Abstract
Osteosarcoma (OS) is a malignant bone tumor that mainly occurs in adolescents. It is accompanied by a high rate of lung metastasis, and high mortality. Recent studies have suggested the important roles of tripartite motif-containing (TRIM) family proteins in regulating various substrates and signaling pathways in different tumors. However, the detailed functional role of TRIM family proteins in the progression of OS is still unknown and requires further investigations. In this study, we found that tripartite motif-containing 22 (TRIM22) was downregulated in OS tissues and was hence associated with better prognosis. In vitro and in vivo functional analysis demonstrated that TRIM22 inhibits proliferation and metastasis of OS cells. Nuclear factor erythroid 2-related factor 2 (NRF2), a redox regulator, was identified as a novel target for TRIM22. TRIM22 interacts with and accelerates the degradation of NRF2 by inducing its ubiquitination dependent on its E3 ligase activity but independent of Kelch-like ECH-associated protein 1 (KEAP1). Further, a series of gain- and loss-of-function experiments showed that knockdown or overexpression of NRF2 reversed the functions of knockdown or overexpression of TRIM22 in OS. Mechanistically, TRIM22 inhibited OS progression through NRF2-mediated intracellular reactive oxygen species (ROS) imbalance. ROS production was significantly promoted and mitochondrial potential was remarkably inhibited when overexpressing TRIM22, thus activating AMPK/mTOR signaling. Moreover, TRIM22 was also found to inhibit Warburg effect in OS cells. Autophagy activation was found in OS cells which were overexpressed TRIM22, thus leading to autophagic cell death. Treatment with N-Acetylcysteine (NAC), a ROS scavenger or the autophagy inhibitor 3-Methyladenine (3-MA) abolished the decreased malignant phenotypes in TRIM22 overexpressing OS cells. In conclusion, our study indicated that TRIM22 inhibits OS progression by promoting proteasomal degradation of NRF2 independent of KEAP1, thereby activating ROS/AMPK/mTOR/Autophagy signaling that leads to autophagic cell death in OS. Therefore, our findings indicated that targeting TRIM22/NRF2 could be a promising therapeutic target for treating OS.
Keywords: Osteosarcoma, TRIM22/NRF2, ROS, Autophagic cell death, Warburg effect
Graphical abstract
Highlights
-
•
TRIM22 inhibits proliferation and metastasis of OS cells.
-
•
TRIM22 interacts with and accelerates NRF2 degradation by inducing its ubiquitination dependent on E3 ligase activity.
-
•
TRIM22 inhibited OS progression through NRF2-mediated intracellular ROS imbalance.
-
•
TRIM22 inhibits OS progression by promoting NRF2 degradation, thereby activating ROS/AMPK/mTOR/Autophagy signaling.
1. Introduction
Osteosarcoma (OS) is a type of bone cancer which begins in primary mesenchymal cells that mainly form long bones. It has a high fatality rate worldwide. Because of lung metastasis and drug resistance, the 5-year survival rate of OS is still at a low level despite the development of treatments including surgery and chemotherapy in last decades [[1], [2], [3]]. The overall survival probability of OS individuals with metastasis or recurrence is currently low (10–20%) due to limited therapeutic approaches [4,5]. Therefore, it's important and urgent to explore the potential mechanism in OS progression as well as seek comprehensive and effective approaches for treating OS.
Tripartite motif-containing (TRIM) protein family is known as a subfamily of RING type E3 ubiquitin ligase family [6]. Besides the RING finger domain, TRIM family proteins also consist of one or two zinc-binding motifs, named B-boxes and a related coiled-coil region. Several studies have reported the functional role of TRIM family proteins in many biological processes, including transcriptional regulation, cell proliferation, metastasis, apoptosis and tumorigenesis [7,8]. Ubiquitylation is one of the most post-translational modifications in modulating various biological processes including cell cycle, cellular signaling, DNA repair, protein stability control and carcinogenesis [[9], [10], [11]]. Majority of oncogenic or tumor-inhibitory proteins are regulated by post-translational modification, mostly by ubiquitin-proteasome system. Therefore, identifying and clarifying the functional role of E3 ligases are frontiers in tumor biology. Nevertheless, relevant studies in OS are rare and deserve further investigations.
TRIM22 is a member of the TRIM family proteins which was first identified as an IFN-induced protein as well as a transcriptional target of TP53 [12,13]. Several proteins involved in certain biological processes, including IκBα, NS5A and NOD are found to be the substrates of TRIM22 [[14], [15], [16]]. Ji et al. reported that TRIM22 could activate NF-κB signaling in GBM by degrading IκBα [14]. Another study has shown that TRIM22 could suppress the expression level and replication of HBV gene dependent on the nuclear-located RING domain [15]. However, the detailed functional role of TRIM22 in OS and its underlying mechanism is still unclear.
This present study identifies TRIM22 as a functional E3 ligase which is downregulated in OS and hence correlated with better prognosis. Our results showed that TRIM22 inhibits OS progression by interacting with, and accelerating the degradation of NRF2 independent of KEAP1, thus activates ROS/AMPK/mTOR/Autophagy signaling. This study reveals a novel role of TRIM22/NRF2 axis in OS progression and the underlying mechanism. Therefore, targeting TRIM22/NRF2 axis may be a promising therapeutic target for treating OS.
2. Results
2.1. TRIM22 is downregulated in OS and correlated with a better prognosis
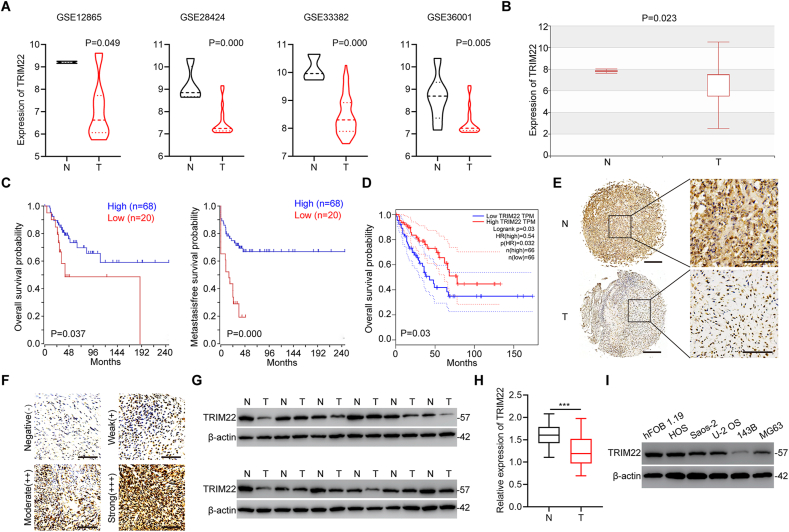
Expression of TRIM22 was first examined in four published datasets obtained from OS samples deposited in GEO (GSE12865, GSE28424, GSE33382 and GSE36001). The analyzed data showed that TRIM22 is significantly downregulated in tumor specimens compared with non-tumor tissues (Fig. 1a). The results from the Human Cancer Metastasis Database also confirmed the downregulation of TRIM22 in OS tissues (Fig. 1b). Based on the online database, Kaplan-Meier analysis demonstrated that patients with a high level of TRIM22 expression in OS had a much better prognosis (Fig. 1c). Data from GEPIA online database also showed that patients with high TRIM22 expression in sarcoma had a better prognosis (Fig. 1d). In addition, the downregulation of TRIM22 protein expression was confirmed combining immunohistochemistry (IHC) staining with OS tissue microarray (TMA) (Fig. 1e, f). Next, Western blot assays were performed in twelve randomly selected pairs of tumor tissues as well as corresponding non-tumor tissues and further confirmed the decrease of TRIM22 protein expression in OS tissues (Fig. 1g). Downregulation of TRIM22 mRNA level in OS specimens was also validated by qRT-PCR in forty paired tumor specimens and corresponding non-tumor specimens (Fig. 1h). Moreover, Western blot analysis was used to determine TRIM22 expression in OS cell lines including HOS, Saos-2, U-2 OS, 143B and MG63 cells and the normal human osteoblast cell line hFOB 1.19 cell. Consistently, TRIM22 protein expression levels were found to be decreased in OS cells lines (Fig. 1i). Therefore, these results indicated that TRIM22 might play an inhibitory role in OS progression and is associated with better prognosis.
Fig. 1.
TRIM22 is downregulated in OS and correlated with a better prognosis. (a) The expression of TRIM22 in tumor tissues and normal non-tumor tissues from GSE12865, GSE28424, GSE33382 and GSE36001 (unpaired two-tailed Student's t-test). (b) The expression of TRIM22 from the Human Cancer Metastasis Database (unpaired two-tailed Student's t-test, http://hcmdb.i-sanger.com/index, HCMDB). (c) Kaplan-Meier analysis of the overall survival and metastasis-free survival from an online database (http://hgserver1.amc.nl/cgi-bin/r2/main/cgi). (d) The correlations between TRIM22 expression and overall survival in the GEPIA database (http://gepia.cancer-pku.cn/index.html). (e) Representative IHC staining of TRIM22 in OS tissues and paired non-tumor tissues. Scale bar, 100 μm. (f) Expression of TRIM22 by IHC staining of OS tissues. Scale bar, 100 μm. (g) The expression of TRIM22 was determined in twelve pairs of OS tissues and matched non-tumor tissues by Western blot. (h) The expression of TRIM22 was examined in forty pairs of OS tissues and matched non-tumor tissues by qRT-PCR (n = 40, ***P<0.001, unpaired two-tailed Student's t-test). (i) The expression of TRIM22 in OS cell lines and hFOB 1.19 cell line was evaluated by Western blot. IHC = immunohistochemistry.
2.2. Inhibitory effects of TRIM22 on proliferation of OS cells in vitro and in vivo
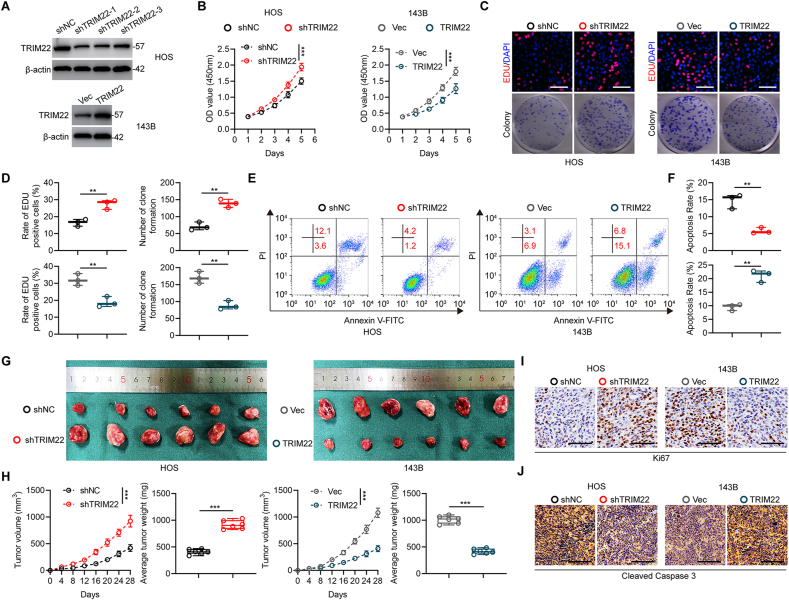
According to the expression profiles of TRIM22 in OS cell lines, the HOS cells with high expression of TRIM22 were used to silence TRIM22 by lentivirus-mediated transfection of sh-TRIM22. Results showed that sh-1 exhibited the highest silence efficiency and were hence chosen for further studies (Fig. 2a). In addition, TRIM22 was stably overexpressed in low TRIM22-expression 143B cells (Fig. 2a). Results from CCK-8 assays indicated that cell proliferation was markedly promoted after silencing TRIM22, whereas it was inhibited after overexpressing TRIM22 in OS cells (Fig. 2b). The EdU assay and colony formation assay were further carried out and confirmed that silencing of TRIM22 promoted cell proliferation whereas overexpression of TRIM22 inhibited cell proliferation significantly in OS cells (Fig. 2c, d). Furthermore, the flow cytometric analysis of cell apoptosis indicated that silencing of TRIM22 in HOS cells inhibited cell apoptosis while overexpression of TRIM22 induced cell apoptosis in 143B cells (Fig. 2e, f).
Fig. 2.
Inhibitory effects of TRIM22 on proliferation of OS in vitro and in vivo. (a) Western blot analysis of TRIM22 expression after transfection in HOS and 143B cells. (b) CCK-8 assays in HOS and 143B cells with TRIM22 knockdown or overexpression, respectively (n = 3, ***P<0.001, two-way ANOVA with post hoc test). (c, d) EdU assays and colony formation assays in HOS and 143B cells with TRIM22 knockdown or overexpression, respectively and quantification of the EdU assay and colony formation assay (n = 3, **P<0.01, unpaired two-tailed Student's t-test). Scale bar, 200 μm. (e, f) Flow cytometric analysis and quantification of apoptosis in HOS and 143B cells with TRIM22 depletion or overexpression, respectively (n = 3, **P<0.01, unpaired two-tailed Student's t-test). (g) Representative images of xenograft tumors after subcutaneous injection of OS cells with TRIM22 knockdown or overexpression and corresponding controls 28d after inoculation. (h) Time course of growth and tumor weight of OS xenografts (n = 6, ***P<0.001, two-way ANOVA with post hoc test (tumor volume) and unpaired two-tailed Student's t-test (average tumor weight)). (i) IHC staining of Ki67 of tumor sections. Scale bar, 100 μm. (j) IHC staining of cleaved caspase 3 of tumor sections. Scale bar, 100 μm. CCK-8 = cell counting kit-8; EdU = 5-ethynyl-2′-deoxyuridine; IHC = immunohistochemistry.
Transfected HOS and 143B cells were subcutaneously injected into nude mice to further investigate the role of TRIM22 in OS development in vivo. As presented in Fig. 2g and h, the volume and weight of tumors in TRIM22 knockdown group were significantly promoted as compared with those in negative control group. In contrast, overexpression of TRIM22 reduced xenografted tumor growth, size and weight in vivo. Furthermore, Ki67 staining showed an increase in the rate of proliferation in TRIM22 knockdown xenografts and a decrease in the rate of proliferation in TRIM22 overexpression xenografts (Fig. 2i). In addition, by staining cleaved caspase 3 which is a marker for detecting cell apoptosis, it was found that the apoptosis rate decreased in TRIM22 knockdown xenografts and increased in TRIM22 overexpression xenografts (Fig. 2j). In conclusion, these results demonstrated that TRIM22 plays an essential role in inhibiting proliferation and promoting apoptosis in OS in vitro and in vivo.
2.3. Inhibitory effects of TRIM22 on metastasis of OS cells in vitro and in vivo
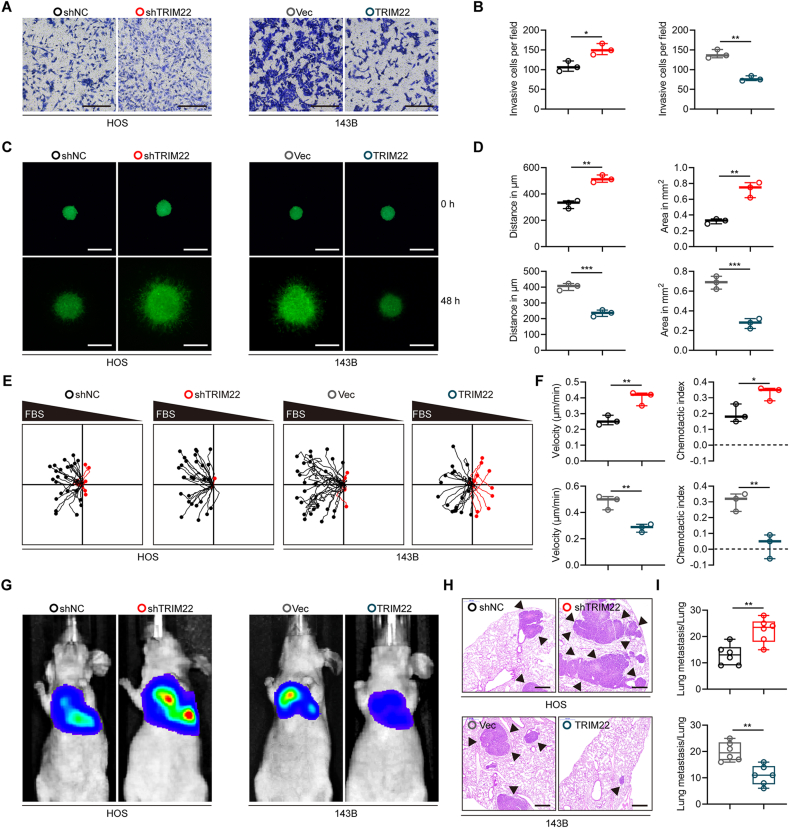
A series of functional experiments were performed to further investigate whether TRIM22 was associated with metastasis of OS cells. The transwell invasion assay revealed that silencing TRIM22 significantly enhanced invasiveness of HOS cells whereas overexpressing TRIM22 attenuated invasiveness of 143B cell (Fig. 3a, b). The results of 3D tumor spheroid cell-invasion assay, also validated that TRIM22 could significantly inhibit invasiveness of OS cells (Fig. 3c, d). Furthermore, the effects of TRIM22 on OS cells were evaluated using a microfluidic migration chamber with stable, shear-minimized linear chemo-attractive or chemo-repulsive gradients as previously described [17,18]. Attracted by FBS, OS cells moved toward the source of FBS. As shown in Fig. 3e, f, knockdown of TRIM22 significantly promoted the migration and directionality to FBS in HOS cells compared with the negative control. However, the TRIM22-upregulated 143B cells exhibited a lower directed migration because they were less sensitive to the FBS gradient.
Fig. 3.
Inhibitory effects of TRIM22 on metastasis of OS in vitro and in vivo. (a, b) Effects of TRIM22 on invasion in OS cells in vitro using transwell invasion assay and quantification of transwell invasion assay (n = 3, *P<0.05, **P<0.01, unpaired two-tailed Student's t-test). Scale bar, 200 μm. (c, d) Representative images of 3D tumor spheroid cell-invasion assay in transfected HOS and 143B cells and quantification of 3D tumor spheroid cell-invasion assay (n = 3, **P<0.01, ***P<0.001, unpaired two-tailed Student's t-test). Scale bar, 500 μm. (e, f) Migration plots of OS cells in a gradient of FBS. The starting point of every track was normalized to the position x = 0 and y = 0. Mean velocities of OS cells and mean chemotaxis indices of OS cells were quantified (n = 3, *P<0.05, **P<0.01, unpaired two-tailed Student's t-test). (g) Representative images of lung metastasis models in nude mice after tail injection of HOS and 143B cells with TRIM22 knockdown or overexpression, respectively. (h) H & E staining of pulmonary metastatic nodules. Scale bar, 500 μm. (i) Quantification of lung metastatic foci (n = 6, **P<0.01, unpaired two-tailed Student's t-test). FBS = fetal bovine serum; H & E = hematoxylin-eosin.
To evaluate the functional role of TRIM22 in metastasis of OS cells in vivo, we injected stably-transfected OS cells into nude mice via the tail vein to generate a pulmonary metastasis model. After six weeks, lung metastasis was remarkably enhanced in TRIM22 knockdown group, but was suppressed in TRIM22 overexpression group compared with the corresponding control groups (Fig. 3g). Treated mice were sacrificed then and lung sections were stained with hematoxylin and eosin (H&E), which further confirmed the previously observed results (Fig. 3h, i). Therefore, these results demonstrated that TRIM22 attenuated OS metastasis in vitro and in vivo.
2.4. TRIM22 binds to and degrades NRF2
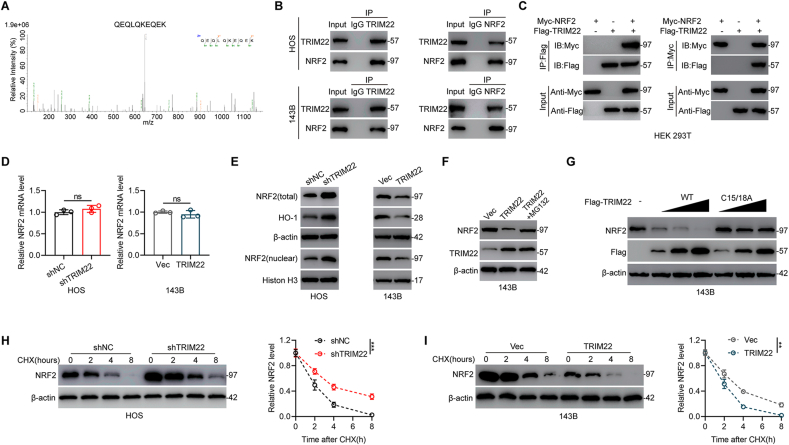
To further investigate the underlying mechanism by which TRIM22 regulates OS proliferation and metastasis, immunoprecipitation coupled with mass spectrometry (IP/MS) was carried out to determine which protein interacts with TRIM22. NRF2, which is well-recognized to play an essential role in redox biology, was then identified as a protein which interacts with TRIM22 (Fig. 4a). Co-immunoprecipitation (Co-IP) analysis in OS cells was performed to confirm the findings. As shown in Fig. 4b, NRF2 was precipitated by TRIM22 and reverse Co-IP confirmed that TRIM22 could also be precipitated by NRF2 in OS cells. Furthermore, in HEK 293T cells, it was found that Flag-tagged TRIM22 co-precipitated with Myc-tagged NRF2 and Myc-tagged NRF2 co-precipitated with Flag-tagged TRIM22 efficiently (Fig. 4c). NRF2 contains six conserved Neh domains (Neh1-Neh6). To further determine the binding domain of NRF2 interacting with TRIM22, a series of truncated Myc-tagged NRF2 were constructed (Fig. S1a). As shown in Fig. S1b, Co-IP assays showed that fragments of NRF2 containing amino acids 205–433 could interact with TRIM22, which indicated that TRIM22 binds to the Neh6 domain of NRF2. Therefore, these results support the interaction between TRIM22 and NRF2.
Fig. 4.
TRIM22 binds to and degrades NRF2. (a) IP/MS analysis indicated that NRF2 interacts with TRIM22. (b) Detection of endogenous protein interactions between TRIM22 and NRF2 in OS cells lysates. (c) Detection of exogenous protein interactions between TRIM22 and NRF2 in HEK 293T cells. Flag-tagged TRIM22 and Myc-tagged NRF2 plasmids were transfected into HEK 293T cells. (d) NRF2 mRNA levels in OS cells in indicated groups (n = 3, ns, no significance, unpaired two-tailed Student's t-test). (e) Western blot analysis of NRF2 and HO-1 protein levels in transfected OS cells. (f) Evaluation of NRF2 protein levels in 143B cells overexpressed with TRIM22 with or without proteasome inhibitor MG132 treatment. (g) Increasing amounts of Flag-tagged TRIM22 (WT or C15/18A mutant) were transfected and the expression levels of NRF2 and Flag-tagged TRIM22 were detected by Western blot in 143B cells. (h) Detection of NRF2 protein levels in shNC and shTRIM22 transfected HOS cells in the presence of CHX (10 μg/ml) for indicated time point (n = 3, ***P<0.001, two-way ANOVA with post hoc test). (i) Detection of NRF2 protein levels in 143B cells with overexpression of TRIM22 or corresponding control in the presence of CHX (10 μg/ml) for the indicated time points (n = 3, **P<0.01, two-way ANOVA with post hoc test). IP/MS = Immunoprecipitation coupled with mass spectrometry; CHX = cycloheximide.
Since TRIM22 interacts with NRF2, the influence of silencing and overexpressing TRIM22 on NRF2 expression levels in OS cells were then investigated. Silencing and overexpression of TRIM22 significantly increased and decreased NRF2 protein levels, respectively, but had no influence on NRF2 mRNA levels (Fig. 4d, e). These results indicate that TRIM22 regulates NRF2 at the protein level but not at the mRNA level. We further determined whether TRIM22 inhibits the transcriptional activity of NRF2. As shown in Fig. S2a, knockdown of endogenous TRIM22 in HOS cells resulted in significantly increased mRNA expression levels of several known NRF2 transcription targets including NQO1, GCLC, GCLM and HO-1. In contrast, overexpression of TRIM22 led to decreased mRNA expression levels of NRF2 target genes (Fig. S2b). Also, silencing or overexpression of TRIM22 also significantly increased or decreased the protein levels of HO-1 (Fig. 4e). Further, addition of the proteasome inhibitor MG132 reversed the decrease in NRF2 protein levels after overexpressing TRIM22 (Fig. 4f). Experiments to investigate whether NRF2 is degraded by TRIM22 were also conducted in this study. It was found that NRF2 protein levels were significantly decreased after overexpression of TRIM22, while not influenced by overexpression of a catalytically inactive C15/18A variant of TRIM22 (Fig. 4g). Then, we explored the effect of depletion or overexpression of TRIM22 on the protein stability of endogenous NRF2 in the presence of protein synthesis inhibitor cycloheximide (CHX). It was shown that silencing TRIM22 significantly suppressed NRF2 degradation whereas overexpression of TRIM22 markedly promoted NRF2 degradation (Fig. 4h, i).
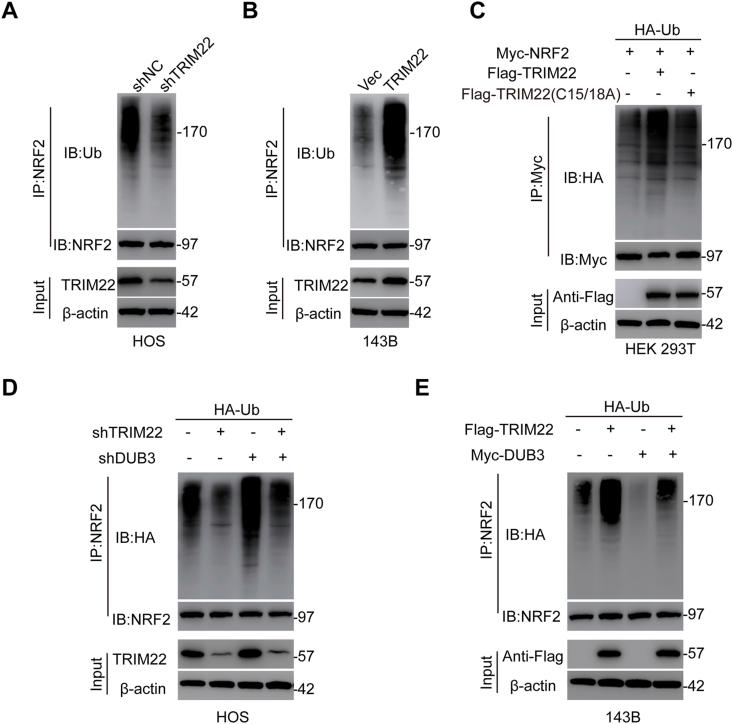
The association of the TRIM22 ability to degrade NRF2 with its activity as a E3 Ub ligase mediating protein ubiquitination was also investigated in this study. Compared with shNC, depletion of TRIM22 significantly decreased NRF2 ubiquitination in HOS cells (Fig. 5a). However, in 143B cells, overexpression of TRIM22 significantly increased ubiquitination of NRF2 (Fig. 5b). TRIM22-regulated NRF2 ubiquitination was further confirmed by co-transfecting the Flag-TRIM22 (either WT or C15/18A mutant), Myc-NRF2 as well as HA-Ub in HEK 293T cells. It was found that TRIM22 overexpression increased NRF2 ubiquitination while transfection with C15/18A mutant TRIM22 showed little effect on NRF2 ubiquitination (Fig. 5c). These results suggested that TRIM22 regulates the stability of NRF2 by regulating its proteasomal degradation via polyubiquitination. It has been reported that deubiquitinating enzyme 3 (DUB3) function as a deubiquitinase to regulate NRF2 protein stability at the posttranslational level [19]. Then we surmised that TRIM22 may exert an antagonistic role to counteract DUB3-mediated deubiquitylation of NRF2. To further prove this, TRIM22 together with DUB3 were knocked down as well as overexpression of TRIM22 together with DUB3 were also performed to assess the ubiquitination and expression of NRF2. As shown in Fig. 5d, knockdown of TRIM22 decreased NRF2 ubiquitination whereas silencing DUB3 increased NRF2 ubiquitination. Silencing TRIM22 alongside knockdown of DUB3 abolished the decreased ubiquitination caused by TRIM22 inhibition. On the other hand, overexpression of TRIM22 increased NRF2 ubiquitination whereas upregulation of DUB3 decreased NRF2 ubiquitination. However, overexpression of TRIM22 together with DUB3 attenuated the upregulated ubiquitination of NRF2 due to TRIM22 overexpression (Fig. 5e). Generally, these findings indicated that TRIM22 regulates NRF2 stability by counteracting DUB3-mediated ubiquitination in OS cells.
Fig. 5.
Effects of TRIM22 on ubiquitination of NRF2. (a) Evaluation of endogenous NRF2 ubiquitination in HOS cells transfected with shNC or shTRIM22. (b) Evaluation of endogenous NRF2 ubiquitination in 143B cells with overexpression of TRIM22 or corresponding control. (c) Evaluation of exogenous NRF2 ubiquitination in HEK 293T cells co-transfected with Flag-tagged TRIM22 (WT) or Flag-tagged TRIM22 (C15/18A), HA-tagged Ub and Myc-tagged NRF2. (d) HOS cells were co-transfected with shTRIM22, shDUB3 as well as HA-Ub, and then the NRF2 ubiquitylation linkage was assessed. (e) 143B cells transfected with Flag-TRIM22, Myc-DUB3 as well as Ub, and then NRF2 ubiquitylation linkage was assessed.
2.5. TRIM22 decreases NRF2 stability independent of KEAP1
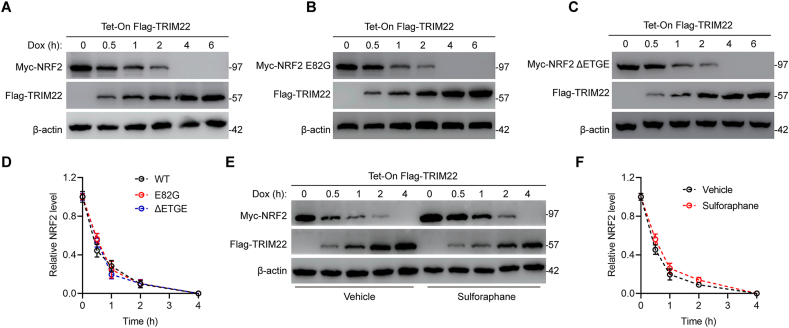
It's well-recognized that KEAP1 interacts with NRF2 and promotes NRF2 ubiquitination. To further evaluate whether TRIM22-mediated NRF2 proteasomal degradation was regulated by KEAP1, a HEK 293T TetON cell line with doxycycline-inducible expression of a Flag-tagged TRIM22 was established. As expected, in response to doxycycline, the expression of Flag-TRIM22 was increased while the expression of NRF2 was decreased (Fig. 6a). These results further confirmed above results that TRIM22 promotes proteasomal degradation of NRF2. Next, mutations within the Neh2 domain of NRF2, which were demonstrated to disrupt the high affinity KEAP1-binding site as well as inhibit interaction with NRF2, were generated to further determine whether the degradation of NRF2 by TRIM22 was affected by KEAP1. Interestingly, the results showed that both the E82G and the ΔETGE variant of NRF2 couldn't counteract the degradation of NRF2 (Fig. 6b–d). Additionally, NRF2 inducer sulforaphane which modifies critical cystine residues of KEAP1 to inhibit the ubiquitination of NRF2 was used. Prior to TRIM22 induction, sulforaphane could increase the expression of NRF2. However, in response to TRIM22, sulforaphane failed to prevent the degradation of NRF2 (Fig. 6e). The degradation rate of NRF2 in response to TRIM22 was similar in the absence and presence of sulforaphane (Fig. 6f). Collectively, these results indicated TRIM22 promotes NRF2 proteasomal degradation independent of interaction between NRF2 and KEAP1.
Fig. 6.
TRIM22 promotes KEAP1-independent NRF2 proteasomal degradation. (a) Myc-tagged NRF2 was expressed in HEK 293T Tet-On Flag-TRIM22 cells with doxycycline (Dox) inducible expression of Flag-TRIM22. Cells were exposed to Dox as indicated. Cell lysates were evaluated by immunoblotting with anti-Myc and anti-Flag. (b–c) Plasmids encoding NRF2 variant deficient for KEAP1 binding including E82G or ΔETGE were transfected and cell lysates were evaluated by immunoblotting with anti-Myc and anti-Flag. (d) Quantification of Myc-NRF2 expression level in a-c (n = 3, ns, no significance, two-way ANOVA with post hoc test). (e) Cells were exposed to a vehicle or the NRF2 inducer sulforaphane for 6h prior to Dox administration and cell lysates were evaluated by immunoblotting with anti-Myc and anti-Flag. (f) Quantification of Myc-NRF2 expression level in e (n = 3, ns, no significance, two-way ANOVA with post hoc test). Dox = doxycycline.
2.6. NRF2 is upregulated and negatively correlated with TRIM22 in OS
The correlation between NRF2 and TRIM22 was investigated because NRF2 is a substrate of TRIM22. The expression level of NRF2 protein was examined in twelve randomly selected pairs of OS specimens and corresponding non-tumor specimens by Western blot. The results of this study showed that NRF2 expression is increased in OS specimens (Fig. S3a). Furthermore, a significant negative correlation between the expression of TRIM22 and NRF2 protein was found using Spearman correlation test (Fig. S3b). Based on an online database, Kaplan-Meier analysis demonstrated that patients with a high level of NRF2 expression in OS have a much worse prognosis (Fig. S3c). Moreover, NRF2 expression was demonstrated to be increased in OS cell lines (Fig. S3d). Further, a series of in vitro functional experiments were performed then to investigate the function of NRF2 in OS progression. NRF2 was overexpressed in HOS cells while silenced in 143B cells. The results showed that overexpression of NRF2 in HOS cells promoted malignant phenotypes including proliferation, migration and invasion while downregulation of NRF2 in 143B cells showed the opposite effects (Figs. S4a–h). Therefore, these results indicated that NRF2 is negatively correlated with TRIM22 and may act as an oncogenic protein.
2.7. TRIM22 promotes OS cells proliferation and metastasis through interacting with and degrading NRF2 in vitro and in vivo
The role of NRF2 in the mechanism by which TRIM22 inhibits the progression of OS was also investigated through the knockdown of lentiviral-mediated NRF2 in TRIM22-silenced HOS cells and overexpression of NRF2 in TRIM22-upregulated 143B cells. Several in vitro functional rescue experiments were performed as described above. Results showed that the increased malignant phenotypes including proliferation, migration and invasion in TRIM22-silenced HOS cells were reversed when NRF2 was knocked down, whereas NRF2 overexpression diminished the decreased malignant phenotypes in TRIM22-upregulated 143B cells (Fig. S5).
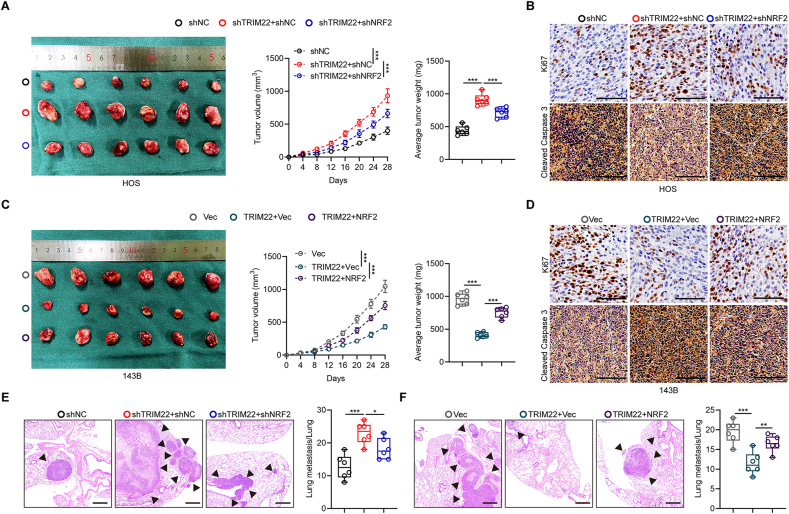
Xenografts were also generated in nude mice to investigate if NRF2 is an essential downstream substrate for TRIM22 which inhibits the development of OS. It was found that the ectopic downregulation of NRF2 in HOS cells counteracted the upregulated volume and weight of the xenograft tumors when depletion of TRIM22 (Fig. 7a). Conversely, overexpression of NRF2 in 143B cells abolished the decreased malignant effects when overexpressing TRIM22 (Fig. 7c). Results from Ki67 and cleaved caspase 3 staining validated these results (Fig. 7b, d). In pulmonary metastatic models, silencing NRF2 abolished promotion in lung metastasis by TRIM22 knockdown in HOS cells while overexpression of NRF2 in 143B cells significantly counteracted the inhibitory-metastasis effect of TRIM22 overexpression on pulmonary metastasis (Fig. 7e, f). Overall, these results revealed that NRF2 is the downstream target of TRIM22 and mediates the role of TRIM22 in inhibiting OS malignant progression.
Fig. 7.
TRIM22 inhibits OS cells proliferation and metastasis through interacting with and degrading NRF2 in vivo. (a) Representative images of tumors and quantification of tumor volumes and tumor weights in nude mice bearing HOS cells in indicated groups (n = 6, ***P<0.001, two-way ANOVA with post hoc test (tumor volume) and one-way ANOVA with post hoc test (average tumor weight)). (b) IHC staining of Ki67 and cleaved caspase 3 of tumor sections in indicated groups. Scale bar, 100 μm. (c) Representative images of tumors and quantification of tumor volumes and tumor weights in nude mice bearing 143B cells in indicated groups (n = 6, ***P<0.001, two-way ANOVA with post hoc test (tumor volume) and one-way ANOVA with post hoc test (average tumor weight)). (d) IHC staining of Ki67 and cleaved caspase 3 of tumor sections in indicated groups. Scale bar, 100 μm. (e, f) H & E staining and quantification of pulmonary metastatic nodules in indicated groups (n = 6, *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA with post hoc test). Scale bar, 500 μm. IHC = immunohistochemistry; H & E = hematoxylin-eosin.
2.8. TRIM22/NRF2 cascade inhibits OS progression and Warburg effect via ROS/AMPK/mTOR/-mediated autophagy activation
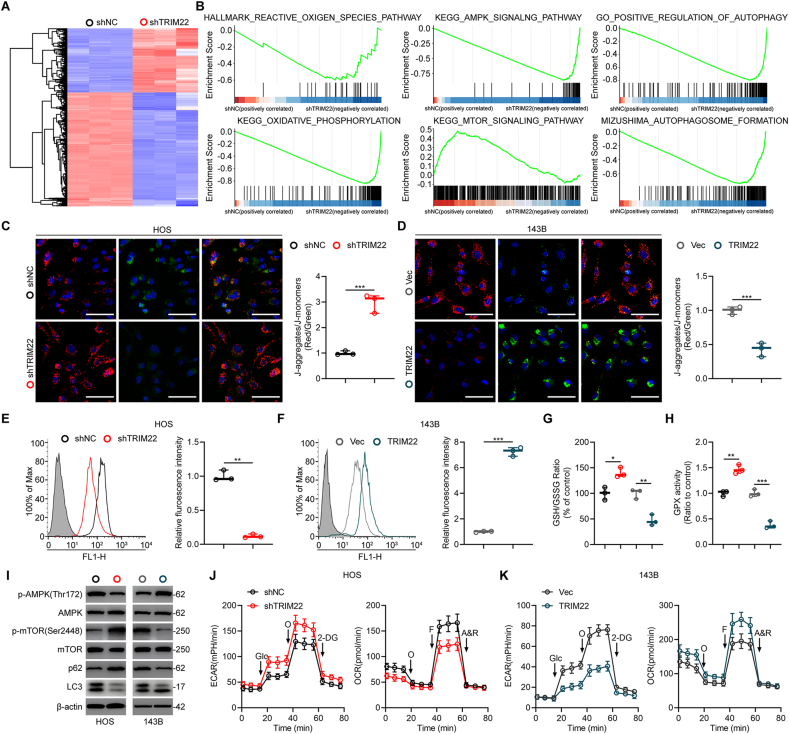
To further explore the mechanism of TRIM22 in inhibiting OS progression, transcriptome analysis by high-throughput RNA sequencing (RNA-Seq) was performed in HOS cells transfected with shNC and shTRIM22. As shown in Fig. 8a, 451 DEGs were upregulated while 892 DEGs were downregulated in TRIM22-downregulated HOS cells. Subsequently, gene set enrichment analysis (GSEA) was performed and results showed that the expression of TRIM22 was markedly correlated with ROS production, AMPK signaling activation and mTOR signaling inhibition, suggesting a potential regulatory role of TRIM22 in ROS/AMPK/mTOR signaling (Fig. 8b). NRF2 is a widely-accepted gene which regulates redox biology and inhibits ROS production. Ongoing studies are investigating the potential applications of regulating ROS levels in the tumor microenvironment as a tumor treatment approach since maintaining low ROS levels in tumor preserves malignancy [20]. Taken these into consideration, it was speculated that TRIM22 might lead to ROS accumulation, thus inhibiting OS progression via degrading NRF2. First, JC-1 staining was used to evaluate the mitochondrial potential. The results from the staining showed that knockdown of TRIM22 in HOS cells significantly increased the mitochondrial potential (Fig. 8c). In contrast, overexpression of TRIM22 in 143B cells markedly decreased the mitochondrial potential and this indicated mitochondrial damage (Fig. 8d). Flow cytometry was used to detect intracellular ROS. Results showed that TRIM22 could lead to ROS production in OS cells (Fig. 8e, f). Additionally, reduced glutathione (GSH)/oxidized glutathione (GSSG) ratio and glutathione peroxidase (GPX) activity were further evaluated. We found that silence of TRIM22 in HOS cells significantly increased GSH/GSSG ratio and enhanced GPX activity while overexpression of TRIM22 in 143B cells showed the opposite effects (Fig. 8g, h). Several studies have suggested that AMPK signaling is extremely sensitive to oxidative stress and can be phosphorylated by ROS production [[21], [22], [23], [24]]. The activation of AMPK could result in the inhibition of mTORC1 and thus activating autophagy. Also, our GSEA results indicated ROS/AMPK/mTOR signaling is inactivated when silencing of TRIM22. Western blot analysis confirmed that overexpressing TRIM22 could increase the expression level of phosphorylated AMPK protein while decreased the expression level of phosphorylated mTOR protein, whereas the silencing of TRIM22 showed the opposite results (Fig. 8i). In contrast, overexpression of NRF2 decreased the expression level of phosphorylated AMPK protein while increased the expression level of phosphorylated mTOR protein, whereas downregulation of NRF2 showed the opposite results (Fig. S4i).
Fig. 8.
Effects of TRIM22 on ROS production, mitochondrial potential and Warburg effect in OS cells. (a) Heatmap of DEGs of HOS cells transfected with shNC and shTRIM22. (b) GSEA was used to identify the distribution of genes in the ROS-related, AMPK signaling-related, mTOR signaling-related and autophagy-related pathway gene set of HOS cells transfected with shNC and shTRIM22. (c, d) Mitochondrial potential was assessed and quantified by JC-1 staining in HOS and 143B cells (n = 3, ***P<0.001, unpaired two-tailed Student's t-test). Scale bar, 100 μm. (e, f) Detection and quantification of ROS levels in HOS and 143B cells by flow cytometry (n = 3, **P<0.01, ***P<0.001, unpaired two-tailed Student's t-test). (g, h) Evaluation of GSH/GSSG ratio and GPX activity in HOS and 143B cells (n = 3, *P<0.05, **P<0.01, ***P<0.001, unpaired two-tailed Student's t-test). (i) Western blot analysis of AMPK/mTOR signaling and LC3 as well as p62 protein levels in HOS and 143B cells. (j, k) Detection of ECAR and OCR in HOS and 143B cells (n = 3, two-way ANOVA with post hoc test). DEGs = differentially expressed genes; GSEA = gene set enrichment analysis; ROS = reactive oxygen species; GSH = reduced glutathione; GSSG = oxidized glutathione; GPX = glutathione peroxidase; ECAR = extracellular acidification rate; OCR = O2 consumption rate.
It has been noted that the Warburg effect, which was identified as the phenomenon that cancer cells mainly produce energy through aerobic glycolysis rather than oxidative phosphorylation (OXPHOS), contributes to the survival and apoptosis-inhibition of cancer cells. Interestingly, AMPK is metabolic sensor which contributes to maintain cellular energy homeostasis. It is regarded as a key player in the shift of Warburg effect in cancer cells. Therefore, this study further determined whether the TRIM22 could regulate the Warburg effect in OS. Depletion of TRIM22 resulted in an increase in the glycolytic rate (ECAR) while a decrease in the mitochondrial respiration (OCR) in HOS cells (Fig. 8j). Contrarily, overexpression of TRIM22 inhibited the Warburg effect in 143B cells (Fig. 8k). The functional role of NRF2 in the Warburg effect in OS cells was also investigated and results showed that NRF2 promoted the Warburg effect (Figs. S4j and k). These results indicated that TRIM22 contributed to the inhibition of Warburg effects in OS.
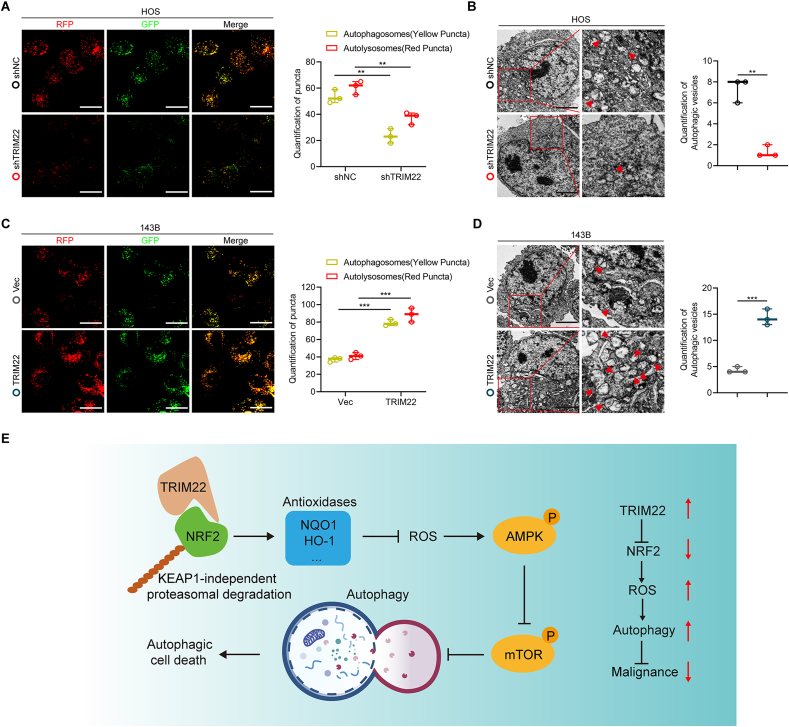
The activation of AMPK/mTOR signaling has been demonstrated to be associated with autophagy activation [25]. Also, GSEA results showed that the expression of TRIM22 was significantly correlated with autophagy pathway components, suggesting a potential regulatory role of TRIM22 in autophagy regulation (Fig. 8b). Thus, this study further evaluated the functional role of TRIM22 in the regulation of autophagy. First, to evaluate the formation of autophagy flux, OS cells were transfected with GFP-mRFP-LC3. It was found that the dot accumulation of GFP-mRFP-LC3 was significantly decreased when ablation of TRIM22 in HOS cells under a confocal microscopy (Fig. 9a). The autophagosomes were detected by transmission electron microscope (TEM), confirmed the aforementioned results (Fig. 9b). Moreover, that Western blot analysis detected the expression of LC3 and p62 also confirmed that inhibition of autophagy when silencing TRIM22 (Fig. 8i). Contrarily, overexpression of TRIM22 in 143B cells markedly promoted the activation of autophagy (Fig. 8, Fig. 9i). Also, NRF2 was found to inhibit autophagy in OS cells (Fig. S4i, l-o). Overall, these results indicated that TRIM22 promotes autophagy dependent on ROS/AMPK/mTOR signaling in OS cells.
Fig. 9.
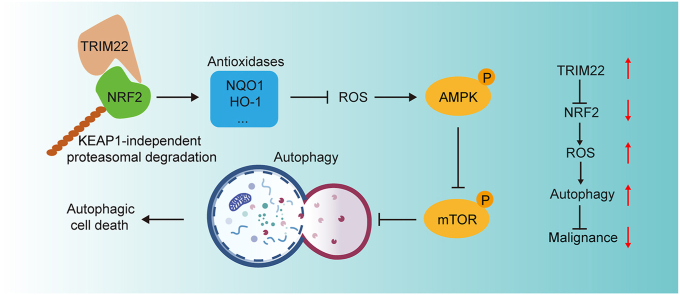
Effects of TRIM22 on autophagy in OS cells. (a) HOS cells transfected with GFP-mRFP-LC3 lentivirus were monitored, and the cellular puncta were evaluated. Scale bar, 20 μm (n = 3, **P<0.01, one-way ANOVA with post hoc test). (b) TEM was applied to determine the autophagic microstructure of transfected HOS cells (n = 3, **P<0.01, unpaired two-tailed Student's t-test). Scale bar, 2 μm. (c) 143B cells transfected with GFP-mRFP-LC3 lentivirus were monitored, and the cellular puncta were evaluated (n = 3, ***P<0.001, one-way ANOVA with post hoc test). Scale bar, 20 μm. (d) TEM was applied to determine the autophagic microstructure of transfected 143B cells. Scale bar, 2 μm (n = 3, ***P<0.001, unpaired two-tailed Student's t-test). (e) Potential underlying mechanism by which TRIM22/NRF2 axis inhibits OS progression through ROS/AMPK/mTOR/Autophagy signaling.
2.9. Inhibition of ROS or autophagy abolished the functional role of TRIM22 in inhibiting OS progression
To determine the role of TRIM22/NRF2 axis in the regulation of ROS/AMPK/mTOR and autophagy activation in OS cells, TRIM22-overexpressing 143B cells were treated with the ROS scavenger, NAC or the autophagy inhibitor 3-MA. The ROS/AMPK/mTOR signaling and autophagy was significantly inhibited by treatment of TRIM22-overexpressing 143B cells with NAC (Figs. S6a–c). Meanwhile, Treatment of the cells with NAC markedly abolished the decreased malignant phenotypes in TRIM22-overexpressing 143B cells (Figs. S6d–f). Moreover, treatment with 3-MA also significantly reversed the downregulated malignant behaviors in TRIM22-overexpressing 143B cells (Fig. S7). Overall, these results further suggested that TRIM22/NRF2 axis inhibits OS progression through ROS/AMPK/mTOR/Autophagy signaling.
3. Discussion
Using a series of loss- and gain-of-function in vitro and in vivo experiments, this present study indicate the role of TRIM22 in inhibiting OS progression and malignancy. Firstly, TRIM22 was found to be significantly downregulated in OS specimens and the expression level of TRIM22 was correlated with better outcomes. Various functional experiments were used to evaluate the role of TRIM22 in proliferation, apoptosis, migration and invasion of OS cells. TRIM22 was shown to positively inhibit the malignant phenotypes by destabilizing and promoting the degradation of NRF2, which is a well-known redox controller. A series of rescue functional experiment were conducted to confirm the TRIM22/NRF2 axis in inhibiting the progression of OS. ROS production and mitochondrial potential was evaluated and the role of TRIM22/NRF2 in regulation of ROS in OS was confirmed. It was finally found that TRIM22 could also inhibit the Warburg effect in OS cells and promote autophagic cell death by activating ROS/AMPK/mTOR/Autophagy signaling (Fig. 9e).
Osteosarcoma is the most common malignant bone tumor which mainly occurs in adolescents. Early lung metastasis is common and suggests a poor prognosis in patients suffered from OS [26,27]. Therefore, the underlying mechanism in OS development should be better investigated to develop effective therapeutic approaches for treating OS with metastasis. Several studies have revealed the important role of TRIM family proteins in various tumors [8]. For instance, TRIM14 was found to promote chemoresistance and EMT process via stabilizing DVL2 and ZEB2 in GBM respectively [28,29]. TRIM26 was demonstrated to inhibit HCC progression by degrading ZEB1 [30]. TRIM31 was shown to promote HCC progression through accelerating ubiquitination and degradation of TSC1-TSC2 complex [31]. Interestingly, Zhang et al. reported that TRIM24 enhances stemness and invasiveness through upregulating SOX2 in GBM [32] while another study found that TRIM24 could suppresses the development of HCC [33]. This indicates that even if the same TRIM family protein may exert opposite functions by targeting different proteins in diverse diseases. However, the functional role of TRIM family proteins in OS have been rarely investigated. In our study, TRIM22 was found to be significantly downregulated in OS specimens and associated with better outcome in patients. A series of in vitro and in vivo functional experiments demonstrated that TRIM22 inhibits tumor growth and metastasis in OS. However, the potential mechanism by which TRIM22 inhibits OS progression has not been fully investigated.
Next, IP/MS was performed to explore the underlying mechanism and protein-to-protein interaction underlying the functional role of TRIM22. NRF2 was identified as a potential protein that interacts with TRIM22 in OS cells. NRF2, which is well-known as an important transcription factor for regulating redox homeostasis, is promising in treating some diseases [34,35]. NRF2 expression and activation is regulated transcriptionally, post-transcriptionally as well as post-translationally. Under homeostatic conditions, NRF2 transcriptional and protein levels are at a low level. Three E3 ubiquitin ligase complexes, named KEAP1-CUL3-RBX1, β-TrCP-SKP1-CUL1-RBX1 as well as HRD1, regulate NRF2 ubiquitination and proteasomal degradation while KEAP1-regulated NRF2 degradation is the main regulatory way [36,37]. A previous study reported that KEAP1 could be deubiquitinated and stabilized by USP15 and thus promotes NRF2 degradation [38]. Zhang et al. reported that DUB3 interacts with and stabilized NRF2 [19]. In this study, it was demonstrated that TRIM22 interacts with NRF2 and promotes NRF2 ubiquitination and degradation independent of KEAP1. Moreover, our findings demonstrated that TRIM22 regulates the degradation of NRF2 by counteracting DUB3-mediated deubiquitylation of NRF2 in OS cells. However, there is need to investigate whether TRIM22 could cooperate with or competing with DUB3 or other unknown proteins to regulate the stability of NRF2. Moreover, it has been reported that NRF2 contains a KEAP1-independent degron within the Neh6 domain which is directly phosphorylated by GSK3β leading to increased NRF2 proteasomal degradation regulated by β-TrCP-SKP1-CUL1-RBX1 ubiquitin ligase complex [39,40]. In addition, once phosphorylation of the Neh6 domain increased, the interaction between the β-TrCP substrate adapter and NRF2 is evidently promoted. Interestingly, our results showed that TRIM22 interacts with the Neh6 domain of NRF2. Therefore, further studies are warranted to investigate whether the interaction between TRIM22 and NRF2 is regulated by phosphorylation of the Neh6 domain and specific phosphorylation events which potentially mediate the effect of TRIM22 on NRF2 degradation. Also, the involvement of β-TrCP in TRIM22-promoted NRF2 degradation still requires further investigation.
ROS is important signaling molecules and is involved in the transmission of information via a variety of signaling pathways in cells [41,42]. Excessive ROS production induces cancer cells autophagy and autophagic death by activating AMPK as well as other signaling pathways, hence inhibiting the initiation and progression of tumors [20,24]. For instance, Liu et al. reported that BDH2 inhibits tumor progression through ROS/PI3K/AKT/mTOR signaling [43]. Tang et al. showed that silencing HSP60 could suppress the progression of GBM by ROS/AMPK/mTOR signaling [44]. Another study demonstrated that salinomycin could promote autophagic cell death through increasing ROS production and hence activating PI3K/AKT/mTOR and ERK/p38 MAPK signal pathways [45]. NRF2 and its downstream target genes including HO-1 and NQO1 classically regulates ROS production. It's well-recognized that NRF2 is high-upregulated in majority of tumors. Therefore, cancer cells employ the cytoprotective ability of NRF2 to create a microenvironment which facilitates the survival of cancer cells [46,47]. Under a toxic threshold of high expression of NRF2 in tumors, ROS is balanced to avoid cell apoptosis or death in cancer cells. Several studies have proven that inhibiting NRF2 pathway could inhibit the progression of tumors and enhance the therapeutic effects of chemotherapeutic drugs [37]. The results of our study also showed that using ROS scavenger NAC significantly reversed the decreased malignant phenotypes in TRIM22-overexpressing OS cells. The findings suggested that overexpression of TRIM22 could lead to the degradation of NRF2 and hence increases ROS production as well as activates AMPK/mTOR/Autophagy signaling in OS.
Several previous studies suggest that autophagy plays an essential role in various biological processes. However, the exact functional role of autophagy is still controversial in tumor biology [[48], [49], [50]]. On the one hand, many studies have shown the cytoprotective role of autophagy in the development of malignant tumors via promoting cancer cell survival, cell proliferation, EMT as well as drug resistance. On the other hand, other studies have shown that autophagy can inhibit tumorigenesis and metastasis due to autophagic cell death in cancer cells. Therefore, autophagy may play crucial roles in the development of malignant tumors by promoting the survival of cancer cells or contributing to the apoptosis or death of cancer cells. Although our findings suggested that TRIM22 can lead to autophagic cell death in OS cells, the functions of autophagy in the development of malignant tumors should be more comprehensively investigated in tumor biology.
In conclusion, this study showed that TRIM22 inhibits OS progression by interacting with and promoting the ubiquitination and degradation of NRF2 independent of KEAP1. TRIM22/NRF2 axis further activated ROS/AMPK/mTOR/Autophagy signaling and lead to autophagic cell death in OS. Therefore, these findings indicated that targeting TRIM22/NRF2 axis may be a promising therapeutic target for treating OS.
4. Materials and methods
4.1. Clinical specimens and cell lines
OS tissues and matches of non-tumor normal tissues were collected during surgery. The OS tissue specimens were quickly frozen in liquid nitrogen and stored at −80°C until RNA and protein extraction. Human OS cell lines including HOS, Saos-2, U-2 OS, 143B and MG63, normal human osteoblast cell line hFOB 1.19 as well as HEK 293T cells were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China) and cultured according to the established protocol. The stable TRIM22-knockdown HOS cells, stable NRF2-overexpression HOS cells, stable NRF2-knockdown 143B cells and stable TRIM22-overexpression 143B cells were established according to the manufacturer's descriptions. The HEK 293T Tet-ON Flag-TRIM22 advanced stable cell line and NRF2 variants were generated accordingly. Indicated cell culture medium was supplemented with 1 μg/μL of doxycycline (Dox) to induce Flag-TRIM22 protein expression.
4.2. Cell counting kit-8 assay (CCK-8), colony-formation assay, 5-ethynyl-2′-deoxyuridine (EdU) assay and cell apoptosis assay
These functional in vitro experiments for testing cell proliferation and apoptosis in OS cells were conducted as described in our previous studies [51,52].
4.3. Invasion assay, 3D tumor spheroid cell-invasion assay and chemotaxis assay
Invasion of OS cells was measured using 24-well BD Matrigel invasion chambers (BD Biosciences) according to our previous studies [51,52]. Further, 3D tumor spheroid cell-invasion assay was performed as described in our previous studies [51,52]. The stable transfected OS cells were cultured at a density of 20,000 cells/mL in a 96-well ultralow adherence plate (#7007, Costar). OS cells were then induced to aggregate into a multicellular spheroid after 96 h followed by adding Matrigel to the wells. The motion of these multicellular spheroid cells was then observed under a fluorescence microscopy after 48 h.
Time-lapse imaging of established OS cells was performed as described previously [17,18]. OS cells were seeded into the microfluidic device (Ibidi). A stable linear-gradient of conditioning molecules was formed across the microfluidic device by supplying a medium of 10% fetal bovine serum (FBS) to the source reagent channel. A real-time video microscopy (Zeiss Cell Discoverer 7) was then used to image the osteosarcoma cells. Image J (National Institutes of Health, USA) was used to track cell migration. Thirty migratory cell tracks were imaged and recorded per trial. The migration track of each cell was plotted after normalizing the start point to x = 0 and y = 0. The cell chemotactic index was defined as the ratio of net distance of cell migration in the direction of gradient to the total length of cell track.
4.4. qRT-PCR
TRIzol reagent (Invitrogen, USA) was used to extract total RNAs from OS cells and the frozen tissues. The PrimeScript RT Master Mix Kit (TaKaRa Bio lnc., Japan) was used to perform mRNA reverse transcription according to the manufacturer's descriptions. The SYBR Green PCR Kit (TaKaRa Bio lnc.) was then used to carry out qRT-PCR on an ABI7900 fast real-time PCR system (Applied Biosystems, USA) and β-actin was used as the internal control.
4.5. Autophagosome detection by GFP-mRFP-LC3 and transmission electron microscopy (TEM)
Autophagosome was evaluated using a GFP-mRFP-LC3 lentivirus (Obio, China). Location and quantitation of autophagosomes were then observed under a confocal microscopy (LSM710, Zeiss, Germany). Autophagosomes were labeled red and green (yellow fluorescence) whereas autophagic lysosomes were labeled red. Transmission electron microscope (TEM) was performed and images were taken under an electron microscope (FEI Tecnai, USA).
4.6. Measurement of oxidative phosphorylation and glycolysis
A Seahorse XF96 Metabolic Flux Analyzer (Seahorse Biosciences, USA) was used to measure the extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) in OS cells in the indicated groups according to the manufacturer's instructions. Briefly, 3 × 104 cells in indicated groups were seeded into each well of a Seahorse XF96 cell culture microplate. Extracellular acidification rate was assessed through sequential injection of 10 mM glucose, 1 mM oligomycin and 80 mM 2-deoxyglucose (2-DG). The oxygen consumption rate was determined through sequential addition of 1 mM ATP synthase blocker oligomycin, 1 mM mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone and 2 mM complex I and III inhibitors antimycin A and 2 mM rotenone. Data quantification was carried out using XFe Wave software (Seahorse Biosciences) according to protocol of the manufacturer.
4.7. ROS and mitochondrial membrane potential evaluation
ROS Assay Kit (Beyotime, China) was used to determine ROS levels using 2′,7′-dichlorofluorsecein-diacetate (DCFH-DA) by flow cytometry analysis. Mitochondrial membrane potential was evaluated using JC-1 Assay Kit (Beyotime, China) and quantified by aggregate-to-monomer (red/green) fluorescence intensity ratio.
4.8. Detection of GSH/GSSG ratio and GPX activity
The ratio of reduced glutathione (GSH)/oxidized glutathione (GSSG) was detected by GSH and GSSG Assay Kits (Beyotime, S0053) according to the manufacturer's protocols. Glutathione peroxidase (GPX) activity was evaluated by a Glutathione Peroxidase Assay Kit (Beyotime, S0056) according to the manufacturer's protocols.
4.9. Immunohistochemistry (IHC)
IHC staining of 100 human OS samples in tissue microarray (TMAs) were performed using anti-TRIM22 and -NRF2 antibodies. The immunostaining results were examined by two independent researchers. All tissues were fixed in 4% paraformaldehyde and embedded in paraffin according to the established protocol. The paraffin was then cut into 4-μm thick sections and incubated overnight with the primary indicated antibodies followed by the secondary antibody incubation.
4.10. RNA sequence (RNA-Seq)
RNA-Seq analysis was performed by Genminix Information Co., Ltd., (Shanghai, China). Total RNAs from HOS cells transfected with shNC and shTRIM22 (n = 3/group) were extracted. Quality RNA samples were converted into cDNA libraries. The purified products were enriched with 12–15 cycles of PCR to create the final cDNA library. Libraries were then sequenced on the Illumina Hiseq X Ten following the manufacturer's protocols. Fold changes >1.5 and P < 0.05 represented differentially expressed genes (DEGs).
4.11. Western blot
Cells was lysed using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime). Equivalent amounts of protein were separated by SDS-PAGE, transferred to PVDF membranes (Merck-Millipore), blocked with 5% BSA and incubated with specific primary antibodies at 4 °C overnight. Corresponding secondary antibodies were then incubated with membranes for 2 h, and the enhanced chemiluminescence reagent (Merck-Millipore) was then used to visualize the reacting bands. Semi-quantification of the protein bands density was carried out using Image J software.
4.12. Immunoprecipitation (IP)
RIPA lysis buffer containing protease inhibitors (Beyotime) was used to collect and lyse OS cells and HEK 293T cells. The protein concentrations were determined using a BCA protein assay kit (Thermo Fisher Scientific, USA). Cell lysates were precleared with protein A/G-agarose beads (Santa Cruz Biotechnology, USA) for 1 h and immunoprecipitated with the indicated antibodies at 4 °C overnight. After that, the lysates were collected and incubated with protein A/G-agarose beads for 2 h and the immunocomplexes were washed five times with RIPA lysis buffer and the bound proteins were then eluted by boiling and subjected to SDS-PAGE for Western blot analysis.
4.13. In vivo ubiquitination assays
Anti-NRF2 antibodies were used to immunoprecipitate the endogenous NRF2 and then ubiquitin antibodies were used for immunoblotting to evaluate endogenous NRF2 ubiquitination. Ubiquitination of exogeneous NRF2 was evaluated after transfection of Flag-tagged TRIM22 (WT or C15/18A mutant), Myc-tagged NRF2 and HA-tagged ubiquitin into HEK 293T cells. Lysate proteins were precipitated and determined by immunoblotted with specific antibodies.
4.14. Animal experiments
All animal work was done in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Nanjing Medical University. Four-week-old nude mice (BALB/c nude mice) were obtained from the Animal Model Institute of Nanjing University (Nanjing, China) and used for in vivo tumor growth assay. The nude mice were randomly divided into several groups according to experimental requirements (n = 6). Stably transfected OS cells (2 × 106 cells) in 100 μL medium were subcutaneously injected into the nude mice. For tumor metastasis assay, medium containing 2 × 106 cells were injected through caudal vein. Development of metastases was imaged and evaluated using the IVIS200 imaging system (Caliper Life Science, USA). The investigators were blinded to the experimental groups and outcome evaluations.
4.15. Statistical analysis
Data are presented as mean ± SD and contain at least three independent biological replicates. Statistical analysis was performed using GraphPad Prism version 8 (GraphPad Software, USA). For two-group comparisons, the unpaired two-tailed Student's t-test was performed; for comparisons between more than two groups, one-way or two-way ANOVA followed by Tukey's post-hoc test was performed. A P-value < 0.05 was considered as statistically significant.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was sponsored by Shanghai Sailing Program (Grant No. 21YF1457000).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102344.
Contributor Information
Weihua Cai, Email: caiwhspine@sina.com.
Jianling Bai, Email: baijianling@njmu.edu.cn.
Xuhui Zhou, Email: zhouxuhui@smmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gianferante D.M., Mirabello L., Savage S.A. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017;13(8):480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14(11):722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 3.Ritter J., Bielack S.S. Osteosarcoma. Ann Oncol. 2010;21 doi: 10.1093/annonc/mdq276. Suppl 7vii320-325. [DOI] [PubMed] [Google Scholar]
- 4.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison D.J., Geller D.S., Gill J.D., Lewis V.O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 6.Vunjak M., Versteeg G.A. TRIM proteins. Curr. Biol. 2019;29(2):R42–R44. doi: 10.1016/j.cub.2018.11.026. [DOI] [PubMed] [Google Scholar]
- 7.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Hatakeyama S. TRIM proteins and cancer. Nat. Rev. Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 9.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19(1):59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 10.Rong Y., Fan J., Ji C., Wang Z., Ge X., Wang J., et al. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W., Ge X., Zhou Z., Jiang D., Rong Y., Wang J., et al. Deubiquitinase USP18 regulates reactive astrogliosis by stabilizing SOX9. Glia. 2021;69(7):1782–1798. doi: 10.1002/glia.23992. [DOI] [PubMed] [Google Scholar]
- 12.Obad S., Brunnstrom H., Vallon-Christersson J., Borg A., Drott K., Gullberg U. Staf50 is a novel p53 target gene conferring reduced clonogenic growth of leukemic U-937 cells. Oncogene. 2004;23(23):4050–4059. doi: 10.1038/sj.onc.1207524. [DOI] [PubMed] [Google Scholar]
- 13.Gongora C., Tissot C., Cerdan C., Mechti N. The interferon-inducible Staf50 gene is downregulated during T cell costimulation by CD2 and CD28. J. Interferon Cytokine Res. 2000;20(11):955–961. doi: 10.1089/10799900050198390. [DOI] [PubMed] [Google Scholar]
- 14.Ji J., Ding K., Luo T., Zhang X., Chen A., Zhang D., et al. TRIM22 activates NF-kappaB signaling in glioblastoma by accelerating the degradation of IkappaBalpha. Cell Death Differ. 2021;28(1):367–381. doi: 10.1038/s41418-020-00606-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao B., Duan Z., Xu W., Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology. 2009;50(2):424–433. doi: 10.1002/hep.23011. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Zhang B., Wei M., Xu Z., Kong W., Deng K., et al. TRIM22 inhibits endometrial cancer progression through the NOD2/NFkappaB signaling pathway and confers a favorable prognosis. Int. J. Oncol. 2020;56(5):1225–1239. doi: 10.3892/ijo.2020.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D.Y., Sun N.H., Lu Y.P., Hong L.J., Cui T.T., Wang C.K., et al. GPR124 facilitates pericyte polarization and migration by regulating the formation of filopodia during ischemic injury. Theranostics. 2019;9(20):5937–5955. doi: 10.7150/thno.34168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayakawa K., Ohkawa Y., Yoshizaki S., Tamaru T., Saito T., Kijima K., et al. Macrophage centripetal migration drives spontaneous healing process after spinal cord injury. Sci. Adv. 2019;5(5) doi: 10.1126/sciadv.aav5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q., Zhang Z.Y., Du H., Li S.Z., Tu R., Jia Y.F., et al. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26(11):2300–2313. doi: 10.1038/s41418-019-0303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poillet-Perez L., Despouy G., Delage-Mourroux R., Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015:4184–4192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faubert B., Boily G., Izreig S., Griss T., Samborska B., Dong Z., et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metabol. 2013;17(1):113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y., Wang J., Guo S., Pourteymour S., Xu Q., Gong J., et al. Non-lethal sonodynamic therapy facilitates the M1-to-M2 transition in advanced atherosclerotic plaques via activating the ROS-AMPK-mTORC1-autophagy pathway. Redox Biol. 2020:32101501. doi: 10.1016/j.redox.2020.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong D., Zhang Z., Chen L., Huang W., Zhang F., Wang L., et al. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020:36101600. doi: 10.1016/j.redox.2020.101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y., Hu X., Liu Y., Dong S., Wen Z., He W., et al. ROS signaling under metabolic stress: cross-talk between AMPK and AKT pathway. Mol. Cancer. 2017;16(1):79. doi: 10.1186/s12943-017-0648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsey B.A., Markel J.E., Kleinerman E.S. Osteosarcoma overview. Rheumatol Ther. 2017;4(1):25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna C., Fan T.M., Gorlick R., Helman L.J., Kleinerman E.S., Adamson P.C., et al. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin. Cancer Res. 2014;20(16):4200–4209. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng S., Cai X., Li Y., Jian X., Zhang L., Li B. Tripartite motif-containing 14 (TRIM14) promotes epithelial-mesenchymal transition via ZEB2 in glioblastoma cells. J. Exp. Clin. Cancer Res. 2019;38(1):57. doi: 10.1186/s13046-019-1070-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Tan Z., Song L., Wu W., Zhou Y., Zhu J., Wu G., et al. TRIM14 promotes chemoresistance in gliomas by activating Wnt/beta-catenin signaling via stabilizing Dvl2. Oncogene. 2018;37(40):5403–5415. doi: 10.1038/s41388-018-0344-7. [DOI] [PubMed] [Google Scholar]
- 30.Li X., Yuan J., Song C., Lei Y., Xu J., Zhang G., et al. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo P., Ma X., Zhao W., Huai W., Li T., Qiu Y., et al. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene. 2018;37(4):478–488. doi: 10.1038/onc.2017.349. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L.H., Yin Y.H., Chen H.Z., Feng S.Y., Liu J.L., Chen L., et al. TRIM24 promotes stemness and invasiveness of glioblastoma cells via activating Sox2 expression. Neuro Oncol. 2020;22(12):1797–1808. doi: 10.1093/neuonc/noaa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang S., Minter L.C., Stratton S.A., Yang P., Abbas H.A., Akdemir Z.C., et al. TRIM24 suppresses development of spontaneous hepatic lipid accumulation and hepatocellular carcinoma in mice. J. Hepatol. 2015;62(2):371–379. doi: 10.1016/j.jhep.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016:711624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013:145–149. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34(1):21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villeneuve N.F., Tian W., Wu T., Sun Z., Lau A., Chapman E., et al. USP15 negatively regulates Nrf2 through deubiquitination of Keap1. Mol. Cell. 2013;51(1):68–79. doi: 10.1016/j.molcel.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004;279(30):31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 40.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/beta-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a keap1-independent manner. Mol. Cell Biol. 2011;31(6):1121–1133. doi: 10.1128/Mcb.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brieger K., Schiavone S., Miller F.J., Jr., Krause K.H. Reactive oxygen species: from health to disease. Swiss Med. Wkly. 2012:142w13659. doi: 10.4414/smw.2012.13659. [DOI] [PubMed] [Google Scholar]
- 42.Liu W., Tang P., Wang J., Ye W., Ge X., Rong Y., et al. Extracellular vesicles derived from melatonin-preconditioned mesenchymal stem cells containing USP29 repair traumatic spinal cord injury by stabilizing NRF2. J. Pineal Res. 2021;71(4) doi: 10.1111/jpi.12769. [DOI] [PubMed] [Google Scholar]
- 43.Liu J.Z., Hu Y.L., Feng Y., Jiang Y., Guo Y.B., Liu Y.F., et al. BDH2 triggers ROS-induced cell death and autophagy by promoting Nrf2 ubiquitination in gastric cancer. J. Exp. Clin. Cancer Res. 2020;39(1):123. doi: 10.1186/s13046-020-01620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H., Li J., Liu X., Wang G., Luo M., Deng H. Down-regulation of HSP60 suppresses the proliferation of glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci. Rep. 2016:628388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim K.Y., Park K.I., Kim S.H., Yu S.N., Park S.G., Kim Y.W., et al. Inhibition of autophagy promotes salinomycin-induced apoptosis via reactive oxygen species-mediated PI3K/AKT/mTOR and ERK/p38 MAPK-dependent signaling in human prostate cancer cells. Int. J. Mol. Sci. 2017;18(5) doi: 10.3390/ijms18051088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Homma S., Ishii Y., Morishima Y., Yamadori T., Matsuno Y., Haraguchi N., et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res. 2009;15(10):3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 47.Lignitto L., LeBoeuf S.E., Homer H., Jiang S., Askenazi M., Karakousi T.R., et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell. 2019;178(2):316–329. doi: 10.1016/j.cell.2019.06.003. e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh S.S., Vats S., Chia A.Y., Tan T.Z., Deng S., Ong M.S., et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37(9):1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 49.Eskelinen E.L. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011;11(4):294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Morselli E., Galluzzi L., Kepp O., Vicencio J.M., Criollo A., Maiuri M.C., et al. Anti- and pro-tumor functions of autophagy. Biochim. Biophys. Acta. 2009;1793(9):1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y., Liu W., Tang P., Jiang D., Gu C., Huang Y., et al. miR-624-5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J. Exp. Clin. Cancer Res. 2019;38(1):488. doi: 10.1186/s13046-019-1491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge X., Liu W., Zhao W., Feng S., Duan A., Ji C., et al. Exosomal transfer of LCP1 promotes osteosarcoma cell tumorigenesis and metastasis by activating the JAK2/STAT3 signaling pathway. Mol. Ther. Nucleic Acids. 2020:21900–21915. doi: 10.1016/j.omtn.2020.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.