Abstract
Autotrophic ammonia oxidizer (AAO) populations in soils from native, tilled, and successional treatments at the Kellogg Biological Station Long-Term Ecological Research site in southwestern Michigan were compared to assess effects of disturbance on these bacteria. N fertilization effects on AAO populations were also evaluated with soils from fertilized microplots within the successional treatments. Population structures were characterized by PCR amplification of microbial community DNA with group-specific 16S rRNA gene (rDNA) primers, cloning of PCR products and clone hybridizations with group-specific probes, phylogenetic analysis of partial 16S rDNA sequences, and denaturing gradient gel electrophoresis (DGGE) analysis. Population sizes were estimated by using most-probable-number (MPN) media containing varied concentrations of ammonium sulfate. Tilled soils contained higher numbers than did native soils of culturable AAOs that were less sensitive to different ammonium concentrations in MPN media. Compared to sequences from native soils, partial 16S rDNA sequences from tilled soils were less diverse and grouped exclusively within Nitrosospira cluster 3. Native soils yielded sequences representing three different AAO clusters. Probes for Nitrosospira cluster 3 hybridized with DGGE blots from tilled and fertilized successional soils but not with blots from native or unfertilized successional soils. Hybridization results thus suggested a positive association between the Nitrosospira cluster 3 subgroup and soils amended with inorganic N. DGGE patterns for soils sampled from replicated plots of each treatment were nearly identical for tilled and native soils in both sampling years, indicating spatial and temporal reproducibility based on treatment.
Nitrification, the microbial oxidation of ammonium to nitrate, can lead to significant nitrogen (N) losses from ecosystems by producing potentially mobile forms of N. In most systems, chemolithotrophic bacteria contribute more to nitrification than do heterotrophic microorganisms (8), which do not use reduced N compounds as electron donors (36). Although autotrophic nitrification is carried out in two steps by two distinct groups of bacteria, the ammonia oxidizers and nitrite oxidizers, the former is responsible for the rate-determining first step (16). Autotrophic ammonia oxidizers (AAOs) thus play a key role in determining whether systems retain or lose N. All known terrestrial AAOs fall within a monophyletic assemblage of the β subdivision of the class Proteobacteria (β-proteobacteria) and are represented by the genera Nitrosomonas and Nitrosospira (6), which comprise at least seven phylogenetically distinct clusters (27). The close phylogenetic relationship among β-proteobacterial AAOs (33) contrasts with the diverse phylogeny of microorganisms known to carry out other N cycle processes such as ammonification and denitrification.
Nitrification rates vary widely in soils and are thought to be controlled principally by ammonium concentration, temperature, moisture, and oxygen (18). Little is known about how AAO population structure affects nitrification or how environmental factors affect AAO population structure. Nitrate concentrations, net nitrification, and most-probable-number (MPN) estimates of AAOs are generally lower in undisturbed soils than in agricultural soils (24), which has led to hypotheses that grassland and forest soils suppress AAO populations (17). Reported estimates of soil AAO numbers based on standard MPN methods may be misleading, however, because AAO outgrowth depends on ammonium concentrations in MPN media (2, 30). Despite low net nitrification, significant gross nitrification has also been measured in some forest (25) and grassland (35) soils, indicating that nitrifier populations in these soils are extant but poorly characterized. Changes in AAO populations that result from agricultural disturbance, therefore, may contribute to the high net nitrification rates observed in agricultural soils.
N fertilization and tillage constitute two key components of agricultural disturbance. Studying the effects of N fertilization and tillage on native AAO populations supports ecological approaches to reducing agricultural N losses, which can be as high as 60% of the fertilizer N applied (19). The objective of our study was to compare AAO populations in soils with different tillage and N fertilization histories at the long-term ecological research (LTER) site at the Kellogg Biological Station (KBS) near Kalamazoo, Mich. (20). The combination of N fertilization and tillage, which reduces soil carbon through organic-matter oxidation (15), may make energy substrate availability more favorable for autotrophic nitrifiers than for heterotrophs (36). We therefore expected soils from native and tilled plots to provide different habitats for AAOs and to yield significant differences in AAO population size and structure. Soils from adjacent plots undergoing succession following long-term tillage were also analyzed to evaluate effects of tillage cessation on AAO populations.
We compared AAO population structures in LTER soils by using PCR amplification of microbial community DNA with primers specific for 16S rRNA genes (rDNA) of β-AAOs and close relatives (13). Clone libraries of PCR products from each soil treatment were constructed for partial sequencing and phylogenetic analysis and were used in hybridization tests with group-specific probes (27). We also compared AAO populations on the basis of their denaturing gradient gel electrophoresis (DGGE) patterns following PCR amplification with the primers of Kowalchuk et al. (10). We estimated AAO population sizes by using MPN media (24) containing various concentrations of (NH4)2SO4 to address possible differences in population sensitivity to ammonium (30). Previous 16S rDNA studies of indigenous soil AAO populations have compared acidic and neutral agricultural soils (26, 27), seaward and inland coastal dune sands (10), wetland soils (11), and agricultural soils with and without amendments of pig slurry (5). This study represents the first comparison of AAO populations in native and tilled soils. To discern relationships between soil disturbance regimens and AAO populations, we interpreted our molecular and microbiological results in the light of soil management histories and field data from the KBS LTER Web site (7a, 20).
MATERIALS AND METHODS
Soils, treatments, and soil properties.
Soils were sampled from plots at the National Science Foundation KBS LTER site near Kalamazoo, Mich. (20). This site was established to study ecological interactions affecting agricultural productivity, nutrient availability, and biotic diversity in ecosystems representative of the upper midwestern United States. Soils at this site are classified as Typic Hapludalfs (33a) belonging to the Kalamazoo and Oshtemo soil series (fine, loamy, mixed, mesic).
Two replicate plots from conventionally tilled (CT) and never-tilled, native (NTS) treatments were sampled in July 1994 and August 1995. Two historically successional plots (HTS), as well as their internal N-fertilized (HTS-N) microplots (5 by 5 m), were sampled in August 1995. The CT, NTS, and HTS treatments, which are respectively designated treatments 1, 8, and 7, are more fully described in the LTER Web pages (7a). Main treatment plots are 100 m2 in area. Approximate distances between sampled plots within treatments were 900, 500, and 100 m, for the CT, HTS, and NTS treatments, respectively. CT and HTS plots were established in 1989 on soil that had been in a small-grain–soybean–corn rotation for approximately 100 years. Starting in 1989, the HTS plots were allowed to become revegetated with extant successional flora while the CT plots were maintained in crop rotations. CT plots supported corn and soybeans in alternating years from 1989 to 1994, with wheat introduced as a third rotation crop in 1995. Corn was planted in 1993, soybeans were planted in 1994, and wheat was planted in 1995. These plots were conventionally tilled (annual moldboard plowing, disking, and cultivation) and treated with prescribed applications of herbicides and fertilizers. Ammonium nitrate was broadcast in a single application at rates of 124 kg of N ha−1 for the corn crop and 84 kg of N ha−1 for wheat. HTS-N microplots were fertilized with 125 kg of N ha−1 broadcast in July of each year. Native plots (NTS) were established on adjacent areas of grass vegetation that had been maintained by annual mowing following clearing of the native deciduous forest in 1956.
Soils were sampled to a 10-cm depth with a 2.5-cm corer (14 g of fresh soil per core). For each plot, samples were composited from 20 soil cores. All samples were stored at 4°C until they were analyzed in the laboratory. Gravel and other debris were removed by hand, and soils were mixed inside plastic bags by shaking and kneading. Soil water contents were determined from replicate subsamples dried at 110°C for 48 h. All results are based on soil dry weights.
Total C, microbial biomass C, and direct microscopic counts were determined on composited samples from each replicate main plot in 1994 and 1995 as part of the KBS LTER data collection program. Carbon analysis was performed on oven-dried, ground samples in a Carlo Erba NA 1500 series 2 nitrogen-carbon analyzer (Fisons Instruments, Beverly, Mass.). Microbial biomass C content was measured by the chloroform-fumigation-incubation method (7). Direct microscopic counts were made by staining soil smears with 5-(4,6-dichlorotriazin-2-yl)aminofluorescein (Sigma Chemical Co., St. Louis, Mo.) and obtaining random digitized images of the smears under epifluorescence microscopy (1) with a charge-coupled device camera (Princeton Instruments, Trenton, N.J.). Images were transferred to a Power Macintosh 7100/66 and displayed for counting by using IP Lab Spectrum software (Signal Analytics Corp., Vienna, Va.). Relevant field level data on soil C, N, and microbial biomass from the KBS LTER Web site (7a) are compiled in Table 1.
TABLE 1.
KBS LTER data on soil C, N, and microbial biomass for CT, HTS, and NTS treatmentsa
| Treat-ment | Total C (%)b | Microbial-biomass C (μg g−1)c | Direct microscopic counts (109 cells g of soil−1)
|
Soil pH | NO3− N (μg g of soil−1)d
|
NH4+ N (μg g of soil−1)d
|
Net nitrification in field (μg g−1 day−1)e | % of available NH4+ N converted in fieldf | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| July 1994 | August 1995 | 1994 | 1995 | 1994 | 1995 | ||||||
| CT | 0.81 ± 0.17a | 186 ± 81a | 0.9 ± 0.04a | 1.9 ± 0.15b | 6.8 | 4.2 ± 2.3a | 1.9 ± 1.17a | 2.7 ± 1.1a | 2.6 ± 1.5a | 0.19 ± 0.15 | 68 |
| HTS | 1.1 ± 0.21a | 230 ± 118a | 1.6 ± 0.03b | 2.5 ± 0.12bc | NDg | 0.7 ± 0.8a | 0.6 ± 0.42a | 4.1 ± 1.3a | 4.5 ± 2.1a | 0.12 ± 0.15 | 30 |
| NTS | 1.97 ± 0.17b | 415 ± 114b | 1.9 ± 0.05b | 3.0 ± 0.13c | 5.4 | 0.7 ± 1.0a | 0.5 ± 0.35a | 10.7 ± 3.3b | 7.7 ± 2.1a | 0.17 ± 0.17 | 21 |
Means and standard deviations from all replicate plots for each treatment are shown. There were six replicate plots for CT and HTS and four replicate plots for NTS. Values followed by different letters indicate significant differences of at least P < 0.05. These data are from the KBS LTER Web site (7a).
Measured April and August 1994.
Measured August 1995.
Means of monthly data from May through November 1994 and April through November 1995.
Means of monthly measurements of net nitrification over 21 days from May through October 1994 and May through September 1995.
Percentage based on the net increase in NO3− N after 21 days divided by the sum of the initial NH4+ N pool plus the amount of inorganic N mineralized during incubation. Inorganic-N measurements before and after incubation were used to calculate percentages.
ND, not determined.
MPN enumeration.
MPN counts of AAOs were determined with different ammonium concentrations by using a microtechnique procedure (21). Soil samples were dispersed for 1 min in a Waring Blendor with 100 mM sodium phosphate buffer (pH 7.0), and coarse particles were allowed to settle for 1 min. Soil suspensions were then serially diluted in three separate 96-well microtiter plates containing ATCC Medium 929 (American Type Culture Collection, Manassas, Va.) with 0.05, 0.5, and 10 g of (NH4)2SO4 liter−1. Confidence intervals (P < 0.05) were determined on the basis of twofold dilutions and eight wells per dilution (37). Inoculated plates were double sealed with Parafilm, wrapped in humidified plastic bags, and incubated at 25°C in the dark for 2 months before testing of aliquots from the wells for nitrate and nitrite. Nitrite was analyzed by using modified Griess-Ilosvay reagents (24). Nitrate was analyzed with Szechrome NB reagent (Polysciences, Inc., Warrington, Pa.) prepared in accordance with the manufacturer’s instructions. Fifty microliters of MPN medium was added to the well of a plate and mixed with 250 μl of the Szechrome reagent. After 15 min, the color of the test mixture was compared to that of nitrate standards ranging from 0.1 to 100 g of NO3− N liter−1. Negative controls consisted of uninoculated MPN media in microtiter plates held for 8 weeks.
DNA extraction from soils.
Microbial community DNA was extracted (38) from 5-g (fresh weight) samples of soil. DNA was separated from soil humic substances by subjecting the crude extract to electrophoresis in 0.8% (wt/vol) low-melting-point agarose (Gibco BRL, Gaithersburg, Md.). The DNA bands were excised from the gels, and the agarose was dissolved with agarase (Boehringer Mannheim Corp., Indianapolis, Ind.). The DNA mixture was concentrated and washed twice with distilled water in Centricon-100 ultracentrifugal filters (Amicon, Inc., Beverly, Mass.). DNA concentrations and purities were determined at 260, 280, and 230 nm with a Hewlett-Packard 8452A spectrophotometer (Hewlett-Packard Co., Sunnyvale, Calif.).
PCR with 16S rDNA primers.
Purified soil DNA was amplified by using the 16S rDNA primers of McCaig et al. (13). The β-AMOf and β-AMOr primers correspond to positions 141 to 161 and 1301 to 1320 of Escherichia coli rDNA, respectively (12). PCR was carried out in 50-μl reaction volumes with a Perkin-Elmer GeneAmp PCR System 9600 (Perkin-Elmer, Foster City, Calif.) by using a hot-start procedure to reduce nonspecific amplification. Each reaction mixture contained 10 ng of template DNA, 4 pmol of each primer, and 1 U of Taq polymerase (Perkin-Elmer) in final concentrations of 2.5 mM MgCl2 and 0.12 mM deoxyribonucleoside triphosphates in PCR buffer. Positive controls contained 10 ng of Nitrosomonas europaea genomic DNA as a template. Negative controls contained dilutions of processed agarose gel slices. PCR conditions were as follows: initial denaturation at 94°C for 2 min; 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min. Four to five duplicate reaction mixtures were amplified at a time, and the PCR products were pooled to reduce potential bias.
Cloning, sequencing, and phylogenetic analysis of PCR products.
Pooled mixtures of PCR products were concentrated in a Microcon-100 unit (Amicon, Inc., Beverly, Mass.) and isolated by agarose gel electrophoresis. Gel slices containing the PCR products were excised, and DNA was purified by using QIAEX II reagents (Qiagen, Inc., Chatsworth, Calif.) before washing and concentration in Microcon-100 units. PCR products were ligated into the pGEM-T vector by using T4 ligase (Promega, Corp., Madison, Wis.). Epicurean coli XL1-Blue supercompetent cells (Stratagene, Inc., La Jolla, Calif.) were prepared and transformed with the ligation mixtures in accordance with the manufacturer’s directions. Plasmid DNA preparations were obtained from clones by using the Wizard Minipreps kit (Promega Corp.). Since the primers of McCaig et al. (13) can generate amplification products from β-proteobacteria other than AAOs, clones were analyzed by T tracking (26). In the T-track screening, cloned inserts were partially sequenced with the 536r 16S rDNA sequencing primer, [35S]dATP, and dideoxythymidine terminators to generate single-lane band patterns (T tracks) in sequencing gels for comparison with patterns from AAOs in the database (27). However, T tracking did not screen out nonspecific inserts from a cluster of sequences which group basal to the β-subgroup ammonia oxidizer assemblage (β-deep group of sequences; see Table 4). Similar sequences have also been retrieved from soils sampled in Scotland (26, 27), sediments (14), and freshwater samples (26). Such sequences fall outside the β-AAO radiation (unpublished observations), so they were not included in the phylogenetic analysis.
TABLE 4.
Numbers of probe-positive clones in libraries from soil community DNA in 1994 and 1995
| Yr and treatment | Total no. of clones tested | % of clones hybridizing with any AAO probe | % of clones hybridizing with probe for deep-branched non-AAO clade |
|---|---|---|---|
| 1994 | |||
| CT | 281 | 75 | 0 |
| NTS | 672 | 7 | 14 |
| 1995 | |||
| CT | 994 | 8 | 0.6 |
| NTS | 962 | 0.5 | 6.0 |
| HTS | 826 | 1 | 0.5 |
| HTS-N | 882 | 5 | 1.4 |
Sequencing and phylogenetic analysis were performed on 25 clones from the 1994 samples. Clones from tilled and native soils were designated KZOO_H and KZOO_D, respectively. Sequences (approximately 320 bases corresponding to E. coli positions 180 to 500) were obtained manually and aligned with other available 16S rDNA AAO sequences (27). Distance matrix analysis was performed by using the Kimura correction (9) and neighbor joining (22) with PHYLIP 3.5 (3). The ARB sequence management system (29) was used to generate bootstrap support values from 100 parsimony–maximum-likelihood analyses (32) of the aligned sequences.
Colony blots and probe hybridizations.
For both sampling years, several hundred clones were hybridized with oligonucleotide probes designed to detect 16S rDNA sequences from specific clades within the β-subgroup ammonia oxidizers (27). In addition, the probe β-Deep_446 (CTAATGACGGTACTAC) was designed to specifically hybridize to the group of deep-branched β-proteobacterium-like sequences that clustered basal to the ammonia oxidizer radiation. Cloned DNA from fresh colonies was transferred (23) and cross-linked to Hybond N+ membranes (Amersham Corp., Arlington Heights, Ill.) by using a Stratalinker (Stratagene, Inc.). Clones containing PCR inserts with known sequences were included in the clone libraries during membrane preparation to serve as positive and negative hybridization controls. Group-specific oligonucleotide probes (described in reference 27) were used in colony blot hybridizations.
Probes were end labeled with [γ-32P]ATP (Du Pont NEN Biotechnology Division, Wilmington, Del.) by using T4 polynucleotide kinase (Stratagene, Inc.). The 32P-labeled probes were added to hybridization solution (23) to obtain activities of approximately 1 μCi ml−1. Membranes containing DNA from clone libraries were prehybridized in QuikHyb solution (Stratagene, Inc.) for 1 to 2 h in glass hybridization tubes in a Techne hybridization oven (Techne, Inc., Princeton, N.J.). Membranes were hybridized with 32P-labeled probes and washed under the conditions described in reference 27 and placed in film cassettes for exposure to autoradiogram film (Kodak, Inc., Rochester, N.Y.). The probe β-Deep_446 was hybridized and washed at 42°C.
DGGE.
Soil DNA extracts from both sampling years were subjected to PCR-DGGE analysis. Template concentrations of 20 ng per PCR amplification were used with the primers CTO178f-GC and CTO637r (10). This primer pair generates a fragment containing 459 bp of rDNA sequence and a 38-bp GC clamp. These primers are more specific than those used to generate clone libraries and do not amplify sequences clustering within the β-Deep group of 16S rDNA from other known β-proteobacteria (10). The PCR amplifications were carried out with the Expand High Fidelity PCR System (Boehringer Mannheim, Mannheim, Germany) in 50-μl volumes. DGGE was carried out by the methods of Kowalchuk et al. (10) in 8% polyacrylamide gels (38 to 50% denaturant) with a D-Gene system (Bio-Rad Laboratories, Hercules, Calif.). DGGE gels were run in 0.5× TAE (23) and stained following electrophoresis with ethidium bromide to visualize DNA bands. Blots were prepared from the gels by electrophoretic transfer (10) for hybridization with the following 32P-labeled probes: β-AO233, specific for all β-subgroup AAOs; Nsp436, specific for the genus Nitrosospira; and NspCL3_454, specific for a Nitrosospira subgroup provisionally named Nitrosospira cluster 3 (26).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the cloned sequences used are U56606 to U56633.
RESULTS
MPN counts.
MPN counts from native soils were significantly lower (P < 0.05) than MPN counts from tilled, unfertilized successional, and fertilized successional soils at ammonium concentrations of 10 and 2,000 ppm (Table 2). With ammonium at 100 ppm, MPN counts from native soils were lower but not significantly different from the MPN counts of other soils. For all soils, MPNs were lowest in the 2,000-ppm ammonium medium but this difference was significant only for the native soils. Hence, culturable AAOs in native soils appeared to be more sensitive to 2,000-ppm ammonium than did AAOs in the other soils. Six years of revegetation in the successional plots did not result in a significant reduction in culturable AAO populations. MPN counts were higher in fertilized than in unfertilized successional soils, but the differences were not significant (Table 3).
TABLE 2.
Mean MPN estimatesa
| Treatment | 10 ppm | 100 ppm | 2,000 ppm |
|---|---|---|---|
| CT | 130 (74–212)a | 93 (52–163)a | 63 (27–130)a |
| NTS | 13 (7–21)b | 46 (20–96)ab | 0.19 (0.11–0.31)c |
| HTS | 60 (28–121)a | 170 (60–408)a | 53 (25–106)a |
| HTS-N | 140 (50–310)a | 270 (74–668)a | 71 (36–135)a |
Mean numbers (103) of AAOs per gram of dry soil in ATCC Medium 929 containing 0.05, 0.5, and 10 g of (NH4)2SO4 liter−1, which corresponds to NH4+ N at 10, 100, and 2,000 ppm, respectively, are shown. Values in parentheses represent MPN confidence limits (P = 0.05) for twofold dilutions and eight replicates per dilution. Values followed by different letters are significantly different (P < 0.05) between treatments.
TABLE 3.
Mean DNA yields from replicate plots of tilled, native, and successional soils
| Treatment | Mean DNA yield (μg of g soil−1)a ± SD
|
|
|---|---|---|
| 1994 | 1995 | |
| CT | 2.9 ± 0.1a | 12.8 ± 3.8b |
| HTS | NDb | 13.6 ± 2.9b |
| NTS | 13.0 ± 2.8b | 15.7 ± 3.7b |
Values followed by different letters are significantly different (P < 0.05) between treatments. DNA concentrations were calculated from the spectrophotometric A260. Values include Standard deviations based on triplicate subsamples from two replicate plots.
ND, not determined.
Comparative DNA yields from soils.
Mean DNA yields (micrograms per gram of soil) from native soils were significantly higher (P < 0.05) than DNA yields from tilled soils in 1994, when tilled plots were in soybeans (Table 3). No significant differences were observed between DNA yields from tilled, native, and unfertilized successional soils in 1995, when tilled plots were in wheat. Respective ranges of spectrophotometric ratios of absorbance at 260 and 280 nm and 260 and 230 nm were slightly higher for 1994 samples (1.9 ± 0.04 and 2.4 ± 0.05) than for 1995 samples (1.6 ± 0.09 and 1.4 ± 0.12). Both sets of samples, however, yielded equivalent PCR product band intensities from 10 ng of template DNA. There was no evidence that impurities in the DNA extracts inhibited PCR amplification efficiency.
Probe hybridizations of clone blots.
Hybridizations were carried out with several hundred blotted clones by using the β-AO235 and β-Deep_446 probes (for all β-AAOs and deep-branched non-AAOs, respectively). High percentages of nonspecific amplification products (Table 4) were obtained from all soils with the β-AMOf and β-AMOr primers (13), and percentages appeared to be higher when soils contained more microbial biomass. Clone libraries from tilled soils in 1994, for example, contained fewer nonspecific products than in 1995 (Table 4), when direct microscopic counts (Table 1) and total DNA yields (Table 3) were significantly higher. In both years, percentages of clones hybridizing to the all-β-AAO probe were higher in libraries from tilled soils than in those from native soils (Table 4). The converse was true with the probe for deep-branched non-AAOs. In 1995, libraries from fertilized and unfertilized successional soils contained 5 and 1% clones, respectively, which hybridized with the all-β-AAO probe. The percentage of clones hybridizing with the β-Deep probe was slightly higher for unfertilized than fertilized successional soils (Table 4).
Sequence diversity in clone libraries in 1994.
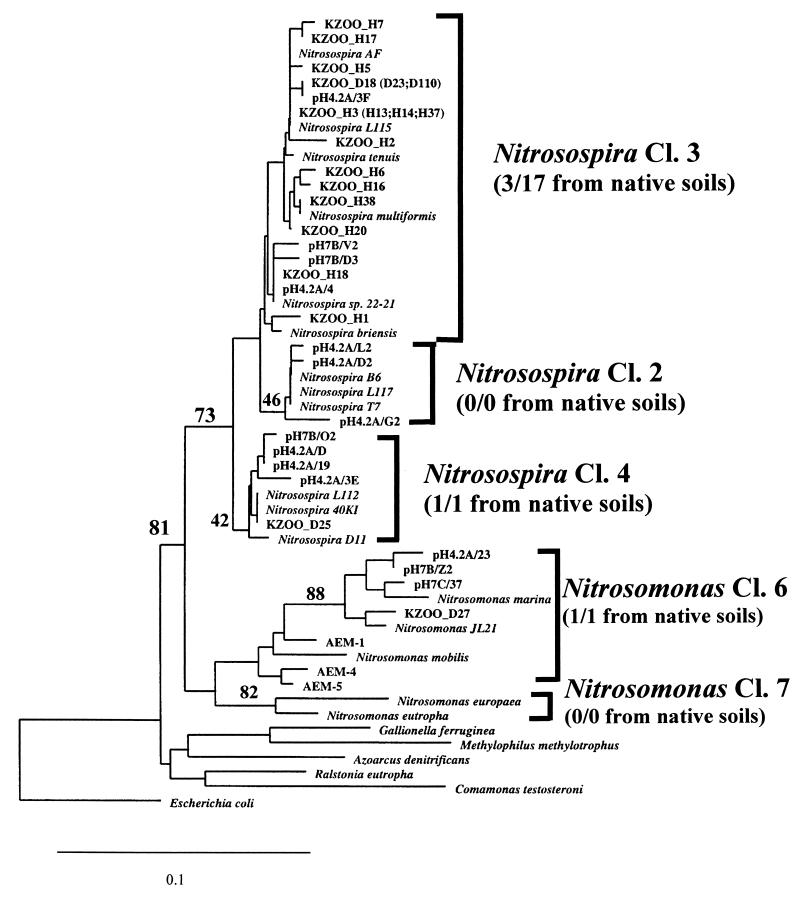
Characteristic AAO motifs in T tracks were exhibited by nearly all (18 of 19) of the clones in libraries from tilled soils. Only 50% (13 of 26) of the clones in native-soil libraries showed the AAO motif, indicating that at least half of the clones contained nonspecific PCR inserts. Partial sequences of 14 clones from tilled-soil libraries and 11 from native-soil libraries were aligned with other AAO and β-proteobacterial sequences in the database, and a phylogenetic tree was generated. Of the 11 clones from native-soil libraries, 6 contained sequences showing affinity with the deep-branching clade. Since there is no reason to assume that the source organisms have the phenotype of autotrophic ammonia oxidation, these sequences were not included in Fig. 1. All AAO-like sequences from tilled-soil libraries (designated KZOO_H) fell within a single clade (Nitrosospira cluster 3). The most dissimilar sequences among these clones differed at 3% of the base positions (of a total 318 bases). The five sequences from native-soil libraries (KZOO_D) were more diverse, being distributed among Nitrosospira cluster 3 (three), Nitrosospira cluster 4 (one), and Nitrosomonas cluster 6 (one). With 73% bootstrap support in this analysis for the genus Nitrosospira as a clade distinct from the genus Nitrosomonas, we concluded that the AAO sequences from native soils were more genetically diverse than those from tilled soils.
FIG. 1.
Neighbor-joining tree (22) showing relationships of partial 16S rDNA sequences from soil communities with reference AAO sequences and those of other selected β-proteobacteria. The scale equals 10% estimated substitutions calculated by the Kimura correction (9). Bootstrap values represent percentages from 100 parsimony–maximum-likelihood analyses (29). Cloned sequences from native and tilled soils are designated KZOO_D and KZOO_H, respectively. Abbreviations representing other AAO sequences are as previously described (27).
DGGE and probe hybridizations.
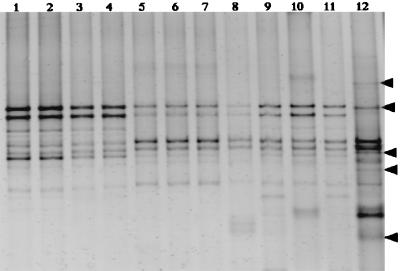
DGGE patterns for soils sampled from replicate plots within each treatment were remarkably similar for tilled and native soils in both sampling years (Fig. 2), indicating spatial and temporal reproducibility based on treatment. Soils from fertilized microplots within the successional treatments also yielded similar DGGE patterns. On the other hand, unfertilized successional soils from each main plot generated a distinctive DGGE banding pattern. Bands unique to HTS replicate block 3, identified by arrows in Fig. 2, suggested a greater potential for heterogeneity in successional treatments.
FIG. 2.
DGGE band patterns of 16S rDNA PCR amplification products obtained with the CTO_PCR primers (10) and community DNA. DGGE gels were stained with ethidium bromide and photographed under UV illumination. Lanes: 1, 3, 5, and 7, PCR products from 1994 samples; 2, 4, 6, and 8, PCR products from 1995 samples; 1 and 2, tilled, replicate plot 5; 3 and 4, tilled, replicate plot 6; 5 and 6, native (never-tilled), replicate plot 3; 7 and 8, native (never-tilled), replicate plot 4; 9 through 12, PCR products from 1995 samples; 9, fertilized successional microplot within replicate plot 1; 10, unfertilized successional, replicate plot 1; 11, fertilized successional microplot within replicate plot 3; 12, unfertilized successional, replicate plot 3. The arrows on the right indicate the locations of unique bands in lane 12.
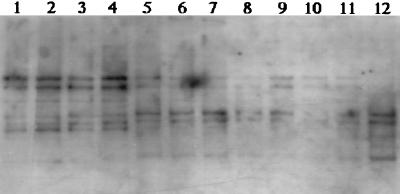
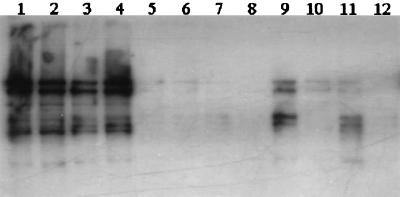
DNA from all lanes in blots of DGGE gels exhibited approximately equivalent hybridization intensities with the Nsp436 probe, which was designed to detect all Nitrosospira sequences (Fig. 3). With NspCL3_454 (for Nitrosospira cluster 3), however, strong hybridization signals were obtained only in lanes containing DNA from tilled and fertilized successional soils (Fig. 4). Hybridization patterns in Fig. 3 and 4 indicated that bands from DNA of native soils and unfertilized successional soils were derived from Nitrosospira DNA but specifically not from cluster 3. Because Nitrosospira cluster 1 has only been found in marine samples, the majority of PCR-DGGE products from these soils were attributed to Nitrosospira cluster 2 or 4.
FIG. 3.
Autoradiogram of a DGGE blot hybridized with a 32P-labeled Nsp436 probe specific for all Nitrosospira sequences (Table 1). The blot contains DNA from the gel shown in Fig. 2, and the lane designations are the same as in Fig. 2.
FIG. 4.
Autoradiogram of a DGGE blot hybridized with a 32P-labeled NspCL3_454 probe for Nitrosospira cluster 3 (Table 1). The blot contains DNA from the gel shown in Fig. 2, and the lane designations are the same as in Fig. 2.
Measurements of other soil and microbial properties.
Results of molecular analyses of AAO population differences were complemented with other data on soil and microbial properties. Total C, microbial-biomass C, and direct microscopic counts were significantly higher in native than in tilled soils (Table 1). Native soils were also characterized by lower pHs, higher ammonium N concentrations, and lower nitrate N concentrations than tilled soils. Although net nitrification rates during field incubations of native and tilled soils did not show significant differences, the percentages of total ammonium N pools (i.e., initial ammonium N plus ammonium N mineralized during incubation) that were converted to nitrate N during 21 days of incubation were higher in tilled soils.
DISCUSSION
Treatments at this LTER site enabled a comparison of AAO populations in adjacent plots of undisturbed, native soils and agricultural soils that had been tilled and fertilized for 100 years. Tilled soils, which had significantly higher nitrate and lower ammonium levels (Table 1), also contained significantly higher numbers of culturable AAOs. Compared to culturable AAOs from native soils, growth of AAOs from tilled soils was less affected by ammonium concentration in the MPN media (Table 2). Tilled soils yielded a higher percentage of 16S rDNA clones hybridizing with β-AAO-specific probes than did native soils (Table 3). Whereas all AAO sequences from tilled soils grouped within Nitrosospira cluster 3, sequences from native soils fell within three different subgroups (Fig. 1). Thus, 16S rDNA sequencing and phylogenetic analysis indicated greater genetic diversity among AAO sequences from undisturbed soils.
The relatively lower genetic diversity of AAO rDNA sequences in tilled soils may have been due to repeated plowing disturbance, which would reduce niche heterogeneity in the soil. Furthermore, repeated N fertilizer additions could result in selection for more ammonium-tolerant and culturable AAOs. When sampling was extended in 1995 to successional treatments, we obtained evidence that such population shifts had occurred in the fertilized microplots. Nitrosospira cluster 3 sequences predominated in DGGE blots from fertilized successional soils but were of lower relative abundance in blots from unfertilized successional soils (Fig. 4). Since DGGE blots from unfertilized successional and native soils hybridized with the all-Nitrosospira probe, these soils apparently contained Nitrosospira cluster 4 (and/or cluster 2), probes for which had not been developed at the time of these experiments. These latter AAO subgroups may be less readily cultured than Nitrosospira cluster 3, which had previously comprised all cultured strains of the genus Nitrosospira (12) until cluster 2 and 4 isolates were recovered from soils in Norway and The Netherlands (10, 34). If less readily cultured AAOs predominate in other grassland, forest, and climax ecosystem soils, this would explain previous observations that such soils contain no or very low numbers of nitrifiers (2, 16).
No strong hybridization was observed between DGGE blots from 1994 native soils and cluster 3 probes (Fig. 4), even though the 1994 clone libraries from these soils yielded three sequences representing cluster 3 (Fig. 1). DGGE blots from native soils, therefore, contained comparatively small amounts of cluster 3 DNA. Despite the fact that DGGE gel patterns were very similar for both tilled and native soils (Fig. 2), only DGGE blots from tilled soils hybridized strongly with probes for Nitrosospira cluster 3 (Fig. 4). These hybridization results can be explained by the fact that bands at identical locations in DGGE gels may contain DNA fragments with different probe target domains. Migration distances of DNA fragments in DGGE gels depend on the melting behavior of their least stable domains. The ca. 500-bp DGGE fragments from tilled and native soils in our study apparently had the same migration-determining domains but different probe target domains. These results are consistent with those of Kowalchuk et al. (10), who concluded that DGGE band locations are not good predictors of phylogenetic position and that probe hybridization tests are needed to confirm the presence of specific AAO subgroups.
The higher diversity observed among AAO rDNA sequences from native soils could reflect the physical heterogeneity than can develop in undisturbed soils. Analysis of soils from successional plots, which had been undisturbed since 1989, enabled us to assess whether AAO populations reverted to those more characteristic of native soils. We observed an apparent progression in the effects of nondisturbance on AAO populations in successional soils. Within 4 years, nitrification activity and nitrate levels declined but numbers of culturable AAOs were not significantly lower than those found in tilled soils (Tables 2 and 3). A similar progression was observed by Stienstra et al. (28), who conducted MPN enumerations and potential-nitrification assays of successional soils after 3, 7, 20, and 46 years of nonfertilization. Significantly lower nitrate levels and potential nitrification activities were observed after 3 years, and further significant reductions were observed in these soils after 7 years. MPN counts of AAOs did not decline until after 20 years. Stienstra et al. (28) concluded that decreasing availability of ammonium in successional soils was the key factor decreasing nitrification activity, and they also suggested that successional soils selected against AAOs having higher pH optima. In native soils at the LTER site, lower pH (Table 2) could also have affected AAO populations.
Similar DGGE banding patterns were obtained from tilled soils in both 1994 and 1995, even though the tilled plots had been planted with different crops (soybeans and wheat). In theory, reproducible DGGE patterns result from the structural similarity of bacterial communities. The presence of the wheat crop thus did not appear to change AAO population structure in tilled soils, even though direct microscopic counts and total DNA yields in 1995 were significantly higher than in 1994 (Tables 1 and 3). These results suggest that AAO population structures exhibited some degree of temporal stability. Furthermore, DGGE patterns appeared to be spatially reproducible, as shown by the similar patterns generated by soils from both replicate plots of each treatment (except for unfertilized successional main plots). Felske and Akkermans (4) also observed spatially consistent DGGE patterns from RNA extracted from soils sampled over an area of several hundred square meters. Nevertheless, artifactual PCR reproducibility may still be caused by selective experimental biases of PCR amplification (31). In our study, we addressed this potential for bias by performing independent molecular analyses with more than one PCR primer set (10, 13). Based on our use of two different molecular approaches in conjunction with MPN data and LTER field measurements, we conclude that our molecular analysis results reflect actual differences in the AAO populations of these soils.
ACKNOWLEDGMENTS
This study was supported in part by NSF grant BIR-9120006 to the Center for Microbial Ecology. Site access and a graduate research grant (LTER grant DEB-02332) were provided by the NSF. Cooperative research with the University of Aberdeen was facilitated by a scientific interchange grant from the U.S. Department of Education.
REFERENCES
- 1.Bloem J, Bolhuis P R, Veringa M R, Weiringa J. Microscopic methods for counting bacteria and fungi in soil. In: Alef K, Nannipieri P, editors. Methods in applied soil microbiology and biochemistry. London, England: Academic Press Ltd.; 1995. pp. 162–173. [Google Scholar]
- 2.Donaldson J M, Henderson G S. A dilute medium to determine population size of ammonia oxidizers in forest soils. Soil Sci Soc Am J. 1989;53:60–62. [Google Scholar]
- 3.Felsenstein J. PHYLIP: phylogeny inference package. Seattle: University of Washington; 1993. [Google Scholar]
- 4.Felske A, Akkermans A D L. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb Ecol. 1998;36:31–36. doi: 10.1007/s002489900090. [DOI] [PubMed] [Google Scholar]
- 5.Hastings R C, Ceccherini M T, Miclaus N, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular biological analysis of ammonia oxidising bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 6.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 7.Horwath W R, Paul E A. Microbial biomass. In: Weaver R W, et al., editors. Methods of soil analysis. Part 2—microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1994. pp. 753–773. [Google Scholar]
- 7a.Kellogg Biological Station Long-Term Ecological Research. Michigan State University Board of Trustees. [Online.] http://lter.kbs.msu.edu. [24 May 1999, last date accessed.]
- 8.Killham K. Heterotrophic nitrification. In: Prosser J I, editor. Nitrification. Oxford, United Kingdom: IRL Press; 1986. pp. 117–126. [Google Scholar]
- 9.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 10.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalchuk G A, Bodelier P L E, Heilig H J G, Stephen J R, Laanbroek H J. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol Ecol. 1998;27:339–350. [Google Scholar]
- 12.Maidak B L, Larsen N, McCaughey N, J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 14.McCaig A E, Phillips C J, Stephen J R, Kowalchuk G A, Harvey S M, Herbert R A, Embley T M, Prosser J I. Nitrogen cycling and community structure of proteobacterial β-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl Environ Microbiol. 1999;65:213–220. doi: 10.1128/aem.65.1.213-220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Post W M, Mann L K. Changes in soil organic carbon and nitrogen as a result of cultivation. In: Bouwman A F, editor. Soils and the greenhouse effect. London, England: John Wiley & Sons, Ltd.; 1990. pp. 401–406. [Google Scholar]
- 16.Prosser J I. Autotrophic nitrification in bacteria. Adv Microbiol Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 17.Rice E L, Pancholy S K. Inhibition of nitrification in climax ecosystems. Am J Bot. 1972;59:1033–1040. [Google Scholar]
- 18.Robertson G P. Factors regulating nitrification in primary and secondary succession. Ecology. 1982;63:1561–1573. [Google Scholar]
- 19.Robertson G P. Nitrogen use efficiency in row-crop agriculture: crop nitrogen use and soil nitrogen loss. In: Jackson L E, editor. Ecology in agriculture. San Diego, Calif: Academic Press; 1997. pp. 347–365. [Google Scholar]
- 20.Robertson G P, Klingensmith K M, Klug M J, Paul E A, Crum J R, Ellis B G. Soil resources, microbial activity, and primary production across an agricultural ecosystem. Ecol Appl. 1997;7:158–170. [Google Scholar]
- 21.Rowe R, Todd R, Waide J. Microtechnique for most-probable-number analysis. Appl Environ Microbiol. 1977;33:675–680. doi: 10.1128/aem.33.3.675-680.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor joining method: a method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schmidt E L, Belser L W. Nitrifying bacteria. In: Page A L, Miller R H, Keeney D R, editors. Methods of soil analysis, part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy; 1982. pp. 1027–1041. [Google Scholar]
- 25.Stark J M, Hart S C. High rates of nitrification and nitrate turnover in undisturbed coniferous forests. Nature. 1997;385:61–64. [Google Scholar]
- 26.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephen J R, Kowalchuk G A, Bruns M-A V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soils by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stienstra A W, Klein Gunnewiek P, Laanbroek H J. Repression of nitrification in soils under a climax grassland vegetation. FEMS Microbiol Ecol. 1994;14:45–52. [Google Scholar]
- 29.Strunk O, Ludwig W. ARB: a software environment for sequence data, 2.1.1. Munich, Germany: Department of Microbiology, Technical University of Munich; 1997. [Google Scholar]
- 30.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 1994;28:1523–1532. [Google Scholar]
- 31.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford D L. PAUP: phylogenetic analysis using parsimony. Champagne-Urbana, Ill.: Illinois Natural History Survey; 1993. [Google Scholar]
- 33.Teske A, Alm E, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.U.S. Department of Agriculture. U.S. soil classification system. U.S. Washington, D.C: Department of Agriculture; 1992. [Google Scholar]
- 34.Utaker J B, Bakken L, Jiang Q Q, Nes I F. Phylogenetic analysis of seven new isolates of ammonia-oxidising bacteria based on 16S rRNA gene sequences. Syst Appl Microbiol. 1995;18:549–559. [Google Scholar]
- 35.Watson C J, Mills C L. Gross nitrogen transformations in grassland soils as affected by previous management intensity. Soil Biol Biochem. 1998;30:743–753. [Google Scholar]
- 36.Wood P M. Nitrification as a bacterial energy source. In: Prosser J I, editor. Nitrification. London, England: Society of General Microbiology; 1989. pp. 39–62. [Google Scholar]
- 37.Woomer P L. Most probable number counts. In: Weaver R W, et al., editors. Methods of soil analysis. Part 2—microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1994. pp. 59–80. [Google Scholar]
- 38.Zhou J, Bruns M-A V, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]