Abstract
The Lactobacillus johnsonii VPI 11088 groESL operon was localized on the chromosome near the insertion element IS1223. The operon was initially cloned as a series of three overlapping PCR fragments, which were sequenced and used to design primers to amplify the entire operon. The amplified fragment was used as a probe to recover the chromosomal copy of the groESL operon from a partial library of L. johnsonii VPI 11088 (NCK88) DNA, cloned in the shuttle vector pTRKH2. The 2,253-bp groESL fragment contained three putative open reading frames, two of which encoded the ubiquitous GroES and GroEL chaperone proteins. Analysis of the groESL promoter region revealed three transcription initiation sites, as well as three sets of inverted repeats (IR) positioned between the transcription and translation start sites. Two of the three IR sets bore significant homology to the CIRCE elements, implicated in negative regulation of the heat shock response in many bacteria. Northern analysis and primer extension revealed that multiple temperature-sensitive promoters preceded the groESL chaperone operon, suggesting that stress protein production in L. johnsonii is strongly regulated. Maximum groESL transcription activity was observed following a shift to 55°C, and a 15 to 30-min exposure of log-phase cells to this temperature increased the recovery of freeze-thawed L. johnsonii VPI 11088. These results suggest that a brief, preconditioning heat shock can be used to trigger increased chaperone production and provide significant cross-protection from the stresses imposed during the production of frozen culture concentrates.
Lactobacillus species are used extensively as starter cultures in a variety of fermentations and as probiotics, which are thought to directly affect the health of the host through such activities as immunostimulation, pathogen exclusion, and maintenance of the normal microflora (reviewed by Sanders [25]). Future uses of lactobacilli will probably include the construction of strains capable of releasing essential nutrients and enzymes, secreting bacteriocins, or displaying epitopes as components in oral vaccines. The general utility of Lactobacillus species in these applications is directly related to their GRAS (Generally Recognized as Safe) status and will be dependent on the availability of cost-effective methods for the production and delivery of viable cultures.
The stresses associated with the production, storage, and distribution of frozen, lyophilized, or spray-dried bacterial culture concentrates can dramatically reduce their viability and activity. Bacteria have evolved complex stress responses to promote their survival under severe conditions. Following a brief heat shock at temperatures above the normal growth range, transient synthesis of a set of highly conserved stress proteins occurs. Among these are molecular chaperones which confer enhanced resistance to elevated temperature (20) and significant cross-protection against other stresses such as osmotic shock (33) and freezing (13). Molecular chaperones bind substrate proteins in a transient noncovalent manner, prevent premature folding, and promote the attainment of the “correct state” in vivo (10).
Among the most abundant bacterial proteins under normal conditions, members of the GroES and GroEL chaperone family are greatly induced by any form of cellular stress that leads to protein denaturation (11). The general importance of these chaperones is emphasized by the fact that in Escherichia coli, GroEL and GroES are required for growth at all temperatures (6). Temperature-sensitive mutations in either gene also lead to a pleiotropic phenotype in which RNA, DNA, and protein synthesis are impaired at high temperature. Approximately 30% of E. coli protein species fail to reach their native form in vivo when GroEL is limiting, indicating that a specific subset of cytoplasmic proteins rely on this chaperone to achieve their native form (12). In addition to its established role in protein folding and assembly, GroEL was recently shown to participate in a complex capable of protecting mRNA from nuclease degradation, suggesting that it plays an additional role as an RNA chaperone (7). The molecular chaperones groES and groEL are typically arranged as an operon, and their translation products are assembled into single or double heptameric rings, respectively (8). In the presence of nucleotide, GroES forms a 1:1 complex with GroEL, which binds the protein substrate, possibly in its central cavity (15). Release is contingent upon ATP-hydrolysis, and multiple cycles of binding and release may be necessary for a protein to reach its native conformation (36).
Increasing the available GroES and GroEL concentration prior to the stresses associated with freezing, lyophilization, or spray-drying may offer additional protection against protein denaturation and produce a more viable and physiologically active product. The purpose of this study was to isolate and characterize the groES/groEL operon of L. johnsonii VPI 11088 (NCK88) as a first step in developing an understanding of the impact of molecular chaperones on stress tolerance.
(The preliminary results of this study were reported at the 96th General meeting of the American Society for Microbiology [34]. After the completion of this study, the groESL operon of Lactobacillus helveticus was published by Broadbent et al. [3] and showed a highly similar operon organization, structural genes, and regulatory elements. These features are noted for comparison throughout this report.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were propagated in Luria-Bertani (LB) broth (24) or brain heart infusion broth (BBL Microbiology Systems, Cockeysville, Md.) and electroporated by the method of Dower et al. (4), and transformants were selected with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) at 50 and 200 μg/ml, respectively, and either 100 μg of erythromycin/ml of brain heart infusion agar or 50 μg of ampicillin/ml of LB agar. PCR-generated groESL fragments were cloned in pT7Blue (Novagen, Madison, Wis.), transformed as above, and selected with 50 μg of ampicillin/ml of LB agar. Lactobacilli were propagated in MRS broth (Difco Laboratories, Inc., Detroit, Mich.) supplemented when necessary with 7.0 μg of erythromycin/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| L. johnsonii VPI 11088 | ATCC 11506 (NCK 88) | Virginia Polytechnic Institute (18) |
| E. coli XL1-Blue MRF=Kan | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F = proAB, lacIqZΔM15 Tn5 (Kanr)] (Con) | Stratagene |
| Plasmids | ||
| pBluescript-II KS(+) | 2.9 kb; lacZ Apr | Stratagene |
| pT7Blue | 3.0 kb; lacZ Apr | Novagen |
| pTRKH2 | 6.9 kb; Emr | 21 |
| pTRK 453 | 5.3 kb; pBluescript-II KS(+) (EcoRV)::2.4-kb NCK88 PCR product; Apr | This study |
| pTRK 454 | 4.5 kb; pT7Blue::1.5-kb NCK88 PCR product; Apr | This study |
| pTRK 455 | 4.1 kb; pT7Blue::1.1-kb NCK88 PCR product; Apr | This study |
| pTRK 475 | 19.0 kb; pTRKH2 (BglII)::12.0-kb NCK88 chromosomal fragment; Emr | This study |
| pTRK 476 | 5.9 kb; pBluescript-II KS(+) (SpeI-ClaI)::3.0-kb pTRK475 fragment; Apr | This study |
DNA isolation, manipulation, and sequencing.
Total Lactobacillus DNA was isolated as previously described (35). Large-scale E. coli plasmid preparations, cesium chloride gradient purification, and standard cloning procedures were performed by the method of Sambrook et al. (24). Nested deletions were carried out with exonuclease III and mung bean nuclease (Stratagene) as described by Zhu and Clark (39). Miniprep plasmid DNA was isolated by the method of Sambrook et al. (24) for clone analysis, and when used for routine sequencing, it was extracted twice with phenol-chloroform and once with chloroform. DNA sequencing was performed with [35S]dATP (New England Nuclear, Boston, Mass.) by the dideoxy-chain termination method of Sanger et al. (26) with Sequenase version 2.0 and the 7-deaza-dGTP kit (United States Biochemical/Amersham Life Science, Cleveland, Ohio). Gel compression artifacts generated in regions containing excessive secondary structure were reduced through the addition of terminal deoxynucleotidyl transferase (United States Biochemical/Amersham Life Science) following termination as described by the manufacturer. The pBluescript-II KS-specific primers T3 and T7 (Stratagene), the pT7Blue-specific primers R-20 and U-19 (Novagen), and numerous groESL-specific primers synthesized by Genosys Biotechnologies, Inc. (The Woodlands, Tex.), were used in the sequencing reactions. The complete DNA sequence was assembled and analyzed by using the PC/Gene software (IntelliGenetics, Inc., Mountain View, Calif.). Sequence comparisons with the database of the National Center for Biotechnology Information (NCBI) were made by using the Basic Local Alignment Search Tool (BLAST) of Altschul et al. (1).
Cloning the groESL operon.
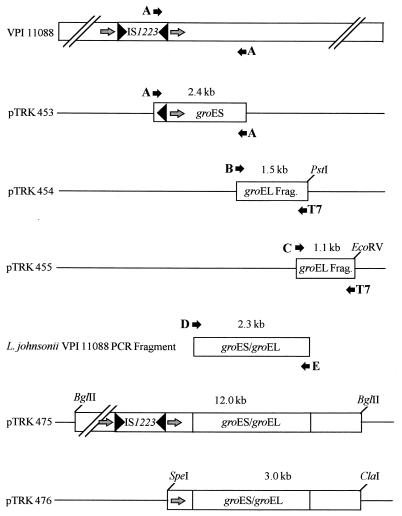
The first 1,296 bp of the groESL operon was amplified as part of a 2.4-kb fragment by PCR with Taq DNA polymerase (Boehringer Mannheim Corp., Indianapolis, Ind.) and one IS1223-specific oligonucleotide (5′-ATGCACCAGGAGAAACG-3′) primer (primer A) (Fig. 1). The fragment was blunt ended with Klenow enzyme (Boehringer Mannheim Corporation) and cloned into EcoRV-digested pBluescript-II KS(+), producing pTRK453 (Fig. 1). The remainder of the operon was cloned in two steps by the single-specific-primer PCR method of Shyamala and Ames (29) as outlined by Ross and Claiborne (23). In the first step, total genomic DNA was digested to completion with PstI and cloned into similarly digested pBluescript-II KS(+). The ligation mix was amplified by PCR with the vector-specific primer T7 and the groESL-specific primer (5′-GATGGTGTTACTATCGCCAA-GAGTATTG-3′) (primer B), and the resultant 1.5-kb fragment was cloned into the T-vector pT7Blue (Novagen), yielding pTRK454. In the second step, total DNA was digested to completion with EcoRV and cloned in similarly digested pBluescript-II KS(+). The ligation mix was again amplified by PCR with the vector-specific primer T7 and the groESL-specific primer (5′-CGTTCAG-CACTTCAAAATGCTGCTTC-3′) (primer C), and the resulting 1.1-kb fragment was cloned into pT7Blue, yielding pTRK455. Oligonucleotide primers designed to anneal near the 5′ end (5′-CACTA-TACAATCCAAGAAAC-3′) (primer D) and 3′ end (5′-GGTCATCACTGTCATCTTC-3′) (primer E) of the chaperone were used to amplify the groESL operon from total L. johnsonii NCK88 DNA. The PCR fragment was gel purified in SeaKem GTG agarose (FMC Bioproducts, Rockland, Maine), labeled with [α-32P]dCTP (New England Nuclear) by using the Multiprime DNA labeling system (United States Biochemical/Amersham Life Science), and used to probe a partial genomic library of NCK88. The library was constructed by completely digesting total DNA with BglII, separating the fragments on a 1.1% NuSieve GTG agarose gel (FMC Bioproducts), transferring only the outer three lanes of the gel to a Magnacharge nylon membrane (Micron Separations, Inc., Westboro, Mass.) by the method of Southern (30), and hybridizing with the groESL probe as described by Le Bourgeois et al. (16). A 1.0-cm band, corresponding in position to the hybridization signal, was excised from the remaining gel, the agarose was digested with α-agarase (New England Biolabs, Beverly, Mass.), and the precipitated DNA was cloned into BglII-digested pTRKH2. The ligation mix was used to electroporate E. coli XL1-Blue MCR′ Kan cells, and 700 transformants were replica plated, transferred to Magnagraph nylon membranes (Micron Separations, Inc.) by the colony lift method used with the Genius kit (Boehringer Mannheim Biochemicals), and hybridized against the same groESL probe used to construct the partial library. The Lactobacillus groESL operon was detected in a single clone (pTRK475) containing a 12.0-kb BglII fragment. A 4.5-kb SpeI-ClaI fragment, hybridizing with both the chaperone and IS1223, was subcloned into pBluescript-II KS(+), producing pTRK476, although a 1.5-kb deletion occurred in the process.
FIG. 1.
Strategy for cloning and sequence analysis of the L. johnsonii VPI 11088 groESL operon. A 2.4-kb fragment containing groES and the 5′ portion of groEL was amplified from the Lactobacillus chromosome by PCR with the IS1223-specific primer A and cloned to yield pTRK453. The remainder of groEL was recovered in two steps by using single-specific-primer PCR (36) and primers B and C, yielding pTRK454 and pTRK455, respectively. Following the assembly of the complete groESL sequence, the chaperone operon was amplified from the chromosome by using primers D and E and used as a probe to recover the genomic copy (pTRK475) from a partial BglII genomic library cloned in pTRKH2. The restriction enzymes SpeI and ClaI were used to subclone the chaperone operon into pBluescript-II KS(+), yielding pTRK476. The gray arrows indicate the 121-bp direct repeats (see the text for details).
Lactobacillus total-RNA isolation.
All Lactobacillus total RNA isolations were performed on 10-ml cultures which were pelleted by centrifugation at 6,058 × g, frozen in a dry ice-ethanol bath and held at −70°C. A 1-ml volume of TRIzol reagent (GibcoBRL, Gaithersburg, Md.) was added to each cell pellet, and the mixture was transferred to 2.0 ml screw-cap microcentrifuge tubes along with a 0.7 g of glass beads (106 μm in diameter) (Sigma Chemical Co., St. Louis, Mo.). The contents of the tubes were homogenized in a Mini-Beadbeater-8 cell disruptor (Biospec Products, Bartlesville, Okla.) for three 1-min cycles (and chilled on ice between the cycles). The phases were separated by centrifugation following the addition of 0.2 ml of chloroform and an additional 15-s homogenization. Total RNA was recovered as specified by the manufacturer.
Determination of the groESL transcription start sites.
L. johnsonii NCK88 was grown at 37°C to an optical density at 590 nm (OD590) of 0.60. Duplicate cultures were either transferred to 55°C or maintained at 37°C for 10 or 20 min, chilled briefly on ice, and collected by centrifugation. Total RNA was isolated as above, and 15 μg was incubated at 65°C for 5 min with 30 U of RNase inhibitor (Boehringer Mannheim Corp.), 2.8 μl of 5× first-strand buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), and 1 pmol of oligonucleotide in a final volume of 14 μl. Separate reactions were conducted with one of three oligonucleotides complementary to positions 775 to 752 (primer B′) (5′-CGGCAATAATTTCGCCCATTTGAG-3′), positions 382 to 360 (primer C′) (5′-GGTGTAATTACTATTGTACAGAC-3′), or positions 275 to 294 (primer A′) (5′-CACTATA-CAATCCAAGAAAC-3′) (see Fig. 2). Annealing was accomplished by allowing the total RNA-oligonucleotide solutions to cool slowly from 65°C to room temperature. The primer extension reactions were carried out for 60 min at 42°C following the addition of 6.0 μl of a master mix containing 75 μCi of [35S]dATP, 500 μM dCTP, 500 μM dTTP, 500 μM dGTP, 50 μM dATP, 33 mM dithiothreitol, 2,000 U of Superscript-II, RNase H− reverse transcriptase (GibcoBRL), and 1× first-strand buffer. The RNA-DNA hybrids were extracted once with phenol-chloroform, precipitated with ethanol, and resuspended in 4 μl of sequencing stop solution. The length of the primer extension product was calculated by comparing mobility in a 6.0% polyacrylamide gel with that of the product of a sequencing reaction generated with the same primer.
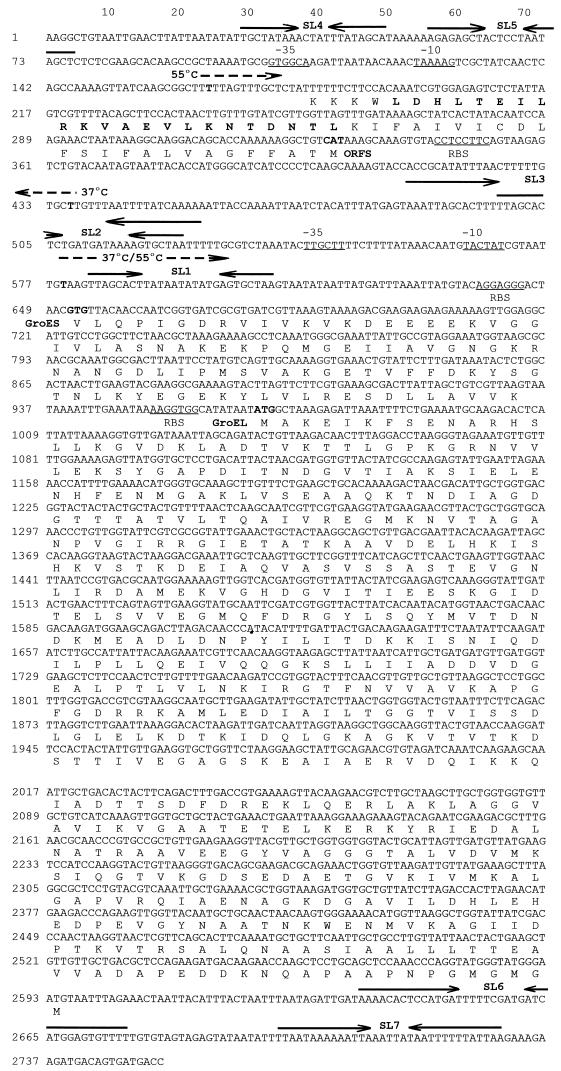
FIG. 2.
Nucleotide sequence of the L. johnsonii groESL chaperone operon. Initiation sites are indicated by boldface type, and the direction of transcription and the temperature at which promoter activity was detected are marked by dashed arrows. The hexamers indicative of promoters are underlined, and the inverted repeats termed CIRCE elements are designated SL1 and SL2. SL2 is overlapped by an additional IR (SL3). The IRs SL4 and SL5 precede the chaperone operon, while SL6 and SL7 may constitute a terminator. Potential ribosome binding sites are underlined, and the corresponding amino acid sequences of GroES, GroEL, and ORFS are indicated near the respective translation start sites. The sequence of ORFS is written 3′ to 5′ relative to the coding strand, and the putative leucine zipper is demarcated in boldface type.
Slot-blot hybridizations of groESL mRNA.
Identical 100-ml volumes of MRS broth were inoculated from an overnight culture incubated at 37°C. After reaching an OD590 of 0.6, the cultures were held at 37°C or shifted to 42, 47, 50, or 55°C. At the initial time point and every 30 min thereafter, a 10-ml sample was collected from each culture, briefly centrifuged, and stored at −70°C. After 150 min of incubation at the elevated temperatures, all the cultures were returned to 37°C for a 30-min recovery and a 10-ml sample was collected as before. Total RNA was isolated from all cell pellets, and volumes of RNA equivalent to 10 μg were alkali denatured, transferred to Zeta Probe blotting membranes (Bio-Rad Laboratories, Richmond, Calif.) with a Bio-Dot SF microfiltration apparatus (Bio-Rad) as specified by the manufacturer, and treated with one UV-auto-cross-linking cycle by using the UV Stratalinker 1800 (Stratagene). Prehybridization and hybridization were carried out at 65°C in 0.5 M NaHPO4 (pH 7.2)–1.0 mM EDTA–7.0% sodium dodecyl sulfate (SDS) with the same [α-32P]dCTP-labeled, PCR-generated probe used in cloning the L. johnsonii groESL operon described above.
Northern blot analysis of groESL mRNA.
Duplicate cultures of NCK88 were grown to an OD590 of 0.6 at 37°C, at which time one culture was shifted to 55°C. Aliquots (10 ml) were collected every 10 min for 1 h. Total RNA was isolated, separated by electrophoresis through a 1.5% agarose–formaldehyde denaturing gel, transferred to Magna Charge nylon membranes (Micron Separations, Inc.) by the method of Sambrook et al. (24), and fixed by UV cross-linking. Prehybridizations (3 h) were carried out at 42°C in 50% formamide–5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–2.5× Denhardt’s reagent, 2.0% SDS–0.1% denatured salmon sperm DNA. The groESL fragments used as probes in the separate hybridization reactions were generated by PCR from total NCK88 DNA by using primer pairs specific for one of three regions in the chaperone operon. Probe B (see Fig. 2), a 107-bp fragment specific for the 5′ region preceding the operon, was generated by using primers spanning bp 275 to 295 (5′-CACTATACAATCCAAG-AAAC-3′) and 381 to 360 (5′-GGTGTAATTACTATTGTACAGAC-3′). Probe C, a 153-bp PCR fragment specific for groES, was amplified by using primers spanning bp 623 to 643 (5′-GATTTAAATTATGTACAGGAG-3′) and 775 to 752 (5′-CGGCAATAATTTCGCCCATTTGAG-3′). Probe D, a 1,274-bp PCR fragment specific for groEL, was amplified by using primers spanning bp 1117 to 1144 (5′-GATGGTGTTACTATCGCCAAGAGTATTG-3′) and 2390 to 2371 (5′-ACTTCTGGGTCTTCATGTTC-3′). The fragments were gel purified, labeled as described above, and used in 16-h hybridization reactions at 42°C. The hybridization solution differed from the prehybridization solution only in the absence of the denatured salmon sperm DNA. The membranes were washed twice at room temperature for 20 min with 100 ml of 2× SSPE–1.0% SDS and once at 55°C for 20 min with the same solution and exposed to X-OMAT autoradiography film (Eastman Kodak Co., Rochester, N.Y.).
Analysis of freeze tolerance in L. johnsonii NCK88 following heat shock.
An overnight culture of NCK88 was used to inoculate 100 ml of MRS broth, which was incubated at 37°C until an OD590 of 0.6 was reached. The cells were pelleted by centrifugation and resuspended in fresh MRS broth. Half of the culture was held at 37°C, and the other half was transferred to a water bath at 55°C. At 0, 15, 30, and 45 min, triplicate 1-ml samples were removed from the 37 and 55°C cultures and placed in an Eppendorf tube. The cells were centrifuged, washed once, and then resuspended in 1 ml of sterile distilled water. Duplicate 100-μl samples were immediately plated on MRS agar for determination of the number of CFU per milliliter. All remaining samples in Eppendorf tubes were then frozen at −20°C and held for exactly 7 days. The samples were then thawed in water at room temperature, diluted in 10% MRS broth, and plated on MRS agar for determination of the number of surviving CFU per milliliter. The percent survival was calculated from the ratio of CFU per milliliter after freezing to CFU per milliliter before freezing. Each reported value was the mean determined from three samples plated in duplicate.
RESULTS
Isolation and sequence analysis of the groESL operon.
During PCR experiments to localize the insertion element IS1223 (35) relative to the genetic determinants for the bacteriocin lactacin F (18), the IS-specific primer A alone generated a ∼2.4-kb fragment, which was cloned into pBluescript-II KS(+) to yield pTRK453 (Fig. 1). Sequence analysis revealed that the insert contained one complete open reading frame (ORF), preceded by a ribosome binding site (GGAGGG), which could encode a protein of 94 amino acids, and a second truncated ORF, also preceded by a ribosome binding site (AGGTGG), which could encode a protein fragment of 110 amino acids. A comparison with the protein database revealed that the 94-residue protein bore significant homology to numerous prokaryotic and eukaryotic GroES proteins while the truncated protein was homologous to numerous GroEL proteins. The pTRK453 clone proved to be unstable and difficult to sequence, with deletions occurring between two inverted-repeat (IR) structures in the promoter region preceding the groES gene. Difficulties with groES instability have been reported previously for both Bacillus subtilis (17) and Lactococcus lactis subsp. lactis (14). Single-specific-primer PCR was used to recover the remainder of the groEL gene as two fragments in pTRK454 and pTRK455 (Fig. 1). The complete nucleotide sequence of the L. johnsonii NCK88 groESL operon was assembled by using the partial sequences derived from the clones pTRK453, pTRK454, and pTRK455. Any ambiguities remaining in the sequence derived from PCR-generated clones were resolved by using the chromosomal groESL recovered in pTRK476. The complete DNA sequence and amino acid translation are presented in Fig. 2.
The 2,753-bp L. johnsonii NCK88 region contained two ORFs encoding putative GroES and GroEL proteins of 94 and 543 residues, respectively. Both ORFs were preceded by ribosome binding sites. Only groES was immediately preceded by a potential promoter region, which was itself bracketed by three sets of inverted repeats (stem-loops) (SL1, SL2, and SL3). The only possible terminator structures detected in the sequence, designated SL6 and SL7 (Fig. 2), followed the second ORF (groEL), providing further evidence that the two ORFs probably comprise an operon. Compared with other chaperonins, the putative L. johnsonii GroEL protein displayed 84, 70, and 64% identity to those of Lactobacillus helveticus, Bacillus subtilis, and Clostridium perfringens, respectively. The putative L. johnsonii GroES protein exhibited 70, 48, and 44% identity to the homologous proteins of L. helveticus, B. subtilis, and C. perfringens, respectively. A third potential ORF (ORFS), transcribed in the opposite direction to groESL (Fig. 2), was located on the noncoding strand. ORFS is preceded by a potential ribosome binding site (AGGAGG) and promoter region and could encode a short protein of only 50 amino acids. A scan for protein sites and signatures (PC/Gene) detected a putative leucine zipper within the short peptide.
During subsequent experiments, the region immediately preceding the copy of IS1223, located upstream from the groESL operon, was sequenced (pTRK475 [Fig. 1]). The area contained a 121-bp, imperfect direct repeat (105 bp conserved) (data not shown) which was also found to precede the groESL operon between nucleotides 34 and 154 (Fig. 2). The 121-bp direct repeats bracket IS1223 and are probably responsible for the deletion of the insertion element during recovery of pTRK476 via homologous recombination.
Heat shock induction of the Lactobacillus groESL operon.
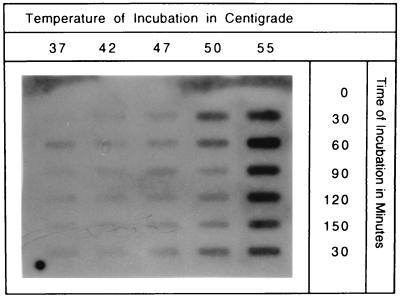
Although transcription of groESL operons is induced by a number of protein-denaturing stresses, chaperone response to heat shock is well documented. To evaluate the heat shock response in lactobacilli and determine the most effective temperature for subsequent groESL induction experiments, slot-blot hybridization was used to analyze total RNA isolated from L. johnsonii cultures following exposures of up to 150 min at temperatures ranging from 37 to 55°C. Based on the strength of the hybridization signal, the strongest expression of groESL in this temperature range occurred at 55°C following a 30- to 60-min exposure (Fig. 3). Additional time and temperature conditions were not evaluated.
FIG. 3.
Heat shock induction of the Lactobacillus groESL operon. Total RNA was isolated from L. johnsonii VPI 11088 following timed exposure to various extremes in temperature and analyzed by slot-blot hybridization. Volumes of RNA equivalent to 10 μg/slot were transferred, fixed, and probed with a groESL fragment spanning nucleotides 275 to 2753. For the final 30-min incubation, all samples were transferred back to 37°C. The temperatures and duration of exposure preceding the final 30-min incubation are indicated as column and row headings, respectively.
Characterization of Lactobacillus groESL transcription activity by Northern blotting.
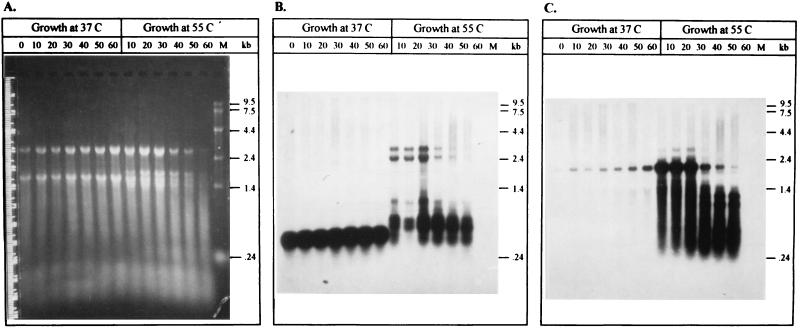
Chaperone transcription activity was evaluated from cultures at 37°C or heat shock conditions (55°C) by probing multiple Northern blots with DNA fragments isolated from specific regions of the groESL operon. Hybridization with probe B, specific for the promoter region (Fig. 4), revealed the presence of a strong transcript of 0.5 kb in the RNA isolated from 37°C samples (Fig. 5B). Following a shift to 55°C, the 0.5-kb transcript signal was diminished and new transcripts of 0.7, 0.8, 2.5, and 3.1 kb appeared. Hybridization of RNA extracted from 37°C cultures with the groES-specific probe C (Fig. 3) revealed a single 2.1-kb transcript (Fig. 5C), which coincides with the expected size of the total groESL transcription product. A shift to heat shock conditions (55°C) clearly increased the strength of expression of the 2.1-kb transcript and produced minor transcripts of 3.1 and 2.5 kb and possible degradation products ranging from 1.2 to 0.5 kb. Hybridization of a duplicate Northern blot with the groEL-specific probe D (Fig. 4) yielded the same hybridization pattern (data not shown), which was expected since groES and groEL are typically transcribed together (reviewed by Gupta [9]).
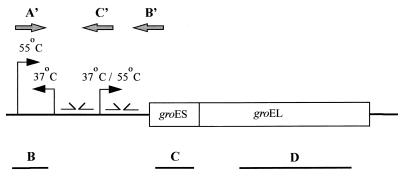
FIG. 4.
Organization of the groESL operon and RNA analysis. Primer extension reactions were carried out with primers A′, B′, and C′. Northern blots of total RNA were hybridized with probe B, targeted against the promoter region, and probes C and D, targeted against groES or groEL, respectively. The initiation sites for the primary transcripts, the temperatures at which they are active, and the IRs comprising the CIRCE elements are represented.
FIG. 5.
Northern blot analysis of Lactobacillus groESL mRNA isolated from cultures maintained under normal or heat shock conditions. Total RNA was isolated from log phase cultures maintained at 37 or 55°C, and 10-μg volumes were separated by electrophoresis through multiple 1.5% agarose–formaldehyde gels and transferred to nylon membranes. Lane 0 contains RNA isolated prior to the temperature shift, lanes 10 to 60 contain RNA isolated at 10-min intervals after the temperature shift, and lane M contains RNA molecular weight markers (Gibco/BRL). (A) Representative agarose-formaldehyde gel which has been stained with ethidium bromide and photographed. (B and C) Northern blots were probed with either a 107-bp fragment specific for the groESL promoter region (B) or a 153-bp fragment specific for groES (C).
Identification of the transcriptional start sites within the groESL promoter region.
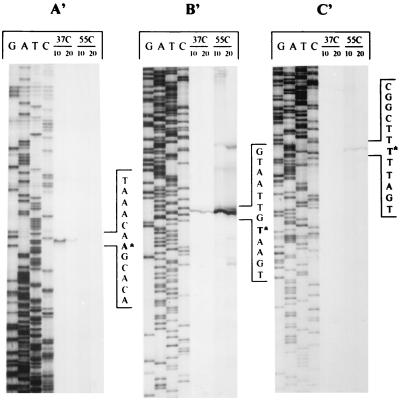
To determine the general characteristics and organization of the Lactobacillus groESL promoter, transcription initiation sites were mapped by primer extension. Total RNA was isolated from log-phase cells following a 10- or 20-min exposure to either 37°C (nonstressed) or 55°C (heat shock) and annealed with one of three primers. The initiation site for the primary groESL transcript was determined by extending from position 775 with primer B′, diagrammatically represented in Fig. 4. Under nonstressed conditions, a moderate level of initiation occurred at nucleotide 579 (Fig. 6B′), which is positioned between two sets of inverted repeats, SL1 and SL2 (Fig. 2). These inverted repeats varied by only 1 bp from a regulatory structure termed CIRCE (40), which is involved in the negative regulation of groESL expression in B. subtilis (37). Transcription markedly increased following a shift in growth temperature to 55°C (Fig. 6B′). Some additional but minor heat shock-dependent transcription activity was detected with primer C′, which anneals between nucleotides 360 and 382. Nucleotide 165, which is located upstream from the putative CIRCE elements, was identified as the 55°C heat shock initiation site (Fig. 6C′).
To confirm the identity of the 37°C (0.5-kb) transcript detected in Northern blots by hybridization with the 5′-specific probe B (Fig. 5B), an extension reaction was performed with primer A′ (Fig. 3), which anneals between nucleotides 275 and 294, complementary to the noncoding strand. A transcription initiation site was identified at nucleotide 436 (Fig. 6A′). The transcript, while clearly visible at 37°C, disappeared following heat shock. Taken together, these data indicate that the 600-bp region preceding groES contains three temperature-sensitive promoters, one of which is divergent (2).
Impact of heat shock on tolerance to freezing.
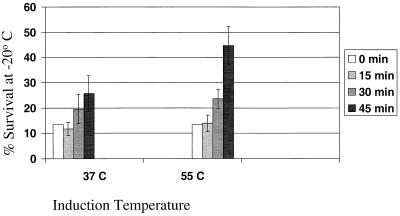
Since the parameters for heat shock induction of the groESL operon had been established, experiments were conducted to evaluate the effect of increased chaperone expression on freezing-stress tolerance of L. johnsonii. Cells exposed to the induction temperature of 55°C showed an approximate 1.5-log-unit reduction in the number of viable cells after the 45-min treatment, compared to the control cells held at 37°C for 45 min, which showed no significant change in population (data not shown). However, the viable cells remaining after 45 min at 55°C showed significantly greater survival after freezing at −20°C for 7 days than did the control cells (Fig. 7). The control culture showed a modest increase in survival from 14 to 26%, which was expected since the culture entered a later stage of growth. On the other hand, exposure to 55°C for 30 and 45 min increased the survival after freezing to 24 and 45%, respectively. The time course for the onset of protection by 55°C preconditioning corresponded to the pattern of heat shock induction of groESL RNA (Fig. 3). While treatment at 55°C has an overall negative impact on cell survival, induction of the chaperone operon in L. johnsonii appears to correlate with the onset of cross-protection to another temperature stress, in this case freezing.
FIG. 7.
Heat shock induction of stability to freezing in L. johnsonii. Cultures were held at either 37 or 55°C for 0, 15, 30, or 45 min before being frozen and stored at −20°C for 7 days. The percent survival was calculated from the ratio of CFU per milliliter determined in the 37 and 55°C pretreated cultures before and after freezing. Triplicate samples were taken at each time point and enumerated in duplicate before and after freezing.
DISCUSSION
The groES/groEL chaperone operon of L. johnsonii was identified adjacent to a copy of IS1223 and characterized. The significance of this genomic arrangement is unknown, but a similar organization has been reported for IS1016 in Neisseria meningitidis (31). PCR-based methods have been used to recover an entire groESL operon (32), as well as groES fragments which have proven difficult to clone from genomic preparations (14, 17). The majority of the 2,753-nucleotide sequence was generated from PCR-derived clones, with the exception of the promoter region. Given the importance of this regulatory region in the bacterial stress response, it was sequenced in its entirety from a genomic clone. Translation of the coding strand revealed the presence of two contiguous ORFs bearing significant homology to ORFs encoding numerous GroES and GroEL proteins. Sequence analysis revealed the presence of a third potential ORF (ORFS), running counter to the chaperone operon, in the region preceding groES. Even though potential transcription and translation signals were detected upstream from ORFS, a BLAST search (1) of the protein database revealed no significant homologies to the putative peptide. A structural analysis detected the presence of a potential leucine zipper at the carboxyl-terminus of the ORFS product. The heptad leucine repeats, characteristic of leucine zippers (21a), are involved in the dimerization of some DNA-binding proteins.
The groESL operons of both Clostridium acetobutylicum (19) and B. subtilis (27), although strongly induced by heat, are preceded by typical vegetative promoters. A highly conserved palindromic structure is found near the promoters of these and other heat shock genes in a wide range of bacteria. These 9-bp IRs, termed CIRCE, are thought to form a stem-loop structure and function as negative cis elements in the heat shock regulation of downstream genes (40). Evidence has been presented suggesting that CIRCE plays a dual role as both a regulatory element, providing for rapid transcript turnover under nonstressed conditions, and a promoter-proximal operator (37). A CIRCE-dependent, negative regulator has been tentatively identified as hrcA in B. subtilis (28, 38) and Caulobacter crescentus (22), respectively, and is transcribed with the genetic components of the dnaK, dnaJ, and grpE chaperone operon. The reported consensus sequence of the CIRCE elements (5′-TTAGCACTC-9N-GAGTGCTAA-3′) is generally present as a single copy near the groES and dnaK promoters. Two potential CIRCE elements, designated SL1 and SL2, were identified in the region immediately preceding the L. johnsonii groESL chaperone operon (Fig. 3).
With the exception of the dnaK operon of L. lactis subsp. lactis (5), where dual CIRCE elements flank the promoter for an hrcA homolog, and the groESL operon of L. helveticus (3), the published sequences of previously isolated CIRCE-based heat shock promoter regions display a single set of IRs. The presence of two CIRCE elements in L. johnsonii, one on either side of the primary promoter, would probably provide a stronger level of negative regulation than could be afforded by a single copy. Since groESL induction experiments are typically conducted at 10°C above the optimum growth temperature, the appearance of two CIRCE elements could explain the relatively high temperature (55°C) required for elevated expression of the groESL operon in L. johnsonii. Repression could be reduced if the effective concentration of the negative regulator, near the dual CIRCE elements, were decreased through degradative processes or simple titration. In either case, the probability of simultaneous binding at both CIRCE elements would be reduced and the likelihood of subsequent promoter activity would be increased. Alternatively, the impact of the two CIRCE elements on promoter inactivation may vary considerably, since the putative negative regulator may display different binding affinities for SL1 and SL2. It is interesting that the inverted repeats that form SL2, occur within a larger pair of inverted repeats (SL3), which could mask the upstream CIRCE element from regulatory control.
Northern blot analysis of total RNA confirmed the heat shock induction of the groESL operon and revealed differential transcript activity. Hybridization experiments with either groES- or groEL-specific probes revealed the presence of a 2.1-kb transcript at 37°C which is greatly induced following heat shock (Fig. 5C). In addition to the 2.1-kb product, which is the expected size of the groESL transcript, heat shock clearly induced minor transcripts of 2.5 and 3.1 kb, suggesting that multiple promoters are active at 55°C. When the 5′ region of the operon was used as a probe, a 0.5-kb transcript was detected in RNA isolated at 37°C (Fig. 5B). The production of this transcript was greatly reduced following heat shock, and multiple transcripts of 3.1, 2.5, 0.8, and 0.7 kb appeared. Several interpretations of these data are possible. During growth under noninducing conditions (37°C), any transcripts initiating from promoters upstream from the groESL operon would probably terminate at or near the CIRCE elements, yielding fragments of 0.5 kb or less. Heat shock-induced release of a negative regulator could prevent premature termination and allow transcription to continue into the chaperone operon. This is consistent with the appearance of 2.5- and 3.1-kb transcripts following induction, which were longer than the expected full-length groESL transcript of 2.1 kb. Alternatively, transcription may initiate at multiple promoters within the 5′ region: one on the noncoding strand under normal growth conditions (37°C) and running in the opposite orientation from that of groESL, and another on the coding strand under heat shock conditions (55°C).
Primer extension revealed the presence of three active promoters in the region preceding the Lactobacillus groESL operon. Primary groESL transcription activity was localized to nucleotide 579, which lies near the base of the putative CIRCE element designated SL1 (Fig. 2). Since the −35 and −10 hexamers preceding this start site are positioned squarely between SL1 and SL2, the binding of a negative regulator to either CIRCE element would probably affect promoter activity. The dramatic increase in transcription activity that accompanied a shift from 37 to 55°C (Fig. 6B) would be expected if a regulatory protein released one or both CIRCE elements following heat shock, allowing RNA polymerase more effective access to the promoter. Heat shock revealed the presence of a second initiation site, 414 bases upstream from the primary one at position 165 (Fig. 6C), which could generate the 2.5-kb transcript observed in Northern blots (Fig. 5B and C). Since activity at this site is triggered by heat stress, it may provide a mechanism for increasing the concentration of chaperone transcripts under extreme conditions. The presence of a heat-sensitive divergent promoter in the region immediately preceding the groESL operon was confirmed when a third transcription initiation site was detected at nucleotide 436, on the noncoding strand (Fig. 6A). ORFS, which immediately followed this start site, contained both a potential ribosome binding site and a leucine zipper. Since some DNA binding proteins are known to contain leucine zipper regions (21a) and the transcription activity of the divergent promoter is heat labile, it is possible that ORFS performs some regulatory function in response to environmental signals such as heat shock. Alternatively, since they have a significant 5′ homology, the divergent transcript could effect chaperone expression by the formation of a partial RNA duplex with any transcripts initiating from the secondary groESL promoter. Both possibilities are highly speculative since a similar divergent promoter has not been detected in other CIRCE-based regulatory regions.
FIG. 6.
Determination of Lactobacillus groESL transcription start sites by primer extension. Total RNA was isolated from log-phase cultures held for 10 or 20 min at either 37 or 55°C. Volumes of RNA equivalent to 40 μg were annealed with one of three primers (listed below), and extension reactions were carried out. The lengths of the extension products were estimated by comparing mobility in a 6.0% polyacrylamide gel. Lanes G, A, T, and C indicate the dideoxy termination lanes. Extension products were generated with RNA isolated from 37 and 55°C cultures incubated for 10 or 20 min. Divergent promoter activity was detected with primer A′ (5′-CACTATACAATCCAAGAAAC-3′), which was designed complementary to the noncoding strand 5′ to groES. Primary and secondary groESL activity were detected by using primers designed to anneal to the coding strand either 3′ (B′) (5′-CGGCAATAATTT-CGCCCA-TTTGAG-3′) or 5′ (C′) (5′-GGTGTAATTACTATTGTACAGAC-3′) to the start of groES.
Heat shock induction of the groESL chaperone operon in L. johnsonii provided some cross-protection to freezing. While protection against other types of stress was not evaluated in this study, it is probable that increasing chaperone concentrations at opportune times can improve the general tolerance of lactobacilli to stresses encountered during production, concentration, storage, and distribution of fermentative or probiotic cultures. In this study, we found that stress protection correlated with the timing and level of expression of the chaperone operon. Determination of optimum conditions for RNA expression over a stress operon provides a means of evaluating and optimizing pretreatment conditions that are likely to provide stress protection, without the need for lengthy empirical determinations that attempt to catalog various pretreatments with culture survival or activity after stress.
This study has revealed that features of the groESL operon are shared between various lactic acid bacteria, notably other Lactobacillus species and Lactococcus lactis. The significant homology between DNA sequences, with similar regulatory elements and induction conditions, indicate that these systems may be widely conserved among the considerably diverse lactobacilli and lactic acid bacteria. In this regard, refined pretreatments that elevate the levels of molecular chaperones are likely to provide enhanced stress tolerance and culture fitness across the many important members of this functional and beneficial group of lactic acid bacteria.
ACKNOWLEDGMENTS
This work was supported by the North Carolina Dairy Foundation; Rhodia, Inc., Madison, Wis.; and the Southeast Dairy Foods Research Center. H. Girgis is a GAANN Biotechnology Fellow at NCSU.
Footnotes
Paper FSR98-38 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Biol Chem. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Beck C F, Warren R A J. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988;52:318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadbent J R, Orberg C J, Wei L. Characterization of the Lactobacillus helveticus groELS operon. Res Microbiol. 1998;149:247–253. doi: 10.1016/s0923-2508(98)80300-8. [DOI] [PubMed] [Google Scholar]
- 4.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton T, Shearman C, Gasson M. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis subsp. lactis. J Gen Microbiol. 1993;139:3253–3264. doi: 10.1099/00221287-139-12-3253. [DOI] [PubMed] [Google Scholar]
- 6.Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgellis D, Sohlberg B, Hartl F U, von Gabain A. Identification of GroEL as a constituent of an mRNA-protection complex in Escherichia coli. Mol Microbiol. 1995;16:1259–1268. doi: 10.1111/j.1365-2958.1995.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 8.Georgopoulos C, Welch W J. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 9.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 10.Hendrick J P, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 11.Hightower L E. Heat shock, stress proteins, chaperones and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 12.Horwich A L, Low K B, Fenton W A, Hirshfield I N, Furtak K. Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- 13.Kaul S C, Obuchi K, Iwahashi H, Komatsu Y. Cryoprotection provided by heat shock treatment in Saccharomyces cerevisiae. Cell Mol Biol. 1992;38:135–143. [PubMed] [Google Scholar]
- 14.Kim S G, Batt C A. Cloning and sequencing of the Lactococcus lactis subsp. lactis groESL operon. Gene. 1993;127:121–126. doi: 10.1016/0378-1119(93)90626-e. [DOI] [PubMed] [Google Scholar]
- 15.Langer T, Pfeifer G, Martin J, Baumeister W, Hartl F-U. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 1992;11:4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Bourgeois P, Lautier M, Mata M, Ritzenhaler P. New tools for the physical and genetic mapping a Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Wong S-L. Cloning and characterization of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3981–3992. doi: 10.1128/jb.174.12.3981-3992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muriana P M, Klaenhammer T R. Conjugal transfer of plasmid-encoded determinants for bacteriocin production and immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987;53:553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narberhaus F, Bahl H. Cloning, sequencing, and molecular analysis of the groESL operon of Clostridium acetobutylicum. J Bacteriol. 1992;174:3282–3289. doi: 10.1128/jb.174.10.3282-3289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neidhardt F C, VanBogelen R A. Heat shock response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. II. Washington, D.C: American Society for Microbiology; 1987. pp. 1334–1345. [Google Scholar]
- 21.O’Sullivan D J, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 21a.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 22.Roberts R C, Toochinda C, Avedissian M, Baldini R, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross R P, Claiborne A. Cloning, sequence and overexpression of NADH peroxidase from Streptococcus faecalis 10C1: structural relationship with the flavoprotein disulfide reductases. J Mol Biol. 1991;221:857–871. doi: 10.1016/0022-2836(91)80180-3. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanders M E. Summary of conclusions from a consensus panel of experts on health attributes of lactic cultures: significance to fluid milk products containing cultures. J Dairy Sci. 1993;76:1819–1828. doi: 10.3168/jds.S0022-0302(93)77514-1. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt A, Schiesswohl M, Völker U, Hecker M, Schumann W. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3993–3999. doi: 10.1128/jb.174.12.3993-3999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shyamala V, Ames G F-L. Genome walking by single-specific-primer polymerase chain reaction: SSP-PCR. Gene. 1989;84:1–8. doi: 10.1016/0378-1119(89)90132-7. [DOI] [PubMed] [Google Scholar]
- 30.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Stevens D S. Transposable elements and migratory DNA in Neisseria meningitidis. Section 59: horizontal gene transfer. Presented at the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. [Google Scholar]
- 32.Tamada H, Ohta T, Hamamoto T, Otawara-hamamoto Y, Yanagi M, Hiraiwa H, Hirata H, Kagawa Y. Gene structure of heat shock proteins 61KDa and 12KDa (thermophilic chaperonins) of thermophilic bacterium PS3. Biochem Biophys Res Commun. 1991;179:565–571. doi: 10.1016/0006-291x(91)91408-5. [DOI] [PubMed] [Google Scholar]
- 33.Völker U, Mach H, Schmid R, Hecker M. Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J Gen Microbiol. 1992;138:2125–2135. doi: 10.1099/00221287-138-10-2125. [DOI] [PubMed] [Google Scholar]
- 34.Walker D C, Klaenhammer T R. Proceedings of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Cloning and molecular characterization of the groESL chaperone operon from Lactobacillus johnsonii, abstr. 1729. [Google Scholar]
- 35.Walker D C, Klaenhammer T R. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J Bacteriol. 1994;176:5330–5340. doi: 10.1128/jb.176.17.5330-5340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman J S, Kashi Y, Fenton W A, Horwich A L. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 37.Yuan G, Wong S-L. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence(CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan G, Wong S-L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6368. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu K Y, Clark J M. Rapid construction of nested deletions of recombinant plasmid DNA for dideoxy sequencing. BioTechniques. 1995;18:222–224. [PubMed] [Google Scholar]
- 40.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]