Abstract
Klebsiella pneumoniae is a globally significant opportunistic pathogen causing healthcare-associated and community-acquired infections. This study examined the epidemiology and the distribution of resistance and virulence genes in clinical K. pneumoniae strains in Kenya. A total of 89 K. pneumoniae isolates were collected over six years from five counties in Kenya and were analyzed using whole-genome sequencing and bioinformatics. These isolates were obtained from community-acquired (62/89) and healthcare-associated infections (21/89), and from the hospital environment (6/89). Genetic analysis revealed the presence of blaNDM-1 and blaOXA-181 carbapenemase genes and the armA and rmtF genes known to confer pan-aminoglycoside resistance. The most abundant extended-spectrum beta-lactamase genes identified were blaCTX-M-15 (36/89), blaTEM (35/89), and blaOXA (18/89). In addition, one isolate had a mobile colistin resistance gene (mcr-8). Fluoroquinolone resistance-conferring mutations in gyrA and parC genes were also observed. The most notable virulence factors were those associated with hyper-virulence (rmpA/A2 and magA), yersiniabactin (ybt), salmochelin (iro), and aerobactin (iuc and iutA). A total of 38 distinct sequence types were identified, including known global lineages ST14, ST15, ST147, and ST307, and a regional clone ST17 implicated in regional outbreaks. In addition, this study genetically characterized two potential hypervirulent isolates and two community-acquired ST147 high-risk clones that contained carbapenemase genes, yersiniabactin, and other multidrug resistance genes. These results demonstrate that the resistome and virulome of Kenyan clinical and hospital environmental K. pneumoniae isolates are diverse. The reservoir of high-risk clones capable of spreading resistance, and virulence factors have the potential to cause unmanageable infection outbreaks with high morbidity and mortality.
Keywords: antimicrobial resistance, virulence, whole-genome sequencing, Kenya, Klebsiella pneumoniae
1. Introduction
Klebsiella pneumoniae is a Gram-negative, rod-shaped ubiquitous bacterium that inhabits soil, water, and sewage ecosystems. It is also found on various human body sites and organ systems, including skin, nose, throat, and intestinal tract, as part of the natural microflora [1]. K. pneumoniae is a prominent member of the Klebsiella pneumoniae species complex that consists of seven species that include Klebsiella pneumoniae, Klebsiella quasipneumoniae subsp. quasipneumoniae, Klebsiella quasipneumoniae subsp. similipneumoniae, Klebsiella variicola subsp. variicola, Klebsiella variicola subsp. tropica, Klebsiella quasivariicola, and Klebsiella africana [2]. The first four species are commonly associated with human infections such as pneumonia, urinary tract infections, soft tissue and wound infections, septicemia, and pyogenic liver abscesses [3].
The World Health Organization (WHO) declared antimicrobial resistance (AMR) as one of the top 10 most serious global public health threats facing humanity [4]. The WHO lists K. pneumoniae as one of the AMR bacteria of concern due to its demonstrated proclivity for developing antimicrobial resistance to many classes of antibiotics such as penicillins, cephalosporins, and quinolones [5,6,7] which are typically used to treat K. pneumoniae infections. This resistance is due to both chromosomal-encoded and plasmid-encoded genes. The abundance of AMR genes carried on plasmids and mobile genetic elements have earned K. pneumoniae its reputation as a “key trafficker” of AMR genes between Klebsiella species and other Enterobacterales, illustrating its importance to AMR spread and development [8]. As carbapenems are considered one of the last resort treatments for multidrug-resistant (MDR) K. pneumoniae, i.e., isolates resistant to three or more drug classes [9], the global increase in carbapenem resistance [10] presents a threat to public health. The situation is aggravated by the isolation of colistin-resistant K. pneumoniae [11] since colistin is used as a last-line antibiotic for treating carbapenem-resistant K. pneumoniae. Global MDR high-risk lineages such as ST14, ST15, ST147, ST307, and ST607 have been identified and have spread rapidly across the globe increasing the need for global AMR surveillance. In Kenya, ST15 and ST17 have been reported in Kilifi [6] and ST14 in Nairobi [12].
In Kenya, several studies have indicated an emergence of the clonal spread of MDR K. pneumoniae and horizontal transfer of AMR genes [6,12,13,14,15]. Extended-spectrum beta-lactamase (ESBL) producing K. pneumoniae isolates have been reported in several locations such as Nairobi, Kisumu, Kisii, Homabay, Migori, Kilifi, and Eldoret [6,13,15]. Increased ESBL-producing Enterobacterales, the most prevalent of which in Kenya are CTX-M-15 and TEM, is associated with the increased use of third generation cephalosporins in the early 2000s [10,13,16,17]. To exacerbate the situation, carbapenem use in Kenya is increasingly leading to the emergence of carbapenem-resistant Enterobacteriaceae (CRE). In 2011, Poirel, et al., were the first to detect CRE in Kenya from urine samples bearing the New Delhi Metallo-β-lactamase (NDM-1) carbapenemase [12]. In 2017, there were reports of K. pneumoniae isolates that encoded blaNDM-1 and blaSPM carbapenemase genes [15] and isolates recovered from bacteremias bearing the pNDM-MAR-like plasmid backbone that has been shown to carry the blaNDM gene [6]. More recently, Musila et al. (2021) identified K. pneumoniae isolates with the blaOXA-181 carbapenemase gene. In addition, K. pneumoniae resistance to aminoglycosides, tetracyclines, chloramphenicol, and fluoroquinolones has also been observed in Kenya, typically associated with ESBL production in MDR [6,10,18]. Aminoglycosides are widely used to treat bacterial infections due to the ease of administration and access in Kenya [19]. Musila et al. (2021) also reported the 16S rRNA methyltransferase genes, rmtF and rmtC, in K. pneumoniae isolates which confer pan-aminoglycoside resistance [18]. Resistance to chloramphenicol and sulfonamides is high in part due to its use as a first-line treatment option for enteric infections such as typhoid and HIV prophylaxis in Kenya [10].
In addition to antimicrobial resistance, K. pneumoniae possesses virulence genes that enhance its ability to cause infections, increase cell fitness, and evade the host immune system. K. pneumoniae colonizes host cells using adhesins such as fimbriae and pili [20]. The production of a robust capsular polysaccharide confers resistance to host immune cells along with the O-antigen portion of the liposaccharide (LPS). K. pneumoniae attacks rival bacteria and eukaryotic cells by injecting potent endotoxins using type VI secretion machinery [21]. Highly virulent K. pneumoniae increases the expression of magA, rmpA, and rmpA2 genes linked to the mucoid phenotype for hypervirulence [22]. Other mechanisms include the use of allantoin for a carbon and nitrogen source, the use of efflux pumps to eject antibiotics [23], and the capture of iron molecules from the host cells using siderophores (aerobactin, salmochelin, yersiniabactin) [24]. Although some studies in Kenya have identified K. pneumoniae lineages ST11, ST15, and ST17, associated with hospital outbreaks [6,14], and the high-risk MDR ST147 [19], few studies have examined virulence genes in Kenyan K. pneumoniae isolates [14].
In Kenya, screening of AMR phenotypes, genes, and sequence typing is done mainly using manual or automatic antimicrobial susceptibility testing (AST) and polymerase chain reaction amplification techniques [12,15,17] These techniques provide limited targeted information and are laborious compared to whole genome sequence data. As such, there is limited information on the STs and extent of AMR genes carried in K. pneumoniae in Kenya [25,26]. Thus, this study combined whole genome sequencing analysis with phenotypic antimicrobial susceptibility testing to identify the AMR genes and gene mutations associated with the phenotypic patterns observed and also to conduct in silico sequence typing of known and novel sequence types (STs).
A clearer understanding of K. pneumoniae sequence types (STs) circulating within the community and healthcare systems across Kenya and the diversity and distribution of AMR and virulence genes would facilitate the monitoring and control of high-risk clones that are potentially hypervirulent and/or multidrug-resistant. Given the public health and clinical importance of K. pneumoniae and the knowledge gaps in Kenya, this study set out to examine K. pneumoniae isolates across Kenya and describe: (1) the sequence types and their distribution across Kenya, (2) the antimicrobial resistance phenotypes, and (3) the AMR and virulence genes present in the K. pneumoniae isolates.
2. Materials and Methods
2.1. Study Site
The bacterial culture, DNA extraction, quantification, and Oxford Nanopore sequencing were conducted in the Kenya Medical Research Institute (KEMRI)—Center for Microbiology Laboratory. Illumina sequencing was performed at the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN), Walter Reed Army Institute of Research (WRAIR). The bioinformatic analysis was conducted using the high-performance computing server at the Department of Emerging Infectious Diseases, USAMRD-Africa in KEMRI, Nairobi.
2.2. Study Samples
The study analyzed hospital environmental and clinical isolates of K. pneumoniae that were collected between May 2015 and March 2020 from eight hospitals in five counties in Kenya as part of an ongoing antimicrobial resistance surveillance study (KEMRI2767/WRAIR 2089) and an environmental study (KEMRI 3482/WRAIR 2416). The clinical samples were urine, wound swabs, and pus collected from consenting patients with suspected bacterial infections. The environmental samples were collected via swabs of high-touch areas in the participating hospitals. Klebsiella pneumoniae identification and antimicrobial susceptibility testing (AST) were performed on the Vitek2® system (bioMérieux, Lyon, France) using the GN-ID and XN05-AST cards. The AST panel consisted of penicillins (piperacillin and ticarcillin/clavulanic acid), monobactam (aztreonam), cephalosporins (cefuroxime, cefuroxime axetil, cefixime, ceftriaxone, and cefepime), carbapenems (meropenem), fluoroquinolones (levofloxacin and moxifloxacin), tetracyclines (tetracycline and minocycline), glycylcycline (tigecycline), phenicol (chloramphenicol), and trimethoprim. The AST results were interpreted according to CLSI guidelines (2018) [27], and isolates were classified as either multidrug-resistant (resistant to three or more drug classes) or non-multidrug resistant and ESBL positive or negative.
2.3. DNA Extraction and Sequencing
A total of 49 K. pneumoniae isolates were inoculated on Mueller Hinton Agar plates and incubated for 24 h at 37 °C. Klebsiella pneumoniae JH930422.1 was included as a positive control. Approximately 14 × 108 cells/mL at OD 600 of overnight bacterial cells were re-suspended in nuclease-free water and centrifuged at 5400× g for 10 min to pellet the cells. Total DNA was then extracted using the DNeasy® UltraClean Microbial Kit (QIAGEN Inc., Hilden, Netherlands) according to the manufacturer’s instructions. DNA purity was determined on the Nanodrop One (ThermoFisher Scientific, Waltham, MA, USA) and quantified on the Qubit dsDNA fluorometer (ThermoFisher Scientific, Waltham, MA, USA). For library preparation, the extracted DNA was end-repaired using the NEBNext® UltraII End Repair/dA-Tailing kit (New England Biolabs, Ipswich, MA, USA) using the manufacturer’s instructions. Ligation Sequencing Kit-LSK109 (Oxford Nanopore Technology, Oxford, United Kingdom), EXP-NBD 104 (1–12), and EXP-NBD 114 (13–24) Native Barcoding kits (Oxford Nanopore Technology, Oxford, UK) were used to barcode each DNA sample and adapters added. The DNA library was prepared and loaded onto a FLO-MIN106 R9.4.1 flow cell for sequencing based on the standard Oxford Nanopore Technology (ONT) 1D-sequencing protocol. The sequencing run was launched on the MinKNOW software (v20.10.3, Oxford Nanopore technology, Oxford, UK). A quality control experiment was conducted to evaluate the Nanopore workflow. This was done by sequencing the lambda phage DNA on the FLO-MIN106 R9.4.1 flow cell for 6 h as per the Lambda DNA control experiment protocol, and the sequences were compared with the reference sequence (NC_001416.1). Guppy software (v4.4.2, Oxford Nanopore Technology, Oxford, UK) was used to basecall and trim the barcodes and adapters from both ends of the reads.
The whole genome sequence reads of 40 additional K. pneumoniae isolates, sequenced on an Illumina MiSeq platform at MRSN-WRAIR as previously described [19], were included.
2.4. De novo Assembly of Raw Reads and Database Querying
The fastQ files of the long reads were filtered to retain only those with a Q-score ≥ 7. The adapters in the short-pair-ended reads were trimmed using Trimmomatic v0.39 [28]. The trimmed Illumina reads were assessed for quality using FastQC v0.11.9 [29] before de-novo assembly using the default Shovill v1.1.0 [30] pipeline settings. Next, the draft assemblies were polished using pilon v1.24 [31]. Finally, the ONT long-reads were de novo assembled using flye assembler v2.8.1 [32] with the plasmid option, followed by one polishing round using medaka v1.3.2 [33]. All polished draft assemblies from Illumina and ONT sequencing were analyzed in the same way. First, the quality was assessed using QUAST [34]. Then, the draft assemblies were queried using the ABRicate v1.0.1 [35] pipeline against CARD [36] to identify AMR genes, VFDB [37] to identify virulence factors, and PlasmidFinder [38] to identify plasmids. The assemblies were queried against the Klebsiella MLST database using the command line mlst v2.19 [39] pipeline to determine the sequence types, while the capsule (K) and O types were determined using the Kleborate pipeline [40] against the Kaptive database [41]. Next, a new ybt-typing scheme updated in the Kleborate [40] pipeline was explored to assign allelic profiles to yersiniabactin genes. Finally, a maximum-likelihood phylogenetic tree was generated using Parsnp v1.2 [42] and NC_009648.1 as the reference genome. The tables were created using flextable [43] (R package), while the circular tree and heatmaps were generated using Interactive Tree of Life (iToL) v6.3.2 [44] tree annotator.
3. Results
3.1. Epidemiological and Clinical Characteristics of K. pneumoniae Isolates
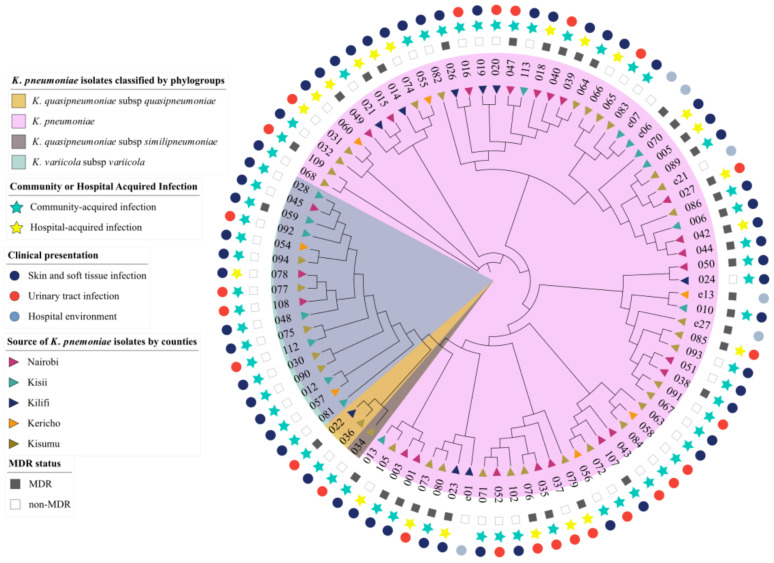
Clinical samples were collected from skin and soft tissue infections (SSTIs) (64%, 57/89), urinary tract infections (UTIs) (29%, 26/89), and the hospital environment (7%, 6/89) from five counties in Kenya: Kisumu (39%, 35/89), Nairobi (26%, 23/89), Kisii (17%, 15/89), Kilifi (10%, 9/89), and Kericho (8%, 7/89). A total of 62 isolates were isolated from community-acquired infections (CAI), while 21 were from healthcare-associated infections (HAI).
Of the four members of the K. pneumoniae Complex identified [2], K. pneumoniae subsp. pneumoniae represented the largest proportion of all isolates at 79% (70/89), with 44 recovered from SSTIs, 20 recovered from UTIs, and 6 from the hospital environment. This phylogroup had the largest number of MDR isolates (39%). The second most represented phylogroup was K. variicola subsp. variicola (18%, 17/89) with 15 isolates recovered from SSTIs and 2 isolated from UTIs. There was only 1 MDR isolate (kkp059) in this category. The least represented phylogroups were K. quasipneumoniae subsp. quasipneumoniae (2%, 2/89) and K. quasipneumoniae subsp. similipneumoniae (1%, 1/89) (Figure 1). Among the isolates from these minor subspecies, kkp022 and kkp034 were isolated from a UTI, while kkp036, the only MDR in this category, was recovered from an SSTI. All K. quasipneumoniae subsp. quasipneumoniae, K. quasipneumoniae subsp. similipneumoniae, and K. variicola subsp. variicola, except for one isolate (kkp078), were from community-acquired infections from different geographical locations (Figure 1). There was no evident geographical clustering of all the phylogroups by county or infection types.
Figure 1.
Circular cladogram showing epidemiological and clinical characteristics of K. pneumoniae isolates (n = 89). The color shading indicates the clustering of the isolates by phylogroups: Klebsiella pneumoniae (pink), Klebsiella quasipneumoniae subsp. similipneumoniae (brown), Klebsiella quasipneumoniae subsp. quasipneumoniae (tan), and Klebsiella variicola subsp. variicola (blue). The triangle symbols in the innermost ring represent the geographic source of the isolates: Nairobi (purple), Kisii (teal), Kilifi (blue), Kericho (orange), and Kisumu (brown). The star symbols represent the isolates recovered from community-acquired (blue) or healthcare-associated infections (yellow). The square symbols represent the multidrug resistance status of the isolates as either multidrug-resistant (black) or non-multidrug-resistant (white). The circle symbols in the outermost ring represent the clinical presentation of the isolates, i.e., skin and soft tissue infections (dark-blue), urinary tract infection (red), and the hospital environment (light-blue).
3.2. Genomic Characteristics of K. pneumoniae Isolates
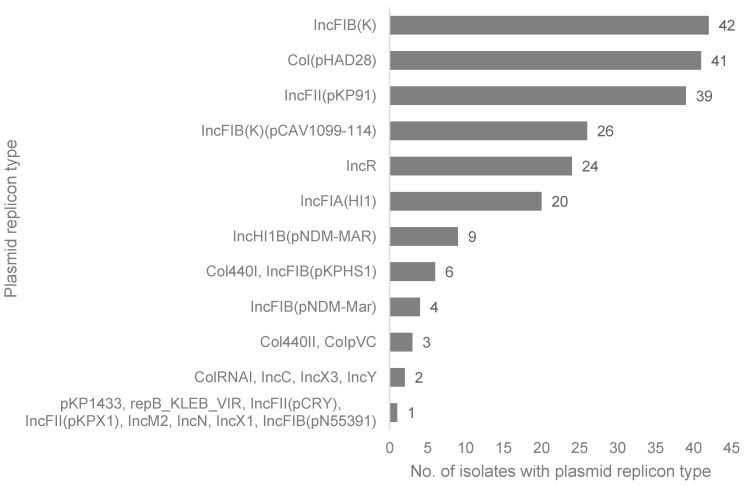
Draft genomes were generated from the 89 isolates: 40 via Illumina short-read sequencing and 49 via MinION-based long-read sequencing. The sizes of the draft genomes ranged from 5.2 to 5.9 Mb with an average G + C content of 57.27%, typical of K. pneumoniae genomes [45] (Table S1). The average N50 for the short and long reads was 226,402 and 5,147,285 base pairs, respectively. The isolates had 0–8 plasmid replicons, averaging 3 per isolate. The highest number of plasmid replicons were in genomes kkp001 (8), kkp018 (8), and kkp0e21 (8) (Table S2), whereas five genomes had no predicted replicons: kkp012, kkp030, kkp070, kkp102, and kkp112. The most abundant plasmid replicon types identified belonged to the Col and Inc family (particularly the IncF type) (Figure 2). The other Inc-like plasmid replicons identified were IncR, IncH, IncX, IncC, IncN, IncM, and IncY. Seven types of Col plasmid replicons, the second most represented type, were identified, dominated by Col(pHAD28) (Figure 2). Other plasmid types identified included pKP1433 and rep_KLEB_VIR in kkp056 in kkp043 genomes, respectively.
Figure 2.
The types and number of plasmid replicons identified in the Kenyan K. pneumoniae isolates (n = 89).
3.3. Multilocus Sequence Types and Distribution of K. pneumoniae Isolates
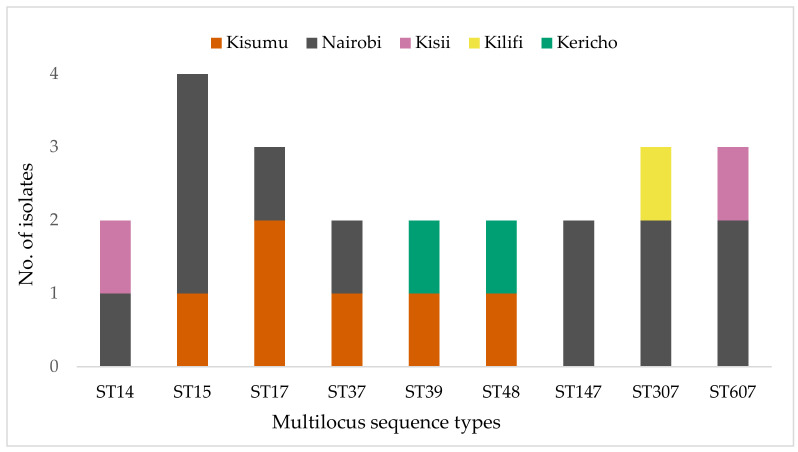
The STs could only be assigned to 57% (51/89) of the isolates. The 37 unassigned isolates were not typeable due to low genome coverage (Table S1). The multilocus sequence types (STs) of the 52 K. pneumoniae isolates were diverse, with 38 different STs identified. STs that were represented by more than one isolate were ST15 (4/52), ST17 (3/52), and ST607 (3/52 each); two isolates each represented the ST14, ST37, ST39, ST48, ST147, and ST307 lineages (Figure 3). The remaining 29 sequence types were represented by a single isolate and were geographically distributed as follows: Kisumu (n = 17), ST20, 45, 101, 198, 336, 391, 751, 1927, 2010, 3717, 3397, 3609, 5594, 5595, 5596, 5598, 5599; Nairobi (n = 6), ST55, 219, 966, 1786, 3692, 5600; Kisii (n = 4),ST25, 711, 3717, 5597; and Kilifi (n = 2), ST364 and 5593. Eight isolates had novel allelic profiles assigned and deposited in the K. pneumoniae MLST database (https://bigsdb.web.pasteur.fr/) (Table 1). There was no evident clustering of STs by infection type or ESBL and MDR status. Kisumu had the highest number of distinct STs (23), followed by Nairobi (12), Kisii (6), Kilifi (3), and Kericho (2). The high-risk lineages, ST14, ST15, ST17, ST307, and ST607, were concentrated in the biggest cities: Kisumu and Nairobi. Global high-risk strains ST14, ST15, ST307, and ST607 were predominantly from the K. pneumoniae.
Figure 3.
Distribution of the most abundant sequence types of the K. pneumoniae isolates by county.
Table 1.
Allelic profiles of K. pneumoniae isolates with novel multilocus sequence types.
| Isolate ID | BIGSdb ID | Species | MLST | gapA | infB | mdh | pgi | phoE | rpoB | tonB |
|---|---|---|---|---|---|---|---|---|---|---|
| kkp022 | 16011 | K. quasipneumoniae | 5593 | 17 | 19 | 39 | 39 | 552 | 21 | 262 |
| kkp034 | 16017 | K. quasipneumoniae | 5594 | 18 | 22 | 56 | 162 | 556 | 13 | 51 |
| kkp075 | 16032 | K. variicola | 5595 | 45 | 18 | 21 | 105 | 455 | 22 | 786 |
| kkp076 | 16033 | K. pneumoniae | 5596 | 2 | 9 | 2 | 1 | 13 | 1 | 787 |
| kkp081 | 16037 | K. variicola | 5597 | 16 | 24 | 21 | 40 | 106 | 17 | 67 |
| kkp090 | 16041 | K. variicola | 5598 | 16 | 18 | 21 | 33 | 55 | 17 | 341 |
| kkp091 | 16042 | K. pneumoniae | 5599 | 294 | 3 | 1 | 1 | 4 | 331 | 4 |
| kkp108 | 16051 | K. variicola | 5600 | 306 | 24 | 21 | 27 | 47 | 22 | 188 |
ID—identity; MLST—multilocus sequence type, the seven housekeeping genes for K. pneumoniae strain typing (gapA—glyceraldehyde 3-phosphate dehydrogenase, infB—translation initiation factor 2, mdh—malate dehydrogenase, pgi—phosphoglucose isomerase, phoE—phosphoporine E, rpoB—beta-subunit of Ribonucleic acid polymerase B, tonB—periplasmic energy transducer (Diancourt, 2005); BIGSdb—K. pneumoniae MLST database.
3.4. Antimicrobial Resistance
3.4.1. Phenotypic Resistance Profiles
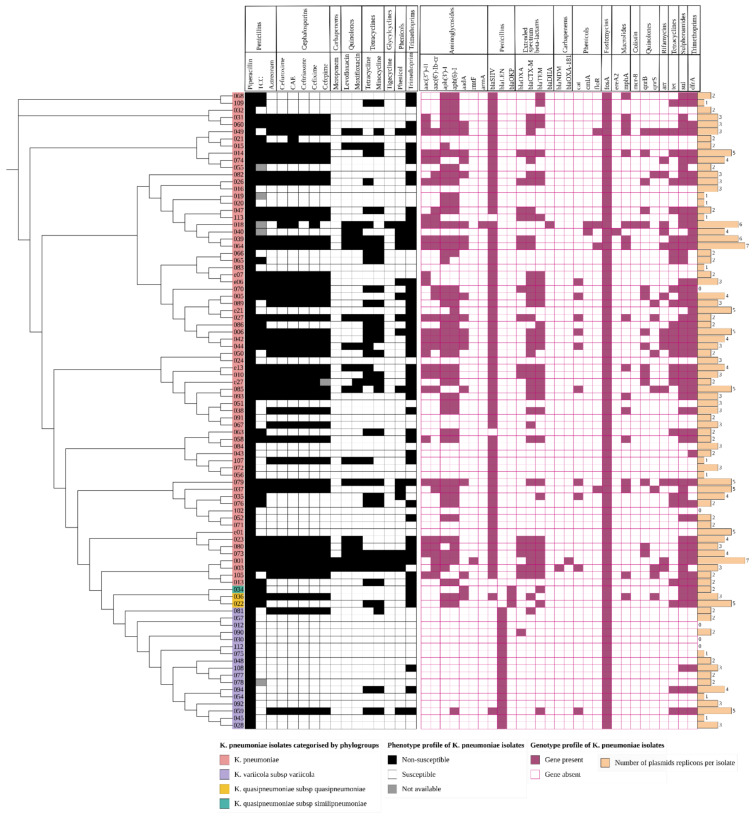
Antibiotic susceptibility tests (AST) performed on the VITEK2® platform (bioMerieux, Lyon, France) (Table S3) identified high levels of non-susceptibility to trimethoprim (57%, 51/89), ticarcillin/clavulanate (49%, 44/89), cefuroxime axetil (49%, 44/89), cefuroxime (48%, 43/89), cefixime (48%, 43/89), ceftriaxone (47%, 42/89), cefepime (46%, 41/89), aztreonam (47%, 42/89), tetracycline (37%, 33/89), and minocycline (34%, 31/89); moderate non-susceptibility to levofloxacin (19%, 17/89), moxifloxacin (20%, 18/89), and chloramphenicol (19%, 17/89); and low levels of non-susceptibility to tigecycline (4%, 4/89) and meropenem (2%, 2/89). All isolates were non-susceptible to piperacillin (Figure 4). Among the dataset, 41% (37/89) were classified as MDR, i.e., resistant to three or more drug classes. A large proportion (47%, 42/89) were ESBL-producing isolates (Figure 4), of which 30 isolates were MDR. One isolate, kkp001, was pan-resistant (resistant to all 16 antibiotics tested in the panel).
Figure 4.
A heatmap of the phenotypic and genotypic profiles and plasmid replicon abundance of K. pneumoniae isolates (n = 89). The cladogram on the left with colored labels on the edge indicates the clustering of the isolates by phylogroups: K pneumoniae (pink), K. quasipneumoniae subsp. similipneumoniae (teal), K. quasipneumoniae subsp. quasipneumoniae (gold), and K. variicola subsp. variicola (grey). The phenotypic profile is represented as an isolate being non-susceptible (black) or susceptible (white) to the antibiotic indicated on the bottom column header (and the drug class it belongs to on the top column header). The grey square indicates an isolate whose phenotypic result was not available. The genotypic profile is represented as a gene present (purple) or absent (white with purple outline). The genes are indicated on the column header and the drug class on the top column header. The beige bar plot represents the number of plasmid replicons found in the isolates; CAE—cefuroxime axetil; TCC—ticarcillin/clavulanate.
3.4.2. Genetic Determinants of Resistance
Comparison of the isolates’ phenotypic and genotypic antimicrobial susceptibility results demonstrated high concordance in the trimethoprim, beta-lactam, and tetracycline antibiotic classes, as demonstrated in Figure 4. In particular, there was strong concordance among the K. pneumoniae isolates between the presence of a blaCTX-M gene and non-susceptibility to beta-lactams and cephalosporins (Figure 4), dfrA gene with trimethoprim non-susceptibility, and tet genes with tetracycline non-susceptibility. Most of the genomes generated from long reads enabled the detection of circularized plasmids bearing AMR genes (Table 2). MDR isolates had more plasmid replicons than the non-MDR isolates which had only 0–3 plasmid replicons (Figure 4).
Table 2.
Antimicrobial resistance and virulence genes identified in the plasmids of the K. pneumoniae isolates; ID—identity.
| Isolate ID | Plasmid ID | Plasmid Replicon | AMR Genes | Virulence Genes |
|---|---|---|---|---|
| kkp001 | kkp001_p002 | IncFII(pKPX1), IncR | aph(3″)-ib, aph(6)-id, CTX-M-15, OXA-181, TEM-181, dfrA, sul2 | |
| kkp005 | kkp005_p002 | IncFIA(HI1),IncR | aac(6′)-ib-cr, aph(3″)-ib, aph(6)-id, CTX-M-15, qnrB, TEM-181, aadA, arr3, CatII, dfrA, qacEdelta1, sul1, sul2 | |
| kkp006 | kkp006_p003 | IncFIA(HI1), IncR | aac(6′)-ib-cr, aph(3″)-ib, aph(6)-id, qnrB, aadA, arr3, CatII, dfrA, qacEdelta1, sul1, sul2, TetD | |
| kkp019 | kkp019_p001 | IncFIA(HI1) | aph(3″)-ib, aph(6)-id | mrkABCDFJ |
| kkp020 | kkp020_p001 | IncFIA(HI1) | aph(3″)-ib, aph(6)-id | mrkABCDFJ |
| kkp024 | kkp024_p002 | IncFIA(HI1) | mrkABCDFJ | |
| kkp026 | kkp026_p001 | IncFIB(K), IncFII(pKP91) | aac(3)-iie, aac(6′)-ib-cr, aph(3″)-ib, aph(6)-id, CTX-M-15, OXA-1, qnrB, TEM-181, dfrA, sul2, TetA | |
| kkp032 | kkp032_p001 | IncFIB(K) | aph(3″)-ib, aph(6)-id | mrkABCDFJ |
| kkp034 | kkp034_p002 | IncY | TEM-181, aadA, dfrA, qacEdelta1, sul1, sul2 | |
| kkp036 | kkp036_p002 | IncN | SHV-134 | |
| kkp037 | kkp037_p001 | IncC, IncFIB(K), IncFII(pKP91) | aac(6′)-ib, aph(3″)-ib, aph(3′)-ia, aph(6)-id, floR, mphA, qacEdelta1, sul1, sul2 | |
| kkp037 | kkp037_p002 | IncX3 | qnrS, SHV-134 | |
| kkp039 | kkp039_p001 | IncFIB(K), IncFII(pKP91) | aadA, dfrA, mphA, qacEdelta1, sul1 | |
| kkp043 | kkp043_p001 | repB_KLEB_VIR | iroBCDN, iucABCD, iutA, rmpA, rmpA2 | |
| kkp043 | kkp043_p002 | IncFIA(HI1) | dfrA, qacEdelta1, sul1 | |
| kkp051 | kkp051_p001 | IncFIB(K)(pCAV1099-114), Col(pHAD28), IncR | aph(3″)-ib, aph(6)-id | |
| kkp052 | kkp052_p003 | IncR | aph(3″)-ib, aph(6)-id, dfrA, qacEdelta1, sul1, sul2 | mrkABCDF |
| kkp066 | kkp066_p001 | IncFIB(K), IncFII(pKP91) | aph(3″)-ib, aph(6)-id, SHV-120, sul2, TetD | |
| kkp068 | kkp068_p001 | IncFIB(K), IncFII(pKP91) | aph(3″)-ib, aph(6)-id, TEM-181, dfrA, mphA, sul2 | |
| kkp083 | kkp083_p001 | IncFIB(K) | astA, iroBDEN | |
| kkp090 | kkp090_p001 | IncFIB(K), IncFII(pKP91) | OXA-926 | |
| kkp094 | kkp094_p002 | IncFIA(HI1) | ant(3″)-iia, dfrA, qacEdelta1, sul1, TetA | |
| kkp108 | kkp108_p002 | IncFII(pKP91) | dfrA, qacEdelta1, sul1 | |
| kkp0e13 | kkp0e13_p001 | IncFII(pKP91) | aac(3)-iie, aac(6′)-ib-cr, aph(3″)-ib, aph(6)-id, CTX-M-15, OXA-1, qnrB, TEM-181, dfrA, sul2, TetA | |
| kkp0e13 | kkp0e13_p002 | IncM2 | aac(3)-IId, CTX-M-15, TEM-181, mphA | |
| kkp0e21 | kkp0e21_p004 | Col(pHAD28) | aph(6)-id, dfrA | |
| kkp0e27 | kkp0e27_p001 | IncFIB(K), IncFII(pKP91) | aph(3″)-ib, aph(6)-id, CTX-M-15, qnrB, TEM-181, dfrA, sul2, TetA | |
| kkp0e7 | kkp0e7_p001 | IncFIB(K), IncFII(pKP91) | aac(3)-iie, CTX-M-15, TEM-181 |
Beta-lactamase resistance genes. In the beta-lactam antibiotic class, genes for penicillinases, ESBLs, and carbapenemases were identified, summarized in Table S4. Each phylogroup demonstrated a distinct chromosomal penicillinase (Table S4), i.e., K. pneumoniae contained blaSHV, K. quasipneumoniae contained blaOKP, and K. variicola contained blaLEN, consistent with previous studies [8]. However, some isolates carried a different additional penicillinase which was observed to have been acquired via a plasmid, e.g., kkp036 (K. quasipneumoniae) obtained a blaSHV-134 from an IncN plasmid (Table 2). There was a high diversity of plasmid-mediated ESBL genes in the isolates: blaCTX-M-15 (40%, 36/89), blaCTX-M-98 (1%, 1/89), blaCTX-M-14 (1%, 1/89), blaTEM181 (39%, 35/89), blaOXA (18/89), and the less common blaLAP2 and blaSCO1. Two MDR isolates had carbapenemase genes: blaNDM1 (kkp003) and blaOXA181 (kkp001). Isolates kkp013 and kkp063 did not demonstrate any β-lactamase genes, and they were unsurprisingly ESBL-negative and non-MDR. Most isolates possessed at least one and at most six β-lactamase genes (Table S4). There were 42 ESBL-positive isolates, and 38 of them had a blaCTX-M co-harbored with 2 or more other types of ESBL genes (Figure 4).
Non-beta-lactamase resistance genes. A previously described [11] colistin-resistant isolate, kkp018, carried an mcr-8, a blaDHA1, and 20 additional AMR genes (Table S4). A total of 67% of the isolates (60/89) carried aminoglycoside-modifying enzymes (AME) genes, i.e., ant(3′) (or aadA) encoding aminoglycoside nucleotidyltransferases; aph(3′), aph(4), and aph(6) encoding aminoglycoside phosphotransferases; rmtF and armA encoding16S rRNA methyltransferases; and aac(3) and aac(6′)-Ib-cr encoding aminoglycoside acetyltransferases (Figure 4, Table S4). The aac(6′)-Ib-cr gene also induces fluoroquinolone resistance. The genes associated with pan-aminoglycoside resistance were found in two isolates: armA (kkp018) and rmtF (kkp001). The genes for efflux pumps associated with aminoglycoside resistance (arnT, crcB, acrD, baeR, cpxA) were well-conserved among all the isolates. In addition, the common MDR efflux pump genes were identified: LptD, CRP, H-NS, KpnEFGH, acrAB, marA, mdtBC, msbA, and ramA.
Three mechanisms for quinolone resistance were detected. First, plasmid-mediated quinolone resistance (PMQR) genes (qnrB or qnrS1) were identified in 26 isolates. Second, chromosomal-encoded efflux pumps (emrR, oqxA, and oqxB) genes were constitutive in all isolates. Third, gene mutations were detected in gyrase A (gyrA)—Ser83Phe, Asp87Aspn, and Ser83Ile—and Deoxyribonucleic acid topoisomerase IV subunit A (parC)—Ser80Ile (Table S6), which are involved in DNA synthesis. A total of 29 isolates (32%, 29/89) had the tetracycline-resistance genes, tetA or tetD, while 15 isolates (17%, 15/89) carried the chloramphenicol resistance genes cat1, catII, and catB3 (encoding chloramphenicol acetyltransferases) as well as floR and cmlA (encoding chloramphenicol efflux pumps). Resistance to sulphonamide and trimethoprim, administered as co-trimoxazole, was mediated by the dfrA trimethoprim resistance gene and sul sulphonamide resistant gene, found in 54 and 55 isolates, respectively. Resistance to other drug classes was conferred by: fosfomycins—fosA; macrolides—mphA, mphE, msrE, and ereA2; rifamycins—arr2 and arr3; and even antiseptics—qacEΔ1 and qacL.
3.5. Virulence Factors Associated with the K. pneumoniae Isolates
Known virulence factors involved in adherence, biofilm formation, capsule synthesis regulation, mucoid phenotype regulation, immune evasion, secretion system, serum resistance, siderophores expression (enterobactin, yersiniabactin, aerobactin, and salmochelin), efflux pump expression, allantoin utilization, and enterotoxin generation were detected among the isolates (Figure 5). The most ubiquitous were the chromosomal genes fim, mrk, and ecp for adherence and biofilm formation, which were present in all isolates except kkp043 which lacked the fim genes. In addition, some isolates carried multiple copies of the mrk genes in plasmids (Table 2). Other genes identified in all the isolates were those for serum resistance factors that determine the ‘O-antigen’ lipopolysaccharide serotype, the immune evasion factors which determine the polysaccharide capsule (K antigen) type, capsule synthesis regulation (rcs), efflux pump expression (acrAB), and enterobactin (ent, fep) (Table S5). In addition, all isolates possessed type VI secretion system loci genes except kkp034.
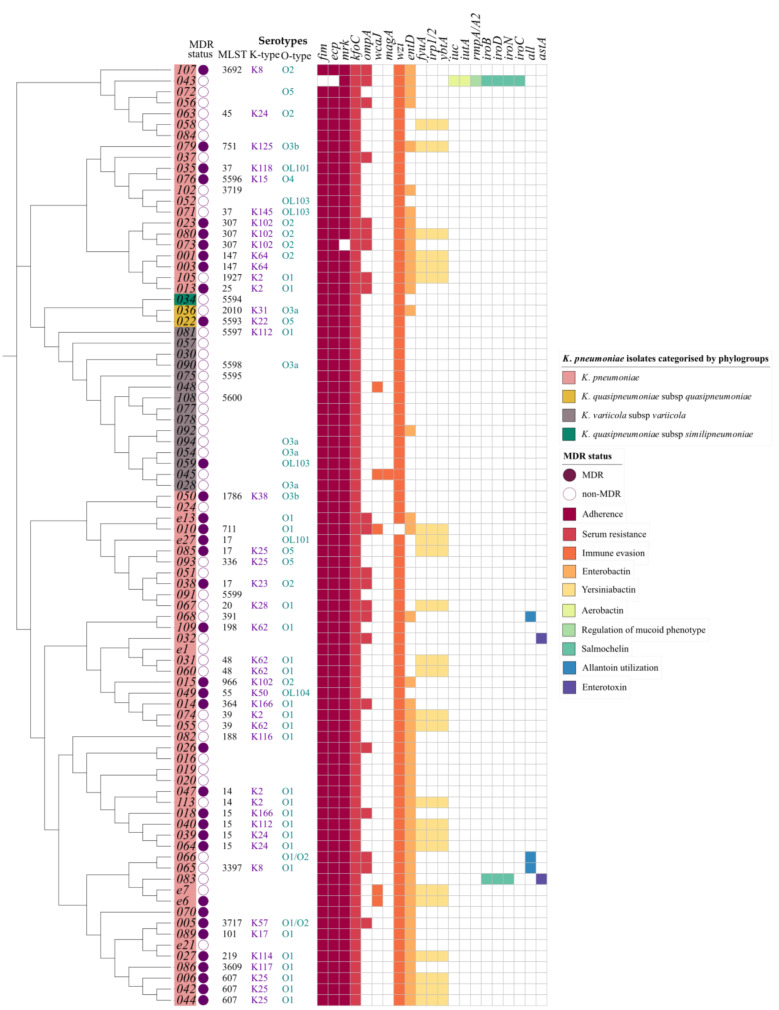
Figure 5.
A heatmap of the multidrug resistance status, ST, serotype, and virulence gene content of K. pneumoniae isolates (n = 89). The cladogram on the left with colored labels on the edge indicates the clustering of the isolates by phylogroups: K. pneumoniae (pink), K. quasipneumoniae subsp. similipneumoniae (blue), K. quasipneumoniae subsp. quasipneumoniae (gold), and K. variicola subsp. variicola (green). The circular symbols represent the multidrug resistance status of the isolates as either multidrug-resistant (purple) or non-multidrug-resistant (white). The serotypes assigned to the isolates are indicated as follows: multilocus sequence type (MLST) (black), capsule type (purple), and O type (teal). The virulence profile is represented as a gene present (color) or absent (white) for factors: adherence (purple), serum resistance (blue), immune evasion (pink), enterobactin (green), yersiniabactin, aerobactin, salmochelin, regulation of mucoid phenotype, enterotoxin and allantoin utilization; blank spaces indicates unassigned serotypes.
This study identified no evident clustering of capsule and lipopolysaccharide types based on geographical locations or clinical presentation. There were 10 different O-loci types identified (O1, O2, O3a, O3b, O4, O5, OL101, OL103, and OL104) in the 57 assigned isolates, whereby O1 (46%, 26/57) and O2 (19%, 11/57) were the most common and clinically significant. There were 24 different K-types in the 45 assigned isolates, and the most abundant were K2 (11%, 5/45), K25 (11%, 5/45), K102 (9%, 4/45), K62 (9%, 4/45), and K24 (7%, 3/45).
The ybt loci, identified in 24/89 isolates, encode for yersiniabactin siderophores found within conjugative transposons in the chromosome. Based on a new typing scheme [46] updated in the kleborate pipeline [40], this study identified four distinct ybt types: ybt14 found within ICEKp5, ybt15 in an ICEKp11, ybt16 in an ICEKp12, and ybt9 within an ICEKp3. In addition, the analysis revealed three isolates (Figure 5) with chromosomally encoded genes for allantoin utilization and two isolates (kkp012 and kkp045) (Figure 5) with magA and K2 capsule types linked to hypervirulence [47].
Several isolates were unique in having plasmid-encoded virulence genes. For example, a hypervirulent isolate (hvKP), kkp043, was identified bearing the repB_KLEB_VIR plasmid containing rmpA and rmpA2 genes, which regulate the expression of the mucoid phenotype, salmochelin (iroBCDN), and aerobactin (iucABCD, iutA). In addition, kkp083 also demonstrated an iroBDEN cluster and a heat-stable enterotoxin (astA) gene carried in an IncF(K)_1 plasmid, in contrast to the chromosomally-bound astA gene in kkp032. This study did not identify MDR hypervirulent isolates with AMR and hypervirulent genes [48]. Furthermore, the MDR isolates did not carry factors associated with hypervirulence, while the hypervirulent isolates were mostly antibiotic susceptible, i.e., they were non-MDR and ESBL-positive (Figure 5).
4. Discussion
This study characterized 89 isolates based on their clinical, geographic, genotypic, and phenotypic characteristics. It was noted that all four recognized phylogroups were represented, with Klebsiella pneumoniae subsp. pneumoniae isolates predominating, consistent with findings from other studies [49,50,51]. Some differences were observed in the characteristics of the phylogroups. For example, Klebsiella variicola subsp. variicola isolates are typically linked to bloodstream infections and UTIs [52,53]; however, those identified in this study were mainly associated with SSTIs (Figure 1) and were largely antibiotic susceptible (Figure 4). In previous studies, Klebsiella quasipneumoniae subsp. similipneumoniae isolates were linked to nosocomial infections such as UTIs [54,55]. Yet in this study, one isolate, kkp034, caused a community-acquired SSTI, potentially indicating a broader distribution of this phylogroup in Kenya than found in other countries, such as sewage in Brazil [56] and a turtle in China [57].
The lineages identified in this study were highly diverse, and most of them were local strains that have not been described in other countries. Globally disseminated high-risk lineages such as ST14, ST15, ST307, and ST607 were identified (Figure 3). These high-risk lineages are bacterial pathogens that easily acquire and disseminate antimicrobial resistance [58]. For example, ST14, ST15, and ST147 have been linked to the spread of carbapenemase resistance genes in many countries [59,60]. In this study, the two extensively drug resistant (XDR) ST147 strains carried a blaOXA-181 (kkp001) and a blaNDM gene (kkp003), respectively, while the MDR ST14/15 strains carried several ESBL genes. Notably, kkp018 (ST15) harbored a mobile colistin-resistant gene, mcr-8, and an AmpC beta-lactamase gene, blaDHA (Figure 4). ST307 and ST607 are emerging strains linked to ESBL infections (Long, et al., 2017), and they were observed in six MDR strains isolated from SSTIs and UTIs (Figure 1 and 5). ST17 is a regional strain that has been implicated in outbreaks in Kilifi [6], Mwanza [61], and Kilimanjaro [62]. This study identified three MDR and ESBL positive ST17 strains, isolated from a community-acquired SSTI, a nosocomial UTI, and a hospital environmental swab (Figure 1 and Figure 5). High-risk clones have also been linked to specific serotypes, e.g., ST607-K25, responsible for a nosocomial outbreak at a neonatal intensive care unit in a hospital in France [63]. Significantly, this study identified three MDR and ESBL-positive ST607-K25 clones associated with SSTIs (Figure 5).
The presence of these high-risk strains in Kenya indicates the significant clinical and public health threat they pose. However, we noticed that this threat was highest in Kisumu and Nairobi, the two largest cities in the country, where most of the global lineages (79%, 11/14) were identified. These cities are travel hubs with large referral hospitals serving patients from a wide geographical area locally and as well as global travelers.
The diverse patient population could explain the concentration of local, regional and global lineages, the great strain diversity, and novel alleles in isolates from the two counties compared to Kisii, Kilifi, and Kericho Counties.
Plasmids are the main vehicle for AMR gene transmission, and in this study, we found that a majority of the AMR genes were carried in plasmids, particularly IncFIB(K) and IncFII(pKP91) (Table 2), which are common among Enterobacterales. The predominant plasmid replicon types belonged to the diverse Incompatibility (Inc) family [64], whose host range is mainly limited to Enterobacterales [65] which are known to have large multi-replicon plasmids [66]. The multi-replicon IncF plasmids contain the FII, FIA, and/or FIB replicons (Table 2 and Table S2) which account for their high abundance (Figure 2). Unsurprisingly, antibiotic susceptible KP isolates (kkp012, kkp030, kkp102, and kkp0112) contained few or no plasmids and were ESBL negative and non-MDR. The MDR kkp070 was exceptional because although it possessed no plasmid replicons, it had several resistance genes integrated into a genetic island in the chromosome. These integration events were not uncommon, as plasmid-associated AMR genes were detected in the chromosome of several isolates: kkp005 and kkp006 carried a blaCTX-M-15 gene, kkp109 had a dfrA14 trimethoprim resistant gene, and kkp001 possessed several AMR genes (aac(6′)-Ib9, blaCTX-M-15, arr-2, and rmtF). The integration of AMR genes into the chromosome is alarming because the resistance is transferred clonally, becomes part of the core genome, and increases the spread and prevalence of non-susceptible K. pneumoniae lineages.
Although the col plasmid family was the second most dominant group, the majority of col-like plasmids did not contain any resistance or virulence genes, except for the abundant colE1 type, Col (pHAD28), which typically carries qnrS1 and other AMR genes [65] as observed in kkp0e21 (Table 2). In addition, col plasmids benefit K. pneumoniae because they produce bacteriocins lethal to rival bacteria. One of the other plasmid families identified was the pK1433 (kkp056) and is associated with the blaKPC2 gene [67] that provides a mechanism for carrying and spreading KPC-type genes. The isolates with plasmids carrying a blaCTX-M gene also carried blaOXA, blaSHV, or blaTEM genes, implying that these resistance genes are transferred together (e.g., ESBL-positive kkp081 and kkp059 isolates possessed blaCTX-M-15, blaSHV-134, and blaTEM181 genes). The most intriguing isolates were the ESBL-positive isolates (kkp015, kkp070, and kkp107) which only contained the blaSHV penicillinase gene. We hypothesize that the ESBL phenotypes in these isolates may have been due to alternative resistance mechanisms such as up-regulation of MDR efflux pumps. The expression of genes encoding MDR efflux pumps is noteworthy because their activity induces resistance of different antibiotic classes non-specifically and can cause phenotypic and genotypic discordance.
Examination of the virulence factors among the K. pneumoniae phylogroups highlighted one main difference. K. quasipneumoniae subsp. similipneumoniae did not demonstrate the Type VI protein secretion system found in the other phylogroups (Table S5); instead, it possessed the Type II secretion as previously noted [68]. Siderophores were the most significant virulence genes detected. The ubiquitous enterobactin scavenges iron from host cells; however, their activity is neutralized by human lipocalin-2 protein [69]. In response, K. pneumoniae uses the more virulent yersiniabactin to bind iron and other heavy metals such as copper to avoid metal toxicity and phagocytosis using reactive oxygen species [70]. According to Holt et al. (2015), acquiring yersiniabactin is usually the first step in accumulating more potent siderophores to make them more invasive. Yersiniabactin genes were found among the K. pneumoniae isolates causing various clinical infections and from diverse geographical locations. Most of them were MDR and ESBL positive, making their infections persistent and antibiotic-resistant and likely contributing to their dominance over the other phylogroups.
Classical hypervirulent K. pneumoniae (hvKP) isolates are generally antibiotic susceptible [71], and they are characterized by having rmpA/A2 and magA virulence factors as well as the K2 capsule type [60]. This study identified one potentially hypervirulent isolate (kkp043), which carried rmpA and rmpA2 genes. In addition, it also carried genes for aerobactin and salmochelin that were carried on a repB_KLEB_VIR plasmid (Table 2). A trade-off between virulence and antibiotic resistance was demonstrated in this potentially hypervirulent isolate (kkp043) as it was non-MDR and ESBL-negative and had only one other plasmid bearing the IncFIA(HI1) replicon, dfrA5, and sul1 genes (Table 2). Additional examples were the two isolates (kkp012 and kkp045) with magA genes which were ESBL-negative and non-MDR. Furthermore, non-MDR K2 strains possessed the ybt operon, which the MDR K2 strains lacked (Figure 5). Despite this, there is growing concern about the rise of MDR hvKP globally [46,72,73,74,75], which cause severe infections with few treatment options [76] emphasizing the significance of monitoring virulence characteristics of K. pneumoniae in Kenya.
The study had several limitations. First, the two sequencing technologies utilized had different strengths and weaknesses, which could introduce bias in the genomic analysis. The draft genomes from the long reads had fewer contigs (<= 12) (Table S1) and were more contiguous, but they had less depth/coverage, while the short reads produced less contiguous draft genomes with greater depth (>40) (Table S1). More contiguity enabled better plasmids reconstruction, while sufficient depth enabled better assignment of multilocus sequence types. Secondly, susceptibility tests were carried out on the Vitek2® platform using a limited number of antibiotics and were not verified using another phenotypic method. Nevertheless, there was good concordance between the phenotypic and genotypic results. Finally, in vivo tests were not conducted to confirm the virulence gene activity, so the virulence results are only predicted.
5. Conclusions
The findings of this study contribute in several ways to our understanding of the genotypic and phenotypic characteristics of Kenyan K. pneumoniae isolates. The multi-center approach provided more nationally relevant data, unlike prior studies with fewer isolates from single sites. These findings describe a K. pneumoniae population with diverse sequence types, highly abundant and diverse resistance, and virulence profiles. In addition, the presence of high-risk clones in the major cities, Nairobi and Kisumu, enhances their transmissibility within and outside the country. Further research should be conducted to correlate the genotypic findings of virulence with phenotypic data, and additional analysis should investigate the genotypic environment of the acquired antimicrobial resistance genes to determine their risk of spread.
Acknowledgments
The authors would like to acknowledge the Kenya Medical Research Institute and the United States Army Medical Research Directorate-Africa staff who carried out the patient sampling, biochemical and antimicrobial susceptibility tests, and the study participants at the participating hospitals. The authors also acknowledge the Walter Reed Army Institute of Research- Multidrug-Resistant Organism Repository and Surveillance Network staff who performed the Illumina sequencing and the Institut Pasteur teams for curating and maintaining the BIGSdb-Pasteur databases at http://bigsdb.pasteur.fr/ (accessed on 25 May 2021).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11050545/s1, Table S1: Genomic and epidemiological characteristics of K. pneumoniae isolates; Table S2: Plasmids replicons present in the K. pneumoniae isolates; Table S3: Phenotypic antimicrobial susceptibility test (AST) results of the K. pneumoniae isolates; Table S4: Antimicrobial resistance genes in the K. pneumoniae isolates; Table S5: Virulence genes identified in the K. pneumoniae isolates; Table S6: Fluoroquinolone mutations identified in the K. pneumoniae isolates.
Author Contributions
Conceptualization, A.M., C.K. (Cecilia Kyany’a), L.M.; methodology, A.M., C.K. (Cecilia Kyany’a), S.K, M.J.M.; visualization, A.M., C.K. (Cecilia Kyany’a), S.K.; writing original draft preparation, A.M., H.J.S., L.M.; supervision, C.K. (Caleb Kibet), J.K., L.M.; formal analysis, A.M.; investigation, A.M.; resources, L.M.; funding, L.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved on 19 April 2021 by the Kenya Medical Research Institute (KEMRI) Scientific and Ethics Review Unit (#2767 and #4107), the Walter Reed Army Institute of Research (WRAIR) Institutional Review Board (#2089), and the U.S. Army Medical Research and Materiel Command, Office of Research Protection, Human Research Protections Office (USAMRMC ORP HRPO) (Log#A-18129).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All genome assemblies are deposited in the NCBI GenBank database under BioProject PRJNA777842. Eight novel allelic profiles were assigned and deposited in the K. pneumoniae MLST database https://bigsdb.web.pasteur.fr/ (accessed on 25 May 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Disclaimer
The material for this publication has been reviewed by the Walter Reed Army Institute of Research, and there is no objection to its publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for the protection of human subjects as prescribed in AR 70-25.
Funding Statement
This research was funded by the Armed Forces Health Surveillance Division, Global Emerging Infections Surveillance Branch, grant number PROMIS ID 20160270153 FY17-20. The study funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gomez-Simmonds A., Uhlemann A.-C. Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella pneumoniae. J. Infect. Dis. 2017;215((Suppl. 1)):S18–S27. doi: 10.1093/infdis/jiw378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 3.Girometti N., Lewis R.E., Giannella M., Ambretti S., Bartoletti M., Tedeschi S., Tumietto F., Cristini F., Trapani F., Gaibani P., et al. Klebsiella pneumoniae Bloodstream Infection: Epidemiology and Impact of Inappropriate Empirical Therapy. Medicine. 2014;93:298–309. doi: 10.1097/MD.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO . National Action Plan on Prevention and Containment of Antimicrobial Resistance, 2017–2022. Regional Office for Africa; Brazzaville, Republic of Congo: 2020. [(accessed on 3 March 2020)]. Available online: https://www.afro.who.int/publications/national-action-plan-prevention-and-containment-antimicrobial-resistance-2017-2022. [Google Scholar]
- 5.Chen L., Mathema B., Chavda K.D., DeLeo F., Bonomo R.A., Kreiswirth B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henson S.P., Boinett C.J., Ellington M.J., Kagia N., Mwarumba S., Nyongesa S., Mturi N., Kariuki S., Scott J.A.G., Thomson N.R., et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int. J. Med. Microbiol. 2017;307:422–429. doi: 10.1016/j.ijmm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawat D., Nair D. Extended-spectrum ß-lactamases in gram negative bacteria. J. Glob. Infect. Dis. 2010;2:263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyres K.L., Wick R.R., Gorrie C., Jenney A., Follador R., Thomson N.R., Holt K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 10.Taitt C.R., Leski T., Erwin D.P., Odundo E.A., Kipkemoi N.C., Ndonye J.N., Kirera R.K., Ombogo A.N., Walson J.L., Pavlinac P.B., et al. Antimicrobial resistance of Klebsiella pneumoniae stool isolates circulating in Kenya. PLoS ONE. 2017;12:e0178880. doi: 10.1371/journal.pone.0178880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyany’a C., Musila L. Colistin Resistance Gene mcr-8 in a High-Risk Sequence Type 15 Klebsiella pneumoniae Isolate from Kenya. Microbiol. Resour. Announc. 2020;9:e00783-20. doi: 10.1128/MRA.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L., Revathi G., Bernabeu S., Nordmann P. Detection of NDM-1-Producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 2011;55:934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apondi O.E., Oduor O.C., Gye B.K., Kipkoech M.K. High Prevalence of Multi-Drug Resistant Klebsiella pneumoniae in a Tertiary Teaching Hospital in Western Kenya. Afr. J. Infect. Dis. 2016;10:89–95. doi: 10.21010/ajid.v10i2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decano A.G., Pettigrew K., Sabiiti W., Sloan D., Neema S., Bazira J., Kiiry J., Onyango H., Asiimwe B., Holden M.T.G. Pan-resistome characterization of uropathogenic Escherichia coli and Klebsiella pneumoniae strains circulating in Uganda and Kenya isolated from 2017–2018. Antibiotics. 2021;10:1547. doi: 10.3390/antibiotics10121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maina D., Revathi G., Whitelaw A.C. Molecular characterization of multidrug-resistant Klebsiella pneumoniae and Escherichia coli harbouring extended spectrum beta-lactamases and carbapenemases genes at a tertiary hospital, Kenya. Microbiol. Med. 2017:32. doi: 10.4081/mm.2017.7076. [DOI] [Google Scholar]
- 16.Bururia J.M., Kinyanjui P.N., Waiyaki P.G., Kariuki S.M. Resistance of Klebsiella Species Isolates From Two Institutions in Nairobi, Kenya, to Commonly Prescribed Antimicrobial Agents. East Cent. Afr. J. Pharm. Sci. 2007;10:22–26. doi: 10.4314/ecajps.v10i1.9757. [DOI] [Google Scholar]
- 17.Maina D., Revathi G., Kariuki S., Ozwara H. Genotypes and cephalosporin susceptibility in extended-spectrum beta-lactamase producing Enterobacteriaceae in the community. J. Infect. Dev. Ctries. 2012;6:470–477. doi: 10.3855/jidc.1456. [DOI] [PubMed] [Google Scholar]
- 18.Maina J., Ndung’U P., Muigai A., Kiiru J. Antimicrobial resistance profiles and genetic basis of resistance among non-fastidious Gram-negative bacteria recovered from ready-to-eat foods in Kibera informal housing in Nairobi, Kenya. Access Microbiol. 2021;3:000236. doi: 10.1099/acmi.0.000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musila L., Kyany’a C., Maybank R., Stam J., Oundo V., Sang W. Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS ONE. 2021;16:e0246937. doi: 10.1371/journal.pone.0246937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcántar-Curiel M.D., Ledezma-Escalante C.A., Jarillo-Quijada M.D., Gayosso-Vázquez C., Morfín-Otero R., Rodríguez-Noriega E., Cedillo-Ramírez M.L., Santos-Preciado J.I., Giron J.A. Association of Antibiotic Resistance, Cell Adherence, and Biofilm Production with the Endemicity of Nosocomial Klebsiella pneumoniae. BioMed Res. Int. 2018;2018:1–9. doi: 10.1155/2018/7012958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbosa V.A.A., Lery L.M.S. Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genom. 2019;20:506. doi: 10.1186/s12864-019-5885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh D.T.N., Kim A.-Y., Kim Y.-R. Identification of Pathogenic Factors in Klebsiella pneumoniae Using Impedimetric Sensor Equipped with Biomimetic Surfaces. Sensors. 2017;17:1406. doi: 10.3390/s17061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni R.T., Onishi M., Mizusawa M., Kitagawa R., Kishino T., Matsubara F., Tsuchiya T., Kuroda T., Ogawa W. The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci. Rep. 2020;10:10876. doi: 10.1038/s41598-020-67820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellington M.J., Ekelund O., Aarestrup F.M., Canton R., Doumith M., Giske C., Grundman H., Hasman H., Holden M.T.G., Hopkins K.L., et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017;23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Sui W., Zhou H., Du P., Wang L., Qin T., Wang M., Ren H., Huang Y., Hou J., Chen C., et al. Whole genome sequence revealed the fine transmission map of carbapenem-resistant Klebsiella pneumonia isolates within a nosocomial outbreak. Antimicrob. Resist. Infect. Control. 2018;7:70. doi: 10.1186/s13756-018-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein M.P. M100-Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 28.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S., Krueger F., Segonds-Pichon A., Biggins L., Krueger C., Wingett S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics; Cambridge, UK: 2010. [Google Scholar]
- 30.Seemann T. Shovill: Faster SPAdes Assembly of Illumina Reads. 2018. [(accessed on 30 June 2021)]. version 1.1.0. Available online: https://github.com/tseemann/shovill.
- 31.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 33.Medaka. Oxford Nanopore Technologies; Oxford, UK: 2021. version 1.3.2. [Google Scholar]
- 34.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. ABRicate. 2022. [(accessed on 30 June 2021)]. version 1.0.1. Available online: https://github.com/tseemann/abricate.
- 36.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carattoli A., Zankari E., Garcìa-Fernandez A., Larsen M., Lund O., Voldby Villa L., Møller Aarestrup F., Hasman H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. mlst. 2021. [(accessed on 30 June 2021)]. version 2.19. Available online: https://github.com/tseemann/mlst.
- 40.Lam M.M.C., Wick R.R., Watts S.C., Cerdeira L.T., Wyres K.L., Holt K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021;12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick R.R., Heinz E., Holt K.E., Wyres K.L. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 2018;56:e00197-18. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.flextable package-RDocumentation n.d. [(accessed on 20 November 2021)]. Available online: https://www.rdocumentation.org/packages/flextable/versions/0.7.0.
- 44.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu P., Li P., Jiang X., Bi D., Xie Y., Tai C., Deng Z., Rajakumar K., Ou H.-Y. Complete Genome Sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a Multidrug-Resistant Strain Isolated from Human Sputum. J. Bacteriol. 2012;194:1841–1842. doi: 10.1128/jb.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam M.M.C., Wyres K.L., Judd L.M., Wick R.R., Jenney A., Brisse S., Holt K.E. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10:77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang C.-T., Chuang Y.-P., Shun C.-T., Chang S.-C., Wang J.-T. A Novel Virulence Gene in Klebsiella pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J. Exp. Med. 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hennequin C., Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. 2016;35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 49.Heinz E., Brindle R., Morgan-McCalla A., Peters K., Thomson N.R. Caribbean multi-centre study of Klebsiella pneumoniae: Whole-genome sequencing, antimicrobial resistance and virulence factors. Microb. Genom. 2019;5:12. doi: 10.1099/mgen.0.000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D., Jenney A., Connor T.R., Hsu L.Y., Severin J., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Musicha P., Msefula C., Mather A.E., Chaguza C., Cain A., Peno C., Kallonen T., Khonga M., Denis B., Gray K.J., et al. Genomic analysis of Klebsiella pneumoniae isolates from Malawi reveals acquisition of multiple ESBL determinants across diverse lineages. J. Antimicrob. Chemother. 2019;74:1223–1232. doi: 10.1093/jac/dkz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imai K., Ishibashi N., Kodana M., Tarumoto N., Sakai J., Kawamura T., Takeuchi S., Taji Y., Ebihara Y., Ikebuchi K., et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: A comparative study, Japan, 2014–2017. BMC Infect. Dis. 2019;19:946. doi: 10.1186/s12879-019-4498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maatallah M., Vading M., Kabir M.H., Bakhrouf A., Kalin M., Nauclér P., Brisse S., Giske C.G. Klebsiella variicola is a frequent cause of bloodstream infection in the stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS ONE. 2014;9:e113539. doi: 10.1371/journal.pone.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elliott A.G., Ganesamoorthy D., Coin L., Cooper M.A., Cao M.D. Complete Genome Sequence of Klebsiella quasipneumoniae subsp. similipneumoniae Strain ATCC 700603. Genome Announc. 2016;4:e00438-16. doi: 10.1128/genomea.00438-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicolás M.F., Ramos P.I.P., De Carvalho F.M., Camargo D.R.A., Alves C.D.F.M., De Morais G.L., Almeida L., Souza R.C., Ciapina L.P., Vicente A.C.P., et al. Comparative Genomic Analysis of a Clinical Isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 Beta-Lactamases Producer Harboring Two Drug-Resistance Plasmids from Southeast Brazil. Front. Microbiol. 2018;9:220. doi: 10.3389/fmicb.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furlan J.P.R., Gallo I.F.L., De Campos T.A., Stehling E.G. Genomic Characterization of a Multidrug-Resistant and Hypermucoviscous/Hypervirulent Klebsiella quasipneumoniae subsp. similipneumoniae ST4417 Isolated from a Sewage Treatment Plant. Microb. Drug Resist. 2020;26:1321–1325. doi: 10.1089/mdr.2019.0417. [DOI] [PubMed] [Google Scholar]
- 57.Li C.F., Tang H.L., Chiou C.S., Tung K.C., Lu M.C., Lai Y.C. Draft genome sequence of CTX-M-type β-lactamase-producing Klebsiella quasipneumoniae subsp. similipneumoniae isolated from a Box turtle. J. Glob. Antimicrob. Resist. 2018;12:235–236. doi: 10.1016/j.jgar.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 58.De Lagarde M., Vanier G., Arsenault J., Fairbrother J.M. High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population. Antibiotics. 2021;10:244. doi: 10.3390/antibiotics10030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giske C.G., Fröding I., Hasan C.M., Turlej-Rogacka A., Toleman M., Livermore D., Woodford N., Walsh T.R. Diverse Sequence Types of Klebsiella pneumoniae Contribute to the Dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 2012;56:2735–2738. doi: 10.1128/AAC.06142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee C.-R., Lee J.H., Park K.S., Jeon J.H., Kim Y.B., Cha C.-J., Jeong B.C., Lee S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mshana S.E., Hain T., Domann E., Lyamuya E.F., Chakraborty T., Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 2013;13:466. doi: 10.1186/1471-2334-13-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonda T., Kumburu H., van Zwetselaar M., Alifrangis M., Mmbaga B.T., Lund O., Kibiki G.S., Aarestrup F.M. Molecular epidemiology of virulence and antimicrobial resistance determinants in Klebsiella pneumoniae from hospitalised patients in Kilimanjaro, Tanzania. Eur. J. Clin. Microbiol. 2018;37:1901–1914. doi: 10.1007/s10096-018-3324-5. [DOI] [PubMed] [Google Scholar]
- 63.Peltier F., Choquet M., Decroix V., Adjidé C.C., Castelain S., Guiheneuf R., Pluquet E. Characterization of a multidrug-resistant Klebsiella pneumoniae ST607-K25 clone responsible for a nosocomial outbreak in a neonatal intensive care unit. J. Med. Microbiol. 2019;68:67–76. doi: 10.1099/jmm.0.000884. [DOI] [PubMed] [Google Scholar]
- 64.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 66.Villa L., García-Fernández A., Fortini D., Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 67.Papagiannitsis C.C., Miriagou V., Giakkoupi P., Tzouvelekis L.S., Vatopoulos A.C. Characterization of pKP1433, a novel KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 340. Antimicrob. Agents Chemother. 2013;57:3427–3429. doi: 10.1128/AAC.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perlaza-Jiménez L., Wu Q., Torres V.V.L., Zhang X., Li J., Rocker A., Lithgow T., Zhou T., Vijaykrishna D. Forensic genomics of a novel Klebsiella quasipneumoniae type from a neonatal intensive care unit in China reveals patterns of colonization, evolution and epidemiology. Microb. Genom. 2020;6:e000433. doi: 10.1099/mgen.0.000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bachman M.A., Lenio S., Schmidt L., Oyler J.E., Weiser J.N. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio. 2012;3:e00224-11. doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaturvedi K.S., Hung C.S., Crowley J.R., Stapleton A.E., Henderson J.P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 2012;8:731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Surgers L., Boyd A., Girard P.M., Arlet G., Decré D. ESBL-Producing Strain of Hypervirulent Klebsiella pneumoniae K2, France. Emerg. Infect. Dis. 2016;22:1687–1688. doi: 10.3201/eid2209.160681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banerjee T., Wangkheimayum J., Sharma S., Kumar A., Bhattacharjee A. Extensively Drug-Resistant Hypervirulent Klebsiella pneumoniae from a Series of Neonatal Sepsis in a Tertiary Care Hospital, India. Front. Med. 2021;8:186. doi: 10.3389/fmed.2021.645955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cejas D., Elena A., Nuñez D.G., Platero P.S., De Paulis A., Magariños F., Alfonso C., Berger M.A., Fernández-Canigia L., Gutkind G., et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019;18:238–242. doi: 10.1016/j.jgar.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Turton J., Davies F., Turton J., Perry C., Payne Z., Pike R. Hybrid Resistance and Virulence Plasmids in “High-Risk” Clones of Klebsiella pneumoniae, Including Those Carrying blaNDM-5. Microorganisms. 2019;7:326. doi: 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan Y., Li Y., Wang G., Li C., Chang Y.-F., Chen W., Nian S., Mao Y., Zhang J., Zhong F., et al. blaNDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob. Resist. Infect. Control. 2019;8:140. doi: 10.1186/s13756-019-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam M., Wyres K.L., Wick R.R., Judd L., Fostervold A., Holt K.E., Löhr I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019;74:1218–1222. doi: 10.1093/jac/dkz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All genome assemblies are deposited in the NCBI GenBank database under BioProject PRJNA777842. Eight novel allelic profiles were assigned and deposited in the K. pneumoniae MLST database https://bigsdb.web.pasteur.fr/ (accessed on 25 May 2021).