly decreased in Ern1S724A/S724A cells after 24 h of high Tm treatment (Fig. 3E). This may indicate similar turnover rate of XBP1s protein in these two types of MEF cells during severe ER stress.
Figure 3.
Effects of S724A mutation upon the dynamic activation of IRE1α in MEF cells.A, MEF cells of the indicated genotypes were treated with Tm at the indicated concentrations for 24 h. Quantitative RT–PCR analysis of Xbp1 mRNA splicing and the mRNA abundance of the indicated genes. B–G, MEF cells were treated with 100 ng/ml or 10 μg/ml Tm for the indicated time intervals. B and E, immunoblot analysis of IRE1α phosphorylation and XBP1s protein. Upper Phos-tag gels with long or short exposure time are shown for the band-shift analysis of IRE1α protein phosphorylation. Tubulin was used as the loading control. C and F, agarose gel analysis of Xbp1 mRNA splicing by RT–PCR. D and G, quantitative RT–PCR analysis of Xbp1 mRNA splicing and the RIDD target Blos1 mRNA. All data are presented as the mean ± SD (n = 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test or two-way ANOVA. IRE1α, inositol-requiring enzyme 1α; MEF, mouse embryonic fibroblast; RIDD, regulated IRE1-dependent decay; Tm, tunicamycin; Xbp1, X-box binding protein 1; XBP1s, spliced XBP1.
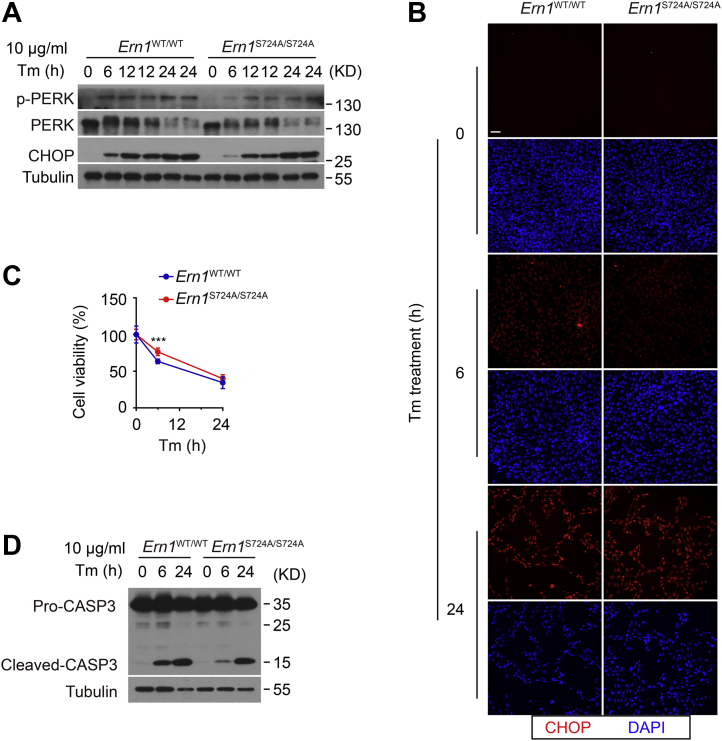
Given the critical role of the IRE1α branch in cell fate control (4), we wondered if this dynamic impairment of IRE1α RNase activation is coupled to cell survival during ER stress. Curiously, at 6 h but not at 24 h following treatment with high Tm at 10 μg/ml, we observed decreased PERK phosphorylation along with lower protein level of the transcription factor CCAAT-enhancer binding protein homologous protein (CHOP) in Ern1S724A/S724A MEF cells (Fig. 4A), which was in accordance with lower accumulation of CHOP protein detected in the nucleus at this particular stage of ER stress (Fig. 4B). Consistently, a significant, though not dramatic, improvement in cell viability, as well as a lower level of the cleaved form of caspase-3, was observed in stressed Ern1S724A/S724A MEF cells at 6 h after Tm treatment (Fig. 4, C and D). This indicates that loss of Ser724 phosphorylation of IRE1α may promote MEF cell survival even in the face of impaired production of XBP1s at early stages of ER stress, likely through attenuating the PERK–CHOP pathway.
Figure 4.
Effects of S724A mutation upon cell survival under ER stress in MEF cells. MEF cells of the indicated genotypes were treated with 10 μg/ml Tm for the indicated time intervals. A, immunoblot analysis of PERK phosphorylation and CHOP protein levels. Tubulin was used as the loading control. B, immunofluorescence staining of CHOP protein along with DAPI staining in MEF cells following Tm treatment. The scale bar represents 50 μm. C, cell viability of Tm-treated MEF cells was determined by CCK8 assay. Shown is the percentage of viability after normalization to their untreated controls. D, immunoblot analysis of cleavage of caspase-3 protein. Tubulin was used as the loading control. Data are presented as the mean ± SD (n = 3 independent experiments). ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test. CCK8, Cell Counting Kit-8; CHOP, CCAAT-enhancer binding protein homologous protein; DAPI, 4′,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; MEF, mouse embryonic fibroblast; PERK, PKR-like endoplasmic reticulum kinase; Tm, tunicamycin.
Next, we considered if inactivation of Ser724 phosphorylation could impact the dynamic activation of IRE1α in a cell type–dependent fashion. Using isolated primary hepatocytes from Ern1WT/WT and Ern1S724A/S724A mice, Phos-tag gel analyses revealed obvious decreases in Tm- or Tg-induced autophosphorylation of IRE1α–S724A protein with less shifting relative to its WT counterpart (Fig. 5A), which is analogous to our observations in MEF cells (Fig. 2A). Under mild ER stress induced by 0.1 μg/ml Tm, we observed overt reductions in autophosphorylation of IRE1α–S724A protein in Ern1S724A/S724A hepatocytes relative to its WT counterpart in Ern1WT/WT hepatocytes (Fig. 5B). Interestingly, Tm treatment induced a time-dependent increase in IRE1α protein level in both hepatocytes (Fig. 5B), which was not observed in MEF cells; and Tm-induced phosphorylation of IRE1α–S724A protein appeared to be higher in Ern1S724A/S724A hepatocytes than that in Ern1S724A/S724A MEF cells, particularly at 24 h after Tm treatment (Figs. 3B and 5B). Whereas Xbp1 mRNA splicing was maintained at similar levels in Ern1WT/WT control hepatocytes after 8 h of Tm treatment, it was decreased to a similar extent (by ∼25%) in Ern1S724A/S724A hepatocytes (Fig. 5C). Unlike our observations in MEF cells (Fig. 3D), S724A mutation resulted in significant reductions in, but not a complete loss of, its RIDD activity for Blos1 mRNA (Fig. 5C). Under severe ER stress induced by 10 μg/ml Tm, we observed similar effects of S724A mutation upon IRE1α autophosphorylation states and XBP1s protein production (Fig. 5D) in association with its RNase activities (Fig. 5E) in Ern1S724A/S724A hepatocytes. Notably, at 24 h after Tm treatment, CHOP protein levels appeared to be elevated in Ern1S724A/S724A hepatocytes (Fig. 5, B and D). However, relative to Ern1WT/WT hepatocytes, no changes in cell viability were detected in Ern1S724A/S724A hepatocytes (Fig. 5F), despite that appreciably higher nuclear accumulation of CHOP protein was found in Ern1S724A/S724A hepatocytes at 24 h following high Tm treatment (Fig. 5G). This may reflect a unique ability of hepatocytes to counteract ER stress–associated cell death without involving the CHOP pathway, which has been frequently shown to mediate apoptosis in other cell types (4).
Figure 5.
Disruption of Ser724phosphorylation attenuates the RNase activation of IRE1α in hepatocytes.A, primary hepatocytes from mice of the indicated genotypes were treated with DMSO control (Con), 10 μg/ml Tm for 4 h, or 1 μM Tg for 2 h. Immunoblot analysis of IRE1α protein, with upper Phos-tag gel analysis of IRE1α phosphorylation. Actin was used as the loading control. B–E, hepatocytes of the indicated genotypes were treated with 100 ng/ml or 10 μg/ml Tm for the indicated time intervals. B and D, immunoblot analysis of IRE1α phosphorylation along with XBP1s and CHOP protein levels. Upper Phos-tag gels are shown for the band-shift analysis of IRE1α protein phosphorylation with long or short exposure time. Tubulin was used as the loading control. C and E, quantitative RT–PCR analysis of Xbp1 mRNA splicing and the RIDD target Blos1 mRNA. F and G, hepatocytes were treated with 10 μg/ml Tm. F, cell viability analysis by CCK8 assay. G, immunoblot analysis of nuclear CHOP protein after 24 h of 10 μg/ml Tm treatment. Lamin A/C was used as the nuclear protein control. All data are presented as the mean ± SD (n = 2 or 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test or two-way ANOVA. CCK8, Cell Counting Kit-8; CHOP, CCAAT-enhancer binding protein homologous protein; DMSO, dimethyl sulfoxide; IRE1α, inositol-requiring enzyme 1α; RIDD, regulated IRE1-dependent decay; RNase, ribonuclease; Tg, thapsigargin; Tm, tunicamycin; XBP1s, spliced X-box binding protein 1.
Taken together, these results from MEF cells and hepatocytes demonstrate an important role of phosphorylation at Ser724 in governing the kinase activity of IRE1α for its further autophosphorylation, which is closely coupled to the activation mode of its RNase activity. Abrogation of phosphorylation at this particular site of the kinase activation loop is likely to cause intrinsic defect in the ability of IRE1α to autophosphorylate sufficiently at additional sites, which are supposed to be needed for its activation to the fullest capacity. Unlike the documented phosphorylation at Ser729 that preferably responds to TLR agonists and primarily regulates its RIDD activity (38), phosphorylation at Ser724 of IRE1α may represent a universal event during its activation upon ER stress, which controls its functional RNase output for both Xbp1 mRNA splicing and RIDD of select RNA substrates. It is currently unclear whether the cell type–selective feature of IRE1α-S724A mutant’s RIDD activity reflects its distinct phosphorylation states exerting an impact upon possible auxiliary factors in the RIDD machinery in different types of cells. In addition, phosphorylation at Ser724 of IRE1α may also exert differing cell type–dependent effects upon cell survival under ER stress.
Abrogation of IRE1α phosphorylation at Ser724 exacerbates ER stress–induced hepatic steatosis
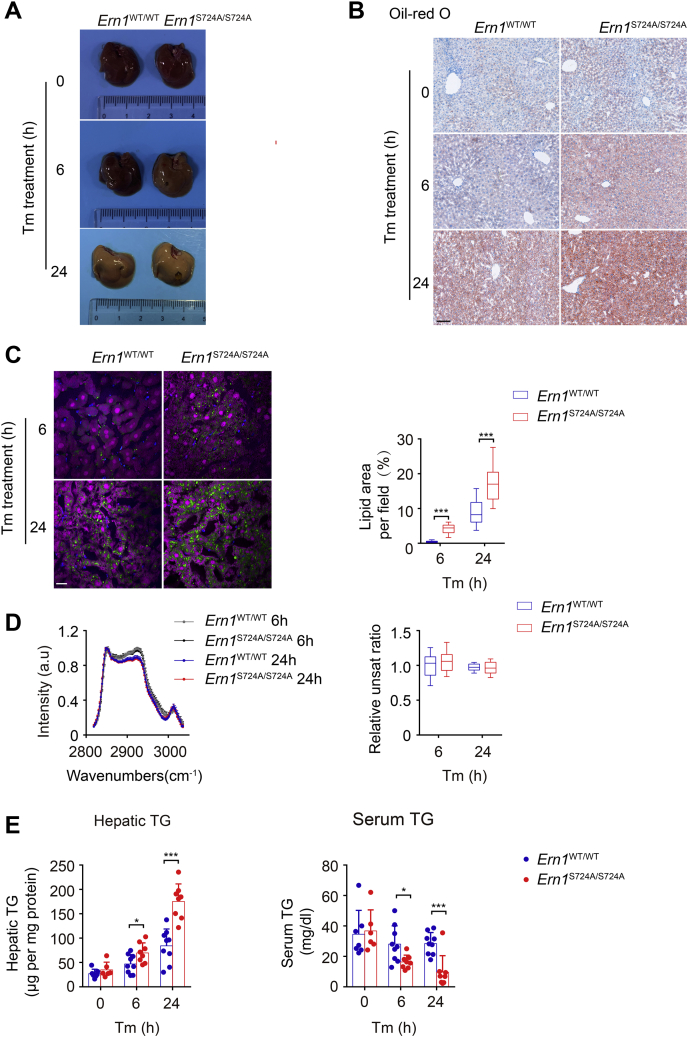
ER stress is a key feature in hepatic steatosis and nonalcoholic fatty liver disease (45, 46, 47), and the IRE1α–XBP1 pathway has been implicated in the control of many aspects of hepatic lipid homeostasis, including de novo lipogenesis, fatty acid β-oxidation, and lipid secretion (43, 48, 49, 50). Given that IRE1α was documented to have a protective role in ER stress–induced liver steatosis (51, 52), we sought to determine the physiological importance of phosphorylation at Ser724 of IRE1α during its activation in the liver in response to chemical ER stress in Tm-treated mice. Abrogation of Ser724 phosphorylation of IRE1α resulted in more severe hepatic steatosis, that is, higher accumulation of liver lipids as determined by Oil-Red O staining, in Ern1S724A/S724A mice than their Ern1WT/WT control group following 6 or 24 h of Tm treatment (Fig. 6, A and B). Consistently, imaging analysis by hyperspectral stimulated Raman scattering (hsSRS) of liver sections (53) also revealed significant elevations in hepatic lipid content (Fig. 5C), but no alterations in the unsaturated ratio of lipids (Fig. 6D) in Tm-treated Ern1S724A/S724A mice relative to Ern1WT/WT mice were revealed. Furthermore, more progressive increases in triacylglycerides (TG) levels were detected in the livers of Ern1S724A/S724A mice than their Ern1WT/WT counterparts following 6 and 24 h of Tm treatment, which was accompanied by higher decreases in their serum TG levels (Fig. 6E). These results indicate that inactivation of Ser724 phosphorylation of IRE1α functionally impairs its ability to regulate hepatic lipid metabolism, leading to higher liver accumulation with lower secretion of neutral lipids, that is, exacerbation of ER stress–induced liver steatosis.
Figure 6.
S724A mutation of IRE1α results in aggravated hepatic steatosis with reduced plasma lipids under ER stress. Male Ern1S724A/S724A mice and their Ern1WT/WT littermates at 2 months of age were injected i.p. with Tm (1 mg/kg body weight) and sacrificed at 6 or 24 h after Tm treatment (n = 6–9 per group). Mice treated with 150 μM dextrose was used as the vehicle control group. A, representative liver images. B, representative images of Oil-Red O staining of livers (n = 3 per group). The scale bar represents 100 μm. C, representative hsSRS images of liver sections from mice at 6 and 24 h after Tm injection. Imaging analysis by the Multivariate Curve Resolution (MCR) algorithm showing chemical distributions of lipid (green), lipofuscin (blue), and protein (magenta) in a field of 200 × 200 μm2 (300 × 300 pixels with dwell time of 10 μs/pixel for imaging). Lipid contents were quantified as areas of lipid signals per field using ImageJ (n = 3 per group). The scale bar represents 20 μm. D, the average hsSRS spectra of hepatic lipids. The Raman peak at 3007 cm−1 represents the vibration from unsaturated =CH stretch. The degree of lipid unsaturation was evaluated by the ratio of Raman intensity at 3007 and 2853 cm−1. E, liver and serum levels of triglycerides. Data represent the mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test. ER, endoplasmic reticulum; hsSRS, hyperspectral stimulated Raman scattering; IRE1α, inositol-requiring enzyme 1α; Tm, tunicamycin.
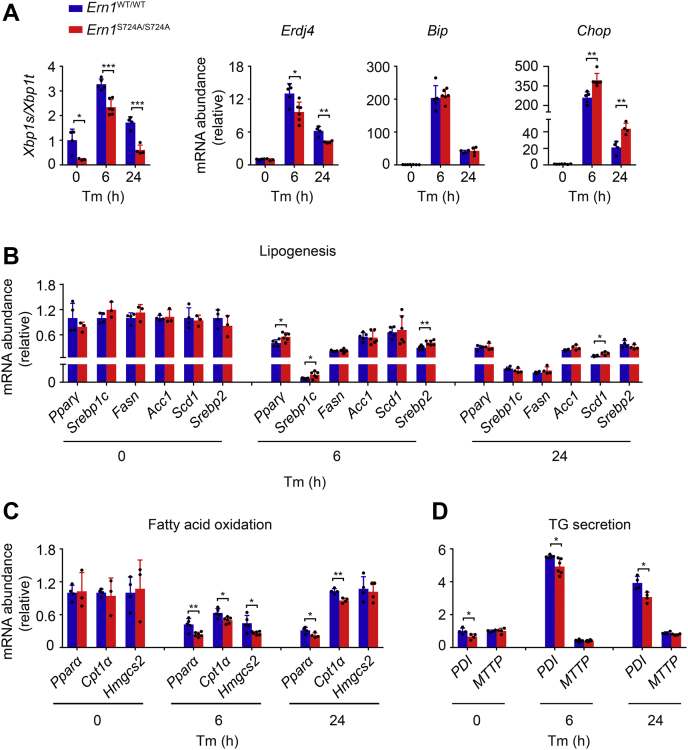
Because hepatic lipid accumulation dynamically involves metabolic pathways including de novo lipogenesis, fatty acid oxidation, as well as lipid secretion and uptake, we next examined whether impairment of IRE1α phosphorylation affected the related gene expression programs in the livers of Ern1S724A/S724A mice. Quantitative RT–PCR profiling revealed significantly blunted induction of Xbp1 mRNA splicing and Erdj4 mRNA expression in Ern1S724A/S724A livers at 6 and 24 h after Tm treatment (Fig. 7A), suggesting a lower RNase activity of the IRE1α–S724A protein in vivo. Surprisingly, while no significant changes were detected in Tm stimulation of binding immunoglobulin protein (Bip) mRNA expression, significantly higher upregulation of Chop mRNA was observed in Ern1S724A/S724A livers (Fig. 7A). In parallel, whereas Tm treatment exerted a suppressive effect upon the expression of key transcriptional regulators and metabolic enzymes in both lipogenesis and fatty acid oxidation (Fig. 7, B and C), we detected significant increases in the expression of lipogenic genes, including peroxisome proliferator–activated receptor gamma (Pparγ), sterol regulatory element–binding transcription factor 1 (Srebp1c), and Scd1 (Fig. 7B), along with significant decreases in that of genes related to fatty acid oxidation such as Pparα and Cpt1α (Fig. 7C), in Ern1S724A/S724A livers at 6 or 24 h after Tm treatment. These data suggest that the lowered RNase activity of IRE1α–S724A protein resulted in decreased Xbp1 mRNA splicing, along with enhanced expression of Chop, which also has been shown to be a key mediator in ER stress–induced suppression of genes related to lipid metabolism in hepatic steatosis (54). Moreover, in the livers of Ern1WT/WT control mice, we observed marked Tm-induced upregulation of PDI (encoding protein disulfide isomerase [PDI]) but not MTTP (encoding microsomal triglyceride–transfer protein [MTTP]) (Fig. 7D), and a significant attenuation of Tm-induced upregulation of PDI was seen in Ern1S724A/S724A livers (Fig. 7D). This blunted expression of PDI, a crucial regulator of TG secretion from the liver, could also be ascribable to the weakened activation of Xbp1 mRNA splicing in Ern1S724A/S724A livers, which is in line with the reported findings (50).
Figure 7.
Effects of S724A mutation of IRE1α upon hepatic gene expression programs related to ER stress and lipid metabolism. Quantitative RT–PCR analysis of the mRNA abundance of the indicated genes in livers of Ern1S724A/S724A mice and their Ern1WT/WT littermates following treatment with Tm or vehicle (n = 3–6 per group). A, the UPR genes. B, lipogenesis. C, fatty acid β-oxidation. D, TG secretion. Data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test. ER, endoplasmic reticulum; IRE1α, inositol-requiring enzyme 1α; TG, triacylglyceride; Tm, tunicamycin; UPR, unfolded protein response.
IRE1α regulates hepatic expression of CHOP and PDI through XBP1s under ER stress
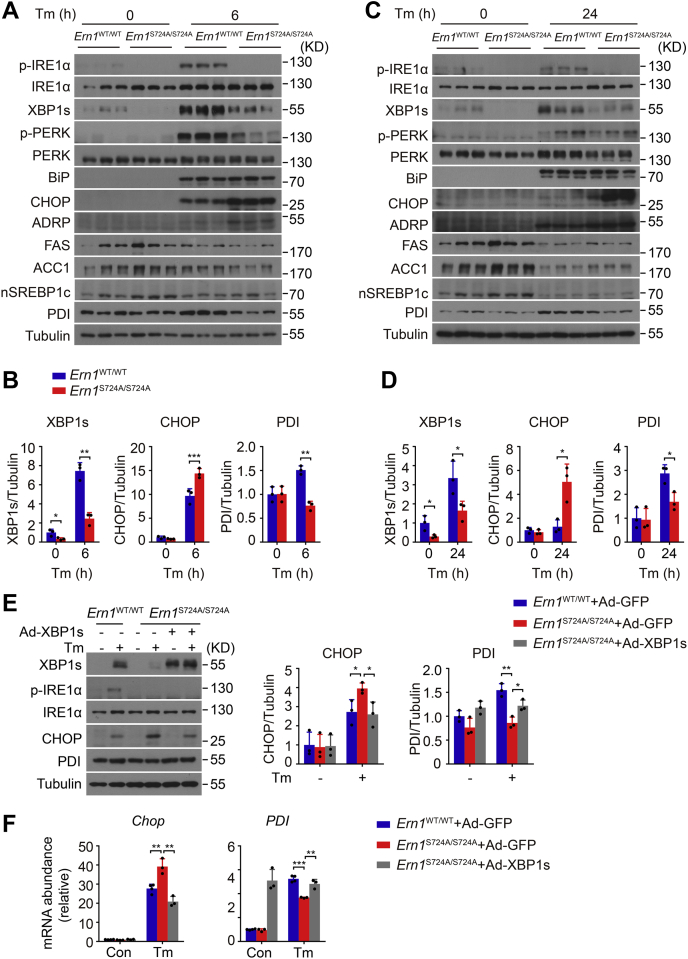
Next, we asked if the dampened production of XBP1s could mechanistically link the insufficient activation of IRE1α to the altered expression of CHOP and PDI in the liver. Consistently, immunoblot analysis showed that abrogation of Ser724 phosphorylation of IRE1α resulted in marked reductions in hepatic XBP1s protein production in Ern1S724A/S724A mice at 6 or 24 h after Tm treatment (Fig. 8, A–D). Interestingly, decreased Tm-induced phosphorylation of PERK but unaltered BiP protein expression was detected, indicating a likely connection between Ser724 phosphorylation of IRE1α and PERK activation. However, robust elevations in CHOP protein levels were seen in Ern1S724A/S724A livers (Fig. 8, A–D), somewhat similar to our observations in primary hepatocytes (Fig. 5, B and D). Thus, such increased CHOP expression was unrelated to the PERK pathway in ER-stressed Ern1S724A/S724A livers. Moreover, we observed significantly reduced PDI protein level along with higher abundance of adipose differentiation–related protein (ADRP), the cytosolic lipid droplet protein marker, but detected no prominent changes in the lipogenic enzymes, fatty acid synthase and acetyl-CoA carboxylase 1 (ACC1), or in the key lipogenic transcription factor SREBP1c, in Ern1S724A/S724A livers relative to their Ern1WT/WT counterparts following Tm treatment (Fig. 8, A–D). These data indicate that in accordance with their mRNA expression levels, deficient production of XBP1s protein was indeed coupled with persistently higher CHOP protein and lower PDI protein expression levels without apparently increasing the ER stress state in the livers of Tm-treated Ern1S724A/S724A mice.
Figure 8.
S724A mutation of IRE1α results in blunted XBP1s production with decreased PDI but increased CHOP expression in ER-stressed livers.A–D, immunoblot analysis of the phosphorylation and expression levels of the indicated proteins in livers of Ern1S724A/S724A mice and their Ern1WT/WT littermates following treatment with Tm or vehicle for 6 h (A and B) or 24 h (C and D). Shown are representative immunoblots for three individual mice per group (A and C), and quantification of XBP1s, PDI, and CHOP protein levels after normalization to tubulin (B and D). E and F, primary hepatocytes from male Ern1S724A/S724A mice and their Ern1WT/WT littermates were infected for 30 h with adenoviruses expressing GFP control or XBP1s protein. Cells were then treated with Tm (100 ng/ml) for 24 h. E, immunoblot analysis of XBP1s protein, IRE1α phosphorylation, as well as PDI and CHOP proteins. Shown also is the quantification of PDI and CHOP protein levels after normalization to tubulin as the loading control. F, quantitative RT–PCR analysis of the mRNA abundance of the indicated genes. Results in (E) and (F) represent three independent experiments. All data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test or two-way ANOVA. CHOP, CCAAT-enhancer binding protein homologous protein; ER, endoplasmic reticulum; IRE1α, inositol-requiring enzyme 1α; PDI, protein disulfide isomerase; Tm, tunicamycin; XBP1s, spliced X-box binding protein 1.
To test whether XBP1s mediates the effects of IRE1α–S724A upon the expression levels of CHOP and PDI, we used adenovirus to overexpress XBP1s protein in primary hepatocytes isolated from Ern1S724A/S724A mice. Similar to our observations in the liver, significant increases in Tm induction of CHOP protein as well as its mRNA expression were detected, and restored expression of XBP1s significantly blunted both in Ern1S724A/S724A hepatocytes (Fig. 8, E and F). In addition, S724A mutation of IRE1α also suppressed Tm-induced elevations in PDI protein as well as its mRNA expression levels, and overexpression of XBP1s reversed both in Ern1S724A/S724A hepatocytes (Fig. 8, E and F). Thus, deficient production of XBP1s, at least in large part, mediated the persistent upregulation of CHOP as well as the downregulation of PDI in hepatocytes, thereby promoting the aggravation of ER stress–induced hepatic steatosis.
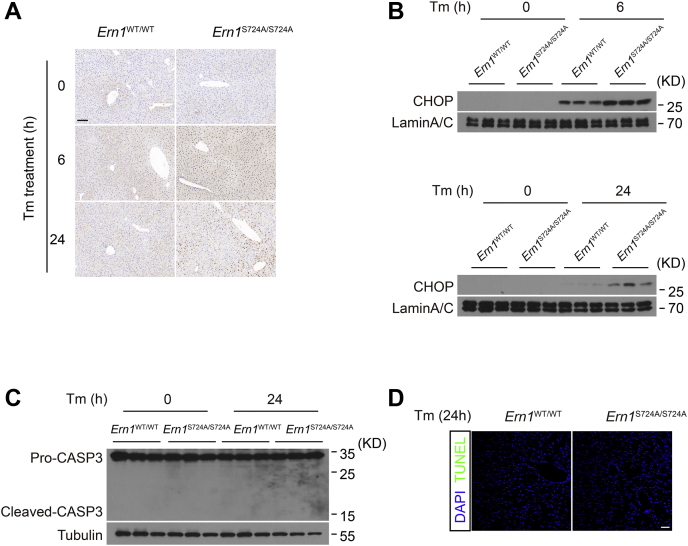
Finally, we went on to examine whether increased hepatic CHOP protein levels were associated with cell death in livers of Tm-treated Ern1S724A/S724A mice. Consistently, immunostaining of liver sections revealed apparently higher intensity of CHOP protein signals (Fig. 9A), and immunoblot analysis showed increased nuclear accumulation of CHOP protein (Fig. 9B), in livers of Ern1S724A/S724A mice after 6 or 24 h of Tm treatment. However, we neither observed detectable levels of the cleaved form of caspase-3 (Fig. 9C) nor found appreciable apoptosis signals by TUNEL analysis, in Ern1S724A/S724A or Ern1WT/WT livers at 24 h following Tm treatment (Fig. 9D). These results further support the notion that CHOP protein may mainly contribute to the dysregulation of hepatic lipid metabolism, rather than mediating cellular apoptosis, during ER stress–induced hepatosteatosis (54).
Figure 9.
S724A mutation of IRE1α has no effect upon apoptosis in ER-stressed livers.A, immunohistochemical staining of CHOP protein in liver sections of Ern1S724A/S724A mice and their Ern1WT/WT littermates following Tm treatment. The scale bar represents 100 μm. B, immunoblot analysis of CHOP protein in liver nuclear extracts. Lamin A/C was used as the nuclear protein control. C, immunoblot analysis of caspase-3 in livers of mice following Tm treatment for 24 h. Tubulin was used as the loading control. D, representative images of TUNEL staining of liver sections after 24 h of Tm treatment. The scale bar represents 50 μm. CHOP, CCAAT-enhancer binding protein homologous protein; ER, endoplasmic reticulum; IRE1α, inositol-requiring enzyme 1α; Tm, tunicamycin.
Discussion
Autophosphorylation of IRE1α has been established as a key step in its functional activation, but it has yet to be fully understood how its phosphorylation states are endogenously regulated and coupled to its RNase outputs in response to various stress states. In the current study, we investigated the role in vivo of phosphorylation at Ser724, one of the three serine residues (Ser724, Ser726, and Ser729) within the kinase activation loop of IRE1α that are known to undergo phosphorylation during its activation upon ER stress (36, 37). Our results revealed that phosphorylation at Ser724 critically governs IRE1α kinase activity for its autophosphorylation at other sites, which presumably enables the full activation of its RNase activity for both Xbp1 mRNA splicing and RIDD of Blos1 mRNA substrate. Furthermore, using Ern1S724A/S724A knock-in mice, we demonstrated in vivo that the activation control by phosphorylation at Ser724 of IRE1α can restrict the severity of ER stress–induced liver steatosis, largely through its RNase-dependent production of XBP1s. Our findings highlight the importance of precisely dissecting the functional consequences in vivo of each individual phosphorylation sites or in combinations, which most likely underlies distinct activation modes of IRE1α under a diversity of stress and pathological conditions.
Phosphorylation at Ser724 has been widely employed for monitoring IRE1α activation during experimental or physiological ER stress conditions. In contrast to the reported phosphorylation at Ser729 of IRE1α (38), whose effect upon its kinase activity remains to be characterized, we found that abrogation of Ser724 phosphorylation resulted in decreased or defective autophosphorylation of IRE1α at certain sites that have yet to be identified. Moreover, these alterations in the phosphorylation state of IRE1α were subsequently connected to its RNase activities, exhibiting possible cell type–specific features with respect to its RIDD outputs. It is also worth noting that Ser724 of IRE1α is not only autophosphorylated but also subjected to phosphorylation by PKA kinase in hepatocytes upon glucagon stimulation (55). Thus, it is conceivable that IRE1α phosphorylation at Ser724 has a key role in governing its activation mode and functional output upon typical ER stress as well as under physiological/pathological stress conditions such as obesity-associated metabolic stress. As IRE1α is known to be complexed with many other protein partners that either regulate its phosphorylation states or perform downstream signaling functions (32, 56), phosphorylation at Ser724 may represent a key step in controlling the overall phosphorylation states of IRE1α, which can in turn not only intrinsically regulate its dimerization/oligomerization for activating its RNase activity toward different substrate RNA targets (57) but also modulate its interactions with other proteins for distinct signaling actions. In this context, it warrants further in-depth investigations with regard to potential stress signal–specific mechanisms in eliciting distinct activation modes of IRE1α through individual or combined phosphorylation events, including those within its kinase activation loop.
Using ER stress–induced hepatic steatosis mouse model, we found that defective activation of IRE1α as a result of S724A mutation caused deficient production of XBP1s, leading to an exacerbation of lipid accumulation in the liver of Tm-treated Ern1S724A/S724A mice. This demonstrates the importance in vivo of phosphorylation at Ser724 of IRE1α for its full activation in counteracting hepatic ER stress, which is consistent with the documented phenotypes in mice with hepatocyte-specific deletion of IRE1α that abolished Xbp1 mRNA splicing (51). Mechanistically, a fully functional IRE1α–XBP1 pathway has been shown to regulate crucial components or regulators of hepatic lipid metabolism, including PDI, PPARα, and CHOP (43, 50, 54). Particularly, given the reported important role of CHOP in mediating ER stress–induced liver steatosis (54) as well as in transcriptionally suppressing the expression of certain master regulators, for example, Pparα, in hepatic lipid metabolism (58), it is likely that CHOP could act as a key contributor in linking the impaired production of XBP1s in Ern1S724A/S724A livers to dysregulated hepatic lipid homeostasis. This is also in line with the reported findings that XBP1s not only downregulates the expression of CHOP (59) but also promotes hepatic lipid secretion through PDI while exerting beneficial metabolic effects upon obesity-associated liver steatosis through suppressing the expression of lipogenic genes (49, 50).
In summary, our study uncovers the functional importance of the phosphorylation at one single serine residue within the kinase activation loop of IRE1α for its dynamic and endogenous activation in response to ER stress. Because IRE1α is a multifunctional protein possessing multiple phosphorylation sites, it requires further detailed molecular dissection of the mechanisms that dictate the activation modes of IRE1α under physiologically and pathologically relevant ER stress conditions such as dietary obesity. This will offer new insights into how dysregulated activation of IRE1α can link ER stress to the pathogenic development of metabolic disorders such as nonalcoholic fatty liver disease.
Experimental procedures
Animals
All animal studies were conducted in strict accordance with the Institutional Guidelines for the humane treatment of animals, with experimental protocols approved by the Committee on Ethics in the Care and Use of Laboratory Animals, College of Life Sciences, Wuhan University. Ern1S724A/S724A knock-in mice were generated by Shanghai Laboratory Animal Co Ltd. For creation of Ern1S724A/S724A knock-in mouse model, a pBR322-based targeting vector was generated to contain the segment from 92,210 to 98,830 of the mouse Ern1 gene (ENSMUSG00000020715) in which the AGT codon for Ser724 residue within exon 17 was substituted with the GCT codon for Ala. Neo cassette flanked by the flippase (FLP) recognition target sequence was then introduced into the targeting sequence between 95,912 and 95,913 of the mouse Ern1 gene. Targeting vector DNA was used to transfect 129SV/EV (SCR012) embryonic stem (ES) cells by electroporation. After selection with the antibiotic G418, 96 clones were screened by PCR analysis to identify recombinant ES clones, using two sets of primer pairs corresponding to the 5′-homology arm (forward: 5′-CTTCGAGCTACTAGATGGCACTCC-3′), the Neo cassette (reverse: 5′-CCGTGCCTTCCTTGACCCTGG-3′; forward: 5′-GGCCTACCCGCTTCCATTGCTC-3′), and the 3′-homology arm (reverse: 5′-GAAGAACTCACAGCAAACATCAG-3′). Two positive clones were identified and then selected for expansion, followed by verification via PCR-based DNA sequencing analysis for the presence of the desired AGT to GCT mutation. Targeted ES cells were subsequently microinjected into 194 C57BL/6J blastocysts for 14 recipients, and three male chimeras were obtained and intercrossed with C57BL/6J females to produce F1 heterozygous mice (n = 8). The Neo selection cassette was then removed by breeding to the FLP mice (129S4/SvJaeSor-Gt [ROSA] 26Sortm1 [FLP1] Dym/J; Jackson Lab), followed by backcrossing of the germline Neo-deleted Ern1S724A/WT mice into the genetic background of C57BL/6J for over nine generations. Ern1S724A/WT mice were then intercrossed to generate homozygous Ern1S724A/S724A mice and Ern1WT/WT littermates. Animals were maintained under a standard humidity- and temperature-controlled environment on a 12 h light/dark cycle, with free access to food and water. For ER stress–induced liver steatosis model, 2-month-old male Ern1S724A/S724A mice and their Ern1WT/WT littermates were injected i.p. with 1 mg/kg body weight of Tm or 150 μM dextrose (vehicle).
Preparation of MEFs
Primary MEFs were isolated from Ern1WT/WT and Ern1S724A/S724A embryos at embryonic day 13.5. Briefly, embryos were minced, trypsinized, and washed with PBS. Primary MEFs were plated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% penicillin–streptomycin. MEFs were then transfected with lentivirus expressing SV40 large T-antigen (kindly provided by Prof Anning Lin at the Shanghai Institutes for Biological Sciences) for immortalization.
Isolation of primary hepatocytes
Primary mouse hepatocytes were isolated from male mice at 8 to 12 weeks of age as previously described (60) with minor modifications. Briefly, collagenase perfusion was performed through the portal vein of anesthetized mice with 50 ml of perfusion buffer (Krebs Ringer buffer containing 3.6 mg/ml glucose, 1 M CaCl2, and 5000 U of collagenase I [Worthington]) at 37 °C. Cells were dispersed, and hepatocytes were collected and plated in collagen-coated plates with Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. Cells were cultured for 8 h before further analysis.
Serum and liver measurements
Hepatic TGs were measured as previously described (61). Briefly, 20 to 30 mg of liver tissues were homogenized in PBS and mixed with CHCl3–CH3OH (2:1, v/v). The organic phase was transferred, air-dried overnight, and resuspended in 1% Triton X-100 in absolute ethanol. Serum and liver TG levels were determined by the Total Triglyceride Kit according to the manufacturer’s instructions (Kehua Bioengineering), and liver TG levels were normalized to the total liver protein.
hsSRS imaging
Liver tissue was fixed in 10% formalin and then embedded in Tissue-Tek optimal cutting temperature compound. Frozen liver tissues sectioned to 30 μm thick using Cryostat (Leica CM 3000) were subjected to hsSRS imaging analysis as previously described (53, 62, 63). Briefly, the spectral focusing–based hsSRS system was used, with a dual-output femtosecond laser (InSight DeepSee; Spectra-Physics) providing pump (800 nm) and Stokes (1040 nm) pulses at an 80 MHz repetition rate. An electro-optical modulator (EO-AM-R-C2; Thorlabs) was used to modulate the Stokes laser at a resonant frequency of 10.5 MHz. The time delay line controlled by a motorized stage was employed, and the microscope (BX51; Olympus) equipped with a water objective (UPLSAPO 60XW; Olympus) was used for laser scanning and imaging. The pump beam was detected by a photodiode (S3994-01; Hamamatsu) with two installed short-pass filters (ET980SP; Chroma), and the SRS signals were acquired by a lock-in amplifier (HF2 LI; Zurich Instruments). The laser power of pump and Stokes beams were set to 50 and 70 mW (measured before galvanometer), respectively.
Oil-Red O staining
Frozen liver sections (10 μm thick) were fixed in formalin and then embedded in Tissue-Tek OCT (Servicebio). Liver sections were stained with freshly prepared Oil-Red O solution (Servicebio) for 10 to 30 min, followed by rinsing with 60% isopropanol. Nuclei were lightly stained with alum hematoxylin (Servicebio).
Immunohistochemistry analysis
For immunohistochemical analysis, MEF cells seeded in 96-well PhenoPlates (PerkinElmer; catalog no.: 6055300) at 20,000 cells/well were treated with Tm and fixed with 4% polyformaldehyde for 15 min at room temperature. Cells were then washed twice with PBS and permeabilized with 0.2% Triton X-100 for 7 min at room temperature. After washing with PBS, cells were incubated with blocking buffer (10% fetal bovine serum) for 1 h. After incubation with CHOP antibody (1:100 dilution; Santa Cruz Biotechnology, Inc; catalog no.: sc-7351) overnight at 4 °C, cells were washed three times with PBS, followed by incubation with Alexa Fluor 488–conjugated secondary antibody (1:500 dilution; Invitrogen, catalog no.: A21202) for 1 h at room temperature. Mounting medium was then added before imaging analysis by the Operetta CLS high-content analysis system (PerkinElmer). For liver tissue sections, samples were blocked with 3% bovine serum albumin for 1 h after antigen retrieval and incubation in 3% H2O2, followed by incubation with CHOP antibody (1:100 dilution; Santa Cruz Biotechnology, Inc; catalog no.: sc-7351) overnight at 4 °C. After washing with PBS, samples were incubated with horseradish peroxidase–conjugated secondary antibody (1:1000 dilution; Servicebio; catalog no.: GB23303) for 1 h at 37 °C prior to incubation with diaminobenzidine tetrahydrochloride. Images were captured by microscopy.
TUNEL and cell viability analyses
Liver sections were analyzed for cell death using the Dead End Fluorometric TUNEL System (Promega; catalog no.: G3250) according to the manufacturer’s instructions. TUNEL signals were visualized by fluorescence microscopy. Cell viability of MEFs and primary hepatocytes was assessed by Enhanced Cell Counting Kit-8 according to the manufacturer’s instructions (Beyotime Biotechnology; catalog no.: C0041).
Adenovirus infection
Recombinant adenoviruses for the overexpression of human IRE1α and its mutant forms, as well as mouse XBP1s, were prepared and used as described previously (44). Primary hepatocytes were infected with adenoviruses at a multiplicity of infection of 20 to 40.
Chemical reagents, antibodies, and immunoblot analysis
Tm (catalog no.: 654380) was purchased from Sigma–Aldrich, and KIRA6 (catalog no.: HY-19708) was from MCE. Antibodies against IRE1α (catalog no.: 3294), p-PERK (catalog no.: 3179), PERK (catalog no.: 3192), BiP (catalog no.: 3177), ACC1 (catalog no.: 3676), PDI (catalog no.: 3501), and caspase 3 (catalog no.:9662) were all purchased from Cell Signaling Technology. Antibodies against phosphorylated IRE1α at Ser724 (p-IRE1α) (catalog no.: AP1146), ADRP (catalog no.: A6276), and CHOP (catalog no.: A0221) were from Abclonal. XBP1s (catalog no.: ab220783) antibody was from Abcam, fatty acid synthase (catalog no.: 610962) antibody was from BD Biosciences, and α-tubulin antibody (T6199) was from Sigma.
For immunoblotting analysis, lysates of liver tissue or cells were prepared by radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl, pH 7.4) containing cOmplete protease-inhibitor cocktail (Sigma). Protein concentrations were measured by Bradford Protein Assay Kit (Thermo Fisher Scientific). Western immunoblotting was performed as previously described (43), and quantification was done using the ImageJ (National Institutes of Health) software.
Phos-tag gel analysis was performed as described (40). Briefly, a 6% SDS-PAGE gel containing 25 mM Phos-tag was prepared according to the manufacturer’s instructions (Phos-tag acrylamide AAL-107; Wako Pure Chemical Industries). IRE1α antibody (catalog no.: 3294; Cell Signaling Technology) was used to detect the phosphorylated and nonphosphorylated forms of IRE1α protein.
For preparation of liver nuclear extracts, frozen livers were homogenized using a Dounce homogenizer in ice-cold homogenization buffer I (pH = 8.0) containing 1% NP-40, 10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 25 mM NaF, 1 mM Na3VO4, 1 mM EDTA, and cOmplete protease-inhibitor cocktail. The homogenates were briefly centrifuged at 450 rpm at 4 °C to remove tissue debris, and the suspension was transferred to a new tube and centrifuged at 12,000 rpm for 10 min at 4 °C. The nuclear pellets were resuspended in buffer II (pH = 8.0) containing 20 mM Hepes, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol, 25 mM NaF, and cOmplete protease-inhibitor cocktail prior to immunoblot analysis using the desired antibodies.
Quantitative RT–PCR analysis
Total RNA was extracted from liver tissue or cells by TRIzol reagent (catalog no.: T9424; Invitrogen), and complementary DNA was synthesized using the RevertAid Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time quantitative PCR was then performed using the ABI Step-one Plus System (Applied Biosystems) with SYBR Green PCR reagents (Applied Biosystems). Gapdh was utilized as the internal control for normalization. The oligonucleotide primers used are as follows:
Mouse Erdj4: forward 5′-ATAAAAGCCCTGATGCTGAAGC-3′ and reverse 5′-GCCATTGGTAAAAGCACTGTGT-3′;
Mouse Xbp1t: forward 5′-TGGCCGGGTCTGCTGAGT CCG-3′ and reverse 5′-GTCCATGGGAAGATGTTCTGG-3′;
Mouse Xbp1s, forward 5′-CTGAGTCCGAATCAGGTGCAG-3′ and reverse 5′-GTCCATGGGAAGATGTTCTGG-3′;
Mouse CHOP: forward 5′-CTGGAAGCCTGGTATGAGGAT-3′ and reverse 5′-CAGGGTCAAGAGTAGTGAAGGT-3′;
Mouse PDI: forward 5′-ACCTGCTGGTGGAGTTCTATG-3′ and reverse 5′-CGGCAGCTTTGGCATACT-3′;
Mouse Bip: forward 5′-ACTTGGGGACCACCTATTCCT-3′ and reverse 5′-ATCGCCAATCAGACGCTCC-3′;
Mouse Gapdh: forward 5′-GGATTTGGCCGTATTGGG-3′ and reverse 5′-GTTGAGGTCAATGAAGGGG-3′;
Mouse Bloc1s1: forward 5′-TCCCGCCTGCTCAAAGAAC-3′ and reverse 5′-GAGGTGATCCACCAACGCTT-3′;
Mouse Pparα: forward 5′-AGAGCCCCATCTGTCCTCTC-3′ and reverse 5′-ACTGGTAGTCTGCAAAACCAAA-3′;
Mouse Cpt1a: forward 5′-CTCCGCCTGAGCCATGAAG-3′ and reverse 5′-CACCAGTGATGATGCCATTCT-3′;
Mouse Hmgcs2: forward 5′-ATATGTGGACCAAACTGACCTGG-3′ and reverse 5′-ACTGTTTTGACAGCCTTGGAC-3′;
Mouse Pparγ: forward 5′-CTCCAAGAATACCAAAGTGC GA-3′ and reverse 5′-GCCTGATGCTTTATCCCCACA-3′;
Mouse Srebp1c: forward 5′-CAGCTCAGAGCCGTGGT GA-3′ and reverse 5′-TTGATAGAAGACCGGTAGCGC-3′;
Mouse Fasn: forward 5′-GGAGGTGGTGATAGCCGGT AT-3′ and reverse 5′-TGGGTAATCCATAGAGCCCAG-3′;
Mouse Acc1: forward 5′-GATGAACCATCTCCGTTGGC-3′ and reverse 5′-CCCAATTATGAATCGGGAGTGC-3′;
Mouse Scd1: forward 5′-AGATCTCCAGTTCTTACACG ACCAC-3′ and reverse 5′-GACGGATGTCTTCTTCCA GGTG-3′.
Mouse Srebp2: forward 5′-CCGCTCTCGAATCCTCTTAT-3′ and reverse 5′-CAGCACCTGACTCCAGTGAC-3′.
Statistical analysis
All data are presented as the mean ± SD. Statistical analysis was conducted using unpaired two-tailed Student’s t test or two-way ANOVA using GraphPad Prism 7.0 (GraphPad Software, Inc). p < 0.05 was considered statistically significant.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Yang Li and Yong Liu conceptualization; Yang Li, J. W., J. D., J. C., and S. H. methodology; S. Y. and P. W. software; Yang Li, S. H., J. W., J. D., J. C., and Z. H. validation; S. Y., P. W., and J. L. formal analysis; Yang Li investigation; Yong Liu resources; Yang Li and S. H. data curation; Yang Li, S. H., and Yong Liu writing–original draft; Yong Liu writing–review & editing; Yang Li, S. H., and J. W. visualization; P. W. and Yong Liu supervision; Yong Liu project administration; P. W., J. L., and Yong Liu funding acquisition.
Funding and additional information
This work was supported by grants from the National Natural Science Foundation of China (grant nos.: 91857204, 31690102, 32021003, and 91739303; to Yong Liu and J. L.) and the Ministry of Science and Technology of China (National Key R&D Program of China; grant no.: 2018YFA0800700; to Yong Liu). Supported also by Fundamental Research Funds for the Central Universities (grant no.: 2042020kf1056; to Yong Liu). P. W. acknowledges the supports from the National Natural Science Foundation of China (grant no.: 62075076), Science Fund for Creative Research Group of China (grant no.: 61421064), and Innovation Fund of the Wuhan National Laboratory for Optoelectronics.
Edited by Ursula Jakob
Footnotes
Present address for Jianli Dai: Abcam Biotech Co Ltd, Hangzhou 310015, China.
References
- 1.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hetz C., Papa F.R. The unfolded protein response and cell fate control. Mol. Cell. 2018;69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Wang M., Kaufman R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 6.Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotamisligil G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J., Ozcan U. Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 2014;289:1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 10.Mori K., Ma W., Gething M.J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 11.Tirasophon W., Welihinda A.A., Kaufman R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Credle J.J., Finer-Moore J.S., Papa F.R., Stroud R.M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 14.Hetz C., Martinon F., Rodriguez D., Glimcher L.H. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J., Liu C.Y., Back S.H., Clark R.L., Peisach D., Xu Z., et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K.P., Dey M., Neculai D., Cao C., Dever T.E., Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali M.M., Bagratuni T., Davenport E.L., Nowak P.R., Silva-Santisteban M.C., Hardcastle A., et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X., Ellis R.E., Lee K., Liu C.Y., Yang K., Solomon A., et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 21.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 22.Han J., Kaufman R.J. Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev. 2017;31:1417–1438. doi: 10.1101/gad.297374.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruggiano A., Foresti O., Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang J., Qi L. Quality control in the endoplasmic reticulum: crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018;43:593–605. doi: 10.1016/j.tibs.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollien J., Weissman J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 27.Hollien J., Lin J.H., Li H., Stevens N., Walter P., Weissman J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Upton J.P., Wang L., Han D., Wang E.S., Huskey N.E., Lim L., et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J.M., Qiu Y., Yang Z.Q., Li L., Zhang K. Inositol-requiring enzyme 1 facilitates diabetic wound healing through modulating microRNAs. Diabetes. 2017;66:177–192. doi: 10.2337/db16-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha H., He Y., Yang L., Qi L. Stressed out about obesity: IRE1alpha-XBP1 in metabolic disorders. Trends Endocrinol. Metab. 2011;22:374–381. doi: 10.1016/j.tem.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S., Xing Y., Liu Y. Emerging roles for the ER stress sensor IRE1alpha in metabolic regulation and disease. J. Biol. Chem. 2019;294:18726–18741. doi: 10.1074/jbc.REV119.007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamu C.E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 34.Welihinda A.A., Kaufman R.J. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- 35.Tirasophon W., Lee K., Callaghan B., Welihinda A., Kaufman R.J. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prischi F., Nowak P.R., Carrara M., Ali M.M. Phosphoregulation of Ire1 RNase splicing activity. Nat. Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang T.K., Lawrence D.A., Lu M., Tan J., Harnoss J.M., Marsters S.A., et al. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol. Cell. 2018;71:629–636.e5. doi: 10.1016/j.molcel.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 38.Tang C.H., Chang S., Paton A.W., Paton J.C., Gabrilovich D.I., Ploegh H.L., et al. Phosphorylation of IRE1 at S729 regulates RIDD in B cells and antibody production after immunization. J. Cell Biol. 2018;217:1739–1755. doi: 10.1083/jcb.201709137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L., Xue Z., He Y., Sun S., Chen H., Qi L. A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi L., Yang L., Chen H. Detecting and quantitating physiological endoplasmic reticulum stress. Methods Enzymol. 2011;490:137–146. doi: 10.1016/B978-0-12-385114-7.00008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh R., Wang L., Wang E.S., Perera B.G., Igbaria A., Morita S., et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae D., Moore K.A., Mella J.M., Hayashi S.Y., Hollien J. Degradation of Blos1 mRNA by IRE1 repositions lysosomes and protects cells from stress. J. Cell Biol. 2019;218:1118–1127. doi: 10.1083/jcb.201809027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao M., Shan B., Liu Y., Deng Y., Yan C., Wu Y., et al. Hepatic IRE1alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARalpha axis signalling. Nat. Commun. 2014;5:3528. doi: 10.1038/ncomms4528. [DOI] [PubMed] [Google Scholar]
- 44.Qiu Y., Mao T., Zhang Y., Shao M., You J., Ding Q., et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baiceanu A., Mesdom P., Lagouge M., Foufelle F. Endoplasmic reticulum proteostasis in hepatic steatosis. Nat. Rev. Endocrinol. 2016;12:710–722. doi: 10.1038/nrendo.2016.124. [DOI] [PubMed] [Google Scholar]
- 46.Lebeaupin C., Vallee D., Hazari Y., Hetz C., Chevet E., Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2018;69:927–947. doi: 10.1016/j.jhep.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Song M.J., Malhi H. The unfolded protein response and hepatic lipid metabolism in non alcoholic fatty liver disease. Pharmacol. Ther. 2019;203:107401. doi: 10.1016/j.pharmthera.2019.107401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrema H., Zhou Y., Zhang D., Lee J., Salazar Hernandez M.A., Shulman G.I., et al. XBP1s is an anti-lipogenic protein. J. Biol. Chem. 2016;291:17394–17404. doi: 10.1074/jbc.M116.728949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B.N., et al. IRE1alpha-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 2012;16:473–486. doi: 10.1016/j.cmet.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K., Wang S., Malhotra J., Hassler J.R., Back S.H., Wang G., et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang S., Yan C., Fang Q.C., Shao M.L., Zhang Y.L., Liu Y., et al. Fibroblast growth factor 21 is regulated by the IRE1alpha-XBP1 branch of the unfolded protein response and counteracts endoplasmic reticulum stress-induced hepatic steatosis. J. Biol. Chem. 2014;289:29751–29765. doi: 10.1074/jbc.M114.565960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu D., Yu Y., Folick A., Currie E., Farese R.V., Jr., Tsai T.H., et al. In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated Raman scattering microscopy. J. Am. Chem. Soc. 2014;136:8820–8828. doi: 10.1021/ja504199s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rutkowski D.T., Wu J., Back S.H., Callaghan M.U., Ferris S.P., Iqbal J., et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao T., Shao M., Qiu Y., Huang J., Zhang Y., Song B., et al. PKA phosphorylation couples hepatic inositol-requiring enzyme 1alpha to glucagon signaling in glucose metabolism. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15852–15857. doi: 10.1073/pnas.1107394108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hetz C., Glimcher L.H. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol. Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Thomas A., Ferri E., Marsters S., Harnoss J.M., Lawrence D.A., Zuazo-Gaztelu I., et al. Decoding non-canonical mRNA decay by the endoplasmic-reticulum stress sensor IRE1alpha. Nat. Commun. 2021;12:7310. doi: 10.1038/s41467-021-27597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chikka M.R., McCabe D.D., Tyra H.M., Rutkowski D.T. C/EBP homologous protein (CHOP) contributes to suppression of metabolic genes during endoplasmic reticulum stress in the liver. J. Biol. Chem. 2013;288:4405–4415. doi: 10.1074/jbc.M112.432344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang C., Wang J.J., Ma J.H., Jin C., Yu Q., Zhang S.X. Activation of the UPR protects against cigarette smoke-induced RPE apoptosis through up-regulation of Nrf2. J. Biol. Chem. 2015;290:5367–5380. doi: 10.1074/jbc.M114.603738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q., Jiang L., Wang J., Li S., Yu Y., You J., et al. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 2009;49:1166–1175. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Shao M., Wu Y., Yan C., Jiang S., Liu J., et al. Role for the endoplasmic reticulum stress sensor IRE1alpha in liver regenerative responses. J. Hepatol. 2015;62:590–598. doi: 10.1016/j.jhep.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 62.Tian S., Li H., Li Z., Tang H., Yin M., Chen Y., et al. Polydiacetylene-based ultrastrong bioorthogonal Raman probes for targeted live-cell Raman imaging. Nat. Commun. 2020;11:81. doi: 10.1038/s41467-019-13784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang B., Yan S., Xiao L., Ji R., Yang L., Miao A.J., et al. Label-free imaging of nanoparticle uptake competition in single cells by hyperspectral stimulated Raman scattering. Small. 2018;14 doi: 10.1002/smll.201703246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.