Abstract
Survival of a nontoxigenic isolate of Escherichia coli O157:H7 at low pH (pH 3.0) was examined over prolonged time periods for each of three population types: exponential-phase cells, stationary-phase cells, and acid-adapted exponential-phase cells. In each population, approximately 5 × 104 CFU ml−1 were detected after a 24-h incubation at pH 3.0. Even after 3 days at pH 3.0, significant numbers of survivors from each of the three populations could be detected. The high level of acid tolerance exhibited by these survivors was found to be quickly lost once they were transferred to conditions which permitted growth to resume, indicating that they were not mutants. Proton flux measurements on the three populations of cells revealed that the initial rates of viability loss at pH 3.0 correlated well with net proton accumulation. Cells showing a high initial rate of viability loss (exponential-phase cells) accumulated protons at the highest rate, whereas resistant populations (adapted or stationary-phase cells) accumulated protons only slowly. Differences in the protein composition of the cell envelope between the three populations were studied by two-dimensional polyacrylamide gel electrophoresis. Complex differences in the pattern of proteins expressed by each population were uncovered. The implications of these findings are discussed in the context of a possible model accounting for acid tolerance in this important food-borne pathogen.
Since the first recognized outbreak in 1982, Escherichia coli O157:H7 has emerged as a serious, potentially life-threatening, human food-borne pathogen (17). Outbreaks involving acidic foods have drawn attention to the acid-tolerant properties of this organism. Acidic foods such as apple cider (34), dry-fermented sausage (9), mayonnaise (35), and yogurt (29) have all been implicated in outbreaks of food poisoning attributed to E. coli O157:H7. In addition to the epidemiological data, survival studies have demonstrated the ability of E. coli O157:H7 to persist in acidic foods (1, 25, 28). Despite these studies, there is no conclusive evidence indicating that this serotype of E. coli is any more acid tolerant than laboratory or commensal strains of the organism. However, acid tolerance in pathogenic strains deserves special attention, as it is likely to play an important role in allowing the organism to survive passage through the low pH of the stomach, thereby increasing the chances of the pathogen establishing infection.
Although there is a substantial body of literature exploring the growth and survival of E. coli O157:H7 in acidic foods, little is known about the molecular basis for acid tolerance in this organism. In the E. coli O157:H7 strains that have been examined so far, it appears that acid tolerance is strongly dependent on growth phase; stationary-phase or starved cultures show high levels of acid tolerance compared to their exponential-phase counterparts (2, 5). Like other enterobacteria, E. coli O157:H7 has been shown to modulate acid tolerance levels in response to changes in extracellular pH, increasing tolerance when the external pH is mildly acid (5). This response, now universally known as the adaptive acid tolerance response (ATR), increases the ability of this pathogen to survive in acidic foods (25). Growth-phase-dependent acid tolerance requires the stress-specific sigma factor RpoS for full induction (9). RpoS is a regulatory factor required for the transcriptional activation of a large number of genes required for tolerance to environmental stress (reviewed in reference 27). Indeed, RpoS has also been shown to be required by E. coli O157:H7 for surviving heat and salt stress and for surviving prolonged storage in dry-fermented sausage (9). The RpoS-regulated genes involved in contributing to acid tolerance remain to be elucidated.
Two other mechanisms have been shown to play a role in acid tolerance in E. coli O157:H7 (also seen in nontoxigenic E. coli strains): the arginine-dependent and glutamate-dependent systems. In the presence of millimolar quantities of either of these amino acids, E. coli strains show an increased ability to resist killing during challenge at low pH. The basis for this protection is not fully understood, but it is thought to result from the intracellular decarboxylation of these amino acids with the concomitant consumption of protons and, therefore, maintenance of intracellular pH (pHi) (26).
In all of the enterobacteria studied, it has become clear that induced acid tolerance, in response to mild acid shock, involves complex changes at the level of protein expression. Protein analyses using two-dimensional (2D) gel electrophoretic approaches have revealed that a large number of proteins are repressed while others are induced under the conditions which trigger the increase in acid tolerance (reviewed in reference 3). In Salmonella typhimurium, a total of 70 proteins show altered expression when cells are exposed to sublethal proton concentrations in the pH range 4.5 to 6.0 (13). A subset of these changes are known to be regulated by RpoS, at least in virulent strains of S. typhimurium (23). Similar complex changes in protein expression are seen when E. coli is exposed to pH 5.0 (19), a pH sufficient to induce adaptive acid tolerance in this organism (16). The functions of these induced proteins in conferring tolerance to low pH remain obscure. It seems clear that improved pHi homeostasis is important for enhanced acid tolerance, at least in S. typhimurium (14). However, recent work on E. coli O157:H7 has shown that the correlation between pHi and cell death does not always hold and that factors other than pHi regulation (perhaps involving protection and/or repair of macromolecules) also determine tolerance to low pH (21).
In this study, we investigated the acid tolerance of a nontoxigenic strain of E. coli O157:H7. Three populations shown to vary with respect to their acid tolerance levels were studied: mid-exponential-phase cells, mid-exponential-phase acid-adapted cells, and stationary-phase cells. In each of the three populations, prolonged survival at pH 3.0 was observed. Even after 3 days at pH 3.0, significant numbers of survivors from each population were detected. Increased acid tolerance was shown to correlate strongly with a decrease in cytoplasmic proton accumulation. In addition, this change in proton permeability was correlated with alterations in the protein composition of the cell membranes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strain used in this study was the nontoxigenic E. coli O157:H7 isolate P1432 (obtained from P. Chapman, PHLS). This strain was isolated from a patient showing symptoms of gastroenteritis. It is negative for the toxin genes as shown by PCR and by a Vero cell culture toxin assay (8a). Strain P400 is an E. coli K-12 isolate (thr leu proA argE thi ara xyl mtl galK lacY supE str) and P460 is an ompA derivative which lacks this outer membrane protein and is defective in conjugation and phage adsorption (32). These strains were obtained from U. Henning, Max Planck Institute, Berlin, Germany. Cultures were grown in tryptic phosphate broth (TPB) (pH 7.0) at 30°C with vigorous aeration. The following three populations of cells were used in this study: (i) mid-exponential-phase cells (optical density at 600 nm [OD600], ≈0.4; 108 CFU ml−1), (ii) mid-exponential-phase cells acid adapted at pH 5.0 for 1 h (OD600, ≈0.4; 108 CFU ml−1), and (iii) stationary-phase cells (overnight culture, OD600, ≈2.0; 5 × 109 CFU ml−1). Under the conditions used in this study, growth rates (μ) were 1.54 and 0.92 h−1 for cultures grown in TPB at pH 7.0 or 5.0, respectively. Values for μ (the reciprical of doubling time in hours) were determined from the slope of the curve obtained by plotting the log10 of OD600 versus time. Strains were maintained at 4°C on brain heart infusion (BHI) agar slopes and stored long term at −80°C in TSB with 7% dimethyl sulfoxide.
Assay of acid tolerance.
Cultures were grown to the appropriate phase of growth in TPB (pH 7.0) at 30°C with vigorous aeration. Cultures were acid challenged by reduction of the medium pH to pH 3.0 with 10 M HCl. Viable cell counts were performed immediately prior to the pH adjustment and at suitable time intervals thereafter. Serial dilutions were performed in 0.1% peptone, and 10 μl of each dilution was spread onto BHI agar plates, which were incubated at 30°C for 24 to 48 h. When small numbers of survivors were expected, volumes of culture between 10 μl and 1 ml were plated directly from the challenge medium. All survival experiments were performed at least three times.
Induction of the ATR.
The ATR was induced by exposing mid-exponential-phase cells to pH 5.0 for 1 h. Cultures were grown to mid-exponential phase as described above and the pH was then reduced to pH 5.0 with 10 M HCl. The culture was incubated at this pH for 1 h prior to acid challenge at pH 3.0. Immediately before acid challenge, cell numbers were determined by viable cell counts (time zero). The acid challenge was performed as described above.
Measurement of growth and loss of acid tolerance.
Cells from each of the three populations defined above were challenged at pH 3.0 for 90 min. A sample was then removed and inoculated (at an inoculum calculated to give approximately 104 viable cells per ml) into fresh TPB at 7.0, and the culture was incubated with shaking at 30°C. At suitable time points thereafter, samples were removed and acid tolerance levels were determined; i.e., the samples were subjected to a 90-min acid challenge at pH 3.0 and the percentage of surviving cells was measured by viable cell counts. Growth was also recorded by determining the number of viable cells at each time point prior to acid challenge. In this way, growth and acid tolerance could be correlated.
Proton flux assay.
Proton flux assays were performed by using a modification of the method of Bender et al. (4). Cells were harvested, washed, and resuspended in 100 mM KCl at a concentration of 30 mg ml−1 (wet weight). Twenty milliliters of this suspension was transferred to a 50-ml beaker and incubated in a circulating water bath at 30°C with continuous stirring. The pH of the suspension was adjusted with either HCl or NaOH until it remained static for 2 min. The suspension was pulsed with 50 μl of 0.5 M HCl, and the pH was recorded continuously at 10-s intervals for 10 min. The initial drop in pH (typically 0.5 pH units) was reversed as protons flowed across the membrane into the cytoplasm. The rate of proton accumulation was calculated directly from the pH increase.
Recovery of membrane proteins.
Cultures were grown to the appropriate phase of growth, and cells were harvested by centrifugation for 10 min at 18,500 × g. The pellets were washed in 50 ml of 20 mM Tris-HCl (pH 7.1), harvested again, and resuspended in 6 ml of 20 mM Tris-HCl–10 mM EDTA (pH 7.1). The cells were sonicated, with cooling on a mixture of ice and ethanol, in bursts of 15 s followed by a cooling period of 45 s. This cycle was repeated seven times. The suspension was then centrifuged at 7,700 × g for 5 min to remove cell debris. The supernatant was retained and transferred to a centrifuge tube. The membrane material was separated from this protein suspension by centrifugation at 39,000 × g for 30 min, and the pellet was washed once in 20 mM Tris-HCl (pH 7.1). After centrifugation at 39,000 × g for 1 h, 800 μl of 20 mM Tris-HCl (pH 7.1) was added to the membrane pellet and this was then stored at −20°C until required for analysis.
Polyacrylamide gel electrophoresis (PAGE).
The total protein content of the membrane fractions was estimated by using a mini polyacrylamide gel (precast Bio-Rad gel). Prior to running the gel, the protein was diluted with an equal volume of sample buffer (0.0625 M Tris-HCl [pH 6.8], 10% [vol/vol] glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS], 5% [vol/vol] 2-mercaptoethanol, 0.002% [wt/vol] bromophenol blue). Equal volumes of the samples to be compared were loaded, and the gels were run at a constant current of 35 mA in a Tris-glycine running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS [pH 8.3]). Gels were stained with Coomassie blue (0.1% brilliant blue in 40% ethanol–7% acetic acid) and destained with 40% ethanol–7% acetic acid. The relative protein concentrations of the samples were assessed visually and were used to normalize the sample loading for the isoelectric focusing (IEF) gels. This method was found to be more reliable than using protein assay kits to determine protein concentration.
2D gel electrophoresis.
2D gels were run on a Pharmacia Multiphor II electrophoresis unit by the method of O’Farrell (30) with modifications described by the manufacturer. Proteins were resolved by using IEF in the first dimension and SDS-PAGE in the second dimension. The IEF gels for the first dimension were 110-mm Immobiline DryStrips with a pH gradient of 4.0 to 7.0 (Pharmacia, Uppsala, Sweden). For the second dimension, ExcelGel XL SDS 8 to 18% gradient polyacrylamide gels (Pharmacia) were used. Prior to running the gels, the protein concentration was standardized by acetone precipitation. The required volume of protein was diluted with 4 volumes of ice-cold acetone. After 5 min, this was centrifuged in a microcentrifuge at 12,500 × g for 10 min. The supernatant was discarded and the pellet was allowed to air dry. The pellet was resuspended in 50 μl of sample buffer (13.5 g of urea, 250 mg of dithiothreitol, 0.5 ml of pharmalyte, 0.13 ml of Triton X-100, 0.05% bromophenol blue). After running in the second dimension, protein spots were stained with Coomassie blue and destained with 40% ethanol–7% acetic acid. Gels were photographed with a Cosmicar Television Lens CCD camera, and the image was captured on computer by using Global Lab Image (Data Translocation, Berkshire, United Kingdom).
RESULTS
Nontoxigenic E. coli O157:H7 (strain P1432) displays an ATR and growth-phase-dependent acid tolerance.
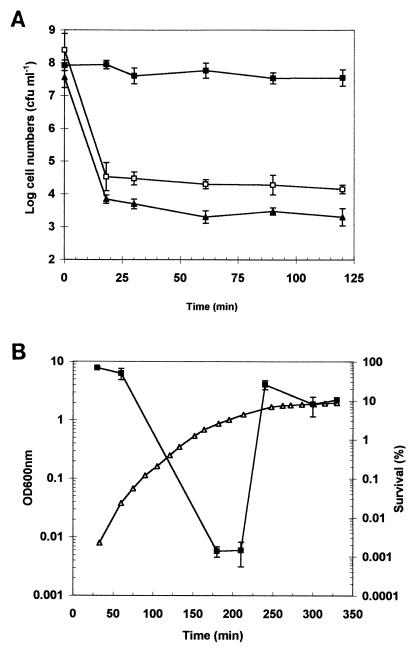
We first investigated whether strain P1432 possessed the ability to alter its acid tolerance levels in response to changing extracellular pH and in response to growth phase. First, cultures of P1342 were grown to mid-exponential phase in TPB (pH 7.0) and either acid adapted (at pH 5.0 for 1 h) or left unadapted. Cultures were then challenged at pH 3.0 and survival rates were recorded. The acid-adapted culture showed a very high level of tolerance to the acid challenge over the time course of the experiment, whereas the unadapted culture was rapidly killed (>1,000-fold reduction in viable cell numbers) within the first 15 min of the assay (Fig. 1A). This result confirms the ability of this strain to induce an ATR. We tested whether this response was dependent on de novo protein synthesis by including the protein synthesis inhibitor chloramphenicol during the adaptive period. The inclusion of chloramphenicol eliminated the ATR, confirming the requirement for protein synthesis for this response (Fig. 1A). Strikingly, the mid-exponential-phase culture showed biphasic death kinetics at pH 3.0, with a small fraction of the population (approximately 5 × 104 CFU ml−1) surviving after the initial period of rapid decline in viability. The pH of the cell-broth mixture was measured over the course of the challenge and was found to remain constant at pH 3.0 (data not shown), ruling out alkalinization of the medium as an explanation of this resistant “tail”. This resistant population was also observed in the chloramphenicol-treated culture, suggesting that it is not due to the induction of an ATR by these cells within the early stages of the acid challenge. It must, therefore, represent cells which are already present in the growing culture. Acid challenges were also performed with mid-exponential-phase cultures at pH 2.5 and 2.0, and similar biphasic death kinetics were observed, though the numbers of survivors detected in the tail were lower, at approximately 104 (standard deviation, ≈70%; n = 3) and 103 CFU ml−1 (standard deviation, ≈120%; n = 3), respectively (data not shown).
FIG. 1.
Adaptive acid tolerance and growth-phase-dependent acid tolerance in E. coli O157:H7 strain P1342. (A) Survival of exponential-phase cells challenged at pH 3.0. Cells were grown to mid-exponential phase in TPB at 30°C and either unadapted (open squares) or adapted at pH 5.0 for 1 h (solid squares) prior to challenge. One culture was adapted in the presence of chloramphenicol (5 μg ml−1) added 15 min prior to adaptation (solid triangles). (B) Acid tolerance measured throughout growth. An overnight culture was used to inoculate TPB (inoculum, 0.5% [vol/vol]), and acid tolerance (percent survival after a 90-min challenge at pH 3.0) was measured at time intervals during growth (solid squares). Growth was recorded by measuring the OD600 (open triangles). For panels A and B, the error bars represent the standard deviations from the means (n = 3).
Acid tolerance levels were also monitored throughout growth. Samples of culture were removed during growth and subjected to an acid challenge (pH 3.0 for 90 min). Tolerance levels were found to be high initially, presumably reflecting the fact that a stationary-phase inoculum was used. Acid tolerance levels dropped dramatically during the exponential phase of growth and then increased rapidly as the culture entered stationary phase (Fig. 1B). Together, these data demonstrate the ability of this nontoxigenic O157:H7 strain of E. coli to alter acid tolerance levels both in response to a mild acidic external pH and upon entry into the stationary phase of growth.
Prolonged acid tolerance of strain P1432.
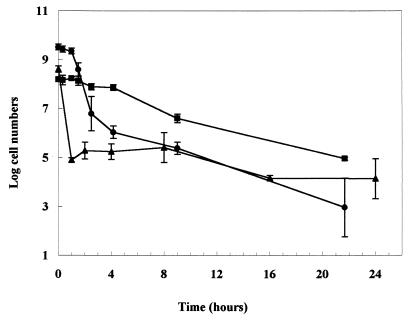
When long-term survival at pH 3.0 was investigated, the death kinetics of mid-exponential-phase cultures were not linear (Fig. 2). Within the first hour of challenge at pH 3.0, there was a rapid decline in numbers to about 105 CFU ml−1. Remarkably, there was no further decline in the number of survivors over the following 24 h. Acid-treated stationary-phase cells exhibited considerably different death kinetics, which were also nonlinear. There was an initial period of acid tolerance for 2 h followed by a more rapid reduction in numbers until about 4 h, after which the numbers declined slowly. After 24 h, the number of surviving cells was similar to that found with mid-exponential-phase cells. If the ATR was induced in mid-exponential-phase cells (by an acid adaptation at pH 5.0 for 1 h prior to the challenge at pH 3.0), a high degree of acid tolerance was detected for about 4 h, after which the numbers declined slowly (Fig. 2). After 24 h, the number of survivors was similar to that found with uninduced cells. Therefore, regardless of the initial physiological state of the cells, similar numbers survived acid treatment at pH 3.0 for 24 h. Incubation at pH 3.0 was continued up to 140 h, and the number of survivors was determined by direct plating. Survivors (between 101 and 104 CFU ml−1) were detected in all three cultures for up to 81 h at the challenge pH (data not shown). These data indicated that a small proportion of the initial population was capable of withstanding extended incubation at pH 3.0.
FIG. 2.
Long-term survival of E. coli O157:H7 (strain P1432) in TPB at pH 3.0. Prior to acid challenge, cultures were grown in TPB (pH 7.0) at 30°C to mid-exponential phase (triangles, nonadapted; squares, adapted at pH 5.0 for 1 h prior to challenge) or stationary phase (circles). The error bars represent the standard deviations from the means (n = 3).
Acid tolerance is lost upon resumption of growth.
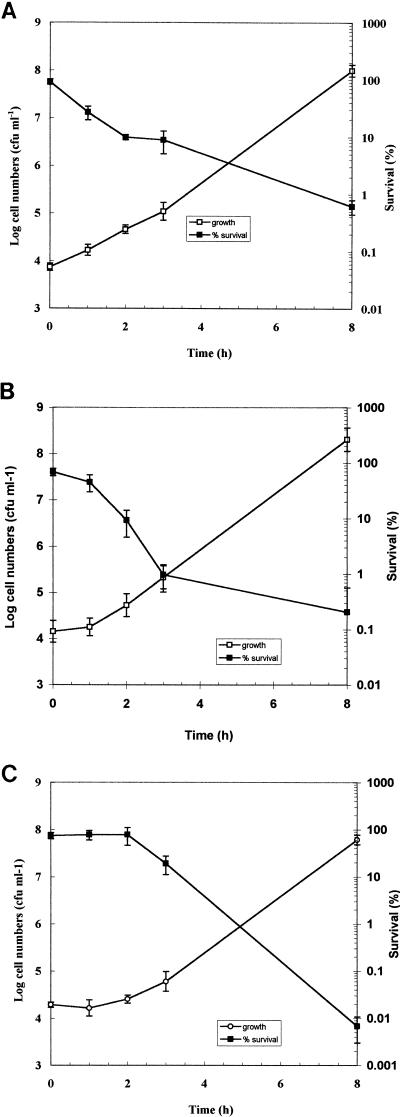
We investigated whether the resistant subset of the three populations, shown in Fig. 1, was lost upon transfer to broth at neutral pH. Growth and acid tolerance (arbitrarily defined as the percentage of surviving cells after a 90-min challenge at pH 3.0) were measured by direct plating to determine if the acid-resistance phenotype was lost upon resumption of growth. Cells were removed from each of the acid-challenged populations after 90 min at pH 3.0 and inoculated into fresh medium to give an inoculum of 104 CFU ml−1. It is clear from the results that there is a very good correlation between growth and loss of acid tolerance (Fig. 3). With the mid-exponential-phase culture, growth began immediately and the percent of survival at pH 3.0 decreased in parallel (Fig. 3A). With ATR-induced cells, there was a lag before growth commenced and this correlated well with the observed loss of acid tolerance (Fig. 3B). When acid tolerance was investigated during outgrowth of stationary phase, there was a lag before acid tolerance decreased and this matched the lag in growth (Fig. 3C). These data indicate that physiological changes that occur during growth lead to the loss of the acid tolerance expressed by these cell populations.
FIG. 3.
Loss of acid tolerance of E. coli O157:H7 strain P1432 upon commencement of growth. After a 90-min challenge at pH 3.0, cells were removed from a mid-exponential-phase culture (A), a mid-exponential-phase culture adapted at pH 5.0 for 1 h (B), or a stationary-phase culture (C) and inoculated in to fresh TPB (pH 7.0). The inoculum was calculated to give approximately 104 viable cells per ml. Samples were taken at various time intervals thereafter, and both acid tolerance (expressed as percent of survival after 90 min at pH 3.0) and growth (expressed as CFU per milliliter) were determined. The error bars represent the standard deviations from the means (n = 3).
Increased acid tolerance correlates with reduced proton accumulation.
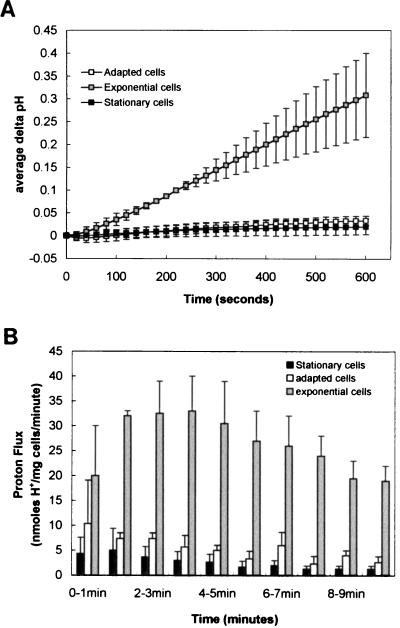
Proton flux assays were performed on each of the three populations to determine the proton accumulation rate of each. Cells suspended in a nonbuffered solution were pulsed with HCl and the subsequent influx of protons was recorded by monitoring the pH of the suspension. Figure 4A shows the average change in pH (measured at 20-s intervals) for mid-exponential-phase, acid-adapted, and stationary-phase cells after the initial pulse of acid. This data was then used to calculate the rate of proton accumulation by these cell populations at 1-min intervals (Fig. 4B). Mid-exponential-phase cells had a proton accumulation rate 5- to 10-fold greater than either acid-adapted or stationary-phase cells. It is important to note that these measurements recorded net proton movement only. Therefore, it was not possible to say whether these observed differences were due to the increased efflux or decreased influx of protons. However, this result indicates that a strong correlation exists between proton permeability and the ability to survive an acidic challenge.
FIG. 4.
Proton flux measurements on cell populations expressing different levels of acid tolerance. Three populations of cells were prepared for analysis as described in Materials and Methods. They were (i) exponential-phase cells without acid adaption (shaded squares and bars), (ii) cells grown to mid-exponential phase and acid adapted at pH 5.0 for 1 h (open squares and bars), and (iii) an overnight culture of stationary-phase cells (solid squares and bars). Proton accumulation measurements were taken over a 10-min period after pulsing cell suspensions with HCl and are shown as either delta pH values (A) or as net proton flux rates in nanomoles of protons per milligram (wet weight) of cells per minute (B).
2D-PAGE analysis of membrane-associated proteins.
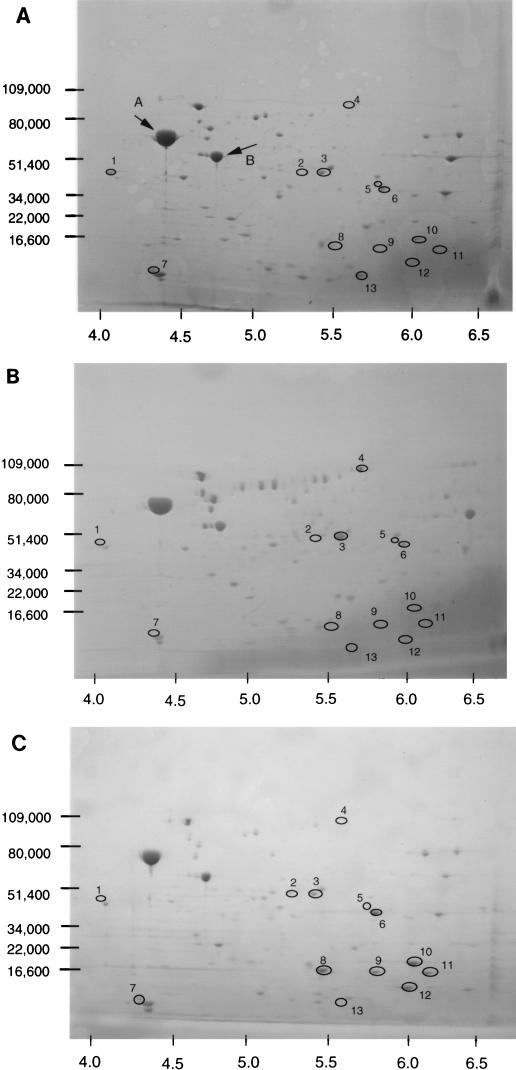
The above result indicated that membrane permeability may be involved in acid tolerance. One factor contributing to the proton flux across the cell envelope could be its protein composition, particularly as the induction of acid tolerance requires de novo protein synthesis. We therefore investigated whether membrane-associated proteins varied depending on the physiological state of the cells. The membrane fraction was isolated from mid-exponential-phase cells, acid-induced mid-exponential-phase cells, and stationary-phase cells of strain P1432. The proteins were separated by 2D-PAGE and analyzed for differences (Fig. 5). To confirm that this procedure successfully recovered membrane proteins, two proteins common to all blots (indicated with arrows labelled A and B in Fig. 5A) were N-terminally sequenced. They showed exact sequence identity with E. coli flagellin (protein A) and the ε subunit of ATP synthase (protein B), indicating that membrane-associated proteins were isolated.
FIG. 5.
2D SDS-PAGE analysis of membrane-associated proteins. Membrane-associated proteins were prepared from cultures of strain P1432 grown to mid-exponential phase without acid adaptation (A), grown to mid-exponential phase and acid adapted at pH 5.0 for 1 h (B), or grown to stationary phase overnight (C). Spots which showed reproducible differences (for each gel, n = 3) in expression are circled and labelled 1 to 13 (see text for details). The positions of the molecular mass standards are indicated in kilodaltons on the vertical axes, and the approximate pI scale is indicated on the horizontal axes.
Mid-exponential-phase acid-induced cells showed increased expression of two membrane-associated proteins (spots 3 and 4, Fig. 5B) compared to unadapted cells. Additionally, a number of proteins were repressed compared to the unadapted control (spots 1, 5, 7, 10, and 13; Fig. 5). Stationary-phase cells showed altered expression of several proteins compared to mid-exponential-phase cells. Six proteins were reproducibly found to be induced (spots 2, 6, 8, 9, 11, and 12; Fig. 5C) while three proteins were found to be repressed (spots 1, 5, and 13; Fig. 5C). Interestingly, three of the proteins which showed altered expression in acid-adapted mid-exponential-phase cells also showed altered expression in stationary-phase cells (spots 1, 5, and 13; Fig. 5). These may represent proteins whose repression is necessary to achieve increased tolerance to low pH in these cell populations.
Attempts were made to identify several of the proteins showing reproducibly altered expression (specifically, spots 2, 8, 9, and 10). Many were N-terminally blocked and so sequence information could not be obtained. However, spot 6 was successfully excised from the gel and analyzed by N-terminal microsequencing. The sequence obtained matched exactly the N-terminal sequence of the outer membrane protein OmpA. We therefore investigated whether an ompA-deficient strain was impaired in its ability to survive an acid challenge. We compared the acid tolerance of the K-12 strain P460 with that of its wild-type parent, P400 (an ompA mutant of E. coli O157:H7 was not available). The ompA-deficient strain (P460) was found to be significantly more sensitive to acid, both in stationary phase (10-fold) and in the mid-exponential phase (1,000-fold) of growth (data not shown). It is possible that the role of this protein in maintaining the structural integrity of the cell envelope (33) contributes in some way to protecting the cell from acidic conditions.
DISCUSSION
Previous studies have demonstrated the ability of E. coli O157:H7 to survive pH 3.0 for 2 to 5 h (2, 5, 8, 15). In this study, we have demonstrated the ability of a nontoxigenic strain of E. coli O157:H7 to survive prolonged exposure to pH 3.0. For each of the three populations of cells investigated in this study (exponential phase nonadapted, exponential phase acid adapted, and stationary phase) significant numbers of survivors can be detected, even after 3 days at pH 3.0, although individually the survival curves are quite different. The well-documented observation (reviewed in reference 3) that stationary-phase and acid-adapted exponential-phase cells show elevated levels of acid tolerance relative to exponential-phase cells also holds true for the strain used in this study. More surprising is the shape of the survival curves. The differences in death kinetics point to the fact that the molecular mechanisms of acid tolerance in acid-adapted cells and in stationary-phase cells are likely to be different. The protection conferred by acid adaptation appears to be greater than that afforded by entry into stationary phase, suggesting that stationary-phase tolerance is not simply the induction of adaptive tolerance via a different signal (i.e., cessation of growth). The exponential-phase culture showed a very pronounced biphasic loss of viability at pH 3.0—a rapid initial decline followed by a prolonged period of survival. It is worth noting that nonlinear death kinetics of this kind have been reported previously for heat-injured E. coli O157:H7 in ground beef or chicken (22). Using a model for nonlinear death, Juneja and colleagues (22) calculated that a subpopulation had a D value of 4.54 min whereas the majority of the population had a D value of 0.61 min at 60°C. These results demonstrate that the tailing phenomenon may not be specific to low-pH-mediated killing. These data also emphasize the need to account for tails when attempting to design processes for the elimination of these organisms from foods.
The nature of the physiological variation contributing to the survival tails remains unclear, but it does not require active protein synthesis during the acid challenge, as the tail is still observed even if chloramphenicol is included during the challenge. Furthermore, even if the acid challenge is lowered to pH 2.0, this tail is still observed, although a smaller percentage of the population survives. Similar data have recently been obtained with toxigenic strains of E. coli O157:H7 by using low-pH challenges in defined media (15). Importantly, Glover and colleagues (15) have also ruled out any role for the stationary-phase sigma factor, RpoS, in contributing to these tails; mutants lacking the rpoS gene were still found to die with marked biphasic death kinetics at low pH. These data suggest that the highly acid-tolerant tail of cells from the exponential-phase culture, which represents 0.1% of the total population, exists naturally within the growing population. It is interesting to note that this is in contrast to the reported requirement for protein synthesis in observing biphasic thermal inactivation of another enteric pathogen, Salmonella enteritidis PT4. In this case, biphasic death kinetics arose as a result of an induced heat shock response occurring during the thermal challenge (20).
Our unpublished observations suggest that this tailing phenomenon is common to many of the strains of E. coli we have examined and is not simply a peculiarity of the strain used in this study. Recent work by Glover and colleagues (15) has also led to the conclusion that nonlinear death kinetics at low pH is a feature common to all pathogenic and commensal strains studied thus far, though the fraction of the initial population surviving as a tail varies considerably from strain to strain. Interestingly, they found that many laboratory strains do not display this phenomenon and speculate that this may be an attribute of strains adept at colonizing the gut. A more detailed analysis, perhaps employing flow cytometry, will be required to elucidate the nature of this population heterogeneity. However, these findings have clear implications for the pathogenic potential of this organism. A population of cells entering the stomach is unlikely to be completely killed if it harbors a subset of cells that are highly acid tolerant, thereby increasing the probability of a successful infection. It is clear, however, that the survival potential of stationary-phase or acid-adapted cells is greater than that of exponentially growing cells over the initial period of acid challenge. Given the relatively short residence time of food in the stomach (approximately 2 h) and the likelihood that only small numbers of bacterial cells will be ingested, these populations are therefore more likely to survive passage through the stomach and subsequently colonize the gut.
Numerous studies have been undertaken in recent years in an attempt to identify factors which contribute to increased acid tolerance in E. coli as well as in other enterobacterial species (reviewed in references 3 and 18). In very few cases have specific mechanisms been identified. Here, we show that a strong correlation exists between increased acid tolerance and decreased permeability of the cell envelope to protons. This result implies that acid-resistant populations (either an acid-adapted exponential-phase population or a stationary-phase population) have an improved ability to maintain pHi. Although it is not yet clear whether this change in permeability is due to an exclusion of protons from the cell or an active efflux of protons from the cell, it seems plausible to suggest that this change in proton flux may directly contribute to the elevated levels of acid tolerance observed. In E. coli, there is evidence that passive influx of protons into the cell, via the outer membrane porin PhoE, may contribute to acid sensitivity (31). A phoE mutant displayed increased levels of acid tolerance, and wild-type cells can be protected from acid killing by the inclusion of polyphosphate during the challenge. Polyphosphate is believed to block the PhoE porin, thereby preventing protons from leaking through the outer membrane (31). Thus, it is clear that proton flux across the cell membrane(s) is an important determinant of acid tolerance in bacteria.
In oral streptococci, permeability to protons is markedly increased by the ATPase inhibitor dicyclohexylcarbodiimide, indicating that proton permeability involves not only passive inflow of protons but also active efflux through the proton-translocating membrane ATPase (4). In Listeria monocytogenes, a gram-positive food-borne pathogen, acid tolerance is dependent upon extracellular pH as well as growth phase (12). Factors which effect the ionic permeability of its membrane have dramatic effects on its tolerance to low-pH stress. For example, nisin, a bacteriocin which has the capacity to dissipate membrane proton motive force, dramatically reduces acid tolerance in both exponential- and stationary-phase cells of this pathogen. The respiratory uncoupler m-chlorophenylhydrazone, which effectively equilibrates the external and internal pHs, has similar effects on acid tolerance (11). These observations underline the essential role of membrane integrity in protecting cells against low external pH.
The changes in membrane permeability observed also correlate with changes in the protein composition of the cell envelope. Several major systems located in the cytoplasmic membrane influence proton circulation. These include K+/H+ and Na+/H+ antiport systems (reviewed in references 6 and 7), the F1F0 proton-translocating ATPase, electron transport chains, and numerous solute-proton symport systems. It may be that altered levels of one or more of these proton translocating systems play a role in enhancing acid tolerance. Further attempts to identify individual protein spots by mass spectrometry or N-terminal microsequencing will be required in order to establish which proteins are likely to be involved.
Recently, it has been shown that acid-adapted E. coli changes the lipid composition of its membranes. Specifically, cells adapted for one doubling at pH 5.0 were found to have elevated levels of cyclopropane fatty acids (8). E. coli cells are also known to increase the cyclopropane fatty acid content of their membranes upon entry into stationary phase (10). Furthermore, the acid tolerance levels of individual strains correlated well with membrane cyclopropane fatty acid content (8). A similar increase in membrane cyclopropane fatty acids was seen when the gram-positive bacterium Clostridium acetobutylicum was grown at an acidic pH (24). It may be that changes in the lipid composition of membranes affect their proton conductance, perhaps reducing the leakage of protons across the membrane when the external proton concentration is high. The changes in the protein composition of the cell membrane described in this contribution may be directly due to changes in membrane lipid composition. It may be that both lipid and protein alterations are necessary to confer elevated levels of protection to the cell in acidic environments. Further studies will be required to determine how each contributes to enabling cells to survive acid stress.
ACKNOWLEDGMENTS
We thank Ian Booth for sharing data prior to publication and for useful discussions. Thanks also to Peter Coote for useful comments on the manuscript.
C.P.O. is supported by a University of Aberdeen ACT(R) medical research fellowship.
REFERENCES
- 1.Abdul-Raouf U M, Beuchat L R, Ammar M S. Survival and growth of Escherichia coli O157:H7 in ground, roasted beef as affected by pH, acidulants, and temperature. Appl Environ Microbiol. 1993;59:2364–2368. doi: 10.1128/aem.59.8.2364-2368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold K W, Kasper C W. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearson S, Bearson B, Foster J W. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 4.Bender G R, Sutton S V W, Marquis R E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin M M, Datta A R. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth I R. Control of proton permeability: its implications for energy transduction and pH homeostasis. FEMS Symp. 1988;44:1–12. [Google Scholar]
- 7.Booth I R, Kroll R G. The preservation of foods by low pH. In: Gould G W, editor. Mechanisms of action of food preservation procedures. Amsterdam, The Netherlands: Elsevier Applied Science; 1989. pp. 119–160. [Google Scholar]
- 8.Brown J L, Ross T, McMeekin T A, Nichols P D. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. J Food Microbiol. 1997;37:163–173. doi: 10.1016/s0168-1605(97)00068-8. [DOI] [PubMed] [Google Scholar]
- 8a.Chapman, P. Personal communication.
- 9.Cheville A M, Arnold K W, Buchrieser C, Cheng C-M, Kaspar C W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cronan J E. Phospholipid alterations during growth of Escherichia coli. J Bacteriol. 1968;95:2054–2061. doi: 10.1128/jb.95.6.2054-2061.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta A R, Benjamin M M. Factors controlling acid tolerance of Listeria monocytogenes: effects of nisin and other ionophores. Appl Environ Microbiol. 1997;63:4123–4126. doi: 10.1128/aem.63.10.4123-4126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis M J, Coote P J, O’Byrne C P. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth phase-dependent acid resistance. Microbiology. 1996;142:2975–2982. doi: 10.1099/13500872-142-10-2975. [DOI] [PubMed] [Google Scholar]
- 13.Foster J W. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J Bacteriol. 1991;173:6896–6902. doi: 10.1128/jb.173.21.6896-6902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster J W, Hall H K. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J Bacteriol. 1991;173:5129–5135. doi: 10.1128/jb.173.16.5129-5135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glover, J., L. Malcolm, F. M. Thomson-Carter, P. Carter, S. Jordan, S. J. Park, and I. R. Booth. Unpublished results.
- 16.Goodson M, Rowbury R J. Habituation to normally lethal acidity by prior growth of Escherichia coli at a sub-lethal acid pH value. Lett Appl Microbiol. 1989;8:77–79. [Google Scholar]
- 17.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli and associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–97. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 18.Hall H K, Karem K L, Foster J W. Molecular responses of microbes to environmental pH stress. Adv Microbiol Physiol. 1995;37:229–272. doi: 10.1016/s0065-2911(08)60147-2. [DOI] [PubMed] [Google Scholar]
- 19.Hickey E W, Hirshfield I N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humpheson L, Adams M R, Anderson W A, Cole M B. Biphasic thermal inactivation kinetics in Salmonella enteritidis PT4. Appl Environ Microbiol. 1998;64:459–464. doi: 10.1128/aem.64.2.459-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan S L, Glover J, Malcolm L, Thomson-Carter F M, Booth I R, Park S F. Augmentation of killing of Escherichia coli O157 by combinations of lactate, ethanol, and low-pH conditions. Appl Environ Microbiol. 1999;65:1308–1311. doi: 10.1128/aem.65.3.1308-1311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juneja V K, Snyder O P, Marmer B S. Thermal destruction of Escherichia coli O157:H7 in beef and chicken: determination of D- and z-values. Int J Food Microbiol. 1997;35:231–237. doi: 10.1016/s0168-1605(96)01237-8. [DOI] [PubMed] [Google Scholar]
- 23.Lee I S, Slonczewski J L, Foster J W. A low-pH-inducible, stationary-phase acid tolerance response in Salmonella typhimurium. J Bacteriol. 1994;176:1422–1426. doi: 10.1128/jb.176.5.1422-1426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LePage C, Fayolle F, Hermann M, Vandecasteele J-P. Changes in the lipid composition of Clostridium acetobutylicum during acetone-butanol fermentation: effects of slovents, growth temperature and pH. J Gen Microbiol. 1987;133:103–110. [Google Scholar]
- 25.Leyer G J, Wang L-L, Johnson E A. Acid adaptation of Escherichia coli O157:H7 increases survival in acidic foods. Appl Environ Microbiol. 1995;61:3752–3755. doi: 10.1128/aem.61.10.3752-3755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Smith M P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowen P C, Hengge-Aronis R. The role of the sigma factor ss (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 28.Miller L G, Kaspar C W. Escherichia coli O157:H7 acid tolerance and survival in apple cider. J Food Prot. 1994;57:460–464. doi: 10.4315/0362-028X-57.6.460. [DOI] [PubMed] [Google Scholar]
- 29.Morgan D, Newman C P, Hutchinson D N, Walker A M, Rowe B, Majid F. Verotoxin producing Escherichia coli O157 infections associated with the consumption of yoghurt. Epidemiol Infect. 1993;111:181–187. doi: 10.1017/s0950268800056880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 31.Rowbury R J, Goodson M. PhoE porin of Escherichia coli and phosphate reversal of acid damage and killing and of acid induction of the CadA gene product. J Appl Bacteriol. 1993;74:652–661. doi: 10.1111/j.1365-2672.1993.tb05199.x. [DOI] [PubMed] [Google Scholar]
- 32.Skurray R A, Hancock R E W, Reeves P. Con− mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J Bacteriol. 1974;119:726–735. doi: 10.1128/jb.119.3.726-735.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape in Escherichia coli: multiple mutants missing the outer membrane lipoprotein and major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele B T, Murphy N, Rance C P. An outbreak of hemolytic uremic syndrome associated with ingestion of fresh apple juice. J Pediatr. 1982;101:963–965. doi: 10.1016/s0022-3476(82)80021-8. [DOI] [PubMed] [Google Scholar]
- 35.Weagant S D, Bryant J L, Bark D H. Survival of Escherichia coli O157:H7 in mayonnaise-based sauces at room and refrigerated temperatures. J Food Prot. 1994;57:629–631. doi: 10.4315/0362-028X-57.7.629. [DOI] [PubMed] [Google Scholar]