Abstract
The chemical composition of each O-antigen subunit in gram-negative bacteria is a reflection of the unique DNA sequences within each rfb operon. By characterizing DNA sequences contained with each rfb operon, a diagnostic serotype-specific probe to Escherichia coli O serotypes that are commonly associated with bacterial infections can be generated. Recently, from an E. coli O157:H7 cosmid library, O-antigen-positive cosmids were identified with O157-specific antisera. By using the cosmid DNAs as probes, several DNA fragments which were unique to E. coli O157 serotypes were identified by Southern analysis. Several of these DNA fragments were subcloned from O157-antigen-positive cosmids and served as DNA probes in Southern analysis. One DNA fragment within plasmid pDS306 which was specific for E. coli O157 serotypes was identified by Southern analysis. The DNA sequence for this plasmid revealed homology to two rfb genes, the first of which encodes a GDP-mannose dehydratase. These rfb genes were similar to O-antigen biosynthesis genes in Vibrio cholerae and Yersinia enterocolitica serotype O:8. An oligonucleotide primer pair was designed to amplify a 420-bp DNA fragment from E. coli O157 serotypes. The PCR test was specific for E. coli O157 serotypes. PCR detected as few as 10 cells with the O157-specific rfb oligonucleotide primers. Coupled with current enrichment protocols, O157 serotyping by PCR will provide a rapid, specific, and sensitive method for identifying E. coli O157.
Enterohemorrhagic Escherichia coli (EHEC) has become the leading cause of hemorrhagic colitis and hemolytic-uremic syndrome in the United States and Canada. The challenge to clinical and food microbiologists is to differentiate E. coli O157:H7 from other commensal E. coli isolates present within the gastrointestinal tract. The transmission of EHEC occurs by the consumption of improperly cooked beef products, person-to-person contact, or drinking of unpasteurized milk or water (14, 15, 28). Dairy and beef cattle have been identified as one reservoir for E. coli O157:H7 (31, 50), although a survey of meat products, including pork, poultry, and lamb, suggests that other animals may also harbor E. coli O157:H7 (18, 39).
Identification of E. coli O157:H7 relies on a combination of biochemical and serological tests. E. coli O157:H7 can be biochemically distinguished from commensal E. coli isolates based on the absence of sorbitol fermentation and β-glucuronidase activity (17). Colony immunoblots or slide agglutination tests with O157 antiserum are subsequently used for confirmation of E. coli isolates as the O157 serotype (17). Current identification schemes, therefore, involve an enrichment step, a screen for sorbitol fermentation, and a final serological confirmation of E. coli O157:H7 (35). The current EHEC isolation and enrichment scheme occasionally results in the report of false positives. Cross-reactivity to O157 polyclonal antisera has been reported for Citrobacter freundii, Yersinia enterocolitica, Pseudomonas maltophilia, Brucella abortus, Brucella melitensis, Escherichia hermanii, Hafnia alvei, Morganella morganii, and Salmonella group N (5, 9, 11). The serological and biochemical basis for the cross-reactivity is the presence of 4-amino-4,6-dideoxy-d-mannopyranosyl as a constituent sugar of the lipopolysaccharide (LPS) (36). Cross-reactivity of O157 antisera to E. hermanii is a particular problem, because the biochemical profile of E. hermanii is similar to that of E. coli O157:H7. Other EHEC O157 strains that are sorbitol negative and H negative have also been identified (24). Although several enteric bacteria exhibit cross-reactivity to O157-typing sera, a thorough analysis of genes necessary for O-antigen synthesis may identify gene sequences unique to E. coli O157:H7.
The chemical composition of each O antigen is a reflection of the unique DNA sequences that are collectively responsible for the synthesis of this polysaccharide. The enzymes involved in the synthesis, polymerization, and attachment of the O-antigen subunits to the core LPS structure are specified by the rfb operon. The number of genes present within this operon varies depending on the sugar compositions and complexities of the O antigens found in the LPSs of Salmonella serovars and E. coli O serotypes (41). The rfb gene cluster has been cloned from a number of gram-negative bacteria, including Salmonella spp. (10, 23, 53), various E. coli O serotypes (25, 42, 44, 46), Shigella spp. (26, 32, 43, 48), Klebsiella pneumoniae (12), Y. enterocolitica (1, 22, 55), Vibrio cholerae (6, 33), and Pseudomonas aeruginosa (19).
Comparisons of DNA sequences within Salmonella rfb operons have identified regions that are conserved and regions that are unique to a given Salmonella serovar (10, 29, 47, 53, 54). Conservation of rfb gene sequences reflects the presence of common sugars present in O antigens in some members of the Enterobacteriaceae family. For example, S. typhimurium, S. paratyphi, and S. typhi produce a common core O-antigen subunit consisting of rhamnose, galactose, and mannose. It is therefore not surprising that at the genetic level the rfb operon is similar in S. typhimurium, S. paratyphi, and S. typhi. In fact, the genes rfbBCAD, which are responsible for the synthesis of TDP-rhamnose, not only are found in Salmonella serovars Typhimurium, Paratyphi, and Typhi but also have been identified in other rfb operons encoding proteins in which rhamnose is a constituent sugar of the O antigen (38). Serological differences observed among S. typhimurium, S. paratyphi, and S. typhi, however, reflect the presence of the unique dideoxy sugars abequose, paratose, and tyvelose, respectively (41). The rfb genes, which specify the synthesis of these unique sugars, have proven to be useful as DNA-based markers for identification and differentiation of Salmonella serogroups A, B, C2, and D (30). The O157 rfb operon also contains gene sequences that specify enzymes that are either unique to E. coli O157 serotypes (7) or evolutionarily divergent enough to serve as molecular markers for E. coli O157 (this study). In this study, a region within the E. coli O157:H7 rfb operon which was specific to E. coli O157 was identified from Southern analysis. DNA sequence analysis revealed similarity to two previously identified rfb genes from Y. enterocolitica and V. cholerae. Sufficient difference in the nucleotide sequence was present to design oligonucleotides specific to E. coli O157 serotypes. E. coli O157 serotypes were identified by PCR with these primers. PCR was able to detect as few as 10 E. coli O157 cells.
MATERIALS AND METHODS
Bacterial strains.
The E. coli O157 serotypes examined in this study were isolated from several sources, including humans and animals. The E. coli O157 serotypes in this study included H7 and other H serotypes. The additional E. coli O serotypes O11, O26, O55, and O111, as well as Salmonella enteriditis and S. typhimurium, were used to test the specificity of O157 gene probes. Also included in this test were M. morganii, H. alvei, and C. freundii (5), bacteria that exhibit cross-reactivity with polyclonal O157 antisera.
Creation of an E. coli O157:H7 genomic library and screening of the library for O-antigen-positive clones.
Genomic DNA from E. coli O157:H7 ATCC 31350 was isolated by cetyltrimethylammonium bromide-NaCl-proteinase K extraction (3) and further purified by CsCl-ethidium bromide gradient ultracentrifugation (40). Chromosomal DNA from E. coli O157:H7 was partially digested with the restriction enzyme Sau3AI under conditions that yielded large fragments of DNA (30 to 40 kb) (37). The DNA fragments were ligated into the BamHI site of pcos2EMBL (37). The ligated DNA was packaged into λ phages with an in vitro λ phage extract (Gigapack II XL; Stratagene, La Jolla, Calif.) and introduced into competent E. coli K-12 by λ-mediated transduction.
The library was screened with polyclonal O157-specific antiserum (Difco, Detroit, Mich.) by the procedure of Morona et al. (33). Nitrocellulose filters were incubated overnight at room temperature with the primary antibody diluted 1/2,000. An anti-rabbit immunoglobulin M–alkaline phosphatase conjugate (Sigma, St. Louis, Mo.), diluted 1/1,000, was subsequently applied to the filters and incubated at room temperature for 30 min. The development of the blots was done with Fast Red TR/Napthol AS-MX (Sigma).
O157-positive E. coli K-12 cosmid clones were checked for antigen expression prior to every small- or large-scale extraction procedure for plasmid DNA or LPS by slide test agglutination with polyclonal O157 antiserum. Cosmid DNA was isolated from E. coli K-12 by sodium doedecyl sulfate lysis (40) and purified by CsCl-ethidium bromide gradient ultracentrifugation.
E. coli O157-antigen-specific sera included rabbit polyclonal O157-specific antiserum (Difco Laboratories), which was used in the initial screen of the E. coli O157:H7 genomic library, and a monoclonal O157-specific antibody, 8-9H (PerImmune Inc., Rockville, Md.). The monoclonal antibody specifically recognizes the O-antigen epitope unique to the O157 serotype in E. coli. This antibody was used to confirm O-antigen-positive cosmid clones identified from the initial screen with polyclonal O157 antiserum.
Characterization of the LPS from E. coli O157-positive cosmid clones.
LPS was extracted from 1.5-ml overnight cultures by proteinase K treatment (21). The LPS samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5% polyacrylamide resolving gel, 4% separating gel) at 75 V. Polysaccharide bands were visualized on the gels by using the periodate-silver staining method (45). For Western analysis, gels were transferred onto nitrocellulose membranes by electroblotting for 1 h at 100 V according to the manufacturer’s specifications for the MiniTrans Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, Calif.). The membranes were prepared for Western analysis by the same procedure described for colony blotting.
Southern analysis.
Chromosomal DNA (1 μg) was digested with 3 U of selected restriction enzymes at 37°C for 2 h. The DNA was separated on a 0.5% agarose gel (1× Tris-borate-EDTA buffer [40] and ethidium bromide [5 μg/ml]) and transferred to a nylon membrane (40). Single-stranded DNA was cross-linked to membranes by using UV light (Fisher Biotech UV Crosslinker, optimal cross-linking setting). Probes were labeled with digoxigenin (DIG)-labeled deoxynucleoside triphosphates by random primer extension (Genius 3; Boehringer Mannheim, Indianapolis, Ind.). DNA probes were prepared from pDS300 subclones as follows. Plasmid DNA was cut with the appropriate restriction enzyme(s), which excised the insert from the plasmid. The DNA was separated on a 0.5% agarose gel (1× Tris-acetate-EDTA buffer [TAE] (40), and the DNA insert was extracted from the gel and purified by using a GenElute Agarose Spin Column (Supelco, Bellefonte, Pa.). The DNA was labeled with DIG-tagged dioxynucleoside triphosphates by using the Genius 3 system. Procedures for DNA-DNA hybridizations and detection were performed as specified in the protocol for the Genius 3 kit. The annealing temperature for hybridizations and washes was 68°C (40).
Strategy for subcloning and identification of E. coli O157-specific DNA probes.
Cosmid pDS300 was digested with a variety of restriction enzymes, including EcoRI, EcoRV, HindIII, PstI, and PvuII. DNA was separated on a 0.5% agarose gel (1× TAE) (40), and DNA bands that corresponded in size to E. coli O157-specific DNA fragments that were identified from Southern blots of E. coli and Salmonella genomic DNAs were extracted from the agarose gel. DNA was extracted from agarose gel slices by using the Supelco GenElute Agarose Spin Column. Extracted DNA was ligated to plasmid DNA pZERO (Invitrogen, San Diego, Calif.) by using T4 DNA ligase (Promega, Madison, Wis.). Competent E. coli TOP10F′ (OneShot TOP10F′; Invitrogen) was transformed with ligated DNA (20), plated on L agar containing Zeocin (200 μg/ml), and incubated overnight at 37°C. The plasmid pZERO is a cloning vector which selects for recombinants via the disruption of ccdB, a lethal gene in E. coli TOP10F′ cells (4). Plasmid DNA was isolated from transformants by alkaline lysis (8). Each DNA insert was subsequently tested for E. coli O157 specificity by Southern analysis.
DNA sequencing.
A plasmid, pDS306, was identified from Southern analysis as being E. coli O157 specific. Approximately 5 μg of plasmid DNA was submitted to the Molecular Genetics Instrumentation Facility at the University of Georgia for sequencing. An ABI automated sequencer for double-stranded DNA sequencing was used to sequence the plasmid insert. Primer walking was used to generate the complete DNA sequence. DNA sequences were entered into the National Center for Biotechnology Information (34) Blast search program, which was queried for any homology between the query nucleotide sequence and reported gene sequences (2).
PCR.
One hundred nanograms of E. coli chromosomal DNA served as the template in 10 μl of PCR mix. This reaction mix consisted of 0.2 mM deoxynucleotides, 2.0 mM MgCl2, 1× PCR buffer (50 mM Tris, pH 7.4), bovine serum albumin (0.25 mg/ml), 50 pmol of forward (O157PF8 [CGTGATGATGTTGAGTTG]) and reverse (O157PR8 [AGATTGGTTGGCATTACTG]) PCR primers, and 0.5 U of Taq DNA polymerase (Boehringer Mannheim). The PCR reaction mix was drawn up into 10-μl-capacity thin-walled capillary tubes, and the ends were heat sealed. The capillary tubes were placed in a Rapidcycler (Idaho Technology, Idaho Falls, Idaho) (52). The program parameters for the hot-air thermocycler were (i) 94°C for 0 s, (ii) 55°C for 0 s, and (iii) 72°C for 15 s for 30 cycles (the thermocycler quickly heats the capillary tubes to 94° and 55°C and holds the reaction temperature at 72°C before repeating the cycle). Thirty cycles of PCR were completed in 10 min. DNA products from PCR were analyzed by gel electrophoresis. DNA was separated on 1.5% agarose–1× TAE gels at 70 V. A 100-bp ladder (Promega) served as a molecular size standard for determining molecular sizes of PCR products. PCR products were purified by using a WIZARD Magic PCR column (Promega). DNA was sent to the Molecular Genetics Instrumentation Facility for double-stranded DNA sequencing. Oligonucleotides serving as forward and reverse PCR primers were also used as primers for sequencing double-stranded DNA.
To test the sensitivity of PCR, E. coli O157:H7 ATCC 31350 cells were serially diluted in 1 ml of saline, starting with 109 CFU/ml. A 10-μl aliquot from each 10-fold dilution was spotted, in triplicate, onto 1% sorbitol–MacConkey agar and incubated overnight at 37°C to enumerate the cells. Each dilution was boiled for 10 min, and the samples were centrifuged at 12,000 rpm for 30 min. A 100-μl aliquot was taken from each sample, diluted 1/10 in saline, and stored at −20°C. One microliter of boiled cells from each dilution served as the template in PCRs with DIG-labeled nucleotides (Boehringer Mannheim) included in the PCR mixtures. A control lacking template was included with each reaction as a check for PCR contamination. The DNAs were separated on 1.5% agarose gels (1× TAE plus 5 μl of ethidium bromide per 100 ml) and transferred to nylon membranes (40). Membranes were blocked with casein, probed with anti-DIG antibody–alkaline phosphatase conjugate, and developed by using 4-nitroblue tetrazolium chloride–5-bromo-4-chloro-3-indoyl phosphate solution as outlined in the Genius 3 kit (Boehringer Mannheim).
PCR was applied to the detection of E. coli O157 in material where the organism may be found. We chose milk and bovine feces for this purpose. An overnight culture of E. coli O157 (1.4 × 109 CFU/ml) was diluted 10-fold in whole milk. Samples were spun at 10,000 × g at room temperature for 15 min. The top fat layer and underlying water were removed, and the pellet was resuspended in distilled water (dH2O). The DNA template was then prepared by boiling the suspension for 10 min and separating the supernatant from milk proteins and cell debris by a brief centrifugation. One microliter of this supernatant served as the template in the PCR. Bovine feces were also inoculated with E. coli O157:H7 and used to determine the ability of the PCR to detect E. coli O157 present in these samples. Two hundred milligrams of bovine feces was added to a 109-CFU/ml culture of E. coli O157:H7. The sample was vigorously vortexed for 5 min. The large insoluble particles present in the sample were removed by a low-speed centrifugation at 3,000 × g for 10 min. The supernatant was boiled for 10 min. The sample was centrifuged again to pellet cell debris. The supernatant was diluted 10-fold in dH2O and served as a template for PCR.
Nucleotide sequence accession number.
Nucleotide and amino acid sequence data have been deposited in the NIH/GenBank database under accession no. AF049343.
RESULTS
Identification of unique gene sequences within a cosmid specifying O157 antigen in E. coli K-12.
The rfb operon from E. coli O157:H7 was successfully cloned into E. coli K-12 host strain LE392. The cosmid library was screened with polyclonal O157-specific antiserum, resulting in the identification of 5 O-antigen-positive clones among the 5,692 clones screened. The O157-specific monoclonal antibody 8-9H reacted with low- and high-molecular-weight LPS from E. coli O157-antigen-positive clones (Fig. 1).
FIG. 1.
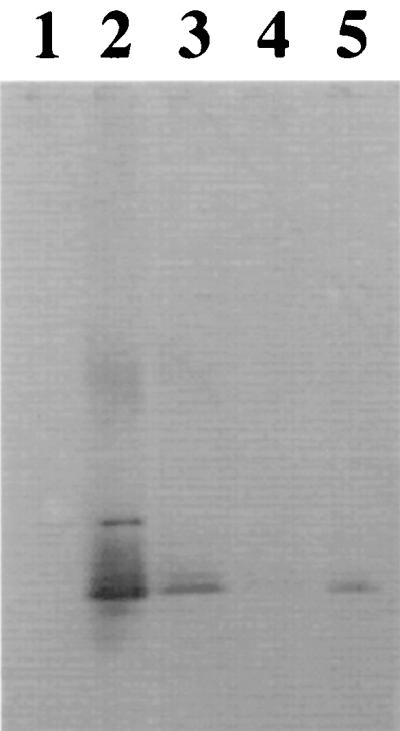
Immunological analysis of LPS produced by O157-antigen-positive clones by Western blotting. Lane 1, S. typhimurium (wild type); lane 2, E. coli O157:H7 ATCC 35150; lane 3, E. coli LE392 with pDS23; lane 4, S. typhimurium rfc; lane 5, E. coli LE392 with pDS300. LPS was separated on 18% acrylamide resolving gels (75 V/gel), electrophoretically transferred to nitrocellulose membranes, and probed with O157-specific monoclonal antibody 8-9H. The primary antibody was detected by using rabbit anti-mouse immunoglobulin G antibody–alkaline phosphatase conjugate and FAST RED alkaline phosphatase substrate.
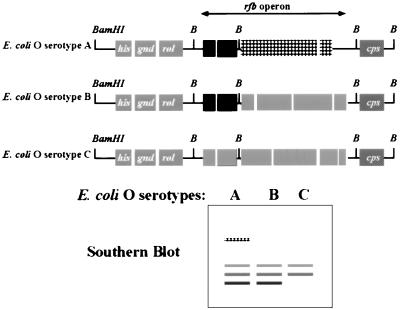
The cosmid DNAs from these O157-positive clones contained DNA inserts of between 30 and 40 kb in size. These DNA inserts probably contain not only the rfb genes necessary for O-antigen synthesis but genes flanking this operon as well. The genetic map of the rfb locus (his-rol-gnd-rfb-cps) is conserved among all E. coli O serotypes and is similar in its genetic organization to the S. typhimurium rfb locus (41). There is also conservation in rfb gene sequences for O serotypes that specify sugars commonly found in different O serotypes (41). Among these conserved gene sequences, we also anticipated finding O-serotype-specific genes that encode glycosyltranserase, reductases, epimerases, isomerases, and O-antigen polymerase, which are collectively responsible for the unique carbohydrate chemistry present in E. coli O157:H7. In order to generate O-serotype-specific DNA probes, we therefore first needed to distinguish the unique O157 DNA fragments from the conserved gene sequences present within the O157-positive cosmid (Fig. 2).
FIG. 2.
Strategy for identifying gene sequences unique to the E. coli O157 rfb operon. Cosmid DNAs from O-antigen-positive clones were used as DNA probes in Southern analysis of the Enterobacteriaciae genome. The genes flanking the rfb operon are highly conserved between E. coli and S. typhimurium. If the DNA inserts contain the rfb operon and genes upstream or downstream of this operon, then Southern analysis will identify similar-size DNA fragments (gray bands with horizontal or diagonal stripes) in all E. coli O serotypes and other enteric bacteria included in the study. The rfb operons will also contain genes whose products synthesize a sugar common to many O serotypes. The cosmid DNA probes are expected to recognize similar-size DNA fragments (black) present in only those E. coli O serotypes that synthesize a sugar(s) common to both. Finally, DNA sequences within the DNA insert that are unique to a specific E. coli O serotype will recognize only DNA fragments in that O serotype (diamond pattern) when used as a DNA probe in Southern blotting.
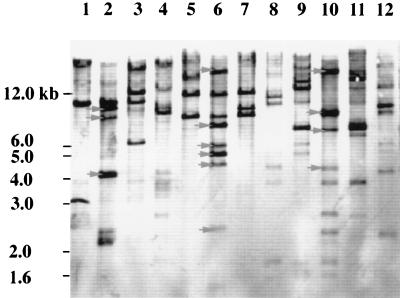
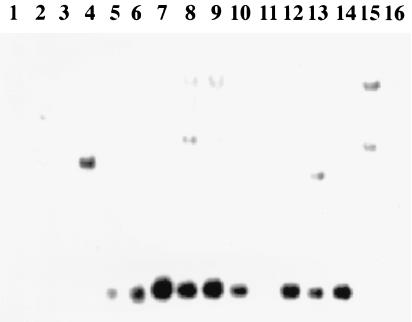
Experiments were therefore initiated to identify the regions within the DNA inserts that are unique to E. coli O157 serotypes. Southern analysis of genomic DNAs from a number of enteric bacteria was performed with the O-antigen-positive cosmids as DNA probes (Fig. 3). With selected restriction enzymes, E. coli O157-specific DNA bands were detected by Southern blotting. E. coli O157-specific bands that ranged in size from 7.5 to 2.3 kb were identified by DNA probe pDS300 with HindIII restriction enzyme digests of E. coli and Salmonella genomic DNAs (Fig. 3). No bands were observed for genomic DNAs of E. hermanii and M. morganii, organisms that cross-react with the polyclonal O157 antiserum (data not shown). Cosmid pDS300 was digested with a number of different restriction enzymes, DNA fragments were separated by gel electrophoresis, and selected DNA bands were extracted from the agarose gel and cloned into DNA sequencing vector pZERO. The DNA fragments that corresponded in size to E. coli O157-specific DNA fragments identified from the first Southern blot were selected. The specificities of the resulting subclones were later assessed by Southern analysis. We identified a number of plasmids with inserts which were specific for E. coli, E. coli O26/O157, and E. coli O157 as DNA probes in Southern analysis. Plasmid pDS306 appeared to be specific for E. coli O157. In Southern analysis, pDS306 identified a 1.9-kb EcoRI DNA fragment in 9 of 10 E. coli O157 serotypes. Although one E. coli O157 isolate did not produce as strong a signal as the other O157 serotypes by Southern blotting, this isolate was weakly positive by PCR (see below). No 1.9-kb EcoRI DNA fragment was observed for other E. coli O serotypes, S. typhimurium, S. enteriditis, and C. freundii under stringent hybridization and wash conditions (Fig. 4).
FIG. 3.
Distribution of E. coli O157 rfb gene sequences present on O-antigen cosmid pDS300 among E. coli O serotypes and S. typhimurium. Lanes 1, 5, and 9, E. coli O26 ATCC 11840; lanes 2, 6, and 10, E. coli O157:H7 ATCC 35150; lanes 3, 7, and 11, E. coli K-12; lanes 4, 8, and 12, S. typhimurium. Chromosomal DNA was digested with restriction enzyme EcoRI (lanes 1 to 4), HindIII (lanes 5 to 8), or PstI (lanes 9 to 12). DNA fragments were separated on 0.5% agarose gels, transferred to nylon membranes, and hybridized with DIG-labeled DNA probe pDS300.
FIG. 4.
Assessing pDS306 as an E. coli O157 serotype-specific DNA probe by Southern blotting. Lane 1, S. typhimurium; lane 2, S. enteriditis; lane 3, E. coli K-12; lane 4, E. coli O26 (ATCC 11840); lane 5, E. coli O157:H7 (ATCC 35150); lane 6, E. coli O157:H7; lane 7, E. coli O157:H7; lane 8, E. coli O157:H11; lane 9, E. coli O157:H16; lane 10, E. coli O157:H42; lane 11, E. coli O157:H7; lane 12, E. coli O157:H7; lane 13, E. coli O157:H7; lane 14, E. coli O157:H7; lane 15, E. coli O55:H7; lane 16, C. freundii. Chromosomal DNA was digested with the restriction enzyme EcoRI. DNA fragments were separated on 0.5% agarose gels, transferred to nylon membranes, and hybridized with DIG-labeled DNA probe pDS306.
The DNA sequence of the insertion in the plasmid pDS306 was determined and exhibited significant homology with reported gene sequences (Table 1). pDS306 has 75 and 69% identity with rfb genes which encode a GDP-mannose dehydratase (Gmd) and WcaG, respectively. The GC content of the E. coli O157 rfb genes was 39%, similar to low GC content present in Salmonella and E. coli rfa and rfb genes (41). The 1.4-kb EcoRI DNA insert present in pDS306 contains two open reading frames (rfbA and rfbB) which correspond in amino acid sequence to a GDP-mannose dehydratase, Gmd (nucleotide positions 1 to 600), and a capsule biosynthesis gene product, WcaG (nucleotide positions 839 to end), respectively.
TABLE 1.
DNA sequence identity between E. coli O157 rfb gene sequences (plasmid pDS306) and reported bacterial gene sequences
| E. coli O157 rfb gene | Gene identified from BLAST search | Function | Identity (%) |
|---|---|---|---|
| ORF1 (rfbA; positions 1 to 601) | Y. enterocolitica O:8 rfb (ORF 13.7) | GDP-mannose dehydratase | 73 |
| V. cholerae rfbD | GDP-mannose dehydratase | 74 | |
| E. coli K-12 cps operon (ORF 0.0) | GDP-mannose dehydratase | 75 | |
| ORF2 (rfbB; positions 837 to 1465) | E. coli K-12 cps operon (ORF 0.9) | Unknown | 69 |
| Y. enterocolitica O:8 rfb (ORF 14.9) | Unknown | 65 |
Specificity and sensitivity of rfbO157-designed primers for detecting E. coli O157 serotypes by PCR.
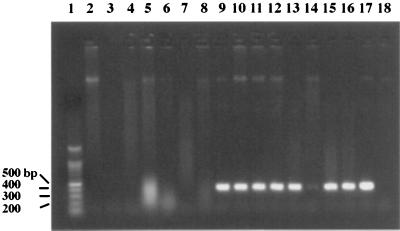
The nucleotide sequence present in the pDS306 DNA insert was searched for potential primer sequences. With primer pair O157PF8 and O157PR8, a 420-bp DNA fragment was observed for all E. coli O157 serotypes examined, while no product was observed for other E. coli O serotypes and C. freundii, an O157 antiserum-positive isolate (Fig. 5). The nucleotide sequences of eight E. coli O157 DNA fragments amplified by PCR with primers O157PF8 and O157PR8 were determined and were found to be similar to the original rfb gene sequence obtained for pDS306. There were a total of only four base pair changes in the nucleotide sequence among the eight E. coli O157 420-bp PCR products (data not shown). We were able to detect as few as 10 E. coli O157:H7 cells by PCR with these primers (Fig. 6).
FIG. 5.
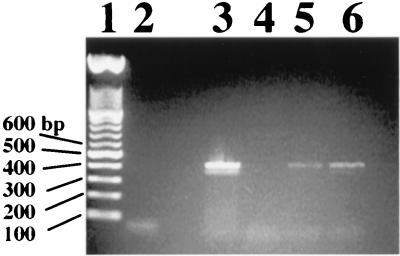
Identification of E. coli O157 serotypes by PCR with E. coli O157 rfbB-specific primers. Lane 1, 100-bp ladder (Promega); lane 2, S. enteriditis; lane 3, S. typhimurium UK1; lane 4, E. coli K-12 LE392 (O16); lane 5, E. coli O11; lane 6, E. coli O18; lane 7, E. coli O26 ATCC 11840; lane 8, E. coli O111; lane 9, E. coli O157:H7; lane 10, E. coli O157:H7; lane 11, E. coli O157:H11; lane 12, E. coli O157:H16; lane 13, E. coli O157:H42; lane 14, E. coli O157:H7; lane 15, E. coli O157:H7; lane 16, E. coli O157:H7; lane 17, E. coli O157:H7; lane 18, C. freundii (O157+). DNA fragments were separated on 1.5% agarose–1× TAE gels at 100 V.
FIG. 6.
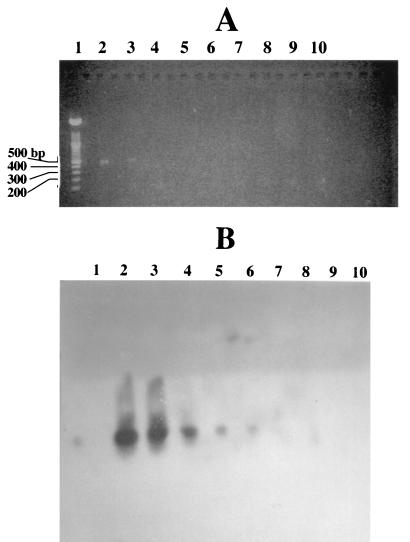
Sensitivity of E. coli O157 rfbB primers in detecting E. coli O157:H7 by PCR. Lanes 1, 100-bp ladder; lanes 2, 10−1 dilution (105 cells); lanes 3, 10−2 (104 cells); lanes 4, 10−3 (103 cells); lanes 5, 10−4 (102 cells); lanes 6, 10−5 (≤10 cells); lanes 7, 10−6 (<10 cells); lanes 8, 10−7 (<10 cells); lanes 9, no DNA; and lane 10, E. coli K-12 LE392. PCR was performed with serial 10-fold dilutions of E. coli O157:H7 ATCC 31350 (109 CFU/ml) and E. coli O157 rfbB-specific primers. DNA fragments were separated on 1.5% agarose–1× TAE gels at 100 V. One microliter of the boiled sample served as the DNA template in the 10-μl PCR mixture. DIG-labeled nucleotides were incorporated to heighten the sensitivity of the PCR. (A) Agarose gel. (B) Nylon membranes probed with anti-DIG antibody.
Application of a PCR test with rfbO157-specific primers to the detection of E. coli O157 in milk and cattle feces.
PCR with this O157-specific rfb primer pair was assessed as to its ability to detect E. coli O157 in samples contaminated with the organism. Whole milk was seeded with E. coli O157 and serially diluted 10-fold. The PCR was able to detect as few as 107 cells/ml in the milk. Similarly, the PCR was also applied to the detection of E. coli O157:H7 in cow feces. A fecal specimen was seeded with 109 CFU/ml. The feces were plated onto MacConkey agar and contained both lactose-positive and -negative colonies. Product from sample diluted 1:10 and 1:100 was observed, while no PCR amplicon was observed in the undiluted fecal sample that was spiked with E. coli O157:H7. The 1:100 dilution represented 107 CFU of E. coli O157:H7 per ml in the sample (Fig. 7).
FIG. 7.
Detection of E. coli O157:H7 in cow feces by PCR. Cow feces was inoculated with 109 CFU of E. coli O157:H7 per ml. Debris was removed by a low-speed centrifugation, and template was prepared for PCR by boiling the supernatant for 10 min. Undiluted samples and samples diluted 10-fold in dH2O served as DNA templates for PCR. Lane 1, 100-bp ladder; lane 2, no-DNA control; lane 3, E. coli O157:H7 positive control; lanes 4 to 6, undiluted cow feces (lane 4) and cow feces diluted 10-fold (lane 5) and 100-fold (lane 6) that were inoculated with 109 CFU of E. coli O157:H7 per ml.
DISCUSSION
In this study, we created an E. coli O157:H7 cosmid library, identified O157-positive cosmid clones, and eventually identified serotype-specific DNA sequences. From the O157 cosmid library, five O157 positive cosmid clones were identified. When an O157-antigen-positive cosmid was used as a DNA probe, the probe failed to recognize the DNA sequences present within the genomes of H. alvei, M. morganii, and C. freundii. These bacteria exhibit cross-reactivity with polyclonal O157 antiserum and a monoclonal antibody that specifically recognizes O157 antigen among the many distinct E. coli O serotypes. An O157-serotype-specific DNA probe was identified. The DNA insert for pDS306 has homology to known rfb and capsule genes present in E. coli K-12 (42), Y. enterocolitica O:8 (55), and V. cholerae (6). The nucleotide sequences of E. coli O157 rfb genes were sufficiently different from those of E. coli K-12 to distinguish E. coli O157 serotypes from other E. coli serotypes and enteric bacteria like Salmonella and Citrobacter by Southern blotting and PCR. The rfb sequence chosen for design of PCR appears to be ideal, since there was no significant divergence in the nucleotide sequences among the E. coli O157 serotypes. This is an ideal target for PCR, since the LPS enzymes responsible for O-antigen synthesis are not subjected to the same selection pressure as surface proteins like H antigen.
By PCR, E. coli O157:H7 was detected in milk and cow feces spiked with the organism. We were able to detect E. coli O157:H7 in milk and feces at 107 cells/ml in samples laced with this organism. This was especially remarkable for the fecal sample, since no additional steps for concentrating the template were included in this procedure. The sensitivity of the PCR in general was increased 1,000-fold by including DIG-labeled nucleotides in the PCR. A PCR–enzyme-linked immunosorbent assay will increase the sensitivity of the test as well as make processing of large samples more tenable (27), and we are currently developing a PCR–enzyme-linked immunosorbent assay just for this purpose. There were PCR inhibitors present in fecal specimens. Feces are notorious for the presence of PCR inhibitors (51). Further DNA purification will eliminate any additional interference with the PCR.
Probe-based methods have revolutionized detection of salmonellae and other important enteric pathogens in clinical and food specimens. The basis for the molecular diagnosis of infectious diseases is the use of selective PCR amplification in the identification of the variable genetic regions unique for a given serotype or pathogenic group like EHEC. A PCR-based detection system has been established for the specific detection of Salmonella serogroups A, B, C2, and D (30). The advantage of this PCR test over current tests focusing on E. coli O157 virulence genes is because of the genetic instability and sporadic distribution observed for these markers among E. coli isolates that are pathogenic or nonpathogenic for humans (13, 16). The strategy outlined in this study will prove beneficial in the generation of new molecular probes for identifying specific pathogenic E. coli O serotypes. Long PCR has already proven to be effective in the cloning and sequencing of the complete rfb operon of E. coli, including E. coli O157 (49). However, by combining long-PCR products of the rfb operon as DNA probes in Southern analysis of various E. coli O serotypes, unique DNA fragments can be quickly identified for sequencing. Instead of bidirectional sequencing of PCR amplicons of 18 kb or greater, we can focus our attention on smaller DNA fragements of 1 to 2 kb, as demonstrated in this study, which will save time and expense in development of new molecular probes. Our work further supports the hypothesis that unique sequences within a pathogenic E. coli O serotype, such as those located within the rfb gene cluster, can be used as specific DNA probes regardless of the biochemical and serological nature of other microorganisms present within the gastrointestinal tract. Even with an initial enrichment, this O157-specific, PCR-based test will detect E. coli O157 serotypes in less time than the current microbiological methods, while eliminating false positives.
ACKNOWLEDGMENTS
This work was supported by USDA grant 9503249 and by a grant from the Emma Winters Foundation.
We acknowledge Heather Thiel, Judy Verzella, and Philip Lee for their assistance with this project. We also thank Peter Brown for his advice and comments on this work and Richard Wilson, Nancy Strockbine, Carol Maddox, and Michael Doyle for providing us with the bacteria used in this study.
REFERENCES
- 1.Al-Hendy A, Toivanen P, Skurnik M. The effect of growth temperature on the biosynthesis of Yersinia enterocolitica O:3 lipopolysaccharide: temperature regulates the transcription of the rfb locus but not the rfa region. Microb Pathog. 1991;10:81–86. doi: 10.1016/0882-4010(91)90068-l. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology, 2nd ed. Current protocols. New York, N.Y: John Wiley and Sons, Inc.; 1992. [Google Scholar]
- 4.Bernard P, Gabant P, Bahassi E M, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 5.Bettelheim K A, Evangelidis H, Pearce J L, Sowers E, Strockbine N A. Isolation of a Citrobacter freundii strain which carries the Escherichia coli O157 antigen. J Clin Microbiol. 1993;31:760–761. doi: 10.1128/jcm.31.3.760-761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik E M, Bunschoten A E, Willems R J, Chang A C, Mooi F R. Genetic organization and functional analysis of the otn DNA essential for cell-wall polysaccharide synthesis in Vibrio cholerae O139. Mol Microbiol. 1996;20:799–811. doi: 10.1111/j.1365-2958.1996.tb02518.x. [DOI] [PubMed] [Google Scholar]
- 7.Bilge S S, Vary J C, Jr, Dowell S F, Tarr P I. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borczyk A A, Lior H, Ciebin B. False positive identifications of Escherichia coli O157 in foods. Int J Food Microbiol. 1987;4:347–349. doi: 10.1016/s0140-6736(87)92065-4. [DOI] [PubMed] [Google Scholar]
- 10.Brown P K, Romana L K, Reeves P R. Molecular analysis of the rfb gene cluster of Salmonella serovar Muenchen (strain M67): genetic basis of the polymorphism between groups C2 and B. Mol Microbiol. 1992;6:1385–1394. doi: 10.1111/j.1365-2958.1992.tb00859.x. [DOI] [PubMed] [Google Scholar]
- 11.Chart H, Okubadejo O A, Rowe B. The serological relationship between Escherichia coli O157 and Yersinia enterocolitica O9 using sera from patients with brucellosis. Epidemiol Infect. 1991;108:77–85. doi: 10.1017/s0950268800049529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke B R, Whitfield C. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: the rfb gene cluster is responsible for synthesis of d-galactan I O polysaccharide. J Bacteriol. 1992;174:4614–4621. doi: 10.1128/jb.174.14.4614-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dev V J, Main M, Gould I. Waterborne outbreak of Escherichia coli O157. Lancet. 1991;337:1412. doi: 10.1016/0140-6736(91)93092-n. [DOI] [PubMed] [Google Scholar]
- 15.Dorn C R. Review of foodborne outbreak of Escherichia coli O157:H7 infection in the western United States. JAMA. 1993;203:1583–1587. [PubMed] [Google Scholar]
- 16.Dorn C R, Scotland S, Smith H R, Willshaw G A, Rowe B. Properties of Vero cytotoxin-producing Escherichia coli serogroup O157:H7 of human and animal origin belonging to serotypes other than O157:H7. Epidemiol Infect. 1989;103:83–95. doi: 10.1017/s0950268800030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle M P. Escherichia coli O157:H7 and its significance in foods. Int J Food Microbiol. 1991;12:289–302. doi: 10.1016/0168-1605(91)90143-d. [DOI] [PubMed] [Google Scholar]
- 18.Doyle M P, Schoeni J L. Isolation of Escherichia coli O157:H7 from retail fresh meats and poultry. Appl Environ Microbiol. 1987;53:2394–2396. doi: 10.1128/aem.53.10.2394-2396.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg J B, Hatano K, Meluleni G S, Pier G B. Cloning and surface expression of Pseudomonas aeruginosa O antigen in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10716–10720. doi: 10.1073/pnas.89.22.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbs M, Reeves P R. Genetic organization and evolution of Yersinia pseudotuberculosis 3,6-dideoxyhexose biosynthetic genes. Biochim Biophys Acta. 1995;1245:273–277. doi: 10.1016/0304-4165(95)00126-3. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O-antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 24.Karch H, Bohm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kido N, Ohta M, Iida K, Hasegawa T, Ito H, Arakawa Y, Komatsu T, Kato N. Partial deletion of the cloned rfb gene of Escherichia coli O9 results in synthesis of a new O-antigenic lipopolysaccharide. J Bacteriol. 1989;171:3629–3633. doi: 10.1128/jb.171.7.3629-3633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 27.Landgraf A, Reckman B, Pingould A. Direct analysis of polymerase chain reation products using enzyme-linked immunosorbent assay techniques. Anal Biochem. 1991;198:86–91. doi: 10.1016/0003-2697(91)90510-z. [DOI] [PubMed] [Google Scholar]
- 28.Lerman V, Cohen D, Gluck A, Ohad E, Sechter I. A cluster of cases of Escherichia coli O157 infection in a day care center in a communal settlement (kibbutz) in Israel. J Clin Microbiol. 1992;30:520–521. doi: 10.1128/jcm.30.2.520-521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D, Verma N K, Romana L K, Reeves P R. Relationships among the rfb regions of Salmonella serovars A, B, and D. J Bacteriol. 1991;173:4814–4819. doi: 10.1128/jb.173.15.4814-4819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luk J M C, Kongmuang U, Reeves P R, Lindberg A A. Selective amplification of abequose and paratose synthase genes (rfb) by polymerase chain reaction for identification of Salmonella major serogroups (A, B, C2, and D) J Clin Microbiol. 1993;31:2118–2123. doi: 10.1128/jcm.31.8.2118-2123.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M L, Shipman L D, Wells J G, Potter M E, Hedberg K, Wachsmuth I K, Tauxe R V, Davis J P, Arnoldi J, Tilleli J. Isolation of Escherichia coli O157:H7 from dairy cattle associated with two cases of haemolytic uraemic syndrome. Lancet. 1986;2(8514):1043. doi: 10.1016/s0140-6736(86)92656-5. [DOI] [PubMed] [Google Scholar]
- 32.McPherson D F, Morona R, Berger D W, Cheah K C, Manning P A. Genetic analysis of the rfb region of Shigella flexneri encoding the Y serotype O-antigen specificity. Mol Microbiol. 1991;5:1491–1499. doi: 10.1111/j.1365-2958.1991.tb00795.x. [DOI] [PubMed] [Google Scholar]
- 33.Morona R, Matthews M S, Morona J K, Brown M H. Regions of the cloned Vibrio cholerae rfb genes needed to determine the Owaga form of the O-antigen. Mol Gen Genet. 1990;224:405–412. doi: 10.1007/BF00262435. [DOI] [PubMed] [Google Scholar]
- 34.National Center for Biotechnology Information. 16 April 1996. [Online.] http://www.ncbi.nlm.nih.gov.
- 35.Okrend A, Rose B E. Isolation and identification of Escherichia coli O157:H7 from meat. USDA-FSIS Laboratory Communication no. 38, revision no. 3. Washington, D.C: Food Safety and Inspection Service, U.S. Department of Agriculture; 1989. [Google Scholar]
- 36.Perry M B, Bundle D R. Antigenic relationships of the lipopolysaccharides of Escherichia hermanii strains with those of Escherichia coli O157:H7, Brucella melitensis, and Brucella abortus. Infect Immun. 1990;58:1391–1395. doi: 10.1128/iai.58.5.1391-1395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poustka A, Rackwitz H, Frischauf A M, Hohn B, Lehrach H. Selective isolation of cosmid clones by homologous recombination in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4129–4133. doi: 10.1073/pnas.81.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajakumar K, Jost B H, Sasakawa C, Okada N, Yoshidawa M, Adler B. Nucleotide sequence of the rhamnose biosynthetic operon of Shigella flexneri 2a and role of lipopolysaccharide in virulence. J Bacteriol. 1994;176:2363–2373. doi: 10.1128/jb.176.8.2362-2373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samadpour M, Ongerth J R, Liston J, Tran N, Nguyen D, Whittam T S, Wilson R A, Tarr P I. Occurrence of Shiga-like toxin-producing Escherichia coli in retail fresh seafood, beef, lamb, pork, and poultry from grocery stores in Seattle, Washington. Appl Environ Microbiol. 1994;60:1038–1040. doi: 10.1128/aem.60.3.1038-1040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:665–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturm S, Jann B, Jann K, Fortnagel P, Timmis K N. Genetic and biochemical analysis of Shigella dysenteriae 1 O antigen polysaccharide biosynthesis in Escherichia coli K-12: structure and functions of the rfb cluster. Microb Pathog. 1986;1:307–324. doi: 10.1016/0882-4010(86)90056-2. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama T, Kido N, Komatsu T, Ohta M, Kato N. Expression of the cloned Escherichia coli O9 rfb gene in various mutants of Salmonella typhimurium. J Bacteriol. 1991;173:55–58. doi: 10.1128/jb.173.1.55-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 46.Valvano M A, Crosa J H. Molecular cloning and expression in Escherichia coli K-12 of chromosomal genes determining the O7 lipopolysaccharide antigen of a human invasive strain of E. coli O7:K1. Infect Immun. 1989;57:937–943. doi: 10.1128/iai.57.3.937-943.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma N K, Reeves P R. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989;171:5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viret J, Cryz S J, Jr, Lang A B, Favre D. Molecular cloning and characterization of the genetic determinants that express the complete Shigella serotype D (Shigella sonnei) lipopolysaccharide in heterologous live attenuated vaccine strains. Mol Microbiol. 1993;7:239–252. doi: 10.1111/j.1365-2958.1993.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells J G, Shipman L D, Greene K D, Sowers E G, Green J H, Cameron D N, Downes F P, Martin M L, Griffin P M, Ostroff S M, Potter M E, Tauxe R V, Wachsmuth I K. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wittwer C T, Fillmore G C, Hillyard D R. Automated polymerase chain reaction capillary tubes with hot air. Nucleic Acid Res. 1989;17:4353–4357. doi: 10.1093/nar/17.11.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyk P, Reeves P. Identification and sequence of the gene for abequose synthetase, which confers antigenic specificity on group B salmonellae: homology with galactose epimerase. J Bacteriol. 1989;171:5687–5693. doi: 10.1128/jb.171.10.5687-5693.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang S, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, Toivanen P, Skurnik M. The gene cluster directing O-antigen biosynthesis in Yersinia enterocolitica serotype O:8: identification of the genes for mannose and galactose biosynthesis and the gene for the O-antigen polymerase. Microbiology. 1996;142:277–288. doi: 10.1099/13500872-142-2-277. [DOI] [PubMed] [Google Scholar]