Abstract
Microbial community composition associated with benzene oxidation under in situ Fe(III)-reducing conditions in a petroleum-contaminated aquifer located in Bemidji, Minn., was investigated. Community structure associated with benzene degradation was compared to sediment communities that did not anaerobically oxidize benzene which were obtained from two adjacent Fe(III)-reducing sites and from methanogenic and uncontaminated zones. Denaturing gradient gel electrophoresis of 16S rDNA sequences amplified with bacterial or Geobacteraceae-specific primers indicated significant differences in the composition of the microbial communities at the different sites. Most notable was a selective enrichment of microorganisms in the Geobacter cluster seen in the benzene-degrading sediments. This finding was in accordance with phospholipid fatty acid analysis and most-probable-number–PCR enumeration, which indicated that members of the family Geobacteraceae were more numerous in these sediments. A benzene-oxidizing Fe(III)-reducing enrichment culture was established from benzene-degrading sediments and contained an organism closely related to the uncultivated Geobacter spp. This genus contains the only known organisms that can oxidize aromatic compounds with the reduction of Fe(III). Sequences closely related to the Fe(III) reducer Geothrix fermentans and the aerobe Variovorax paradoxus were also amplified from the benzene-degrading enrichment and were present in the benzene-degrading sediments. However, neither G. fermentans nor V. paradoxus is known to oxidize aromatic compounds with the reduction of Fe(III), and there was no apparent enrichment of these organisms in the benzene-degrading sediments. These results suggest that Geobacter spp. play an important role in the anaerobic oxidation of benzene in the Bemidji aquifer and that molecular community analysis may be a powerful tool for predicting a site’s capacity for anaerobic benzene degradation.
In recent years, the potential for anaerobic in situ biodegradation of monoaromatic hydrocarbons has received increasing attention (e.g., references 4, 14, 24, and 37). Petroleum contamination of aquifers is common in the United States and other industrialized countries, with sources of contamination including leaking underground fuel storage tanks, leachate from landfills, and surface spills of petroleum products (42, 47). Although petroleum is comprised of a complex mixture of hydrocarbons, monoaromatic hydrocarbons (benzene, toluene, ethylbenzene, and xylenes [collectively known as BTEX]) are of particular concern. BTEX compounds are the most water-soluble constituents of petroleum and have a relatively low sediment-water coefficient (11, 51). As a result, they are highly mobile in aquifers and are the most prevalent petroleum constituents in contaminated groundwater. Among BTEX compounds, benzene is of the most concern because it is highly toxic and is a known carcinogen, and therefore it is a U.S. Environmental Protection Agency “priority pollutant.”
Although BTEX compounds are rapidly degraded under aerobic conditions (40, 49), aerobic biodegradation is self-limiting due to the low aqueous solubility of oxygen and the rapid depletion of oxygen that occurs in reduced environments (49). At the onset of anaerobic conditions, Fe(III) is generally the most abundant terminal electron acceptor in aquifer sediments (23). Therefore, biodegradation of BTEX compounds under Fe(III)-reducing conditions has the potential to be an effective natural attenuation process (25). However, anaerobic biodegradation of some BTEX compounds, particularly benzene, remains poorly understood. To date, no organism capable of anaerobic benzene oxidation has been isolated, and investigations of anaerobic benzene degradation under various geochemical conditions often report conflicting results (24).
Recent studies in a petroleum-contaminated aquifer in Bemidji, Minn., provided evidence for benzene mineralization under in situ Fe(III)-reducing conditions (4). However, the capacity for benzene oxidation coupled to Fe(III) reduction was not detected at the other petroleum-contaminated aquifers that were evaluated. Furthermore, at the Bemidji aquifer, anaerobic benzene oxidation under in situ conditions was only found within a portion of the Fe(III)-reducing zone (4). Although there were no apparent correlations between benzene oxidation and geochemical parameters (5), there was a strong correlation between anaerobic benzene oxidation and the abundance of members of the family Geobacteraceae (4). This family includes the only organisms known to couple the oxidation of several monoaromatics, including toluene, with Fe(III) reduction (10, 30). The results suggested that the microbial community composition in the Bemidji aquifer may have played a key role in anaerobic benzene degradation and that Geobacteraceae, in particular, were associated with benzene-degrading activity (4). However, past studies have not evaluated the diversity of other microbial groups with molecular methods nor did they evaluate which microorganisms within the family Geobacteraceae are associated with the capacity for anaerobic benzene degradation. Therefore, no conclusions have been drawn regarding which species of Geobacteraceae are associated with benzene-oxidizing activity and whether other, unrelated, phylogenetic groups can also play a role in benzene degradation.
The purpose of this study was to use a multifaceted approach, including biodegradation studies, molecular phylogenetic approaches, lipid analysis, and culturing studies, to analyze the microbial community associated with benzene oxidation at the Bemidji aquifer. The results suggest that a narrow phylogenetic group of organisms within the Geobacter cluster (22) play an important role in the benzene-oxidizing community.
MATERIALS AND METHODS
Study site and sample collection.
Sediment samples were collected from the U.S. Geological Survey Groundwater Toxics Site at Bemidji, Minn. This site is located in a remote forested area and was contaminated in 1979 when a pipeline transporting crude oil burst (19). An anaerobic contaminant plume containing BTEX compounds has formed downgradient from the crude oil source. The contaminated portion of the aquifer is composed of fairly homogeneous, sandy to coarse-grained sediments (19), making it an ideal site for groundwater contaminant research. In addition, considerable information is available regarding contaminant composition, hydrogeological conditions, and geochemical conditions at this site (43).
Samples were collected in 1996 and 1997 as previously described (4) from several locations along the contaminant gradient and from an uncontaminated site. Sites IR-1, IR-2, and IR-3 were sediments in which Fe(III) reduction was the terminal-electron-accepting process (TEAP), with site IR-1 being closest to the contaminant source. Sites IR-1, IR-2, and IR-3 correspond to sites 97-1, 97-2, and 97-3, respectively, as identified in a previous study at this site (4). Sediments 96-1 are Fe(III)-reducing sediments collected in 1996. Methanogenic sediments were sampled closer to the source of contamination and uncontaminated sediments were recovered from a nearby site just outside the contaminant plume. At each site, sediments from a depth of approximately 9 to 10 m were collected aseptically from drilling cores (34) and were held anoxically on ice during overnight transport to the laboratory. Sediment samples were homogenized and divided into aliquots for subsequent manipulations in an N2-filled glove bag. For enrichment cultures, 1 to 2 g of sediment was added to pressure tubes for later dilution as described below. For phospholipid fatty acid (PLFA) and molecular analyses, 100 g of sediment was transferred to sterile conical tubes and stored at −80°C. The TEAP at each of these sites was confirmed with geochemical and activity measurements as previously described (4, 5). Sediments from sites IR-3 and 96-1 mineralized benzene under in situ conditions (5). Anaerobic oxidation of benzene was measured with [14C]benzene as previously described (4).
Enrichment cultures.
Enrichment cultures of benzene-oxidizing Fe(III) reducers were developed with inocula collected from site 96-1. Cells were released from sediment particles and diluted in Fe(III) reducer medium as previously described (4, 29), except that 140 μM benzene was used as the electron donor. Briefly, the medium contained poorly crystalline Fe(III) oxide (30 mM) as an electron acceptor, 4 mM nitrilotriacetic acid as an Fe(III) chelator, and 1.3 mM Fe(II) chloride as a reductant. Initial dilution tubes also contained 0.1% pyrophosphate in order to release cells bound to sediment particles (7). Most-probable-number (MPN) tubes were incubated at 25°C for 2 months prior to enumeration. Utilization of benzene was determined by monitoring benzene removal in the headspace of inoculated tubes relative to uninoculated controls. Benzene concentrations were determined by gas chromatography. Benzene mineralization was also monitored by adding [14C]benzene ([U-14C]benzene, 58.2 mCi/mmol; Sigma Radiochemicals, St. Louis, Mo.) to enrichment cultures and measuring the production of 14CO2 by gas proportional counting as previously described (26). The highest positive dilutions were enriched for Fe(III) reducers capable of degrading benzene by the re-addition of benzene and continued monitoring of benzene removal. Active enrichments were transferred to fresh Fe(III)-reducing media prior to phylogenetic analyses.
16S rDNA phylogenetic analysis of enrichment cultures.
The phylogeny of the dominant organisms in benzene-oxidizing enrichment cultures was investigated by using 16S ribosomal DNA (rDNA) sequence analysis. Prior to DNA extraction, cultures were treated with oxalic acid in order to chelate and remove Fe(III), which inhibits Taq polymerase (48). Then, 15 ml of 300 mM filter-sterilized oxalic acid was added to 5 ml of enrichment culture and mixed thoroughly. Cells were collected by centrifugation at 5,000 × g for 5 min, washed in 1 ml of 30 mM NaHCO3, and resuspended in 250 μl of 30 mM NaHCO3. The resulting cell suspensions were subjected to three freeze-thaw cycles at −70 and 65°C and then extracted with phenol, phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform-isoamyl alcohol (24:1) (39). A 5-μl portion of the resulting aqueous phase was used as a template for PCR with primers 338F-GC (the complement of EUB338) (1) and 907R (21). Genomic DNA was also extracted from pure cultures of Geobacter chapellei, Geobacter sulfurreducens, Escherichia coli, Desulfococcus multivorans, and Desulfuromusa acetexigens (6) for use as reference DNA in PCR and denaturing gradient gel electrophoresis (DGGE). PCR mixtures containing 50 mM KCl, 10 mM Tris-Cl (pH 8.3), 1.5 mM MgCl2, 200 μM each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), 0.6 μM each primer, and 400 ng of bovine serum albumin (BSA) per μl in a total volume of 100 μl were assembled and treated with UV for 10 min. A total of 2.5 U of AmpliTaq Taq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) and template DNA were then added to each reaction. Amplification was performed in a GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer Cetus) with an initial denaturation at 94°C for 90 s; 30 cycles of 94°C for 30 s, 40°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. PCR products were analyzed by agarose gel electrophoresis by standard techniques (39).
The 16S rDNA amplicons were resolved by DGGE with a 50 to 80% denaturant gradient (where 100% is equivalent to 7 M urea and 40% formamide) (35) on a 7% acrylamide gel in 1× TAE (40 mM Tris-acetate [pH 7.4], 20 mM sodium acetate, 1 mM EDTA). DGGE gels were run for 16 h at 65 V and then were stained with ethidium bromide (39). Resolved PCR products were visualized by UV transillumination. Isolated bands were excised and pulverized with a sterile mortar and pestle, and DNA was eluted overnight in 50 μl of 0.1 M Tris (pH 8.0) at 4°C. Partial 16S rDNAs were then reamplified from excised bands, as described above, and analyzed by a second DGGE in order to ensure that heteroduplexes were resolved. Once again, isolated bands were excised, and 16S rDNA was reamplified as described above, except that no GC clamp (35) was incorporated into primer 338F. These PCR products were purified by using Wizard PCR Preps (Promega, Madison, Wis.) and sequenced by using Dye Deoxy Terminator Cycle Sequencing (Perkin-Elmer Cetus) and an ABI 373A automated sequencer (Applied Biosystems, Foster City, Calif.) at the Michigan State University Sequencing Facility or the University of Massachusetts Department of Microbiology.
Phylogenetic diversity of sediment microbial communities.
Detailed phylogenetic analyses were focused on the uncontaminated site and the site where Anderson et al. (4) demonstrated that benzene oxidation occurred under in situ Fe(III)-reducing conditions (site IR-3). The 16S rDNA phylogeny of dominant members of the sediment microbial community was analyzed by using PCR-DGGE and sequence analysis. DNA was extracted from triplicate 500-mg samples of homogenized sediment by using the FastDNA soil extraction kit (Bio 101, Vista, Calif.) as described by the manufacturer, except that cell lysis was carried out with a Mini-BeadBeater (Biospec Products, Inc., Bartlesville, Okla.) at maximum speed for 1 min. DNA was eluted from Bio 101 cartridges with 50 μl of sterile Milli-Q water. For analysis of the diversity of Geobacteraceae, partial 16S rDNA was amplified from triplicate sediment DNA extractions with primers 338F-GC and Geo825R. PCR conditions were as described above for primers 338F and 907R except that PCR mixtures were not treated with UV and the PCR temperature profile included touchdown primer annealing (12) from 65 to 55°C (decreasing 0.5°C per cycle) for 20 cycles, followed by 10 cycles at 55°C. Geobacteraceae 16S rDNA amplicons were analyzed by DGGE on a 55 to 70% denaturant gradient, and individual bands were excised, checked for heteroduplexes, reamplified, and sequenced as described above. For analysis of the dominant members of the bacterial community, partial 16S rDNAs were amplified from triplicate sediment DNA samples by using primers 338F-GC and 907R as described above. In order to improve detection of faint bands in subsequent DGGE analyses, 200 μl of the resulting PCR product from each sediment DNA sample was concentrated by lyophilization. Concentrated PCR products were then resolved by DGGE and reamplified as described above, except that gel denaturant gradients consisted of 30 to 70%, 50 to 70%, and 60 to 80% denaturant.
MPN-PCR analysis of sediment Geobacteraceae.
MPN-PCR analysis of Geobacteraceae 16S rDNA at the methanogenic site was conducted as previously described (4).
Sequence analysis.
16S rDNA sequences were checked for potential chimeras with the Ribosomal Database Project (RDP) CHECK_CHIMERA program and by determining the secondary structure of selected sequences. Sequences were then aligned with closely related 16S rRNA sequences from GenBank and the RDP. Unambiguously aligned base positions were used to calculate Jukes-Cantor distances and construct phylogenetic trees with the maximum-parsimony and least-squares methods.
PLFA analyses.
PLFAs were extracted from sediment samples by adding 142.5 ml of methylene chloride-methanol-phosphate buffer (monobasic, pH 7.0) (1:2:0.8) to 70 g (wet weight) of sediment, followed by sonication for 2 min and incubation for 3 h at room temperature. The extractant was then separated from the sediment by centrifugation and transferred to a clean tube. The remaining sediment was washed with 37.5 ml of methylene chloride, and the resulting extractant was combined with the previous fraction. Then, 37.5 ml of sterile distilled water was added to the extractant and, after thorough mixing, the organic and aqueous phases were separated by centrifugation. The lower (organic) phase was removed by pipetting it to a clean tube and then dried under a stream of N2. Total lipid was fractionated as described by Guckert et al. (17). PLFA were then transesterified into methyl esters (17) prior to identification and quantification by gas chromatography-mass spectrometry (38).
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under accession numbers AF104267 to AF104299.
RESULTS
Distribution of benzene mineralization potential and MPN-PCR of Geobacteraceae.
Anaerobic benzene mineralization under in situ conditions was negligible at the uncontaminated site and in the methanogenic sediments, with less than 6% of the [14C]benzene added recovered as 14CO2 after 85 days. This result contrasted with the mineralization of more than 60% of the added benzene in sediments from IR-3 and was comparable to the very slight benzene mineralization that was also observed at IR-1 and IR-2 (4). MPN-PCR analysis indicated that there were 2.2 × 105 ± 7.8 × 104 (mean ± the standard deviation) copies of Geobacteraceae 16S rDNA g−1 in the methanogenic sediments. This finding compared with previously reported estimates of Geobacteraceae 16S rDNA of 1.48 × 103 ± 1.04 × 103 copies g of sediment−1, 9.24 × 104 ± 3.49 × 104 copies g of sediment−1, and 9.24 × 104 ± 3.49 × 104 copies g of sediment−1 in the background site and at sites IR-1 and IR-2, respectively, but it was significantly lower than the 1.50 × 107 ± 6.72 × 106 copies g of sediment−1 for Geobacteraceae previously found at site IR-3 (4).
PLFA analysis of microbial community.
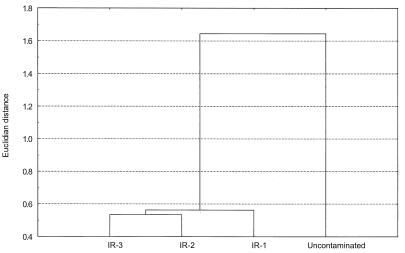
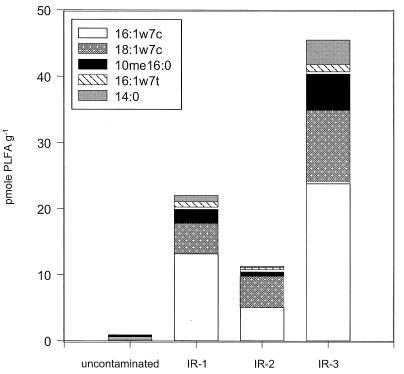
PLFA concentrations were highest at site IR-3 (170.6 pmol g−1), decreased at sites IR-2 and IR-1 (32.2 and 65.0 pmol g−1, respectively), and lowest at the uncontaminated site (7.9 pmol g−1). PLFA data from the methanogenic site were omitted due to heavy petroleum contamination at this site that appeared to carry over into the PLFA assays. Hierarchical cluster analysis of the sediment PLFA profiles (Fig. 1) indicated that microbial community composition at the uncontaminated site was distinct from that at sites where Fe(III) reduction was the dominant TEAP. PLFA profiles from the Fe(III)-reducing sites were similar and exhibited a higher diversity of PLFA than that observed at the uncontaminated site. There were, however, substantial differences between the PLFA profiles extracted from each Fe(III)-reducing site. The distribution of Geobacter species in the sediments was estimated by evaluating the five PLFAs that are most abundant in known Geobacter species (37a), with the exclusion of the ubiquitous saturated PLFAs 16:0 and 18:0. This analysis indicated that the concentration of Geobacter indicative PLFAs was highest at site IR-3, lower at sites IR-2 and IR-1, and lowest at the uncontaminated site (Fig. 2). However, these results must be viewed with caution because the PLFAs used in this analysis are not unique to Geobacter spp. but are also present in other gram-negative bacteria (45).
FIG. 1.
Hierarchical cluster analysis of PLFAs extracted from sediment samples from IR-1, IR-2, IR-3, and the uncontaminated site.
FIG. 2.
Distribution of Geobacteraceae-indicative PLFA functional groups at sites IR-1, IR-2, and IR-3 and the uncontaminated site.
Phylogenetic diversity of sediment microbial community.
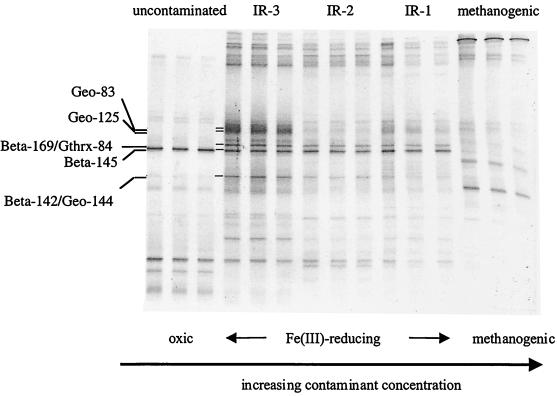
PCR-DGGE analysis of triplicate sediment samples indicated that DGGE patterns from each site were consistent and reproducible (Fig. 3). The DGGE patterns were clearly resolved with fewer than 25 bands per lane (Fig. 3 and 4). The use of several different ranges of denaturant gradients improved the DGGE analysis and provided more-detailed focus on separate segments of the community with each different gradient (Fig. 4).
FIG. 3.
DGGE profiles of bacterial 16S rDNA fragments retrieved from Bemidji sediments along the contaminant gradient. Triplicate DNA extractions were analyzed from each site. A 60 to 80% denaturant gradient range was used.
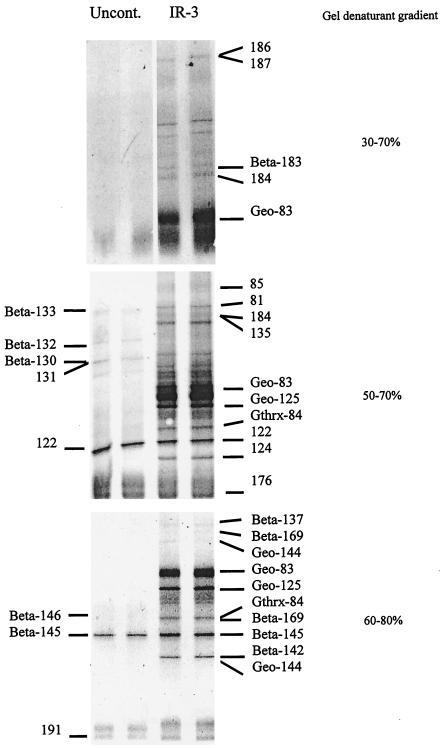
FIG. 4.
DGGE analysis of 16S rDNA fragments amplified from the uncontaminated site (Uncont.) and benzene-oxidizing site IR-3 with primers that target 16S rDNAs of most bacteria in several different denaturant gradient ranges.
Detailed PCR-DGGE and sequence analysis was focused on site IR-3, the most active site of anaerobic benzene degradation (4), and the uncontaminated site. There were marked differences in the DGGE profiles of bacterial 16S rDNA from the two sites (Fig. 4). The diversity of dominant bacteria appeared to be higher at site IR-3, as was evident from the fact that, regardless of the denaturant gradient, far more bands were present in DGGE profiles from site IR-3 than at the uncontaminated site.
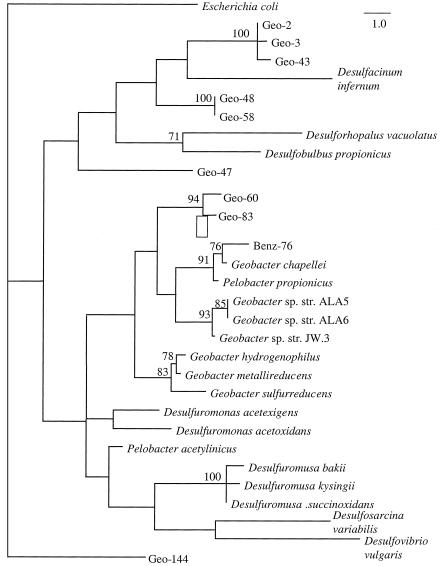
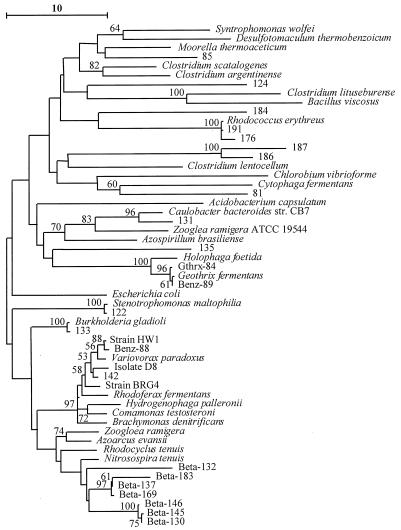
While the two sites shared several 16S rDNA fragments that were either identical or extremely closely related (Fig. 4), the majority of sequences were unique to each site. One of the most conspicuous differences between these sites was band Geo-83, which was the predominant band at site IR-3 and was not detected at the uncontaminated site. Comparative sequence analysis placed Geo-83 within the Geobacter cluster, with its closest relative being G. chapellei (Fig. 5). Other 16S rDNA sequences detected in DGGE profiles from site IR-3, but not at the uncontaminated site, included bands Geo-125 and Geo-144 within the Geobacteraceae family (Fig. 5); Gthrx-84, most closely related to Geothrix fermentans; several beta proteobacteria (Beta-137, Beta-169, and Beta-183, most closely related to Azoarcus evansii, and Beta-142, most closely related to an unidentified toluene degrader, isolate D8 [33]); and several sequences (sequences 81, 85, 124, and 184) which appear to represent novel lineages (Fig. 6).
FIG. 5.
16S rRNA phylogenetic tree of Geobacteraceae and delta proteobacterial sequences from the uncontaminated site and site IR-3 and from benzene-oxidizing enrichment cultures. Phylogenetic trees were constructed by using maximum-parsimony methods with 100 bootstrapped data sets. A total of 366 base positions were considered in the analysis. Bootstrap values of greater than 50 (of 100 trees) are shown adjacent to the nodes. The scale bar is in fixed nucleotide substitutions per 100 sequence positions. A similar tree topology was observed for trees constructed by using least-squares methods (data not shown).
FIG. 6.
Phylogenetic analysis of 16S rRNA sequences from the uncontaminated and benzene-oxidizing sites and from enrichment cultures. Phylogenetic trees were constructed by using maximum-parsimony methods with 100 bootstrapped data sets. Bootstrap values (of 100 trees) that are greater than 50 are shown adjacent to the nodes. A total of 402 base positions were considered in the analysis. The scale bar is in fixed nucleotide substitutions per 100 sequence positions. A similar tree topology was observed for trees constructed by using least-squares methods (data not shown).
Comparison of DGGE patterns from all five sites along the petroleum contaminant gradient at Bemidji indicated major differences in microbial community structure between the uncontaminated site, the Fe(III)-reducing sites (sites IR-3, IR-2, and IR-1, in order of increasing contamination), and the methanogenic (most heavily contaminated) site (Fig. 3). While the overall DGGE patterns did not differ drastically within the Fe(III)-reducing zone, several differences between site IR-3 and sites IR-1 and IR-2 were apparent. These included a much higher intensity of the bands representing Geo-83 and Geo-125 at site IR-3, as well as the presence of several faint bands at sites IR-1 and IR-2 that were not recovered from site IR-3. Bands representing Beta-145 and Gthrx-84 were clearly evident at all of the Fe(III)-reducing sites.
Phylogenetic diversity of Geobacteraceae in sediment community.
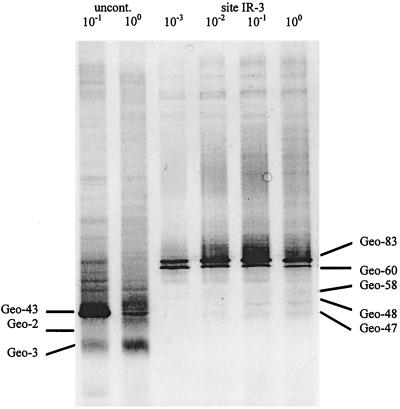
Because of the apparent enrichment of Geobacteraceae in the Fe(III)-reducing sediments that were most active in benzene degradation, the diversity of the Geobacteraceae was examined in more detail with the primer sets designed to specifically target Geobacteraceae. The DGGE patterns from the uncontaminated and benzene-oxidizing sediments revealed marked differences in their Geobacteraceae populations, with no shared sequences apparent. Two 16S rDNA fragments (Geo-83 and Geo-60) were consistently found in PCR products from the benzene-oxidizing sediments from site IR-3 (Fig. 7). Geo-83 was identical in sequence to one of the predominant sequences retrieved from site IR-3 with the less-specific bacterial primers. Like Geo-83, Geo-60 was most closely related to G. chapellei and Pelobacter propionicus. Geo-83 and Geo-60 were the dominant PCR products under several different analysis conditions, including extraction of DNA by a freeze-thaw method (see Materials and Methods), amplification by a seminested PCR protocol (with primers 8F [44] and Geo825R followed by primers 338F-GC and Geo-825R), and 10−5 dilution of template DNA prior to amplification (data not shown). In addition to these two predominant bands, 16S rDNA amplified from site IR-3 contained several bands of much lower intensity (Geo-58, Geo-48, and Geo-47), which were not members of the Geobacter cluster. Phylogenetic analysis of these sequences suggested that they were deeply branching members of the delta subclass of the proteobacteria and indicated that they probably represent undescribed genera (Fig. 5). However, especially because the current analysis is limited to about 300 base positions, it is difficult to infer phylogenetic relationships of sequences with no known close relatives.
FIG. 7.
DGGE analysis of 16S rDNA fragments amplified from the uncontaminated (uncont.) site and the benzene-oxidizing site (IR-3) by using Geobacteraceae-targeted primers.
In contrast to the results from site IR-3, the DGGE patterns from the uncontaminated zone varied considerably among triplicates or when sediment DNA was diluted prior to amplification (Fig. 7). 16S rDNA sequences retrieved from the uncontaminated site with Geobacteraceae-targeted primers (Geo-2, Geo-3, and Geo-43) fell within a tight phylogenetic cluster that appeared to fall outside of the Geobacteraceae family (Fig. 5). These sequences were distinct from those retrieved from site IR-3 and may represent deeply branching delta proteobacteria (Fig. 5).
Benzene-oxidizing enrichment culture.
A sediment-free enrichment culture which reduced Fe(III) in a medium with benzene as the sole electron donor and poorly crystalline Fe(III) oxide as the sole electron acceptor was established with sediments from site 96-1. Site 96-1 is analogous to site IR-3 in that it was determined to be within the Fe(III)-reducing zone of the aquifer and to actively degrade benzene under in situ conditions (4). Benzene-degrading enrichment cultures could not be established with sediments from the uncontaminated site. When [14C]benzene was added to the enrichment culture 14CO2 was produced over time, with ca. 25% of the [14C]benzene added to cultures being mineralized to 14CO2 within 25 days. Thus, it was clearly demonstrated that this enrichment could anaerobically oxidize benzene.
DGGE analysis of 16S rDNA retrieved from benzene-oxidizing enrichment cultures revealed three distinct bands (data not shown), with the most intense band (Benz-76) representing an organism closely related to G. chapellei (Fig. 5). Comparative sequence analysis revealed that one of the remaining bands (Benz-88) was closely related to Variovorax paradoxus HW1 (20) and an unidentified toluene degrader, D8 (33), while the second remaining band (Benz-88) was closely related to G. fermentans.
DISCUSSION
The results from molecular phylogenetic analysis of sediments and enrichment cultures, as well as the PLFA analysis, demonstrate that anaerobic benzene mineralization under Fe(III)-reducing conditions is associated with increased numbers of Geobacter spp. This finding might have been predicted from previous pure-culture studies because Geobacter spp. are the only organisms available in culture that are known to oxidize aromatic compounds with the reduction of Fe(III) (10, 22, 25, 28). However, it is generally considered that the most environmentally significant microorganisms are not readily recovered in laboratory cultures (2). Furthermore, the Geobacter spp. currently in culture are not known to metabolize benzene. Therefore, it was not necessarily expected that organisms closely related to the Geobacter spp. that are available in pure culture would predominate in the zone of active benzene degradation at Bemidji. These results, and the fact that a Geobacter species was a dominant organism in a benzene-degrading, Fe(III)-reducing enrichment culture that was established from the zone of benzene degradation, suggest that it may be possible to culture the environmentally significant microorganisms from the Fe(III)-reducing zone of petroleum-contaminated aquifers.
Comparative studies with MPN-PCR and PLFA analyses.
The Bemidji aquifer provides the only site as yet described in which benzene is anaerobically degraded under in situ Fe(III)-reducing conditions (4, 5). Although benzene oxidation coupled to Fe(III) reduction was previously reported in other aquifer sediments (31, 32), in those cases benzene degradation only took place when the availability of Fe(III) was increased with chelators or humic substances that serve as electron shuttles to Fe(III) (27). Other aromatic hydrocarbons, including toluene and naphthalene, are also degraded under Fe(III)-reducing conditions at the Bemidji site (5). However, the degradation of these hydrocarbons is not localized at only one site as is benzene degradation. Since attempts to obtain pure cultures of benzene-degrading Fe(III) reducers have as yet been unsuccessful, it was considered that comparative studies between sites that were geochemically similar but differed in the capacity to degrade benzene might reveal microorganisms specifically associated with anaerobic benzene degradation.
Results of MPN-PCR studies with PCR primers specific for microorganisms in the family Geobacteraceae reported here and previously (4) indicated that the benzene-degrading sediments from site IR-3 had significantly more Geobacteraceae than did the other sediments. The PLFA analysis also suggested that there was an enrichment of Geobacteraceae at site IR-3. However, the PLFA data further indicated that there was a general increase in the microbial population at site IR-3, suggesting that, in addition to Geobacteraceae, other microorganisms were also enriched at this site. Therefore, in order to more fully explore the microbial populations that might be involved in benzene degradation, the microbial community was analyzed in more detail with DGGE.
Comparison of overall bacterial diversity with DGGE.
Although PCR-DGGE analysis has several potential limitations (13, 18, 50), this approach was clearly effective in identifying the major shifts in the microbial community in the Bemidji aquifer. In an attempt to avoid biases, primers that targeted bacteria were employed under low-stringency conditions at annealing temperatures with a minimum number of amplification cycles. It seems likely from analysis of triplicate sediment DNA extractions that any PCR biases inherent in this approach were consistent between samples and thus differences in DGGE profiles between sites reflected major differences in the composition of the environmental DNA templates.
The fairly simple DGGE patterns obtained with domain-level primers suggests that relatively few species dominated at each of the Bemidji sites, especially at the uncontaminated site. This conclusion is also supported by community PLFA profiles, which exhibited relatively low diversity compared to surficial soils. The apparent simplicity of these subsurface microbial communities is not surprising given the low availability of organic carbon and nutrients generally found in subsurface sediments (3, 5, 16). Other studies of phylogenetic diversity in subsurface microbial communities have also indicated relatively low levels of diversity (8, 36) compared to surficial soil communities (9, 15, 41).
The DGGE profiles indicated that the microbial community composition differed markedly among the aerobic, Fe(III)-reducing, and methanogenic zones of the aquifer. In addition to different bands predominating in samples from each TEAP zone, more bands were recovered from all of the contaminated sediments than from the uncontaminated sediments. These results suggest that different microorganisms have a selective advantage in different TEAP zones and that the BTEX contaminants stimulated the growth of microorganisms that were not dominant members of the microbial community in the uncontaminated sediments. It is likely that dominant microorganisms that grew in response to the BTEX input were involved in BTEX degradation. However, organic matter produced by the BTEX degraders could provide substrates for the growth of lesser quantities of other organisms.
Enrichment of Geobacter spp. in the zone of benzene degradation.
In accord with the MPN-PCR results with Geobacteraceae primers, the DGGE analysis of the sediments with bacterial PCR primers suggested that the IR-3 sediments in which benzene was actively degraded were enriched with Geobacteraceae. Several Geobacteraceae bands such as Geo-83 and Geo-125 (which was almost identical to Geo-83) had much higher intensity at site IR-3 than at site IR-1 or IR-2, and these bands were not detected in sediments from the uncontaminated or methanogenic sites. Geo-83 and the closely related sequences represent a distinct group in the Geobacter cluster within the family Geobacteraceae (Fig. 5). A more detailed evaluation of the diversity of Geobacteraceae with primers specific for this family further demonstrated the enrichment of the Geobacter cluster at site IR-3 and suggested that these organisms were not present in the uncontaminated sediments. The only sequences that were recovered from the uncontaminated sediments with Geobacteraceae-targeted primers were not closely related to known Geobacteraceae spp.
The finding that there is a specific enrichment of Geobacter spp. in the zone of benzene degradation is of interest because Geobacter spp. are the only organisms in pure culture that have been reported to oxidize aromatic compounds to carbon dioxide (28). Although no pure cultures have been found to degrade benzene, as reported here, a Geobacter sp. closely related to the sequences that predominated in the benzene degradation zone was a dominant organism in the benzene-oxidizing, Fe(III)-reducing enrichment culture (Benz-76). A benzene-degrading enrichment culture could only be established with sediments from the zone of benzene degradation and not from the uncontaminated sediments, which lack the Geobacter spp. that predominate in the zone of benzene degradation. Interestingly, a Geobacter cluster organism, FeR, was also the dominant member of toluene-degrading enrichment cultures established with sediments from the Bemidji site (4). Comparison of FeR to the Geobacteraceae sequences described here suggested that FeR was closely related to, but distinct from, Benz-76 (sharing 94 of 102 base positions).
Another sequence that was consistently recovered from the benzene-degrading sediments but not the uncontaminated sediments was Gthrx-84, which is most closely related to G. fermentans. G. fermentans is capable of completely oxidizing acetate to carbon dioxide with Fe(III) serving as the sole electron acceptor but, in contrast to several Geobacter species, it does not use aromatic compounds (28). A sequence closely related to G. fermentans was recovered from the benzene-oxidizing, Fe(III)-reducing enrichment culture, but the DGGE band for this sequence was not as intense as the Geobacter band. Thus, it is not known if this organism was involved in the benzene degradation or survived on the products of the Geobacter sp. in the enrichment that is hypothesized to have been the benzene degrader. Previous studies employing a dilution-to-extinction culturing method demonstrated that G. fermentans was a numerically dominant acetate-oxidizing Fe(III) reducer in uncontaminated sediments from Bemidji, as well as in sediments from the benzene-degrading, Fe(III) reduction zone (4). In contrast to the pervasiveness of Geothrix spp., Geobacter spp. were only numerically significant in the zone of benzene degradation in those culturing studies. While Gthrx-84 was not detected in the uncontaminated sediments, this sequence was found at comparable band intensities in all of the sediments in which Fe(III) reduction was the TEAP. The fact that the Gthrx-84 sequence was not uniquely enriched in the zone of benzene degradation suggests that it is not specifically involved in benzene degradation at site IR-3. In a similar manner, although the sequence Beta-169 was recovered from site IR-3 but not from the uncontaminated site, the intensities of the Beta-169 band were similar in all three Fe(III)-reducing sites, suggesting that it was not uniquely associated with benzene degradation.
The sequence Beta-142 that was also found at site IR-3 but not at the uncontaminated site is most closely related to V. parodoxus HW1 (20) and an unidentified toluene degrader, strain D8 (33). V. parodoxus is an obligate aerobe (46), and although they have not been characterized, strains HW1 and D8 were also grown under aerobic conditions (20, 33). Therefore, it seems unlikely that these organisms are capable of aromatic degradation under anaerobic, Fe(III)-reducing conditions. In addition, Beta-142 was detected at other Fe(III)-reducing sites, albeit with lower band intensities (Fig. 3), indicating that its presence is not unique to the benzene-oxidizing site, IR-3. However, a sequence closely related to V. parodoxus was recovered from the benzene-oxidizing, Fe(III)-reducing enrichment culture along with a Geobacter sequence and a Geothrix sequence. Thus, the possibility that an organism(s) closely related to V. parodoxus is involved in anaerobic benzene degradation cannot be eliminated. Similarly, it is possible that anaerobic benzene degradation is carried out by a microbial consortium that includes Geobacter sp., Geothrix sp., and a relative of V. parodoxus, all of which were detected in both the benzene-oxidizing sediments and the enrichment.
In summary, although comparative analysis of 16S rDNA sequences does not allow definitive determination of which microorganisms are responsible for a specific metabolism in the environment, the results suggest that specific Geobacter spp. are associated with the capacity for benzene degradation in this petroleum-contaminated aquifer. Evidence supporting this conclusion includes (i) the significant increase in Geobacteraceae within the zone of benzene degradation; (ii) specific enrichment of a tight phylogenetic cluster of Geobacter spp. in the zone of benzene degradation that is not found in the uncontaminated sediments; (iii) the fact that the genus Geobacter contains organisms known to be able to oxidize aromatic compounds with the reduction of Fe(III); and (iv) the finding that Geobacter spp. were dominant organisms in a benzene-oxidizing, Fe(III)-reducing enrichment culture established with benzene-degrading sediments from this aquifer. This is an important finding because, to date, the specific enrichment of Geobacter spp. is the only variable which has been found to be associated with the capacity for anaerobic benzene degradation at this site. A variety of other geochemical parameters were not predictive of the potential for benzene degradation (5). Thus, these results demonstrate that phylogenetic studies of contaminated aquifer sediments can yield insights into the microorganisms associated with contaminant degradation and raise the possibility that phylogenetic analyses will be useful for predicting the potential for anaerobic benzene degradation at other sites.
ACKNOWLEDGMENTS
This research was supported by the National Science Foundation (grant DEB-9523932) and the American Petroleum Institute.
We thank G. Delin and W. Larson for help in sample collection and O. Snoeynbos West, C. V. Gaw Van Praagh, and T. Magnuson for technical advice.
REFERENCES
- 1.Amann R I, Binder B, Chisholm S W, Olsen R, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson R T, Lovley D R. Ecology and biogeochemistry of in situ groundwater bioremediation. Adv Microb Ecol. 1997;15:289–350. [Google Scholar]
- 4.Anderson R T, Rooney-Varga J N, Gaw C V, Lovley D R. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum-contaminated aquifers. Environ Sci Technol. 1998;32:1222–1229. [Google Scholar]
- 5.Anderson, R. T., and D. R. Lovley. Naphthalene and benzene degradation under Fe(III)-reducing conditions in petroleum-contaminated aquifers. Bioremediation, in press.
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 7.Balkwill D L, Ghiorse W C. Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl Environ Microbiol. 1985;50:580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boivin-Jahns V, Ruimy R, Bianchi A, Daumas S, Christen R. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol. 1996;62:3405–3412. doi: 10.1128/aem.62.9.3405-3412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coates J D, Phillips E J P, Lonergan D J, Jenter H, Lovley D. Isolation of Geobacter species from diverse sedimentary environments. Appl Environ Microbiol. 1996;62:1531–1536. doi: 10.1128/aem.62.5.1531-1536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman W E, Munch J W, Streicher R P, Ringhand H P, Kopfler F C. Identification and measurement of components in gasoline, kerosene, and no. 2 fuel oil that partition into the aqueous phase after mixing. Arch Environ Contam Toxicol. 1984;13:171–178. [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eganhouse R P, Dorsey T F, Phinney C S, Westcott A M. Processes affecting the fate of monoaromatic hydrocarbons in an aquifer contaminated by crude oil. Environ Sci Technol. 1996;30:3304–3312. [Google Scholar]
- 15.Felske A, Wolterink A, Van Lis R, Akkermans A D. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghiorse W C, Wilson J T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–172. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- 17.Guckert J B, Ringelberg D B, White D C, Hanson R S, Bratina B J. Membrane fatty acids as phenotypic markers in the polyphasic taxonomy of methylotrophs within the proteobacteria. J Gen Microbiol. 1991;137:2631–2641. doi: 10.1099/00221287-137-11-2631. [DOI] [PubMed] [Google Scholar]
- 18.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 19.Hult M F. Ground-water contamination by crude oil at Bemidji, Minnesota, research site. U.S. Geological Survey Toxic Waste—Ground-Water Contamination Study 84-4188. U.S. Geological Survey Water-Resources Investigations Report. Washington, D.C: U.S. Geological Survey; 1984. [Google Scholar]
- 20.Kamagata Y, Fulthorpe R R, Tamura K, Takami H, Forney L J, Tiedje J M. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1997;63:2266–2272. doi: 10.1128/aem.63.6.2266-2272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane D, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonergan D J, Jenter H L, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley D R. Dissimilatory metal reduction. Annu Rev Microbiol. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 24.Lovley D R. Potential for anaerobic bioremediation of BTEX in petroleum-contaminated aquifers. J Ind Microbiol. 1997;18:75–81. [Google Scholar]
- 25.Lovley D R, Baedecker M J, Lonergan D J, Cozzarelli I M, Phillips E J P, Siegel D I. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature. 1989;339:297–299. [Google Scholar]
- 26.Lovley D R, Chapelle F H, Woodward J C. Use of dissolved H2 concentrations to determine the distribution of microbially catalyzed redox reactions in anoxic ground water. Environ Sci Technol. 1994;28:1205–1210. doi: 10.1021/es00056a005. [DOI] [PubMed] [Google Scholar]
- 27.Lovley D R, Coates J D, Blunt-Harris E L, Phillips E J P, Woodward J C. Humic substances as electron acceptors for microbial respiration. Nature. 1996;382:445–448. [Google Scholar]
- 28.Lovley D R, Coates J D, Saffarini D A, Lonergan D J. Dissimilatory iron reduction. In: Winkelman G, Carrano C J, editors. Iron and related transition metals in microbial metabolism. Amsterdam, The Netherlands: Harwood Academic Publishers; 1997. pp. 187–215. [Google Scholar]
- 29.Lovley D R, Giovannoni S J, White D C, Champine J E, Phillips E J P, Gorby Y A, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 30.Lovley D R, Lonergan D J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl Environ Microbiol. 1990;56:1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovley D R, Woodward J C, Chapelle F H. Rapid anaerobic benzene oxidation with a variety of chelated Fe(III) forms. Appl Environ Microbiol. 1996;62:288–291. doi: 10.1128/aem.62.1.288-291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley D R, Woodward J C, Chapelle F H. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature. 1994;370:128–131. doi: 10.1038/370128a0. [DOI] [PubMed] [Google Scholar]
- 33.Moller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy F, Herkelrath W N. A sample-freezing drive shoe for a wire line piston core sampler. Ground Water Monit Remediation. 1996;16:86–90. [Google Scholar]
- 35.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen K, Arlinger J, Hallbeck L, Pettersson C. Diversity and distribution of subterranean bacteria in groundwater at Oklo in Gabon, Africa, as determined by 16S rRNA gene sequencing. Mol Ecol. 1996;5:427–436. doi: 10.1111/j.1365-294x.1996.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 37.Reinhard M, Shang S, Kitanidis P K, Orwin E, Hopkins G D, Lebron C A. In situ BTEX biotransformation under enhanced nitrate- and sulfate-reducing conditions. Environ Sci Technol. 1997;31:28–36. [Google Scholar]
- 37a.Ringelberg, D. B. Unpublished data.
- 38.Ringelberg D B, Townsend G T, Deweerd K A, Suflita J M, White D C. Detection of the anaerobic dechlorinating microorganism Desulfomonile tiedjei in environmental matrices by its signature lipopolysaccharide branched-long-chain hydroxy fatty acids. FEMS Microbiol Ecol. 1993;14:9–18. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Thomas J M, Ward C H. In situ biorestoration of organic contaminants in the subsurface. Environ Sci Technol. 1989;23:760–766. [Google Scholar]
- 41.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Environmental Protection Agency. Underground motor fuel storage tanks: a national survey, NTIS PB 86-216512. U.S. Washington, D.C: Environmental Protection Agency; 1986. [Google Scholar]
- 43.U.S. Geological Survey. 15 December 1998, posting date. References. [Online.] http://wwwmn.cr.usgs.gov/bemidji/index/html. [3 February 1999, last date accessed.]
- 44.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson S G. Gram-negative bacteria. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. New York, N.Y: Academic Press, Inc.; 1988. pp. 299–323. [Google Scholar]
- 46.Willems A, De Ley J, Gillis M, Kersters K. Comamonadaceae, a new family encompassing the acidovorans rRNA complex, including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969) Int J Syst Bacteriol. 1991;41:445–450. [Google Scholar]
- 47.Wilson B, Smith G B, Rees J F. Biotransformations of selected alkylbenzenes and halogenated aliphatic hydrocarbons in methanogenic aquifer material: a microcosm study. Environ Sci Technol. 1986;20:997–1002. doi: 10.1021/es00152a005. [DOI] [PubMed] [Google Scholar]
- 48.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson L P, Bouwer E J. Biodegradation of aromatic compounds under mixed oxygen/denitrifying conditions: a review. J Ind Microbiol Biotechnol. 1997;18:116–130. doi: 10.1038/sj.jim.2900288. [DOI] [PubMed] [Google Scholar]
- 50.Wintzingerode F V, Gobel U F, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 51.Zurcher F, Thuer M. Rapid weathering processes of fuel oil in natural waters: analyses and interpretations. Environ Sci Technol. 1978;12:838–845. [Google Scholar]