Abstract
An alkane-degrading, sulfate-reducing bacterial strain, AK-01, was isolated from an estuarine sediment with a history of chronic petroleum contamination. The bacterium is a short, nonmotile, non-spore-forming, gram-negative rod. It is mesophilic and grows optimally at pH 6.9 to 7.0 and at an NaCl concentration of 1%. Formate, fatty acids (C4 to C16) and hydrogen were readily utilized as electron donors. Sulfate, sulfite, and thiosulfate were used as electron acceptors, but sulfur, nitrite, and nitrate were not. Phenotypic characterization and phylogenetic analysis based on 16S rRNA gene sequence indicate that AK-01 is most closely related to the genera Desulfosarcina, Desulfonema, and Desulfococcus in the delta subdivision of the class Proteobacteria. It is phenotypically and phylogenetically different from strains Hxd3 and TD3, two previously reported isolates of alkane-degrading, sulfate-reducing bacteria. The alkanes tested to support growth of AK-01 had chain lengths of C13 to C18. 1-Alkenes (C15 and C16) and 1-alkanols (C15 and C16) also supported growth. The doubling time for growth on hexadecane was 3 days, about four times longer than that for growth on hexadecanoate. Mineralization of hexadecane was indicated by the recovery of 14CO2 from cultures grown on [1-14C]hexadecane. Degradation of hexadecane was dependent on sulfate reduction. The stoichiometric ratio (as moles of sulfate reduced per mole of hexadecane degraded) was 10.6, which is very close to the theoretical ratio of 12.25, assuming a complete oxidation to CO2. Anaerobic alkane degradation by sulfate reducers may be a more widespread phenomenon than was previously thought.
Alkanes are major components of petroleum fuels, which can be commonly found in contaminated environments, and numerous studies on their biodegradability have been conducted. Earlier investigations concentrated on processes occurring under aerobic condition (4), whereby the initial attack of the aliphatic hydrocarbon chain is generally mediated by monooxygenases. Detailed studies on the enzymology and genetics of specific systems are also well documented (26, 28). In contrast, little is known about the biodegradation of alkanes under anoxic conditions, where oxygen-initiated reactions cannot occur. Cases of alkane degradation under different reducing conditions were sparsely reported in enrichment or microcosm studies over the past decades (6, 8, 10, 11, 15, 16, 27). While degradation by pure cultures of Desulfovibrio under sulfate-reducing conditions has been reported in the early literature (9, 18, 22), these cultures were not preserved and have not been further studied (2). Even now, little is known about the degrading organisms and the degradation mechanism(s). The scarcity of pure bacterial cultures available for detailed studies, the generally slow anaerobic processes, and the requirement for more elaborate microbiological techniques may have hindered the progress in this area.
A key publication by Aeckersberg et al. in 1991 (2) reported the isolation of a sulfate-reducing bacterium which, under rigorous conditions, was shown to degrade and grow on alkanes under strictly anoxic conditions. Later, Rueter et al. (23) reported the isolation of a new type of thermophilic, sulfate-reducing bacterium from the sediment of Guaymas Basin (Gulf of California, Mexico) and demonstrated its ability to perform anaerobic alkane degradation. These reports have again generated interest in this area, and the acquisition of the pure isolates opens opportunities for more detailed investigations on the degradation processes. In this paper, we report the isolation and characterization of a novel alkane-degrading, sulfate-reducing bacterium.
MATERIALS AND METHODS
Initial culturing conditions.
The alkane-degrading bacterial strain AK-01 was isolated from the active sulfate-reducing enrichment cultures established previously with petroleum-contaminated sediments collected from the Arthur Kill, N.Y., a hydrocarbon-impacted intertidal waterway between Staten Island and New Jersey (24). The medium used for maintaining the enrichment cultures and isolating the bacterium contained, per liter of deionized water, the following: NaCl, 23 g; KCl, 1.3 g; MgCl2 · 6H2O, 1 g; CaCl2 · 2H2O, 0.1 g; NH4Cl, 0.5 g; KH2PO4, 0.2 g; Na2SO4, 1.42 g; NaHCO3, 2.5 g; Na2S · 9H2O, 0.5 g; vitamin solution, 1 ml; and trace element solution, 5 ml. The vitamin solution was modified from that of Widdel and Bak (31) and contained, per liter of deionized water, the following: vitamin B12, 1 mg; d-(+)-biotin, 20 mg; folic acid, 20 mg; nicotinic acid, 50 mg; p-aminobenzoic acid, 50 mg; calcium d-(+)-pantothenate, 50 mg; pyridoxine HCl, 100 mg; riboflavin, 50 mg; thiamine, 50 mg; and thioctic acid, 50 mg (final pH adjusted to 7). The trace element solution contained, per liter of 0.01 N HCl, the following: CoCl2 · 6H2O, 6 g; CuCl2, 30 mg; FeCl2 · 4H2O, 0.3 g; H3BO3, 1.14 g; MnCl2 · 4H2O, 4 g; Na2MoO4 · 2H2O, 0.5 g; NiCl2 · 6H2O, 0.3 g; ZnCl2, 0.42 g. All of the above medium components except NaHCO3, Na2S · 9H2O, and the vitamin solution were added to deionized water and then deoxygenated by bubbling with a stream of N2-CO2 gas (70:30) for 1 h. The medium was dispensed into serum bottles, crimp-sealed with butyl rubber stoppers, and autoclaved. A filter-sterilized stock solution containing 1 M NaHCO3 and 67 mM Na2S · 9H2O was then added to the autoclaved medium at 3% (vol/vol). The vitamin solution was also filter sterilized and added to the medium. The final pH of the medium was about 6.8. Medium for the serial dilution procedures was further amended with 0.1 g of yeast extract per liter.
Pure culture growth and maintenance.
After the pure culture of strain AK-01 was obtained, the medium used for routine maintenance and most characterization experiments was the same as the one described above, except that another trace element solution modified from that of Widdel and Bak (31) was used. Each liter of medium received 1 ml of the modified trace element solution, which contained, per liter of 0.1 N HCl, the following: CoCl2 · 6H2O, 190 mg; CuCl2 · 2H2O, 2 mg; FeCl2 · 4H2O, 1.5 g; H3BO3, 6 mg; MnCl2 · 4H2O, 100 mg; Na2MoO4 · 2H2O, 36 mg; NiCl2 · 6H2O, 24 mg; and ZnCl2, 70 mg. Yeast extract (0.1 g/liter) was also added except when stated otherwise. In addition, the pH and NaCl concentration of this mineral salt medium were varied to test for their effects on growth of the strain on hexadecane. The pH was finally set at 7.0 (by addition of sodium hydroxide) and the NaCl concentration was set at 10 g/liter after these conditions were shown to support optimal growth. The incubation temperature was 30°C except when stated otherwise. Growth was monitored routinely by measuring sulfate loss instead of increase in optical density (OD) over time due to interference from water-insoluble substrates like alkanes and long-chain fatty acids. The sulfate reduction rates thus determined were used to estimate the doubling times of the bacterium cultured under specific conditions. (The direct correlation between the amount of sulfate loss and cell growth was confirmed by a supplementary experiment which simultaneously monitored the OD540 and sulfate concentration of a culture of AK-01 grown on octanoate, a water-soluble fatty acid.)
Stock cultures of Desulfovibrio gigas (ATCC 19364) and Desulfobacter curvatus (ATCC 43919) were obtained from the American Type Culture Collection, and a stock culture of Desulfosarcina variabilis (DSM 2060) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Cultures were routinely maintained in media recommended by their respective sources or in the same mineral salt medium used for strain AK-01. Formate (for D. gigas), acetate (for D. curvatus), and benzoate (for D. variabilis) were used as growth substrates. Cultures were incubated at 30°C or room temperature in the dark and without shaking.
A stock culture of strain Hxd3, an alkane-degrading sulfate reducer previously isolated by Aeckersberg et al. (2), was obtained from DSMZ (DSM 6200). This strain had been tentatively named Desulfobacterium oleovorans based on its physiological characteristics (2). Recently, the authors indicate that its placement in this genus may not be appropriate in the light of its 16S rRNA phylogeny (3). Medium 517 recommended by DSMZ was modified as suggested by Friedrich Widdel (30) for the cultivation of the bacterium. It contained, per liter of deionized water, the following: NaCl, 17.6 g; MgCl2 · 6H2O, 3 g; CaCl2 · 2H2O, 0.22 g; NH4Cl, 0.22 g; KH2PO4, 0.18 g; Na2SO4, 2 g; NaHCO3, 2.5 g; Na2S · 9H2O, 0.12 g; vitamin solution, 1 ml; trace element solution, 1 ml; and selenite-tungstate solution, 1 ml. The vitamin solution was the same as that described above except for the following amendments (per liter of deionized water): vitamin B12, 50 mg; Na2HPO4, 0.577 g; and KH2PO4, 0.015 g (final pH = 7.0). The trace element solution was the same as that described above for maintaining strain AK-01, except the following changes (per liter of 0.1 N HCl): FeCl2 · 4H2O, 4.5 g; and ZnCl2, 140 mg. The selenite-tungstate solution contained, per liter of 0.01 N NaOH, the following Na2SeO3, 2 mg; Na2WO4 · 1.5H2O, 4 mg. All of the above medium components, except NaHCO3, Na2S · 9H2O, and the vitamin solution, were added to deionized water and then deoxygenated by bubbling with a stream of N2-CO2 gas (70:30) for 1 h. The medium was dispensed into serum bottles, which were then crimp-sealed with butyl rubber stoppers and autoclaved. A filter-sterilized stock solution containing 1 M NaHCO3, 18 mM Na2S · 9H2O, and 325 mM NaOH was then added to the autoclaved medium at 3% (vol/vol). The vitamin solution was also filter sterilized and added to the medium at 0.1% (vol/vol). The final pH of the complete medium was 7.2. The cultures were incubated at 30°C in the dark and without shaking.
Organic substrates were generally sterilized separately before being added to the autoclaved medium. Water-soluble substrates were added as filter-sterilized concentrated stock solutions. Liquid hydrocarbons like alkanes and 1-alkenes were also added with filter sterilization. Concentrated solutions of sodium salts of long-chain fatty acids (C11 to C16) were autoclaved in sealed bottles and melted in a boiling-water bath before being added to the medium. Only the solid 1-pentadecanol and 1-hexadecanol were added to the medium before it was autoclaved.
Isolation.
Initial enrichments established as described previously (24) were subcultured by transferring a 5% inoculum to fresh medium with hexadecane (0.1 ml per 50 ml of medium) as the sole carbon source. Growth of the subcultures, as indicated by sulfate reduction, was consistently observed upon refeeding with sulfate when it was exhausted (with hexadecane in excess). The subcultures were then serially diluted 10-fold into tubes containing medium plus hexadecane (0.1 ml per 15 ml of medium) and yeast extract (0.1 g/liter). Utilization of hexadecane was indicated by sulfate loss and increase in cell number in the culture medium. Sulfate loss of less than 3% of the total and no cell growth were observed in the tubes with yeast extract only (no hexadecane). In each dilution series, the most highly diluted medium showing growth was further serially diluted in the same manner. After three consecutive series of dilution, taking place over 1 year, cultures apparently with a single morphotype (short rods) were obtained. The purity of the culture was verified by subculturing it into the mineral salt medium containing 5 mM lactate, pyruvate, formate, or butyrate and other complex media including tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.) brain heart infusion (Difco) and nutrient broth (Difco) prepared under anoxic conditions. (Agar shake tubes containing hexadecane as the substrate were also inoculated with cultures obtained by serial dilution, but no visible colony was found over 6 months of incubation.)
Cell surface hydrophobicity.
The cell surface hydrophobicity of AK-01 was determined by an adaptation of the bacterial adherence to hydrocarbons test (21). AK-01 was cultured on hexadecane for 1 month to the stationary phase. Cells were collected by centrifugation, resuspended in about 2 ml of the residual medium to give a concentrated suspension, and used without further washing. Various amounts of the cell concentrate were then added to sealed glass tubes, each containing 4 ml of the mineral salt medium under anoxic conditions. The diluted cell suspensions in the tubes had initial OD540 of 0.3 to 0.6 as determined with a spectrophotometer (Spectronic 20; Bausch & Lomb, Rochester, N.Y.). To each tube, 1 ml of hexadecane (previously deoxygenated by bubbling with N2) was then added. The tubes were vortexed for 2 min and allowed to stand for 30 min for phase separation. The OD540 of the aqueous phase of each tube was again measured. A hydrophobicity index, defined as the percentage of total cells that adhered to hexadecane, was calculated for each tube as follows: 100% × (initial OD540 − final OD540)/initial OD540.
Detection of desulfoviridin.
The presence of desulfoviridin in strain AK-01 was tested by the method of Postgate (20). Cells of AK-01 grown on octanoate (2 mM) were collected by centrifugation, resuspended in deionized water, and lysed by adding drops of 2 N NaOH. The cell lysate was then examined under UV light (365 nm) for red fluorescence. Cultures of Desulfovibrio gigas and Desulfobacter curvatus were used as positive and negative controls, respectively.
G+C content.
The G+C content of the DNA of AK-01 was determined by a high-pressure liquid chromatography (HPLC) method modified from that of Mesbah et al. (14) with salmon sperm DNA as a standard. Eppendorf tubes each containing 50 μl of DNA sample (80 μg per ml) were heated in a boiling-water bath for 2 min and then immediately chilled in an ice bath. To each sample, 100 μl of sodium acetate buffer (30 mM; pH 5.3), 10 μl of zinc sulfate (20 mM), and 6 μl of P1 nuclease (1 mg per ml of sodium acetate buffer; equivalent to 340 U per ml) were added. All tubes of the reaction mixture were first incubated at 37°C for 2.5 h. After that, 10 μl of calf intestinal mucosa alkaline phosphatase (100 U per ml of 0.1 M Tris buffer [pH 8.1]) was added and the tubes were further incubated for 6 h. After incubation, the debris was spun down at 13,500 × g for 10 min in a microcentrifuge and the supernatant was stored frozen at −20°C until analyzed by HPLC.
Utilization of electron donors and acceptors.
Utilization of electron donors for growth by strain AK-01 was tested by adding the compounds to the mineral salt medium and then inoculating it with cultures of strain AK-01 pregrown on hexadecane or fatty acids. Sodium salts of fumarate (5 mM), lactate (5 mM), malate (5 mM), pyruvate (5 mM), succinate (5 mM), formate (5 mM), acetate (5 mM), propionate (5 mM), isobutyrate (4 mM), 2-methylbutyrate (4 mM), 3-methylbutyrate (4 mM), 2-methylheptanoate (2 mM), benzoate (2 mM), and phenylacetate (2 mM) were added as electron donors. Their concentrations were monitored by HPLC with detection by UV absorption (see “Chemical analysis” below). Butyrate (1 mM), pentanoate (1 mM), hexanoate (1 mM), heptanoate (0.5 mM), octanoate (0.5 mM), nonanoate (0.5 mM), and decanoate (0.5 mM) were monitored by extracting the compounds from acidified samples of the culture media with pentane and analyzing them by gas chromatography with flame ionization detection (GC-FID; see “Chemical analysis” below). Water-insoluble fatty acids of C11 to C16 (0.25 mM each) were extracted from the whole culture (100 ml) with hexane after acidification. The extracts were then evaporated dry and methylated by being heated at 80°C in a mixture of methanol and 6 N HCl (volume ratio, 1:1.18) for 10 min. The resulting fatty acid methyl esters were extracted with hexane and analyzed by GC-FID. Autotrophic growth with hydrogen as the electron donor was tested by preparing cultures in yeast extract-free medium and pressurizing the headspace with hydrogen gas (100%). Growth was monitored by measuring sulfate losses in the culture media by ion chromatography (IC). Autoclaved controls were established for all experiments described above.
Strain AK-01 was tested for growth on alkanes with chain lengths from C6 to C30. The sparingly soluble liquid alkanes (C6 to C9) were added to the base medium at concentrations of 100 to 150 μM. For the insoluble liquid alkanes (C10 to C17), 10 μl was added per 20 ml of medium, equivalent to concentrations of about 2.5 to 3 mM. For the insoluble solid alkanes (C18 and C20), 20 mg was added per 20 ml of medium (2 to 4 mmol/liter). Growth on 1-pentadecene, 1-hexadecene, 1-pentadecanol, and 1-hexadecanol was tested by adding 20 μl or 20 mg of the compounds per 100 ml of medium (0.69 to 0.83 mmol/liter). Growth was indicated by sulfate loss and increase in cell number in the experimental cultures compared to sterile controls.
Utilization of electron acceptors by strain AK-01 was tested by using octanoate (2 mM) as the electron donor and by using a potential electron acceptor provided by one of the following chemicals: potassium nitrate (5 mM), sodium nitrite (2 mM), sodium sulfate (10 mM), sodium sulfite (10 mM), sodium thiosulfate (5 mM), or elemental sulfur (0.2% [wt/vol]; equivalent to 60 mM). Cultures were incubated at 30°C and monitored for loss of nitrate, nitrite, sulfate and thiosulfate by IC. Reduction of sulfite and sulfur was monitored by measuring sulfide concentrations in the culture medium by the methylene blue method (7).
16S rRNA gene sequence determination and phylogenetic analysis.
Strain AK-01 was cultured in mineral salt medium (with hexadecane or butyrate) and TSB (supplemented with 1 mM decanoate). Genomic DNA samples were prepared independently from cultures grown under the three different nutritional conditions by density gradient centrifugation in cesium chloride (19). Strain Hxd3 was grown on stearate, and its genomic DNA was extracted by a miniprep method (25).
The 16S rRNA gene of strain AK-01 was amplified by PCR with a reagent kit from Perkin-Elmer, Norwalk, Conn. (product N801-0055). In the initial experiment for verifying the purity of the culture, genomic DNA samples independently extracted from the different cultures described above were used as templates. The universal eubacterial primers 27f (5′ AGA GTT TGA TCC TGG CTC AG 3′) and 907r (5′ CCG TCA ATT CCT TTG AGT TT 3′) (12) were used as forward and reverse primers, respectively, resulting in a partial sequence of the gene containing about 900 bp. The sequences determined for each DNA sample were then compared for identity. In the second experiment to obtain the complete 16S rRNA gene sequence, a new reverse primer was developed by aligning the known 16S rRNA sequences of nine different sulfate-reducing bacteria and complementing one of the most highly conserved regions near the end of the aligned sequences. The new primer, SRB-R3, has a sequence of 5′ TAC CTT GTT ACG ACT TCA CC 3′ (equivalent to bp 1488 to 1507 of the Escherichia coli 16S rRNA gene; accession no. E05133) and was used together with the 27f primer. The amplification parameters recommended by the manufacturer of the PCR kit were used in the PCRs for all the above experiments.
The 16S rRNA gene of strain Hxd3 was amplified by PCR with a kit from Clontech, Palo Alto, Calif. (product K1905-Y). Another reverse primer, custom developed by the approach described for SRB-R3, was used together with the forward primer 27f. The new reverse primer, SRB-R1, has a sequence of 5′ CAA CTC TCA TGG TGT GAC GG 3′ (equivalent to bp 1403 to 1422 of the E. coli 16S rRNA gene; accession no. E05133). The amplification parameters recommended by the manufacturer of the PCR kit were used in the PCRs. The PCR products for both strain AK-01 and strain Hxd3 were cleaned up by a purification kit from Qiagen Inc., Chatsworth, Calif. (product 28104), and sequenced by an automated DNA sequencer (Perkin-Elmer ABI, Foster City, Calif.).
The 16S rRNA gene sequence of strain AK-01 was submitted to the FASTA3 Homology Search Engine (version 3.0t76) (19a) based in the European Bioinformatics Institute for similarity search in the EMBL Nucleotide Sequence Database (10a). From the search results, the first 10 most similar sequences which belong to bacteria fully described in the literature were included in the subsequent phylogenetic analysis. Sequences of a previously isolated alkane-degrading sulfate-reducer, strain TD3 (23), and various type species in key genera of the gram-negative sulfate-reducing bacteria (31) were also retrieved from the EMBL database and included in the analysis. The organisms used in the phylogenetic analysis are (accession numbers of their 16S rRNA sequences in parentheses): strain AK-01 (AF141328), strain Hxd3 (AF141881), strain TD3 (X80922), Desulfovibrio desulfuricans (M34113), Desulfomicrobium baculatum (M37311), Desulfobulbus propionicus (M34410), Desulfoarculus baarsii (M34403), Pelobacter acetylenicus (X70955), Pelobacter carbinolicus (X79413), Geobacter metallireducens (L07834), Pelobacter propionicus (X70954), Syntrophus gentianae (X85132), Syntrophus buswellii (X85131), Desulfobacter postgatei (M26633), Desulfobacterium autotrophicum (M34409), Desulfobotulus sapovorans (M34402), Desulfococcus multivorans (M34405), Desulfonema magnum (U45989), Desulfonema limicola (U45990), Desulfosarcina variabilis (M34407), and Escherichia coli (E05133).
The phylogenetic analysis was conducted by computer programs in the Wisconsin Package version 10.0-UNIX developed by the Genetics Computer Group (Madison, Wis.). An alignment of all the sequences included in the analysis was produced by the PILEUP program. A phylogenetic tree was then constructed by using the PAUPSEARCH and PAUPDISPLAY programs based on the neighbor-joining method. A region of about 1,300 bp of the alignment in which all the sequences overlap (with intermittent gaps and uncertain bases omitted) was used in the construction of the phylogenetic tree.
Mineralization of [1-14C]hexadecane.
Strain AK-01 grown on hexadecane was inoculated into fresh medium at 20% (vol/vol) to make a master culture that was 100 ml. Aliquots of 25 ml were then anaerobically dispensed into 30-ml serum bottles, which were sealed with Teflon-coated butyl rubber stoppers and aluminum crimp seals. For sterile controls, a 100-ml culture grown on hexadecane was autoclaved and replicates of the sterilized cultures were prepared as described above. [1-14C]hexadecane (Amersham, Arlington Heights, Ill.) was dissolved in unlabeled hexadecane to 0.084 μCi/μl, of which 2 μl was added to the experimental cultures through a microliter syringe. To each of the sterile controls, 2 μl of labeled hexadecane at 0.125 μCi/μl was added. All cultures were shaken (200 rpm) for 1 h to disperse the hexadecane in the medium and then incubated at 30°C in the dark without shaking. After 78 days of incubation, the entire contents of each bottle were acidified with HCl and purged with N2 gas for 10 min. The purged gas from the cultures was directed to a series of three vials of Oxosol 14C scintillation cocktail (National Diagnostics, Atlanta, Ga.), in which the liberated 14CO2 was trapped. After purging, 2 ml of the acidified culture suspension was added to 10 ml of ReadySafe scintillation cocktail (Beckman Instruments, Inc., Fullerton, Calif.). All the vials were counted in a scintillation counter (model LS 5000 TD; Beckman).
Utilization of hexadecane and hexadecanoate.
The doubling times of AK-01 grown on hexadecane, octanoate and hexadecanoate were estimated by determining the sulfate reduction rates in these cultures. Cultures containing 20 μl of hexadecane per 100 ml of medium (0.68 mmol/liter), 2.5 mM sodium octanoate, or 0.6 mM sodium hexadecanoate as the growth substrate were prepared and monitored for sulfate loss by IC.
Stoichiometry for hexadecane degradation coupled to sulfate reduction was determined by endpoint measurements of hexadecane and sulfate loss in the cultures. Hexadecane (100 μl per liter of medium; 0.34 mmol/liter) and inoculum pregrown on hexadecane (15%) were added to 1-liter bottles of media unamended with yeast extract. These master cultures were shaken at 200 rpm for 5 days to form a homogeneous suspension of dispersed hexadecane and bacterial cells. Replicate cultures were established by transferring aliquots of 100 ml to 125-ml serum bottles, which were then sealed with Teflon-coated rubber stoppers and aluminum crimp seals. The cultures were incubated at 30°C in the dark without shaking. Sterile controls were established by autoclaving the replicate cultures. After 42 days of incubation, five replicates of experimental cultures and four replicates of sterile controls were sacrificed for determination of hexadecane and sulfate. Hexadecane was extracted with 8 ml of pentane from the entire volume (100 ml) of each culture and analyzed by GC-FID. The sulfate concentration of the remaining aqueous medium was determined by IC. The amounts of hexadecane degraded and sulfate reduced were calculated by subtracting the amounts of hexadecane and sulfate remaining in the experimental cultures from those in the sterile controls.
The growth yield on hexadecane was also determined by measuring the amount of protein produced in the replicate experimental cultures. Cells from five replicates of experimental cultures (not extracted for hexadecane with a solvent) were collected by centrifugation, hydrolyzed by heating in 1 N NaOH at 100°C, and then analyzed for protein by using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) with bovine serum albumin as a standard. Initial protein contents contributed by the inocula were estimated by determining the protein contents of replicate cultures used as the inocula.
Dependence of hexadecane degradation on sulfate reduction was determined by endpoint measurement of hexadecane and sulfate loss in cultures in the presence of limiting or unlimiting amounts of sulfate. The same experimental procedures described for the stoichiometry experiment were used except that the media used either were sulfate free or contained an unlimiting amount of sulfate (412 μmol; concentration = 13.8 mM) with respect to the theoretical amount required to completely oxidize the initially added hexadecane. Cultures prepared in the sulfate-free medium contained less than 1 mM sulfate (27.6 μmol) due to carryover from the inoculum. All cultures were incubated for 44 days and sacrificed for analysis of hexadecane and sulfate.
Stoichiometry for hexadecanoate metabolism coupled to sulfate reduction was determined by end-point measurements of hexadecanoate and sulfate loss in the cultures. Cultures (100 ml) with 0.6 mM sodium hexadecanoate and 2% of inoculum (pregrown on hexadecanoate) were prepared in replicate bottles. For sterile controls, the same inoculum was autoclaved before addition. After 14 days of incubation, three replicates of experimental cultures and sterile controls were sacrificed for determination of hexadecanoic acid and sulfate. Hexadecanoic acid was extracted by dichloromethane from each acidified culture, methylated (as described above), and analyzed by GC-FID. The sulfate concentration of the remaining aqueous medium was determined by IC. The growth yield of hexadecanoate was also determined by measuring the amount of protein produced in three replicates cultures as described earlier.
Chemical analysis.
HPLC analysis was performed on a Beckman System Gold liquid chromatograph with detection by UV absorption. Nonaromatic compounds including fumaric, lactic, malic, pyruvic, succinic, formic, acetic, propionic, isobutyric, 2-methylbutyric, and 3-methylbutyric acids were analyzed on a ion-exchange column (ROA; 300 by 7.8 mm; particle size, 8 μm; Phenomenex, Torrance, Calif.) with 0.005 N H2SO4 as the mobile phase (flow rate, 0.6 ml/min) and UV absorption at 210 nm. Benzoic and phenylacetic acids were analyzed on a reverse-phase column (Ultrasphere ODS C18; 250 by 4.6 mm; particle size, 5 μm; Beckman) with methanol-water-acetic acid (60:38:2) as the mobile phase (flow rate, 1 ml/min) and UV absorption at 254 nm. Nucleosides in the digested DNA sample for G+C content determination was analyzed on the same reverse-phase column but with a mobile phase containing 12% methanol in triethylamine phosphate (20 mM; pH 5.1). The flow rate was set at 1.5 ml per min, and UV absorption was measured at 254 nm.
GC analysis of hexadecane, fatty acid methyl esters (C10 to C16), and free fatty acids (C4 to C10) was performed with a gas chromatograph (model 5890 series II; Hewlett-Packard, Wilmington, Del.) fitted with a flame-ionization detector and a DB-WAX column (30 m by 0.25 mm; J&W Scientific, Folsom, Calif.). For analysis of hexadecane, the column temperature was initially set at 50°C for 0.25 min, increased at 25°C per min to 150°C, and maintained there for 1.25 min. For the fatty acid methyl esters, the column temperature was initially set at 50°C for 1 min, increased at 15°C per min to 220°C, and maintained there for 2 min. For the C4 to C6 fatty acids (free), the column temperature was initially set at 50°C for 1 min and then increased at 20°C per min to 200°C. For the C7 to C10 fatty acids and the 2-methylheptanoic acid (free), the column temperature was initially set at 50°C for 1 min, increased at 25°C per min to 200°C, and maintained there for 3.5 min. In all analyses, the injector and detector temperatures were set at 250 and 300°C, respectively. Dodecane was routinely used as an internal standard.
Analysis of sulfate, thiosulfate, nitrate, or nitrite was performed on an ion chromatograph (model DX-100; Dionex Corp., Sunnyvale, Calif.) equipped with an ion-exchange column (IonPac AS9-SC; 4 by 250 mm; Dionex) and a conductivity detector. The eluent contained 2 mM Na2CO3 and 0.75 mM NaHCO3, and the flow rate was at 2 ml/min. Samples of the culture medium were diluted 50-fold in deionized water before analysis.
Nucleotide sequence accession numbers.
The determined 16S rRNA gene sequences of strains AK-01 and Hxd3 were submitted to GenBank under the accession numbers AF141328 and AF141881, respectively.
RESULTS
Isolation of strain AK-01.
Active consortia enriched on hexadecane under sulfate-reducing conditions had been previously established, and their ability to degrade hexadecane by coupling to sulfate reduction was demonstrated (24). The consortia were serially diluted in mineral salt medium with hexadecane under sulfate-reducing conditions, and growth was observed as sulfate reduction and as an increase in cell number in tubes at a dilution of 10−7 or 10−8. Cultures with apparently a single morphotype were obtained after three consecutive dilution series. The purity of the culture was tested by subculturing it in the sulfate-containing mineral salt medium amended with lactate, pyruvate, formate, or butyrate (growth substrates commonly used by a variety of sulfate-reducing bacteria). Ample growth on formate and butyrate and poor growth on lactate and pyruvate were observed. Only one morphotype appeared with each substrate, and it appeared similar to that grown on hexadecane. In addition, when the putative pure culture was inoculated into complex media (TSB, brain heart infusion broth, and nutrient broth), no bacterial growth of any kind was observed after 1 month of incubation, supporting the purity of the hexadecane-degrading culture. The sequences of the 16S rRNA genes amplified from genomic DNA extracted from cultures grown in three different media (the mineral salt medium with hexadecane, the mineral salt medium with butyrate, and TSB with decanoate) were also compared. The sequences determined for the three different cultures contained about 900 bp (with the 27f-907r primer set) and were identical. These results led us to conclude that a pure culture was obtained.
Morphology.
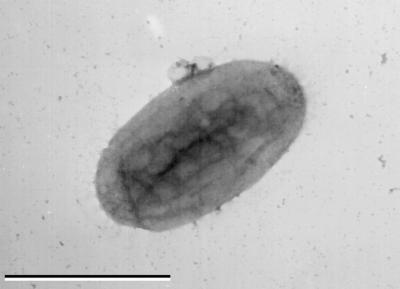
Cells of strain AK-01 are short rods about 1 to 1.5 μm long and 0.5 μm in diameter when grown on hexadecane (Fig. 1). Clumps of dividing cells are commonly seen to cluster closely around droplets of hexadecane under phase-contrast microscopy. On the other hand, cells found dispersed in the aqueous medium do not appear to be actively dividing. When grown on favorable substrates like butyrate and hexadecanoate, cells were typically longer (1.5 to 2 μm), with long unicellular filaments occurring occasionally during log phase of growth. After reaching the late stationary phase, the cells appeared to change from rods to spheres. The cells stained gram negative and showed no motility or sporulation at any stage of growth. Small refractile granules were occasionally seen inside cells (in about 1 or 2 of 10 cells), suggesting the occurrence of storage materials like poly-β-hydroxyalkanoates.
FIG. 1.
Transmission electron photomicrograph of strain AK-01 grown on hexadecane (negatively stained with uranyl acetate). Bar, 1 μm.
Physiology and other characteristics.
Strain AK-01 is mesophilic, with an optimal growth temperature at 26 to 28°C when cultured on hexadecane and 33 to 35°C when cultured on octanoate. The pH optimum for growth on hexadecane is 6.9 to 7.0. Growth on hexadecane was observed at NaCl concentrations of 1 to 60 g/liter, with the optimum at 10 g/liter. (The site from which it was isolated has a salinity of about 20 ppt.) When cultured at 30°C in medium adjusted to the optimal pH and NaCl concentration, doubling times were 3.0, 1.2, and 0.8 days with hexadecane (0.68 mmol/liter), octanoate (2.5 mM), and hexadecanoate (0.6 mmol/liter) as growth substrates, respectively.
Various electron donors and acceptors were tested for utilization by strain AK-01. Hydrocarbons including alkanes and 1-alkenes were utilized for growth by strain AK-01. Growth on alkanes of chain lengths from C13 to C18 was observed within 1 month. Growth on those with longer or shorter carbon chains than this range was not observed within 2 months. The two 1-alkenes (C15 and C16) tested were also utilized. For other nonhydrocarbon aliphatic substrates, the 1-alkanols (C15 and C16), n-saturated fatty acids (C4 to C16), and 2-methylheptanoic acid supported much faster growth than the hydrocarbons did. Oxidation of fatty acids (butyrate and octanoate tested) was complete, and acetate did not accumulate in the cultures. The strain also grew autotrophically on hydrogen with carbon dioxide as the sole carbon source. Growth on acetate, propionate, isobutyrate, 2-methylbutyrate, 3-methylbutyrate, lactate, malate, fumarate, and succinate was relatively poor compared with that on the fatty acids from C4 to C16. No growth on pyruvate, benzoate, and phenylacetate was observed. Sulfate reduction was observed during growth on all utilizable electron donors. Among the tested electron acceptors, sulfate, sulfite, and thiosulfate but not sulfur, nitrite, and nitrate were used for growth on octanoate. Desulfoviridin was not detected in cells of AK-01. The G+C content of its genomic DNA was 57 mol%. Cell surface hydrophobicity was 63 and 74% when the strain was grown to stationary phase on hexadecane and butyrate, respectively.
Table 1 shows the source of isolation, optimal growth temperature, and hydrocarbon degradation properties of strain AK-01 compared to those of strains Hxd3 and TD3, two of the recently reported alkane-degrading sulfate-reducers (1, 23). All three alkane-degrading strains were isolated from environments which are anoxic and chronically exposed to hydrocarbons. While both strains AK-01 and Hxd3 are mesophiles, TD3 is a thermophile that originated from the hydrothermally active Guaymas Basin. AK-01 and Hxd3 utilize a similar range of medium-chain alkanes and are able to use selected 1-alkenes. In contrast, TD3 uses alkanes of shorter chain lengths, with the optimal range from C8 to C12.
TABLE 1.
Comparison of the alkane-degrading sulfate reducers, strains AK-01, Hxd3 and TD3a
| Alkane-degrading bacterial strain | Source | Optimal growth temp (°C) | Alkanes utilized | 1-Alkenes utilizedb |
|---|---|---|---|---|
| AK-01 | Petroleum-contaminated estuarine sediment | 26–28 | C13–C18 | C15, C16 |
| Hxd3 | Precipitates of an oil-water separator in an oil field | 30 | C12–C20 | C14, C16, C17 |
| TD3 | Guaymas Basin sedimentc | 60 | C6–C16d | NDe |
Table 2 summarizes selected taxonomically important characteristics for the gram-negative sulfate-reducing bacteria. Type species for related and key genera (29, 31) are included for comparison to the three alkane-degrading strains, AK-01, Hxd3, and TD3 (1, 23) (see Discussion for details).
TABLE 2.
Characteristics of the alkane-degrading strains AK-01, Hxd3, and TD3 and other type species in various genera of the gram-negative sulfate-reducing bacteriaa
| Bacterial speciesb | Morphology | G+C content (mol%) | Desulfoviridind | Oxidatione | Electron donors for sulfate reductionf
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | For | A | P | FA | L | Fum | M | |||||
| Strain AK-01 | Short rod | 57 | − | C | +* | + | (+) | (+) | 4–16 | (+) | (+) | (+) |
| Strain Hxd3 | Rod | 63 | NR | C | − | − | − | − | 10–18 (4–8) | − | − | − |
| Strain TD3 | Curved rod | NRc | − | C | − | − | − | − | 4–18 | − | − | − |
| Desulfobacteriaceae | ||||||||||||
| Desulfosarcina variabilis | Oval, packages | 51 | − | C | +* | + | (+) | + | 4–14 | + | + | − |
| Desulfonema limicola | Multicellular filament | 35 | + | C | +* | + | (+) | + | 4–14 | + | + | − |
| Desulfococcus multivorans | Sphere | 57 | + | C | − | + | (+) | + | 4–16 | + | − | − |
| Desulfobacterium autotrophicum | Oval | 48 | − | C | +* | + | (+) | (+) | 4–16 | + | + | + |
| Desulfoarculus baarsii | Vibrio | 66 | − | C | − | + | (+) | (+) | 4–18 | − | − | − |
| Desulfobulbus propionicus | Oval | 60 | − | I | + | − | − | + | − | + | − | − |
| Desulfobacter postgatei | Oval | 46 | − | C | − | − | + | − | − | − | − | − |
| Desulfobotulus sapovorans | Vibrio | 53 | − | I | − | − | − | − | 4–16 | + | − | − |
| Desulfovibrionaceae | ||||||||||||
| Desulfovibrio desulfuricans | Vibrio | 59 | + | I | + | + | − | − | − | + | + | + |
| Desulfomicrobium baculatum | Rod | 57 | − | I | (+) | + | − | − | − | + | − | + |
Information for strain AK-01 is from this work. Information for strains Hxd3 and TD3 was adapted from references 1, 2, and 23. Information for all other species was adapted from references 29 and 31. See these references for details.
All except strains AK-01, Hxd3, and TD3 are type species of the respective genera (31).
NR, not reported.
+, present; −, absent.
Extent of organic substrate oxidation. C, complete; I, incomplete.
Abbreviations: For, formate; A, acetate, P, propionate; FA, fatty acids; numbers represent chain lengths (numbers in parentheses indicates poor utilization of those FA); L, lactate; Fum, fumarate; M, malate; *, autotrophic growth. For strain AK-01 only: +, >50% substrate utilized or sulfate reduced within 1 month; (+), >10% substrate utilized or sulfate reduced in 1 to 3 months; −, no substrate utilization or sulfate reduction in 3 months. For other bacteria: +, utilized; (+), poorly utilized; −, not utilized. Note: These data are a compilation and methods vary among studies. Comparison should be made with caution.
Phylogenetic analysis.
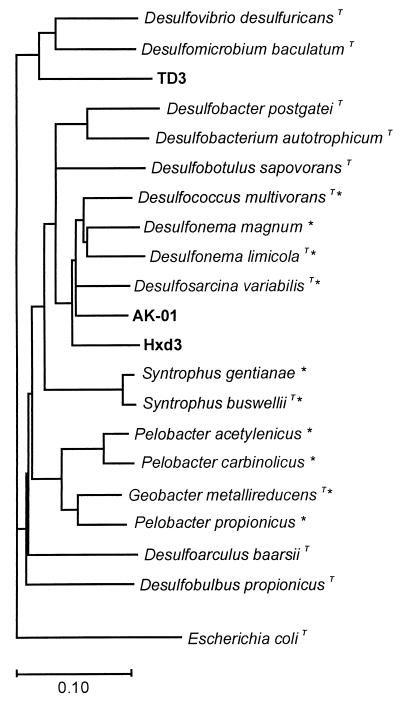
The PCR products obtained by amplification of the 16S rRNA gene of strain AK-01 with the primer set of 27f and SRB-R3 contained approximately 1,500 bp. Those of strain Hxd3 obtained with the primer set of 27f and SRB-R1 contained approximately 1,400 bp. The results that turned up in the similarity search were predominantly 16S rRNA sequences of bacteria that belong to the delta subdivision of the class Proteobacteria. A phylogenetic tree showing the relationship between strains AK-01, Hxd3, TD3 and other selected bacteria is shown in Fig. 2. As illustrated, AK-01 clearly belongs to the delta subdivision of the class Proteobacteria and is most closely related to the type species of the genera Desulfosarcina, Desulfonema, and Desulfococcus. Evolutionary distances (calculated by the Kimura two-parameter method) between strain AK-01 and Desulfosarcina variabilis, Desulfonema limicola, Desulfococcus multivorans, strain Hxd3, and strain TD3 are 9.11, 10.39, 9.35, 10.53, and 21.55 substitutions per 100 bp, respectively.
FIG. 2.
Phylogenetic relationship between the three alkane-degrading strains AK-01, Hxd3, and TD3 and other bacteria in the class Proteobacteria based on 16S rRNA sequence. The tree was constructed from approximately 1,300 aligned bases; T, type species in the genus; ∗, the 10 bacteria with their 16S rRNA sequences most similar to that of strain AK-01 according to the FASTA search.
Degradation of hexadecane coupled to sulfate reduction.
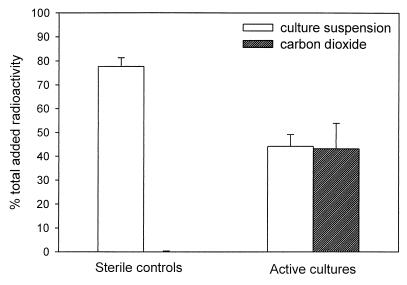
Mineralization of hexadecane to carbon dioxide by strain AK-01 was demonstrated with [1-14C]hexadecane. As shown in Fig. 3, about 40% of the added 14C was recovered as 14CO2 in the experimental cultures after 78 days of incubation. Less than 1% of the added radioactivity was recovered as 14CO2 in the autoclaved controls. Recoveries of the total added radioactivity were about 80%. The incomplete recovery of radioactivity may be due to the long period of incubation needed for this experiment. Despite the presence of their Teflon coating, some sorption of [1-14C]hexadecane by the rubber stoppers may have occurred. Nonetheless, these results indicate that mineralization to carbon dioxide does take place.
FIG. 3.
Formation of 14CO2 in autoclaved and active cultures of strain AK-01 fed with [1-14C]hexadecane after 78 days of incubation. Results are means of triplicate determinations, and error bars represent one standard deviation.
Table 3 shows the stoichiometry and growth yield for hexadecane degradation coupled to sulfate reduction by strain AK-01 compared to those for hexadecanoate degradation. No volatile fatty acids such as acetate and no long-chain fatty acids (free) were found at the end of the experiment in the cultures grown on hexadecane. The measured ratio for hexadecane degradation was 10.6 mol of sulfate per mol of hexadecane, and the growth yield was 7.4 g of protein per mol of hexadecane. The amount of sulfate reduced was 87% of that predicted by the following stoichiometric equation for the complete oxidation of hexadecane to carbon dioxide coupled to sulfate reduction:
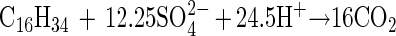 |
1 |
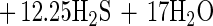 |
When production of cell mass was taken into consideration, equation 1 was modified to include the term of cell mass (using the general formula C5H7O2N). The amount of cell mass produced was indirectly estimated from the measured amount of protein by assuming a 55% (wt/wt) protein content in the cell mass (5). With these assumptions, the revised stoichiometric equation becomes
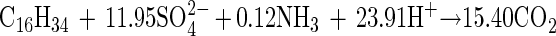 |
2 |
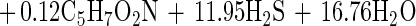 |
Equation 2 predicted a stoichiometric ratio of 11.95 mol of sulfate per mol of hexadecane, and the actual amount of sulfate reduced was 89% of that predicted by the revised equation.
TABLE 3.
Stoichiometry and growth yield of strain AK-01 for hexadecane and hexadecanoate degradation coupled to sulfate reduction
| Growth substrate | Substrate loss (μmol) | Sulfate loss (μmol) | Predicted sulfate lossc (μmol) | Percentage of predictiond | Growth yield (g of protein/mol of substrate) |
|---|---|---|---|---|---|
| Hexadecanea | 20.0 ± 2.7 | 212 ± 33 | 245 | 87 (89) | 7.4 ± 1.2 |
| Hexadecanoateb | 58.7 ± 0.4 | 556 ± 6 | 675 | 82 (86) | 11.1 ± 0.9 |
Results are means of five replicates ± standard deviations.
Results are means of triplicates ± standard deviations.
Obtained by multiplying the number of moles of hexadecane or hexadecanoate lost by the theoretical stoichiometric ratio of 12.25 or 11.5, respectively.
Numbers in parentheses include assimilation of the substrate into cell biomass.
The measured ratio determined for hexadecanoate degradation was 9.47 mol of sulfate per mol of hexadecanoate, and the growth yield was 11.1 g of protein per mol of hexadecanoate. The measured amount of sulfate reduced was 82% of that predicted by the following equation:
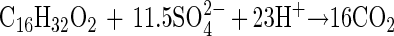 |
3 |
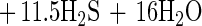 |
This stoichiometric equation 3 was also modified to equation 4 by the same method described above after taking cell biomass production into consideration, with the observed sulfate reduction amounting to 86% of the prediction accordingly:
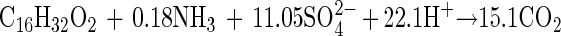 |
4 |
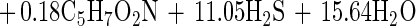 |
Hexadecane degradation by strain AK-01 is also dependent on sulfate reduction. As shown in Table 4, when the sulfate concentration in the medium was unlimiting, 6.6 μmol hexadecane (about 91% of the total) was degraded over 44 days with concomitant sulfate reduction. Only 0.26 μmol of hexadecane (about 3% of the total) was degraded with minimal sulfate reduction under sulfate-limiting conditions.
TABLE 4.
Degradation of hexadecane by strain AK-01 in the presence of unlimiting and limiting amounts of sulfatea
| Initial amt of sulfate (μmol) | Final amt of sulfate (μmol) | Sulfate loss (μmol) | Hexadecane loss (μmol) | % Loss of hexadecane |
|---|---|---|---|---|
| 412 ± 5 | 333 ± 20 | 79 ± 20 | 6.6 ± 0.2 | 91% |
| 27.6 ± 0.4 | 22.6 ± 0.2 | 5.0 ± 0.2 | 0.3 ± 0.3 | 4% |
Amounts of sulfate were deemed unlimiting (412 μmol; 13.8 mM) or limiting (27.6 μmol; 0.92 mM) with respect to the expected sulfate loss determined by multiplying the initial amount of hexadecane (7.5 μmol) by the theoretical stoichiometric ratio of 12.25. Results are means of triplicates ± standard deviations.
DISCUSSION
We have isolated and characterized a sulfate-reducing strain, AK-01, which is able to degrade alkanes under strict anoxic conditions. Its characterization includes both phenotypic and phylogenetic studies, as recommended for the taxonomic examination of the class Proteobacteria (17). Table 2 summarizes a number of phenotypic characteristics of strain AK-01 which are taxonomically important for the gram-negative sulfate-reducing bacteria and are useful for determining the relationship of AK-01 to the other sulfate reducers. Strains Hxd3 and TD3, two of the recently reported alkane-degrading sulfate-reducing bacteria (1, 3, 23), and the type species of key genera in the gram-negative sulfate-reducing bacteria (31) are included for comparison with the new strain. As shown in Table 2, strain AK-01 is different in several aspects from the genus Desulfovibrio in the suggested family Desulfovibrionaceae (31). Desulfovibrio species typically contain desulfoviridin; grow well on lactate, fumarate, and malate but cannot grow on fatty acids; and are incomplete oxidizers of organic substrates. Desulfomicrobium species also characteristically grow well on lactate and malate, incompletely oxidize substrates, and do not utilize fatty acids for growth. In contrast, strain AK-01 does not contain desulfoviridin, grows poorly on lactate and the dicarboxylic acids, but grows very well on fatty acids. Also, substrate oxidation is generally complete.
Strain AK-01 is also physiologically different from the genera Desulfobulbus, Desulfobacter, and Desulfobotulus of the suggested family Desulfobacteriaceae (31). Desulfobulbus species characteristically grow well on lactate and propionate by incomplete oxidation to acetate but cannot utilize any other fatty acids. Desulfobacter species grow effectively on acetate by complete oxidation but do not utilize formate and other fatty acids with longer chains. Desulfobotulus sapovorans, the only species of the genus, utilizes fatty acids (C4 to C16) and lactate with incomplete oxidation to acetate and does not grow on hydrogen, formate, or acetate. In contrast, strain AK-01 is distinctively different from the three genera in these key taxonomic characteristics (see Table 2).
Strain AK-01 is nutritionally similar to species in the genera Desulfoarculus, Desulfobacterium, Desulfococcus, Desulfonema, and Desulfosarcina. AK-01, along with members of these genera, grows well on fatty acids (C4 and above) and is able to utilize formate, acetate, and a variety of dicarboxylic acids. However, AK-01 is indeed morphologically different from the Desulfonema species, which form multicellular filaments, and Desulfosarcina variabilis, which forms cell packets at some growth stage.
The results of the phylogenetic analysis based on 16S rRNA sequences generally agree with the phenotypic characterization but have revealed further details. The 16S rRNA phylogeny (Fig. 2) shows that strain AK-01 is distantly related to the genera in the family Desulfovibrionaceae. Although the strain is phenotypically similar to the genera Desulfoarculus, Desulfobacterium, Desulfococcus, Desulfonema, and Desulfosarcina, this phylogenetic analysis places it closest to the last three genera. In addition, the phylogenetic distances between strain AK-01 and the type species of those three genera (between 9.11 and 10.39) are in the same range as those between the type species themselves (between 9.27 and 10.46). Whether AK-01 should be designated a genus, however, remains to be seen.
Strain AK-01 and the other two alkane-degrading sulfate reducers can also be compared phenotypically and phylogenetically. As shown in Table 2, strains TD3 and AK-01 are both fatty acid utilizers. However, unlike AK-01, TD3 cannot utilize hydrogen, formate, acetate, or propionate as an electron donor. In addition, TD3 is thermophilic while AK-01 is a mesophile. The range of alkanes used by TD3 is also somewhat different from those used by strain AK-01 (Table 1). The 16S rRNA phylogeny places strain TD3 distant from strains AK-01 and Hxd3 (Fig. 2). This is consistent with phylogenetic analyses conducted in other studies, which suggest that the thermophile isolated from a hydrothermally active environment represents a new type of sulfate-reducing bacteria (3, 23).
Strains Hxd3 and AK-01 are both fatty acid utilizers, complete oxidizers (Table 2), and mesophiles and are able to utilize a similar range of alkanes (Table 1). Phylogenetic distance based on 16S rRNA sequences also shows that strain AK-01 is more closely related to strain Hxd3 than to TD3 (Fig. 2). On the other hand, strain Hxd3 is unable to use hydrogen and formate as electron donors. Growth of Hxd3 on the fatty acids from C4 to C8 is poor, in contrast to that of AK-01. Its G+C content of 63% is also substantially higher than that of AK-01 (57%).
An ability to degrade alkanes by strain AK-01 is demonstrated by the formation of radiolabeled CO2 from [1-14C]hexadecane. Utilization of alkanes for growth is shown by cell mass production (as protein produced) upon degradation of hexadecane. The coupling of alkane oxidation to sulfate reduction is indicated by sulfate loss concomitant with hexadecane degradation and the dependence of degradation activity on sulfate reduction. In addition, the amount of sulfate reduced upon hexadecane degradation is reasonably reflected by the stoichiometric equations. The measured ratio of sulfate loss per mole of hexadecane is up to 89% of the predicted ratio when cell mass production is also considered. In previous reports on other anaerobic alkane degraders, the stoichiometric ratio determined for strains Hxd3 and TD3 was 91% of that predicted for complete oxidation of hexadecane (1) and 97% of that predicted for complete oxidation of decane (23).
Although the stoichiometric ratios determined for both hexadecane and hexadecanoate oxidation by strain AK-01 are similar, growth on hexadecane is about four times slower than is growth on hexadecanoate. Similarly, growth of strain Hxd3 on hexadecane is also much slower than that on hexadecanoate and octadecanoate (1, 2). The low water solubility of hexadecane (5.2 × 10−5 mg/liter) may limit its availability, and growth may be impeded by the limited surface area of the hydrophobic hexadecane droplets available for direct contact with the bacterial cells. This is also supported by the observation that actively dividing cells of strain AK-01 are mostly found clumped closely around hexadecane droplets rather than in suspension.
The initial reaction(s) for the oxidation of alkanes might also be a rate-limiting factor for utilization of alkanes. It was observed that cellular fatty acids of strain AK-01 were predominantly C-even when the alkane substrates were C-even and were predominantly C-odd when the alkanes were C-odd (data not shown). The strong impact of the carbon numbers of alkane substrates on those of the predominant cellular fatty acids, although manifested in a different way, can also be observed on strain Hxd3 (3). These observations strongly suggest that alkanes are anaerobically oxidized to fatty acids and are directly assimilated into cellular lipids. The much slower growth of both strains on alkanes than on long-chain fatty acids may therefore be caused by a rate-limiting step(s) in the initial reaction(s) for the anaerobic oxidation of alkanes to fatty acids. The lower growth yield on hexadecane than on hexadecanoate also suggests that the initial reaction(s) may be energy expending.
We report the isolation and characterization of a novel alkane-degrading bacterial strain. As evidenced by this study and those published in the last few years (2, 6, 8, 13, 23), anaerobic alkane degradation by sulfate reducers may be a more widespread phenomenon than was previously thought. Further studies with these pure cultures should provide interesting insights into potentially novel mechanisms for anaerobic alkane metabolism.
ACKNOWLEDGMENTS
We thank Friedrich Widdel for his advice on culturing strain Hxd3, Paula van Schie and Craig Phelps for their advice on the phylogenetic analysis, Beau Ranheim for providing the information on the sampling site, John Grazul for preparing the electron photomicrograph, Maria Rivera and Brian Donovan for technical assistance, and Andreas Naef for translation of the German literature.
This work is supported in part by the Office of Naval Research and the Defense Advanced Research Projects Agency.
REFERENCES
- 1.Aeckersberg F. Anaerober Abbau von Alkanen und 1-Alkenen durch sulfatreduzierende Bakterien. Ph.D. dissertation. Verlag Mainz, Germany: University of Bremen; 1994. [Google Scholar]
- 2.Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991;156:5–14. [Google Scholar]
- 3.Aeckersberg F, Rainey F A, Widdel F. Growth, natural relationships, cellular fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions. Arch Microbiol. 1998;170:361–369. doi: 10.1007/s002030050654. [DOI] [PubMed] [Google Scholar]
- 4.Britton L N. Microbial degradation of aliphatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 89–129. [Google Scholar]
- 5.Brock T D, Madigan M T. Biology of microorganisms. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1991. [Google Scholar]
- 6.Caldwell M E, Garrett R M, Prince R C, Suflita J M. Anaerobic biodegradation of long-chain n-alkanes under sulfate-reducing conditions. Environ Sci Technol. 1998;32:2191–2195. [Google Scholar]
- 7.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 8.Coates J D, Woodward J, Allen J, Philp P, Lovley D R. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl Environ Microbiol. 1997;63:3589–3593. doi: 10.1128/aem.63.9.3589-3593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis J B, Yarbrough H F. Anaerobic oxidation of hydrocarbons by Desulfovibrio desulfuricans. Chem Geol. 1966;1:137–144. [Google Scholar]
- 10.Delaune R D, Hambrick III G A, Patrick W H., Jr Degradation of hydrocarbons in oxidized and reduced sediments. Mar Poll Bull. 1980;11:103–106. [Google Scholar]
- 10a.EMBL Nucleotide Sequence Database. 1997. Release 51, June 1997. FASTA3 Homology Search Engine. [Online.] http://www2.ebi.ac.uk/fasta3/. 31 July 1997, last date accessed.
- 11.Kam C H, Robinson J P. Anaerobic degradation of crude oil. Microbe 86:XIV. Int Congr Microbiol. 1986;14:231. [Google Scholar]
- 12.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–147. [Google Scholar]
- 13.Livingston M R, Lundie L L., Jr . Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Isolation and partial characterization of a sulfidogen that is capable of n-hexadecane degradation, abstr. Q-215; p. 456. [Google Scholar]
- 14.Mesbah M, Premachandran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 15.Moses V, Robinson J P, Springham D G, Brown M J, Foster M, Hume J, May C W, McRoberts T S, Weston A. Proceedings of the 1982 International Conference on Microbial Enhancement of Oil Recovery. 1982. Microbial enhancement of oil recovery in North Sea reservoirs: a requirement for anaerobic growth on crude oil; pp. 154–157. [Google Scholar]
- 16.Muller F M. On methane fermentation of higher alkanes. Antonie Leeuwenhoek. 1957;23:369–384. doi: 10.1007/BF02545890. [DOI] [PubMed] [Google Scholar]
- 17.Murray R G E, Brenner D J, Colwell R R, De Vos P, Goodfellow M, Grimont P A D, Pfennig N, Stackebrandt E, Zavarzin G A. Report of the ad hoc committee on approaches to taxonomy within the Proteobacteria. Int J Syst Bacteriol. 1990;40:213–215. [Google Scholar]
- 18.Novelli G D, ZoBell C E. Assimilation of petroleum hydrocarbons by sulfate-reducing bacteria. J Bacteriol. 1944;47:447–448. [Google Scholar]
- 19.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci, USA. 1998;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postgate J R. A diagnostic reaction of Desulphovibrio desulphuricans. Nature. 1959;183:481–482. doi: 10.1038/183481b0. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. [Google Scholar]
- 22.Rosenfeld W D. Anaerobic oxidation of hydrocarbons by sulfate-reducing bacteria. J Bacteriol. 1947;54:664–665. [PMC free article] [PubMed] [Google Scholar]
- 23.Rueter P, Rabus R, Wilkes H, Aeckersberg F, Rainey F A, Jannasch H W, Widdel F. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulfate-reducing bacteria. Nature. 1994;372:455–458. doi: 10.1038/372455a0. [DOI] [PubMed] [Google Scholar]
- 24.So, C. M., and L. Y. Young. Biodegradation of alkanes by enriched consortia under four different anaerobic conditions. Submitted for publication. [PubMed]
- 25.Song B, Young L Y, Palleroni N J. Identification of denitrifier strain T1 as Thauera aromatica and proposal for emendation of the genus Thauera definition. Int J Syst Bacteriol. 1998;48:889–894. doi: 10.1099/00207713-48-3-889. [DOI] [PubMed] [Google Scholar]
- 26.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 27.Ward D M, Brock T D. Anaerobic metabolism of hexadecane in sediments. Geomicrobiol J. 1978;1:1–9. [Google Scholar]
- 28.Watkinson R J, Morgan P. Physiology of aliphatic hydrocarbon-degrading microorganisms. Biodegradation. 1990;1:79–92. doi: 10.1007/BF00058828. [DOI] [PubMed] [Google Scholar]
- 29.Widdel F. Anaerober Abbau von Fettsäuren und Benzoesäure durch neu isolierte Arten Sulfat-reduzierender Bakterien. Ph.D. dissertation. Göttingen, Germany: University of Göttingen; 1980. [Google Scholar]
- 30.Widdel, F. 1997. Personal communication.
- 31.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]