Abstract
Objectives
The increased infectivity and transmissibility of SARS-CoV-2 variants of concern (VOCs) could cause significant human and economic damage. Hence, understanding their characteristics is crucial to control infection. We evaluated the environmental stability of the Wuhan strain and all VOCs (Alpha, Beta, Gamma, Delta, Omicron BA.1, and Omicron BA.2 variants) on plastic and human skin surfaces and their disinfection efficacy.
Methods
To evaluate environmental stability, residual virus titres on plastic and human skin surfaces were measured over time. Their survival time and half-life were calculated using regression analysis. The effectiveness of ethanol-based disinfectants at different concentrations was determined by in vitro and ex vivo evaluations.
Results
On plastic and skin surfaces, the Alpha, Beta, Delta, and Omicron variants exhibited approximately two-fold longer survival times than the Wuhan strain; the Omicron variants had the longest survival time. The median survival times of the Wuhan strain and the Alpha, Beta, Gamma, Delta, and Omicron (BA.1 and BA.2) variants on human skin surface were 8.6, 19.6, 19.1, 11.0, 16.8, 21.1, and 22.5 h, respectively. The in vitro evaluation showed that the Wuhan strain and the Alpha, Beta, Gamma, Delta, and Omicron (BA.1 and BA.2) variants were completely inactivated within 15 s by 32.5%, 35%, 35%, 32.5%, 35%, 40%, and 40% ethanol, respectively. However, all viruses on human skin were completely inactivated by exposure to 35% ethanol for 15 s.
Conclusions
SARS-CoV-2 VOCs, especially the Omicron variants, have higher environmental stability than the Wuhan strain, increasing their transmission risk and contributing to their spread.
Keywords: SARS-CoV-2, Variants of concern, Environmental stability, Virus survival, Disinfection effectiveness
Introduction
Various SARS-CoV-2 variants have emerged from 2020 to 2021. In particular, those classified as variants of concern (VOCs) can cause significant human and economic damage. Hence, an understanding of their characteristics is crucial for infection control. These VOCs have increased infectivity and transmissibility [1]. In particular, the rapid spread of the Omicron variants has become a serious concern worldwide in 2022 [2,3]. Such increased infectivity and transmissibility can be attributed to several factors, such as increased and prolonged viral shedding from infected individuals, decreased minimum viral load required to establish infection, changes in infection target sites, and increased environmental stability [4,5].
The environmental stability of SARS-CoV-2 has been compared with that of other viruses, such as the SARS-CoV-1 and influenza virus [6,7]. Previous studies suggested that the SARS-CoV-2 Alpha and Beta variants have the same degree of environmental stability [8,9]. Differences in viral stability among all VOCs, including the Omicron BA.1 and BA.2 and Delta variants, have not yet been evaluated and compared. In this study, we improved our previously developed evaluation model and analysed the differences in viral stability and disinfection efficacy between the Wuhan strain and all VOCs.
Methods
Viruses and cells
The SARS-CoV-2 variants analysed in this study included the Wuhan strain (Pango lineage: A, hCoV-19/Japan/TY/WK-521/2019), Alpha variant (Pango lineage: B.1.1.7, hCoV-19/Japan/QK002/2020), Beta variant (Pango lineage: B.1.351, hCoV-19/Japan/TY8-612/2021), Gamma variant (Pango lineage: P.1, hCoV-19/Japan/TY7-501/2021), Delta variant (Pango lineage: B.1.617.2, hCoV-19/Japan/TY11-927/2021), Omicron BA.1 variant (Pango lineage: BA.1, hCoV-19/Japan/TY38-873/2021), and Omicron BA.2 variant (Pango lineage: BA.2, hCoV-19/Japan/TY40-385/2022). All viruses were generously provided by the National Institute of Infectious Diseases (Tokyo, Japan). For virus culture and quantification, VeroE6/TMPRSS2 cells, which express the transmembrane serine protease TMPRSS2, were purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma Aldrich, St Louis, MO, USA). They were supplemented with 5% foetal bovine serum and G418 (Nacalai Tesque, Kyoto, Japan) [10,11]. The viruses were concentrated and purified as follows: 96 h post-infection, the culture medium was harvested and centrifuged for 10 min at 2500 × g at 4°C to eliminate cellular debris. Virions in the supernatant were sedimented through a 20% (w/w) sucrose cushion in phosphate-buffered saline (PBS) via ultracentrifugation at 27,000 rpm for 2.5 h at 4°C using a Beckman SW28 rotor [12,13]. The virus titres were measured using 50% tissue culture infectious dose (TCID50) in VeroE6/TMPRSS2 cells. Three days after inoculation, the cytopathic effect in each well was scored under a microscope, and the TCID50 was calculated.
Construction of skin model to evaluate virus stability and disinfectant effectiveness
Human skin was collected from forensic autopsy specimens that were obtained from the Department of Forensic Medicine of the Kyoto Prefectural University of Medicine. Abdominal skin autopsy specimens from individuals aged 20 to 70 years, which were obtained approximately 1 d after death, were cut into squares with approximate dimensions of 4 cm × 8 cm. Those whose skin was considerably damaged by burning or drowning were excluded. Using the skin-autopsy specimens, an ex vivo model was developed to evaluate the stability of different viruses on the surface of human skin and the effectiveness of different disinfectants against these viruses on the skin [6,14]. The skin from which the panniculus adiposus had been removed was washed with PBS and placed in a culture insert (Corning, Corning, NY, USA) on a membrane with a pore size of 8.0 μm. The culture inserts were placed in six-well plates containing 1.0 mL of DMEM (Sigma-Aldrich).
Evaluation of virus stability on plastic and human skin surfaces
Virus survival was evaluated on plastic (polystyrene plate) and human skin (constructed skin model) surfaces. Virus solutions (4.7 Log10TCID50 in 2 μL PBS) were applied on the surface of plastic or human skin. Each sample was incubated in a controlled environment (25°C, 45–55% relative humidity) for 0–120 h. The virus that remained on the surface was subsequently collected in 1.0 mL of DMEM and titrated [6]. The detection limit for the virus titre remaining on the surface was 0.5 Log10TCID50. Survival time was defined as the time when the virus was no longer detected on the surface. Three independent experiments were performed for each condition, and the results of the residual virus titres on the surfaces were expressed as the mean ± standard error of the mean.
The elapsed time was defined as an explanatory variable (X-axis), and the log virus titre was defined as an explained variable (Y-axis). A linear regression analysis with a logarithmic link function was performed in order to create a regression curve. The measurement limit of the SARS-CoV-2 titre was 0.5 Log10TCID50. The survival times were defined as the X-values when the Y-values of the regression curves were 0.5. The half-life was calculated from the slope of each regression curve when the titres of the virus that remained on the surface were 2.0 and 3.0 log10TCID50 [6,12]. GraphPad Prism 7 (GraphPad, Inc., La Jolla, CA, USA) was used for conducting the statistical analyses.
Evaluation of the effectiveness of alcohol-based disinfectants
The effectiveness of ethanol-based disinfectants was evaluated at different concentrations. The effectiveness of ethanol (EA, Nacalai Tesque) was tested at the following concentrations: 80%, 60%, 50%, 40%, 35%, 32.5%, 30%, 27.5%, 25%, 22.5% and 20% (w/w). Isopropanol (IPA, Nacalai Tesque) was tested at a concentration of 70% (w/w).
First, an in vitro evaluation of the effectiveness of the disinfectant was performed. In a 1500 μL tube, 5 μL of PBS containing virus (final virus titre concentration: 4.5 Log10TCID50/mL) was mixed with 45 μL of various disinfectants for 15 s. The resulting solutions were neutralized with 450 μL of DMEM, and the titres of the remaining viruses were measured [14]. The detection limit for the virus titre concentration was 0.5 Log10TCID50/mL. The EA concentration was defined as an explanatory variable (X-axis), and the log virus titre concentration was defined as an explained variable (Y-axis). Nonlinear regression analyses were conducted using a 4-parameter logistic model [9]. The EA concentration required for log reduction in virus titre concentration to exceed 3.5 in 15 s disinfection reaction (notated as required EA concentration) was defined as the X values when the Y values of the regression curves were 1.0.
Next, the effectiveness of the disinfectants against viruses on human skin was evaluated using the constructed model (ex vivo evaluation). Each solution (5.0 Log10TCID50 in 2 μL PBS) was applied to the human skin surface. Each skin sample was subsequently incubated for 15 min at 25°C with 45%–55% relative humidity in order to completely dry the viral mixture on the skin. Subsequently, 18 μL of disinfectant was applied to each skin sample surface for 15 s, which was then air-dried for 5 min. Afterwards, the remaining viruses on the skin were recovered with 1000 μL of DMEM, and the viral load was measured [14]. The detection limit for the virus titres was 0.5 Log10TCID50. To determine the effectiveness of the disinfectants under each condition, log reductions in virus titres were calculated, normalizing to the PBS control titres. Three independent experiments were performed for each condition, and the results were expressed as the mean ± standard error of the mean.
Ethical considerations
The study protocol, including sample collection procedures, was reviewed and approved by the Institutional Review Board of the Kyoto Prefectural University of Medicine (ERB-C-1593). Written informed consent was obtained from the bereaved families of the study participants.
Results
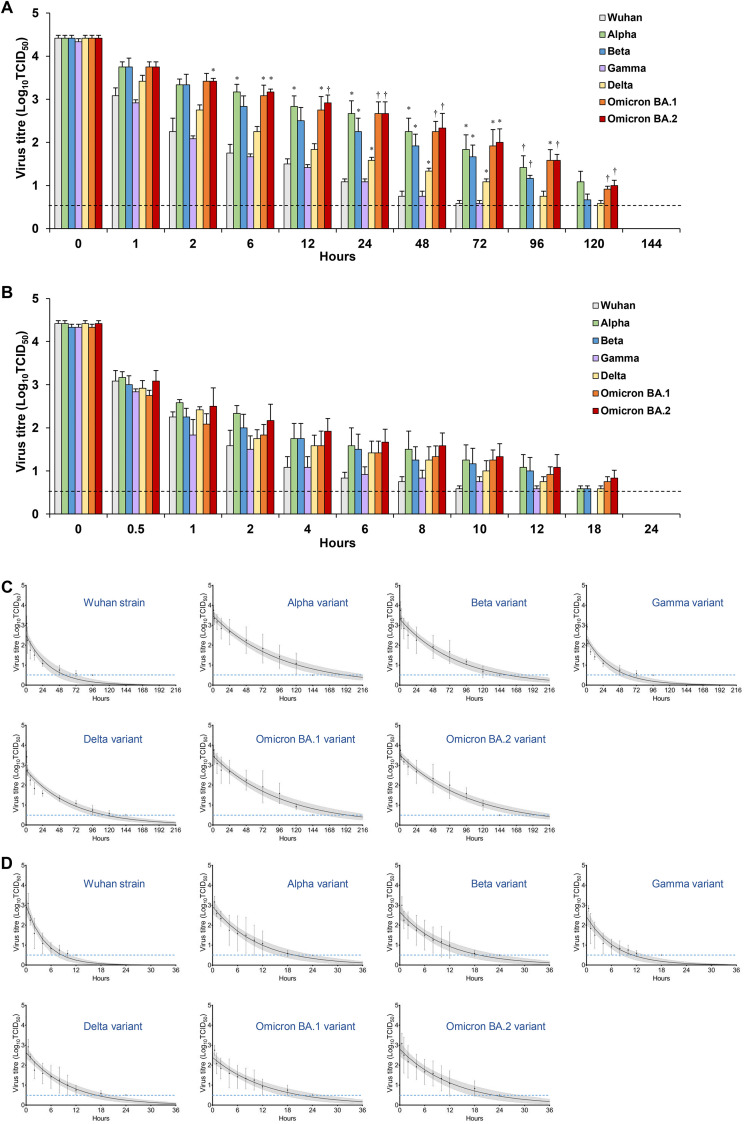
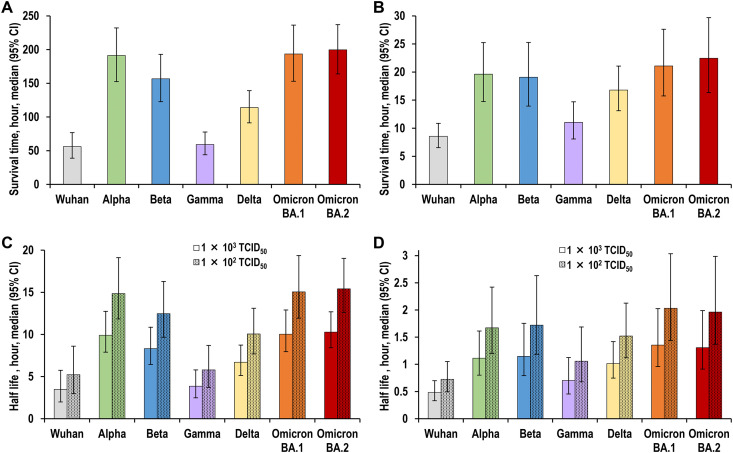
The virus titres that remained on the plastic or human skin surfaces were measured over time. The survival time and half-life were calculated from these titre values using regression analysis (Fig. 1 ). In the plastic surface analysis, the median survival times of the Wuhan strain, Alpha variant, Beta variant, Gamma variant, Delta variant, Omicron BA.1 variant, and Omicron BA.2 variant were 56.0 h (95% confidence interval [CI], 39.0–76.7 h), 191.3 h (95% CI, 152.5–232.1 h), 156.6 h (95% CI, 122.7–192.9 h), 59.3 h (95% CI, 43.9–77.7 h), 114.0 h (95% CI, 91.3–139.1 h), 193.5 h (95% CI, 153.1–236.2 h), and 199.7 h (95% CI, 163.9–237.1 h), respectively (Fig. 2 A and Table 1 ). In the human skin surface analysis, the median survival times of the Wuhan strain, Alpha variant, Beta variant, Gamma variant, Delta variant, Omicron BA.1 variant, and Omicron BA.2 variant were 8.6 h (95% CI, 6.5–10.9 h), 19.6 h (95% CI, 14.8–25.3 h), 19.1 h (95% CI, 13.9–25.3 h), 11.0 h (95% CI, 8.1–14.7 h), 16.8 h (95% CI, 13.1–21.1 h), 21.1 h (95% CI, 15.8–27.6 h), and 22.5 h (95% CI, 16.3–29.7 h), respectively (Fig. 2B and Table 1). The Alpha, Beta, Delta, and Omicron variants had approximately two-fold longer survival times than the Wuhan strain. The Omicron BA.1 and BA.2 variants had similar environmental stability and the longest survival time. Furthermore, the half-life calculations showed the same tendency as the survival time (Fig. 2C and D; Table 1).
Fig. 1.
(A, B) Decrease in residual viral titre on plastic (A) and human skin (B) surfaces over time. Each virus (4.7 Log10TCID50; 50% tissue culture infectious dose) was mixed with 2 μL of phosphate-buffered saline and applied to each surface, which was incubated in a controlled environment (temperature: 25°C, humidity: 45%–55%) for 0–144 h. Viruses on the surface were then recovered in 1 mL of medium and titrated to calculate the titres of the remaining virus. For each condition, three independent experiments were performed, and the results are expressed as the mean ± standard error of the mean. The bars for data below the detection limit were omitted. Dotted horizontal lines represent detection limit titres. The values of residual viral titre on surfaces were evaluated with Student's t-test, and the magnitudes with P < 0.05 were considered significant. ∗P < 0.05 (versus the Wuhan strain). †P < 0.01 (versus the Wuhan strain). (C, D) Stability of viruses on the plastic surface (C) and the human skin surface (D). The elapsed time was defined as an explanatory variable (X-axis), and the log virus titre was defined as an explained variable (Y-axis). Least-squares linear-regression analysis was performed with a logarithmic link function for each virus to generate a regression curve. The upper and lower confidence limits are represented by dotted curves. The dotted horizontal lines represent the detection limit titres. The data shown are expressed as the mean ± standard error of the mean for three independent experiments.
Fig. 2.
(A) Survival times of the various viruses on a plastic surface. (B) Survival times of the various viruses on the surface of human skin. (C) Half-lives of the various viruses on a plastic surface. (D) Half-lives of the various viruses on the surface of human skin. Survival time is defined as the time until the virus can no longer be detected on the surface. All half-lives in the graphs refer to a condition when 2.0 and 3.0 Log10TCID50 (50% tissue culture infectious dose) of virus particles remained on the surface. Data are expressed as the median ±95% confidence interval.
Table 1.
Survival time and half-life of SARS-CoV-2 on the plastic and human skin surface
| Survival time, hour, median (95% CI) |
Half-life, hour, median (95% CI) |
|||||
|---|---|---|---|---|---|---|
| Plastic surface | Skin surface | Plastic surface |
Skin surface |
|||
| 3 (Log10TCID50) | 2 (Log10TCID50) | 3 (Log10TCID50) | 2 (Log10TCID50) | |||
| Wuhan | 56.0 (39.0-76.7) | 8.6 (6.5-10.9) | 3.5 (2.0-5.7) | 5.2 (3.0-8.6) | 0.5 (0.3-0.7) | 0.7 (0.5-1.1) |
| Alpha | 191.3 (152.5-232.1) | 19.6 (14.8-25.3) | 9.9 (7.9-12.7) | 14.9 (11.9-19.1) | 1.1 (0.8-1.6) | 1.7 (1.2-2.4) |
| Beta | 156.6 (122.7-192.9) | 19.1 (13.9-25.3) | 8.3 (6.4-10.9) | 12.5 (9.7-16.3) | 1.2 (0.8-1.8) | 1.7 (1.2-2.6) |
| Gamma | 59.3 (43.9-77.7) | 11.0 (8.1-14.7) | 3.9 (2.5-5.8) | 5.8 (3.7-8.7) | 0.7 (0.5-1.1) | 1.1 (0.7-1.7) |
| Delta | 114.0 (91.3-139.1) | 16.8 (13.1-21.1) | 6.7 (5.1-8.7) | 10.1 (7.7-13.1) | 1.0 (0.8-1.4) | 1.5 (1.1-2.1) |
| Omicron BA.1 | 193.5 (153.1-236.2) | 21.1 (15.8-27.6) | 10.0 (8.0-12.9) | 15.1 (11.9-19.4) | 1.4 (1.0-2.0) | 2.0 (1.4-3.0) |
| Omicron BA.2 | 199.7 (163.9-237.1) | 22.5 (16.3-29.7) | 10.3 (8.4-12.7) | 15.4 (12.6-19.0) | 1.3 (0.9-2.0) | 2.0 (1.4-3.0) |
The elapsed time was defined as an explanatory variable (X-axis), and the log virus titre was defined as an explained variable (Y-axis). A linear regression analysis with logarithmic link function was performed to create a curve of regression. The measurement limit of the SARS-CoV-2 titre was 0.5 Log10TCID50; therefore, the survival time was defined as the X values when the Y values of the regression curves were 0.5. The half-life was calculated from the slope of each regression curve when the titre of virus remaining on the surface was 2.0, and 3.0 Log10TCID50.
The in vitro disinfectant effectiveness evaluation showed that the Wuhan strain and Gamma variant were completely inactivated within 15 s by 32.5% EA (log reduction >4). Meanwhile, the Alpha, Beta, and Delta variants were completely inactivated within 15 s by 35% EA, and the Omicron BA.1 and BA.2 variants were completely inactivated within 15 s by 40% EA.
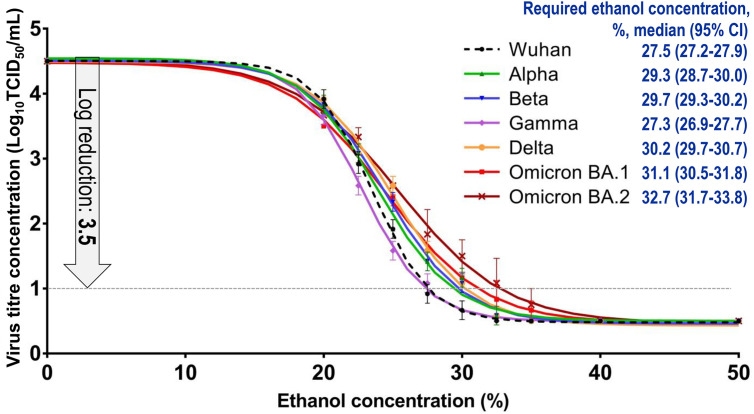
Moreover, the ethanol concentration that was required to achieve a logarithmic decrease of 3.5 in virus titre concentration in a 15 s disinfection reaction (defined as required EA concentration) was calculated. The required EA concentration of the Wuhan strain and the Alpha, Beta, Gamma, Delta, Omicron BA.1, and Omicron BA.2 variants were 27.5%, 29.3%, 29.7%, 27.3%, 30.2%, 31.1%, and 32.7%, respectively (Fig. 3 ).
Fig. 3.
In vitro evaluation of disinfectant effectiveness. The ethanol concentration was defined as an explanatory variable (X-axis), and the log virus titre concentration was defined as an explained variable (Y-axis). The results of the log virus titre concentration are expressed as the mean ± standard error. Nonlinear regression analyses were conducted using a 4-parameter logistic model. The upper and lower confidence limits of the regression curves are omitted. The ethanol concentration required to achieve a logarithmic decrease of 3.5 in virus titre concentration in a 15 s disinfection reaction (notated as the required ethanol concentration) was defined as the X values when the Y values of the regression curves were 1.0.
The Alpha, Beta, Delta, and Omicron variants were slightly more resistant to EA than the Wuhan strain. However, on human skin, an ex vivo evaluation showed that all viruses were completely inactivated after exposure to 35% EA for 15 s (log reduction >4; Table 2 ).
Table 2.
Ex vivo evaluation of disinfectant effectiveness against virus on human skin surface
| Log reduction, mean ± standard error |
|||||||
|---|---|---|---|---|---|---|---|
| Wuhan | Alpha | Beta | Gamma | Delta | Omicron BA.1 | Omicron BA.2 | |
| 70% IPA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 80% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 60% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 50% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 40% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 35% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 |
| 30% EA | >4.00 | >4.00 | >4.00 | >4.00 | >4.00 | 3.67 ± 0.24 | 3.42 ± 0.12 |
| 25% EA | 3.83 ± 0.24 | 2.17 ± 0.24 | 2.25 ± 0.20 | 3.67 ± 0.47 | 2.42 ± 0.31 | 1.67 ± 0.24 | 1.83 ± 0.24 |
| 20% EA | 0.83 ± 0.24 | 0.50 ± 0.35 | 0.58 ± 0.12 | 0.92 ± 0.12 | 0.67 ± 0.12 | 0.33 ± 0.12 | 0.42 ± 0.12 |
The log reduction value was calculated to evaluate disinfectant effectiveness under each condition and was expressed as mean ± standard error. Additionally, the log reduction value of the condition wherein the virus was inactivated below the measurement limit was 4 or more and was expressed as “> 4.00”. EA, ethanol; IPA, isopropanol.
Discussion
The present study reports three major findings as follows: (i) the Alpha, Beta, Delta, and Omicron variants on plastic and skin surfaces had longer survival times than the Wuhan strain; (ii) the Omicron BA.1 and BA.2 variants had similar environmental stability and the longest survival time; and (iii) all viruses on human skin were completely inactivated by exposure to 35 w/w % EA for 15 s, although the Alpha, Beta, Delta, and Omicron variants were slightly more resistant to ethanol than the Wuhan strain.
In 2020, the Wuhan strain's environmental stability was initially reported [6,7,15]. Some studies suggested that the Alpha and Beta variants have the same degree of environmental stability [8,9]. However, no study has directly compared other VOCs with the Wuhan strain. Moreover, the differences in environmental stability between the Wuhan strain and all VOCs, including the Omicron and Delta variants, were previously unknown. Previous studies have shown that the survival times of SARS-CoV-2 on stainless steel, glass, and plastic surfaces do not substantially differ. They also suggested that analysis on a single representative surface can be used in order to compare the environmental stability of different variants [6,16]. Therefore, environmental stability was evaluated on plastic and skin surfaces in this study. Our study showed that on plastic and skin surfaces, the Alpha, Beta, Delta, and Omicron variants exhibited more than twice the survival time of the Wuhan strain. They also maintained their infectivity for more than 16 h on skin surfaces. The Wuhan strain was reported to have higher environmental stability than the influenza virus [6,16], and the environmental stability of these VOCs was even higher than that of the Wuhan strain. This high environmental stability could increase the risk of contact transmission and contribute to the spread of VOC. Additionally, there was no significant difference in the survival times between the Alpha and Beta variants, which had similar environmental stability, consistent with the results of previous studies [8,9]. Hence, future research is needed in order to evaluate the environmental stability of each variant on various surfaces and explore the first variant with a higher environmental stability than the Wuhan strain.
The Omicron variant is currently a major concern because of the rapidly increasing number of infected patients worldwide. The shift in the target site of infection from the lower to the upper respiratory tract and their escape from neutralizing antibodies could be factors causing Omicron variant spread [[1], [2], [3], [4], [5]]. This study also showed that the Omicron variants have the highest environmental stability among VOCs, which might have also contributed to its replacement of the Delta variant and rapid spread. Although the Alpha, Beta, Delta, and Omicron variants showed a slight increase in ethanol resistance, all VOCs on the skin surface were completely inactivated by 15 s exposure to 35% EA. Currently, the World Health Organization recommends the use of EA at concentrations of >52 w/w% (or >60 v/v%) in appropriate infection control (hand hygiene) [17,18]. Therefore, the risk of transmission by VOCs on skin surfaces will be drastically reduced if the appropriate infection control is practised.
This study has some limitations. First, although different sublineages exist for each VOC, we used a single representative in this study. Second, the reason for the higher environmental stability of the Alpha, Beta, Delta, and Omicron variants is unknown, and an evaluation that uses recombinant viruses might identify factors that determine this phenomenon. Third, the survival time and half-life obtained in this environmental stability evaluation might vary depending on the external environment and composition of the body fluid that contains the virus. In this study, in order to accurately analyse the differences in stability between VOCs, the target virus was purified using ultracentrifugation, and the PBS was used as a solvent. Fourth, the relationship between the amount of virus remaining on the surface and the risk of transmission is still unclear. Therefore, it might be reasonable to interpret the survival time in this study as a reference value.
Based on the foregoing results, this study concludes that a higher environmental stability of VOCs, especially the Omicron variants, could increase their transmission risk and contribute to their spread. However, proper hand hygiene practice can reduce such transmission risks.
Author contributions
Study concept and design: RH. Data acquisition: RH, HM, NW, TY, RB, TD. Data analysis and interpretation: RH, YI, and TN. Drafting of the manuscript: RH. Statistical analysis: RH. Secured funding: RH. Administrative/technical/material support: RH, HI. Study supervision: RH and TN.
Transparency declaration
The authors declare no competing financial interests.
Acknowledgments
We thank Editage (www.editage.com) for English language editing. This research was supported by Adaptable and Seamless Technology Transfer Program through Target-driven R&D (ASTEP) from the Japan Science and Technology Agency (JST) [grant number JPMJTR21UE and JPMJTM20PR], JSPS KAKENHI (grant number 21K16326), Mitsubishi Foundation, and Takeda Science Foundation.
Editor: J. M. Hübschen
References
- 1.Tracking SARS-CoV-2 variants. World Health Organization; 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ Available at: [Google Scholar]
- 2.Sars-CoV-2 B. 1.1.529 (Omicron) variant - United States, december 1-8, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1731–1734. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.I. National Center for, D.o.V.D. Centers for Disease Control and Prevention (US) unless a copyright is indicated, information on CDC's sites, blogs, and applications is in the public domain and may be copied and distributed without permission., Atlanta (GA) 2020. Respiratory Diseases, science brief: Omicron (B.1.1.529) variant, CDC COVID-19 science briefs. [Google Scholar]
- 4.Enhancing readiness for Omicron (B.1.1.529) 2021. https://www.who.int/docs/default-source/coronaviruse/2021-12-23-global-technical-brief-and-priority-action-on-omicron.pdf?sfvrsn=d0e9fb6c_8 Technical brief and priority actions for Member States. World Health Organization. Available at: [Google Scholar]
- 5.He X., Hong W., Pan X., Lu G., Wei X. SARS-CoV-2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirose R., Ikegaya H., Naito Y., Watanabe N., Yoshida T., Bandou R., et al. Survival of SARS-CoV-2 and influenza virus on the human skin: importance of hand hygiene in COVID-19. Clin Infect Dis. 2020;73:e4329–e4335. doi: 10.1093/cid/ciaa1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pottage T., Garratt I., Onianwa O., Spencer A., Paton S., Verlander N.Q., et al. A comparison of persistence of SARS-CoV-2 variants on stainless steel. J Hosp Infect. 2021;114:163–166. doi: 10.1016/j.jhin.2021.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister T.L., Fortmann J., Todt D., Heinen N., Ludwig A., Brüggemann Y., et al. Comparable environmental stability and disinfection profiles of the currently circulating SARS-CoV-2 variants of concern B.1.1.7 and B.1.351. J Infect Dis. 2021;224:420–424. doi: 10.1093/infdis/jiab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose R., Itoh Y., Ikegaya H., Miyazaki H., Watanabe N., Yoshida T., et al. Evaluation of the residual disinfection effects of commonly used skin disinfectants against viruses: an innovative contact transmission control method. Environ Sci Technol. 2021;55:16044–16055. doi: 10.1021/acs.est.1c05296. [DOI] [PubMed] [Google Scholar]
- 13.Barcena M., Oostergetel G.T., Bartelink W., Faas F.G., Verkleij A., Rottier P.J., et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci U S A. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirose R., Bandou R., Ikegaya H., Watanabe N., Yoshida T., Daidoji T., et al. Disinfectant effectiveness against SARS-CoV-2 and influenza viruses present on human skin: model-based evaluation. Clin Microbiol Infect. 2021;27:1042.e1–1042.e4. doi: 10.1016/j.cmi.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.L., Chan M.C.W., et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose R., Ikegaya H., Naito Y., Watanabe N., Yoshida T., Bandou R., et al. Reply to gracely. Clin Infect Dis. 2021;73:e854–e856. doi: 10.1093/cid/ciab023. [DOI] [PubMed] [Google Scholar]
- 17.Golin A.P., Choi D., Ghahary A. Hand sanitizers: a review of ingredients, mechanisms of action, modes of delivery, and efficacy against coronaviruses. Am J Infect Control. 2020;48:1062–1067. doi: 10.1016/j.ajic.2020.06.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected. World Health Organization; 2020. https://www.who.int/publications/i/item/10665-331495 Available at: [Google Scholar]