Abstract
SARS-CoV-2 variant detection relies on resource-intensive whole-genome sequencing methods. We sought to develop a scalable protocol for variant detection and surveillance in Paraguay, pairing rRT-PCR for spike mutations with Nanopore sequencing. A total of 201 acute-phase nasopharyngeal samples were included. Samples were positive for the SARS-CoV-2 N2 target and tested with the Spike SNP assay to detect mutations associated with the following variants: alpha (501Y), beta/gamma (417variant/484K/501Y), delta (452R/478K), and lambda (452Q/490S). Spike SNP calls were confirmed using amplicon (Sanger) sequencing and whole-genome (Nanopore) sequencing on a subset of samples with confirmed variant lineages. Samples had a mean N2 Ct of 20.8 (SD 5.6); 198/201 samples (98.5%) tested positive in the Spike SNP assay. The most common genotype was 417variant/484K/501Y, detected in 102/198 samples (51.5%), which was consistent with the P.1 lineage (gamma variant) in Paraguay. No mutations (K417 only) were found in 64/198 (32.3%), and K417/484K was identified in 22/198 (11.1%), consistent with P.2 (zeta). Seven samples (3.5%) tested positive for 452R without 478K, and one sample with genotype K417/501Y was confirmed as B.1.1.7 (alpha). The results were confirmed using Sanger sequencing in 181/181 samples, and variant calls were consistent with Nanopore sequencing in 29/29 samples. The Spike SNP assay could improve population-level surveillance for mutations associated with SARS-CoV-2 variants and inform the judicious use of sequencing resources.
Keywords: SARS-CoV-2, variants, real-time RT-PCR, Paraguay
1. Introduction
Amidst the global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), numerous variants have emerged due to mutations in the positive-sense RNA genome. Variants of concern (VOCs) and variants of interest (VOIs) bear mutations that impact detection, treatment, clinical severity, transmission, and/or immune protection from prior infection or vaccination [1]. Whole-genome sequencing is the reference method for detecting and tracking variants, but this is a time- and resource-intensive process with the capacity varying markedly between regions [2,3,4,5]. It was estimated that 5% of all samples need to be sequenced to reliably detect variants at a prevalence of 0.1–1% [3], yet for many regions, this goal remains unattainable. Limited available sequencing capacity and overwhelming case numbers have motivated the development of real-time reverse transcription PCRs (rRT-PCRs) for the detection and surveillance of specific mutations found among VOCs/VOIs [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20]. For these reasons, our group designed the Spike SNP assay, which is a multiplex rRT-PCR designed to detect specific mutations in the receptor binding motif of the Spike receptor binding domain (RBD; Figure 1) [20].
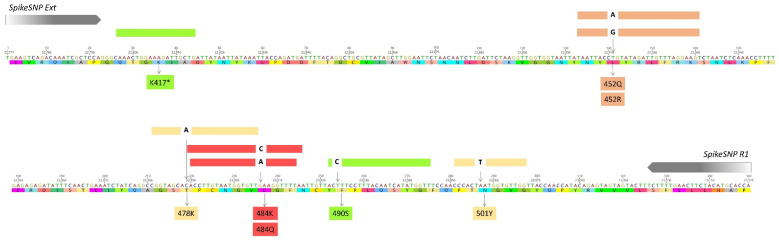
Figure 1.
Graphical display of the 348-base-pair Spike SNP target, including the nucleic acid sequence and amino acid translation of the Wuhan-Hu-1 strain (NC_045512.2). The locations of the forward and reverse primer sequences and all hydrolysis probes are shown. Probe colors correspond to the fluorophores on each probe and the detection channel in the Rotor-Gene Q instrument. Detected nucleotide changes are displayed within the probe, and amino acid mutations are shown below the sequence. The probe for K417 is marked (*), as this is the only probe for which variant sequences result in signal loss and this has only been successfully designed with locked nucleic acid (LNA) bases.
In Paraguay, the first confirmed SARS-CoV-2 infection was reported on 7 March 2020 [21], but 18 months into the pandemic, <0.1% of samples from the country could be sequenced and entered in the GISAID database (www.gisaid.org, accessed 1 March 2022) [22]. Published reports on case series in Paraguay have been limited to descriptive analyses of symptoms and disease outcomes [23,24]. As such, little is known about variants that circulated in the country during the beginning of the pandemic in 2020. However, data from 2021 show that variants originally detected in Brazil, such as the P.1 (gamma variant) and P.2 (zeta) lineages, circulated widely until the middle of that year (GISAID database). Nanopore sequencing on the portable MinION sequencer from ONT (Oxford Nanopore Technologies, Oxford, UK) became available in Asunción, the capital of Paraguay, to detect and monitor SARS-CoV-2 variants for the first time, where this technology provides lower-cost and more-rapid whole-genome sequencing compared to other methods [2,25]. The objective of the current study was to develop an economical and scalable protocol based on the Spike SNP assay and Nanopore sequencing to identify and monitor VOCs/VOIs in acute-phase samples in Paraguay.
2. Methods
2.1. Clinical Samples
Nasopharyngeal (NP) samples were obtained from residual baseline surveillance samples at the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA) Laboratory. Samples were de-identified, aliquoted, and stored at −80 °C until nucleic acid extraction. A convenience set of 201 SARS-CoV-2 positive samples from November 2020 to April 2021 was chosen for this study. Samples tested positive for the envelope gene target of the Charité protocol [26] with a cycle threshold (Ct) < 34 and were collected from different cities in Paraguay from symptomatic (n = 187) and asymptomatic (n = 14) individuals. This study was reviewed and approved by the IICS Scientific and Ethics Committee (P38/2020) and the Emory Institutional Review Board (study 00110736). Patient consent was waived under approval of IICS Scientific and Ethics Committee (P38/2020) due to all samples being de-identified remaining of diagnostics samples.
2.2. Spike SNP Design and Performance
Hydrolysis probes were designed to detect mutations associated with emerging variants of concern: L452Q (CAG > CTG) and F490S (UUU > UCU, C.37, lambda), E484Q (GAA > CAA, B.1.617.1, kappa), T478K (ACA > AAA, B.1.617.2, delta), and a modified T478K containing one mixed base in codon 477 for the detection of omicron. All new probes were designed as described [20], without locked nucleic acids (LNAs). Unmodified probes for 484K and 501Y were also designed and evaluated. New probes were individually evaluated using SARS-CoV-2 genomic RNA in singleplex assays. Probes that provided the most sensitive signal based on the cycle threshold (Ct) value, with preserved specificity, were selected. Subsequently, probes were tested in combination and compared side-by-side with the corresponding singleplex assays. Probe evaluations were performed by utilizing variant control samples that were kindly provided by the National Institutes of Health Variant Task Force and contained the following SARS-CoV-2 variants: alpha, beta, gamma, kappa, lambda, delta, and omicron. Variant control samples were heat-inactivated clinical samples that were prepared in the same manner as all clinical samples and then tested across a range of N2 Ct values from 20–36. Variants and lineages had been confirmed via sequencing (Helix, San Mateo, CA, USA).
Nucleic acids were extracted from all samples on an EMAG instrument (bioMérieux, Durham, NC) from 200 µL of NP swab and eluted in 50 µL. All rRT-PCRs were performed on a Rotor-Gene Q instrument (Qiagen, Germantown, MD, USA) using 20 µL reactions of the Luna Probe One-Step RT-qPCR Kit (New England Biolabs, Ipswich, MA, USA) and 5 µL of nucleic acid eluate. Each sample was tested in a duplex reaction for the SARS-CoV-2 nucleocapsid 2 target and RNase P (N2RP), as well as two different reactions of the Spike SNP assay, including probes for (1) K417, 452R, 484K, and 501Y, as described [20]; and (2) 452Q, 490S, and 484Q. During the study, two probes for 478K were designed, and all samples with 452R were tested in singleplex reactions with the original probe to search for the delta variant. Analytical performance of the second Spike SNP reaction was also evaluated following the inclusion of this probe. As sample collection pre-dated the emergence of omicron, samples were not re-tested with the revised 478K probe that was designed to also detect the mutation in this variant. The N2RP assay was performed as described [20,27]. For full Spike SNP methods, please see the Supplementary Materials. This study was performed in accordance with the STARD 2015 reporting guidelines (accessed 16 January 2022).
2.3. Sequencing
All rRT-PCR products from the first Spike SNP reaction (348-base-pair amplicon) were stored at 4 °C and later shipped to GeneWiz (South Plainfield, NJ, USA) for Sanger sequencing. This was performed to confirm the Spike SNP calls in the current study. The amplicon sequence was considered to be high quality and included for analysis if it achieved a quality score ≥ 40 and had base calls for all probe targets in at least one direction. A quality score ≥ 40, as defined by the company, signifies a mean probability of error ≤ 0.01% for each base call across the amplicon.
A representative subset of 29 samples underwent complete genome sequencing to confirm the variant lineage and identify mutations outside of the Spike SNP amplicon. Samples were selected to include the ancestral strain; alpha, gamma, and zeta variants; and one sample with the L452R mutation. Complete genome sequencing was performed following the Artic Network protocol using the V3 primer scheme [28]. The genomic libraries were prepared and then loaded on an R9.4.1 flow cell (ONT FLO-MIN106) for sequencing with a MinION device (MIN-101B, ONT, Oxford, UK). Basecalling, demultiplexing, and trimming were carried out using the Guppy toolkit integrated into the MinKNOW v.4.3.4 Nanopore software ( ONT, Oxford, UK). Genome assembly and consensus sequences were obtained with Nanopore EPI2ME v.3.4.1(ONT, Oxford, UK) (using Flye and Medaka respectively). The Nextclade tool was used for clade definition and mutation calling [29] and lineages were assigned using the Pangolin web server [30]. Sequences were deposited in the GISAID database.
2.4. Statistics
Basic statistical analyses were performed using GraphPad Prism version 9.0.2 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Spike SNP Optimization
For the initial testing, a Spike SNP assay was developed that contained probes for 452Q, 484Q, and 490S. With the global emergence of B.1.617.2 and the lack of 484Q detection, the 478K probe was incorporated into the assay in place of 484Q. The addition of the original or revised 478K probe had no impact on 452Q or 490S detection (Figure 2), and despite overlapping sequences, the 478K and 484Q probes demonstrated no interference when combined in the same reaction (Figure S1). New probes for 484K and 501Y without LNA bases provided sensitive detection at both sites, though Ct values with the unmodified 484K probe were approximately two cycles later than those generated with the LNA probe (Figure S2). Testing of clinical samples in the current study was performed with the original 484K and 501Y probes to maintain consistency with existing protocols. All primers and probes used in or developed for this study are shown in Table 1, and their respective locations across the amplicon are displayed in Figure 1.
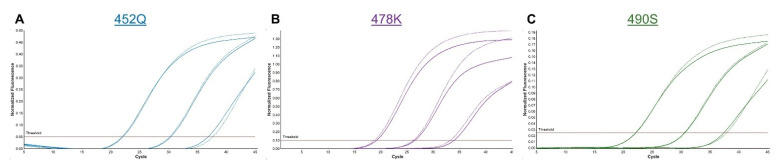
Figure 2.
Addition of the 478K probe did not impact the signal from 452Q or 490S probes. Amplification curves are shown for 100-fold dilutions of C.37 (lambda) and B.1.617.2 (delta) samples tested together in a single run. The results were similar for (A) 452Q and (C) 490S mutations in C.37 with the probe for 478K (solid curves) or without (dotted curves). Results for B.1.617.2 were similar with the 478K probe (B) used in singleplex (dotted curves) or triplex reactions (solid curves). Nonspecific signals were not detected in any channel.
Table 1.
Spike SNP assay primers and probes used in the current study.
| Name | Sequence a | Concentration (nM) b | Location (5′–3′) c |
|---|---|---|---|
| Primers | |||
| SpikeSNP Ext d | TGAAGTCAGACAAATCGCTCC | 400 | 22,777–22,797 |
| SpikeSNP R1 d | TGGTGCATGTAGAAGTTCAAAAG | 400 | 23,103–23,125 |
| LNA Probes | |||
| K417 d | FAM-CAAACTGGA+A+A+G+ATTGCTG-3IABkFQ | 200 | 22,802–22,820 |
| 484K d | Cy5-ACCTTGTAATGGTGT+T+A+AAGGTTTT-3IAbRQSp | 200 | 22,996–23,020 |
| 501Y d | HEX-CCCAC+T+T+ATGGTGTTGG-3IABkFQ | 200 | 23,057–23,073 |
| Unmodified Probes | |||
| 452R d | CFR610-ATAATTACCGGTATAGATTGTTTAGGAAGT-BHQ-2 | 200 | 22,908–22,937 |
| 484K | Q670-CACCTTGTAATGGTGTTAAAGGTTTTAA-BHQ-2 | 200 | 22,995–23,022 |
| 484Q | Q670-CACCTTGTAATGGTGTTCAAGGTTTTAA-BHQ-2 | 200 | 22,995–23,022 |
| 501Y | CFO560-CAACCCACTTATGGTGTTGGTTAC-BHQ-1 | 200 | 23,054–23,077 |
| 452Q | CFR610-ATAATTACCAGTATAGATTGTTTAGGAAGT-BHQ-2 | 200 | 22,908–22,937 |
| 490S | FAM-ACTCTCCTTTACAATCATATGGTTTCC-BHQ-1 | 200 | 23,028–23,054 |
| 478K (original) | CFO560-CGGTAGCAAACCTTGTAATGGTG-BHQ-1 | 200 | 22,987–23,009 |
| 478K (revised) | CFO560-CGGTARCAAACCTTGTAATGGTG-BHQ-1 | 200 | 22,987–23,009 |
Abbreviations: BHQ, black hole quencher; CFO560, CAL Fluor Orange 560; CFR610, CAL Fluor Red 610; Cy5, Cyanine 5; FAM, Fluorescein; IABkFQ/IAbRQSp, Iowa Black quenchers; Q670, Quasar 670; a probe sequences listed 5′-fluorophore-sequence-quencher-3′; “+” before base indicates a locked nucleic acid, bold and underlined base indicates the targeted single nucleotide change; b concentration in the final reaction mixture; c location in Wuhan-Hu-1 complete genome sequence (NCBI Reference Sequence: NC_045512.2); d previously published, presented for clarity.
3.2. SNP Detection in Clinical Samples
Clinical samples were collected between November 2020 and April 2021. A total of 113 participants were female (56.8% of the 199 with gender data available), and the age and day of symptoms at sample collection are shown in Table 2, along with the N2RP and Spike SNP assay results. The distribution of N2 target Ct values is shown in Figure 3 (Ct values provided in Table S1). The most common genotype identified was 417variant/484K/501Y (102/198 samples, 51.5%), which was consistent with B.1.351 (beta) and P.1 (gamma). Large numbers of cases were also detected with a signal for K417 only (64/198, 32.3%), which was consistent with a non-variant/ancestral lineage, and K417/484K (22/198, 11.1%), which was observed in P.2 (zeta), among others. Seven samples (3.5%) had the genotype K417/452R but tested negative with the 478K probe and were thus not consistent with B.1.617.2.
Table 2.
Genotypes detected in the Spike SNP assay, patient demographics, and the percent confirmed using amplicon sequencing.
| Category | n (%) | Age, Years, Mean (SD) a | Day of Symptoms, Mean (SD) b | Ct, Mean (SD) c | Sequencing Confirmed, n/N (%) d |
|---|---|---|---|---|---|
| N2 detected | 201 (100) | 42.6 (16.4) | 4.8 (3.0) | 20.8 (5.6) | ─ |
| Spike SNP detected | 198 (98.5) | 42.5 (16.4) | 4.8 (3.0) | 20.6 (5.3) | 181/181 (100) |
| Genotype e | |||||
| 417var/484K/501Y | 102 (51.5) | 42.8 (14.9) | 4.5 (2.4) | 19.3 (5.0) | 96/96 (100) |
| K417 | 64 (32.3) | 42.6 (18.8) | 5.1 (3.7) | 22.1 (5.4) | 56/56 (100) |
| K417/484K | 22 (11.1) | 37.4 (14.2) | 4.8 (2.7) | 22.2 (5.4) | 20/20 (100) f |
| K417/452R | 7 (3.5) | 54.0 (15.8) | 5.0 (4.6) | 19.4 (4.0) | 7/7 (100) |
| K417/490S | 1 (0.5) | 41 | 4 | 15.2 | 1/1 (100) |
| K417/501Y | 1 (0.5) | 68 | 5 | 19.7 | 1/1 (100) |
| K417/484K/501Y | 1 (0.5) | 19 | 10 | 32.1 | NA |
Abbreviations: var, variant; SD, standard deviation; a age available for 194/201 (96.5%) samples; b day of symptoms available for 136/201 (67.7%) samples; c Ct value in the N2 assay; d number with sequence confirmed (n) over the number of amplicons with high-quality sequence and base calls at each SNP position (N); e displayed as % of Spike SNP positive samples; f one sample had a mixed base (A/G) at the position targeted with the 484K probe.
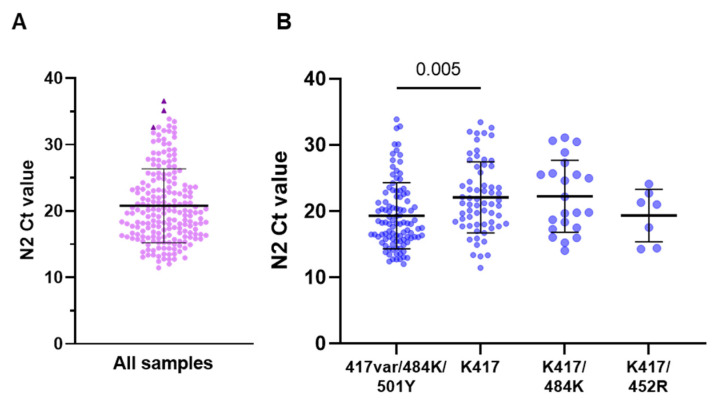
Figure 3.
Distribution of N2 Ct values for (A) all samples and (B) the four most common genotypes detected. (A) N2 Ct values for all tested samples are displayed. Three samples that tested negative in the Spike SNP assay are highlighted (dark triangles) and had N2 Ct values of 32.6, 36.6, and 35.1. (B) N2 Ct values for the four most common genotypes detected. Bars represent mean Ct and standard deviation, and p-values for tested comparisons are displayed above the graph.
Samples with the 417variant/484K/501Y genotype had significantly lower N2 Ct values (mean 19.3, standard deviation 5.0) than all other samples (22.0, 5.4; p < 0.001; data not shown) and samples with K417 only (22.1, 5.4; p = 0.002; Figure 3B). No differences in patient age or day of symptoms were observed based on the genotype (p > 0.05 for all comparisons).
3.3. Amplicon and Whole-Genome Sequencing
High-quality amplicon sequences were obtained from 181 samples (181/201, 90.0%) using Sanger sequencing, and the results confirmed Spike SNP genotype calls for all samples (Table 2). Characteristic findings for samples with 484K, with and without 501Y, are shown (Figure 4). All samples with variant calls at position 417 had the mutation AAG > ACG conferring K417T were observed in P.1. One sample had 484K detected in the Spike SNP assay (genotype K417/484K) but E484 was detected based on the consensus Sanger sequence. On review of the tracing, there was a mixture of G and A bases at the first position of codon 484, which is consistent with the evidence of a mixed viral population in this sample. Notably, this sample had a late Ct value for 484K (38.4) relative to K417 (34.3) when compared to other samples with this genotype (mean Ct difference 0.4, standard deviation 0.2). In addition to mutations targeted in the Spike SNP assay, four samples (2.2%) had a mutation conferring T478R (ACA > AGA) and one sample (0.5%) had a mutation resulting in N501T (AAU > ACU).
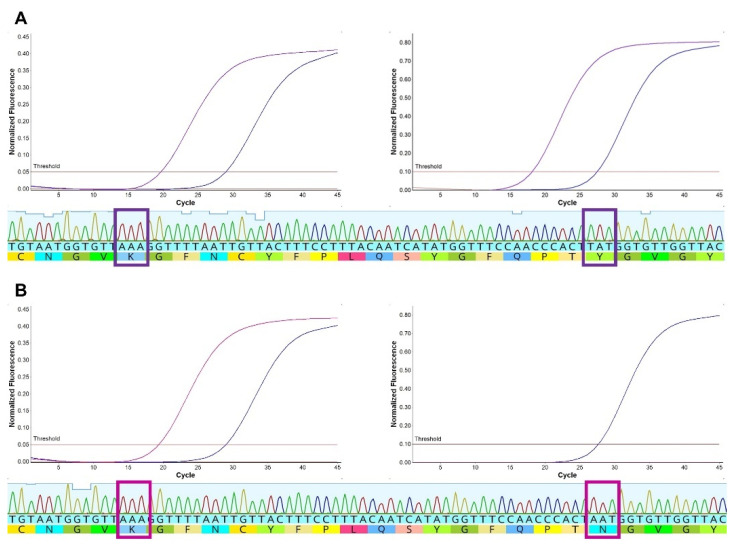
Figure 4.
Spike SNP and amplicon sequencing results were concordant. Amplification curves and amplicon sequence tracings are shown for samples with mutations encoding (A) 484K and 501Y (purple curves) or (B) 484K only with N501 (cranberry). A beta variant control sample is shown for reference (blue curves). Bold squares highlight the SNP mutations in the amplicon sequence.
Whole-genome sequences for 29 samples were obtained using the Artic protocol. Ten different lineages were identified (Table 3) [30]. Nanopore sequencing confirmed Spike SNP genotype calls for all samples. Samples bearing genotype 417variant/484K/501Y belonged to the P.1 lineage and P.1.2 sub-lineage. The genotype of only K417 was found in samples of lineages B.1.1.28, B.1.1.33, B.1.1.277, and N3, all within the 20B Nextstrain clade and lineage B.1.499 (clade 20C) [29]. The P.2 lineage that also belongs to the 20B clade showed the K417/484K genotype. One sample with genotype K417/501Y was classified as B.1.1.7 (alpha). A lineage from 2020 that was still circulating during 2021, namely, A.2.5.2, was found in one sample with genotype K417/452R.
Table 3.
Spike mutations and lineages identified using genome sequencing.
| Sample Code | Spike SNP Genotype |
Nanopore Sequencing | |||
|---|---|---|---|---|---|
| GISAID Accession No. |
Nextstrain CladeWHO Label | Pango Lineage | Spike Deletions/Amino Acid Substitutions a | ||
| PY21-100 | 417var/484K/501Y | EPI_ISL_4071892 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-9 | 417var/484K/501Y | EPI_ISL_2444778 | 20J (V3) gamma |
P.1 | L18F//T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-57 | 417var/484K/501Y | EPI_ISL_4137476 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-58 | 417var/484K/501Y | EPI_ISL_4071897 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S7D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-84 | 417var/484K/501Y | EPI_ISL_4071898 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-16 | 417var/484K/501Y | EPI_ISL_4071893 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-11 | 417var/484K/501Y | EPI_ISL_4071900 | 20J (V3) gamma |
P.1 | L18F/T20N/P26S/D138Y/L189F/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F |
| PY21-90 | 417var/484K/501Y | EPI_ISL_4071899 | 20J (V3) gamma |
P.1.2 | L18F/T20N/P26S/T95I/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/Q675H/T1027I/V1176F/V1228L |
| PY21-150 | K417 | EPI_ISL_2234899 | 20B | B.1.1.33 | D614G |
| PY21-153 | K417 | EPI_ISL_2234891 | 20B | B.1.1.33 | D614G |
| PY21-140 | K417 | EPI_ISL_2234880 | 20B | B.1.1.28 | D614G/V1176F |
| PY21-145 | K417 | EPI_ISL_4084605 | 20B | B.1.1.28 | D614G/V1176F |
| PY21-146 | K417 | EPI_ISL_2234881 | 20B | B.1.1.28 | D614G/V1176F |
| PY21-98 | K417 | EPI_ISL_2444788 | 20B | B.1.1.28 | D614G/V1176F |
| PY21-152 | K417 | EPI_ISL_2234885 | 20B | B.1.1.28 | T22I/D614G/V1176F |
| PY21-144 | K417 | EPI_ISL_4071901 | 20B | B.1.1.28 | D614G/N1135Y/V1176F |
| PY21-43 | K417 | EPI_ISL_4071895 | 20B | B.1.1.28 | A575S/D614G/V1176F |
| PY21-44 | K417 | EPI_ISL_4084604 | 20B | B.1.1.28 | T572I/D614G/V1176F |
| PY21-47 | K417 | EPI_ISL_4071894 | 20B | B.1.1.28 | A575S/D614G/V1176F |
| PY21-148 | K417 | EPI_ISL_2234894 | 20B | B.1.1.28 | V6F/L18F/D614G/V1176F |
| PY21-147 | K417 | EPI_ISL_2234887 | 20B | B.1.1.28 | Q14H/D614G/D1153A/V1176F |
| PY21-154 | K417 | EPI_ISL_2234893 | 20B | B.1.1.277 | D614G/V1176F |
| PY21-52 | K417/484K | EPI_ISL_2444780 | 20B (zeta) |
P.2 | E484K/D614G/V1176F |
| PY21-157 | K417/484K | EPI_ISL_2234879 | 20B (zeta) |
P.2 | L5F/E484K/D614G/A771S/V1176F |
| PY21-143 | K417 | EPI_ISL_2234889 | 20B | N.3 | T76I/D614G |
| PY21-141 | K417 | EPI_ISL_2234883 | 20C | B.1.499 | T478R/D614G |
| PY21-149 | K417 | EPI_ISL_2234897 | 20C | B.1.499 | T478R/D614G |
| PY21-102 | K417/452R | EPI_ISL_4071896 | 19B | A.2.5.2 | L452R/D614G |
| PY21-156 | K417/501Y | EPI_ISL_4084606 | 20I (V1) alpha |
B.1.1.7 | DEL69-70/DEL144N501Y/A570D/D614G/P681H/T716I/S982A/D1118H |
Abbreviations: var, variant; WHO, World Health Organization; a bold text indicates amino acid substitutions detected using the Spike SNP assay.
4. Discussion
Among individuals with a SARS-CoV-2 infection in Paraguay, the Spike SNP assay provided accurate detection of targeted mutations in the RBD and, in conjunction with the employed sequencing protocol, confirmed the predominance of B.1.1.28, P.2, and P.1 in this population between November 2020 and April 2021, as well as the relative absence of B.1.1.7. Given the frequent travel between Paraguay and Brazil, the B.1.1.28 lineage was expected to account for a high proportion of cases in late 2020 and early 2021, to be later replaced by P.1 and P.2. Samples with P.1 had lower Ct values in our population. A similar finding was initially documented in Brazil and likely resulted in increased transmissibility of the gamma variant with the replacement of other lineages [31,32]. The P.1 and P.2 lineages were readily differentiated from one another in the Spike SNP assay, and in a real-world scenario, the N2RP and Spike SNP assays could be performed sequentially to diagnose SARS-CoV-2 infection, identify a set of common mutations, and select samples for downstream sequencing [4,5,20].
For the current study, new probes were generated for mutations that occurred within the target region and are associated with emerging VOCs/VOIs, including delta, lambda, and omicron variants (Figure 1). As new designs do not involve separate primer–probe sets, the assay can be readily modified and reconfigured to include probes that match circulating variants in a particular region. All new probes were designed without LNA bases yet demonstrated similar performances to previous designs (Figure S1). Although LNA bases were shown to be important for the detection of K417 mutations, avoiding their use in other probes improves access to and reduces costs for these oligos. An additional benefit to the current design is that up to four mutations are targeted in a single reaction, which has been found to improve variant classification [9]. Other published rRT-PCR protocols for the detection of a similar number of mutations generate two or more amplicons in single-reaction [12,15,17] or multiple-reaction designs [6,8,14,16], and such protocols may be more difficult to modify in response to the emergence of new variants.
As shown in the current study, the Spike SNP assay can be paired with whole-genome sequencing on a MinION device to provide an economical and tractable molecular protocol for SARS-CoV-2 variant detection. Nanopore sequencing has become a common method worldwide and particularly in locations where the use of short-read sequencing platforms is less feasible. MinION sequencing was well described for SARS-CoV-2 and provides formal confirmation of detected variants and lineages, which cannot be achieved using rRT-PCR alone [4,5,25,31]. However, the Spike SNP assay may provide more sensitive detection of specific mutations than can be achieved with current sequencing protocols. Amplicon sequencing was performed in the current study to confirm Spike SNP calls for assay validation. While this provides less information than whole-genome sequencing, it serves to differentiate B.1.351 from P.1 and identify additional mutations in the receptor-binding motif. In our study, four individuals had the T478R mutation; the whole genome of two of them was sequenced and assigned to the B.1.499 lineage. This lineage circulated widely in Argentina during 2020, but the mutation T478R was not reported. While this mutation is distinct from T478K found in the B.1.617.2 (delta) and B.1.1.529 (omicron), mutations at position 478 have developed in vitro and confer decreased antibody neutralization, indicating that such mutations may benefit the virus through immunity escape [33,34]. Finally, one individual was identified with a possible mixed infection or a minor variant bearing 484K [35,36]. Such infections do not appear to cause more severe clinical disease [35], but this case highlights one potential benefit of rRT-PCR surveillance if key mutations can be detected that are not observed in the consensus sequence.
The degree of multiplexing possible in the Spike SNP assay is limited by the number of channels available in common real-time PCR instruments (4–5), as well as the defined 348-base-pair target region. Based on the results of testing for 478K and 484Q in a single reaction, overlapping probes did not impact the performance (Figure S1). However, multiplexed detection of key mutations outside of the Spike SNP target region would require additional primer–probe sets and assay optimization. A limitation of the current study was the utilization of retrospective convenience samples to evaluate the testing protocol. Although new data on SARS-CoV-2 variants in Paraguay were generated, we cannot calculate variant prevalence from these data.
In conclusion, the Spike SNP assay provided accurate detection of mutations that were associated with VOCs/VOIs in Paraguay. This can be implemented in SARS-CoV-2 testing protocols to triage samples for whole-genome sequencing in the MinION platform or direct amplicon sequencing, which provides additional information. The assay can be implemented in any laboratory performing rRT-PCR to improve the surveillance for these mutations and inform the judicious use of scarce sequencing resources.
Acknowledgments
The authors thank the staff at the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción, and collaborating laboratories: Letizia Carpinelli (CYRLAB), Fátima Ovando (Hospital de Clínicas, UNA), Graciela Riera (MeyerLab), and Stefan Goertzen (Hospital Loma Plata) for their support during the study period and the COVID-19 pandemic in Paraguay. We also thank members of the Variant Task Force at Emory University, namely, Aunradha Rao, Leda Bassit, and Morgan Greenleaf, for the provision of samples with characterized variant strains for assay evaluation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14050873/s1, File S1: Spike SNP Performance; Table S1: Spike SNP and N2RP line data for 201 clinical samples tested from Paraguay; Figure S1: 478K and 484Q probes do not interfere despite overlapping sequences; Figure S2: Performance of unmodified hydrolysis probes compared to probes that include LNA bases.
Author Contributions
Conceptualization, M.M., F.C. and A.R.; Data curation, M.M., A.R. and J.J.W.; Formal analysis, M.M., L.M., A.R. and J.J.W.; Funding acquisition, M.M., A.R. and J.J.W.; Investigation, M.M., P.-V.N., M.S., F.C., A.V., L.F., M.E.G., L.E.R., C.C.D.A., J.F., J.O., F.d.P., L.M., E.N., A.R. and J.J.W.; Methodology, M.M., P.-V.N., M.S., F.C., A.V., L.F., M.E.G., L.E.R., C.C.D.A., J.F., J.O., F.d.P., L.M., E.N., A.R. and J.J.W.; Project administration, M.M. and A.R.; Resources, A.R.; Supervision, M.M.; Writing—original draft, M.M., A.R. and J.J.W.; Writing—review and editing, M.M., P.-V.N., M.S., F.C., A.V., L.F., M.E.G., L.E.R., C.C.D.A., J.F., J.O., F.d.P., L.M., E.N., A.R. and J.J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funding from Consejo Nacional de Ciencia y Tecnología with FEEI support, CONACYT, Paraguay, PINV20-239 (M.M.); National Institutes of Health grants U54 EB027690 02S1, U54 EB027690 03S1, U54EB027690 03S2, and UL1TR002378; and the Doris Duke Charitable Foundation, Clinical Scientist Development Award 2019089 (J.J.W.).
Institutional Review Board Statement
This study was reviewed and approved by the IICS Scientific and Ethics Committee (P38/2020) and the Emory Institutional Review Board (study 00110736).
Informed Consent Statement
Patient consent was waived under approval of IICS Scientific and Ethics Committee (P38/2020) due to all samples being de-identified remaining of diagnostics samples.
Data Availability Statement
All supporting data necessary for interpretation are provided herein. Complete genome sequences were deposited in GISAID and accession numbers are shown in Table 3. Amplicon sequence tracings are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Centers for Disease Control and Prevention SARS-CoV-2 Variant Classifications and Definitions. [(accessed on 2 September 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Concern.
- 2.Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vavrek D., Speroni L., Curnow K.J., Oberholzer M., Moeder V., Febbo P.G. Genomic surveillance at scale is required to detect newly emerging strains at an early timepoint. medRxiv. 2021 doi: 10.1101/2021.01.12.21249613. [DOI] [Google Scholar]
- 4.Pan American Health Organization . Recommendations for Reporting and Notification of Sars-Cov-2 Variants of Concern and Variants of Interest. Pan American Health Organization; Washington, WA, USA: 2021. [Google Scholar]
- 5.World Health Organization . Guidance for Surveillance of Sars-Cov-2 Variants. World Health Organization; Geneva, Switzerland: 2021. [Google Scholar]
- 6.Banada P., Green R., Banik S., Chopoorian A., Streck D., Jones R., Chakravorty S., Alland D. A simple RT-PCR melting temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. J. Clin. Microbiol. 2021;59:e00845-21. doi: 10.1128/JCM.00845-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedotto M., Fournier P.E., Houhamdi L., Levasseur A., Delerce J., Pinault L., Padane A., Chamieh A., Tissot-Dupont H., Brouqui P., et al. Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant. J. Clin. Virol. 2021;139:104814. doi: 10.1016/j.jcv.2021.104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blairon L., Cupaiolo R., Piteus S., Beukinga I., Tre-Hardy M. The challenge of screening SARS-CoV-2 variants of concern with RT-qPCR: One variant can hide another. J. Virol. Methods. 2021;297:114248. doi: 10.1016/j.jviromet.2021.114248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camp J.V., Buchta C., Jovanovic J., Puchhammer-Stockl E., Benka B., Griesmacher A., Aberle S.W., Goerzer I. RT-PCR based SARS-CoV-2 variant screening assays require careful quality control. J. Clin. Virol. 2021;141:104905. doi: 10.1016/j.jcv.2021.104905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heijnen L., Elsinga G., de Graaf M., Molenkamp R., Koopmans M.P.G., Medema G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799:149456. doi: 10.1016/j.scitotenv.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brusaferro S., Brandtner D., Fasanella A., et al. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021;197:117104. doi: 10.1016/j.watres.2021.117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norz D., Grunwald M., Olearo F., Fischer N., Aepfelbacher M., Pfefferle S., Lutgehetmann M. Evaluation of a fully automated high-throughput SARS-CoV-2 multiplex qPCR assay with built-in screening functionality for del-HV69/70- and N501Y variants such as B.1.1.7. J. Clin. Virol. 2021;141:104894. doi: 10.1016/j.jcv.2021.104894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tierling S., Kattler K., Vogelgesang M., Pfuhl T., Lohse S., Lo Porto C., Schmitt B., Nastasja S., Salhab A., Smola S., et al. Rapid base-specific calling of SARS-CoV-2 variants of concern using combined RT-PCR melting curve screening and SIRPH technology. Open Forum Infect. Dis. 2021;8:ofab364. doi: 10.1093/ofid/ofab364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega-Magana N., Sanchez-Sanchez R., Hernandez-Bello J., Venancio-Landeros A.A., Pena-Rodriguez M., Vega-Zepeda R.A., Galindo-Ornelas B., Diaz-Sanchez M., Garcia-Chagollan M., Macedo-Ojeda G., et al. RT-qPCR assays for rapid detection of the N501Y, 69–70del, K417N, and E484K SARS-CoV-2 mutations: A screening strategy to identify variants with clinical impact. Front. Cell. Infect. Microbiol. 2021;11:672562. doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogels C.B.F., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., Goes de Jesus J., et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19:e3001236. doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., Jean S., Eltringham R., Madison J., Snyder P., Tu H., Jones D.M., Leber A.L. Mutation-specific SARS-CoV-2 PCR screen: Rapid and accurate detection of variants of concern and the identification of a newly emerging variant with spike L452R mutation. J. Clin. Microbiol. 2021;59:e0092621. doi: 10.1128/JCM.00926-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Miller J.A., Verghese M., Sibai M., Solis D., Mfuh K.O., Jiang B., Iwai N., Mar M., Huang C., et al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J. Clin. Microbiol. 2021;59:e0085921. doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaniv K., Ozer E., Shagan M., Lakkakula S., Plotkin N., Bhandarkar N.S., Kushmaro A. Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B.1.1.7 and Beta, B.1.351) detection and quantification in wastewater. Environ. Res. 2021;201:111653. doi: 10.1016/j.envres.2021.111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelyas N., Pabbaraju K., Croxen M.A., Lynch T., Buss E., Murphy S.A., Shokoples S., Wong A., Kanji J.N., Tipples G. Precision response to the rise of the SARS-CoV-2 B.1.1.7 variant of concern by combining novel PCR assays and genome sequencing for rapid variant detection and surveillance. Microbiol. Spectr. 2021;9:e0031521. doi: 10.1128/Spectrum.00315-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babiker A., Immergluck K., Stampfer S.D., Rao A., Bassit L., Su M., Nguyen V., Stittleburg V., Ingersoll J.M., Bradley H.L., et al. Single-amplicon, multiplex real-time RT-PCR with tiled probes to detect SARS-CoV-2 spike mutations associated with variants of concern. J. Clin. Microbiol. 2021;59:e014462. doi: 10.1128/JCM.01446-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ministerio de Salud Pública y Bienestar Social Primer Caso del Nuevo Coronavirus en El Paraguay. [(accessed on 1 September 2021)]; Available online: https://www.mspbs.gov.py/portal/20535/primer-caso-del-nuevo-coronavirus-en-el-paraguay.html.
- 22.Ministerio de Salud Pública y Bienestar Social. [(accessed on 25 August 2021)]; Available online: https://www.mspbs.gov.py/index.php.
- 23.Sobarzo P., Rolón López J.C., Narváez Serra P.F., López Cañete S.A. Clinical characteristics of the first 60 patients with SARS CoV-2 admitted to the National Hospital for the period June–August 2020. Rev. Virtual Soc. Parag. Med. Int. 2021;8:69–77. doi: 10.18004/rvspmi/2312-3893/2021.08.01.69. [DOI] [Google Scholar]
- 24.Vargas-Correa A., Mereles E.F., Segovia C.N., Gimenez A.A., Santacruz L., Ojeda M.L., Kunzle E.H., Samudio M. Clinical-epidemiological characteristics of patients confirmed with COVID-19 from the Department of Alto Paraná, Paraguay. Rev. Salud Pública Parag. 2021;11:54–61. doi: 10.18004/rspp.2021.junio.54. [DOI] [Google Scholar]
- 25.Brinkmann A., Ulm S.L., Uddin S., Forster S., Seifert D., Oehme R., Corty M., Schaade L., Michel J., Nitsche A. AmpliCoV: Rapid whole-genome sequencing using multiplex PCR amplification and real-time Oxford nanopore MinION sequencing enables rapid variant identification of SARS-CoV-2. Front. Microbiol. 2021;12:651151. doi: 10.3389/fmicb.2021.651151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brunink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A., Hussaini L., Kraft C.S., Pinsky B.A., Anderson E.J., et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quick J. nCoV-2019 Sequencing Protocol v3 (LoCost) V.3. [(accessed on 1 February 2021)]. Available online: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye.
- 29.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naveca F.G., Nascimento V., de Souza V.C., Corado A.L., Nascimento F., Silva G., Costa A., Duarte D., Pessoa K., Mejia M., et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021;27:1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 33.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francisco R.D.S., Jr., Benites L.F., Lamarca A.P., de Almeida L.G.P., Hansen A.W., Gularte J.S., Demoliner M., Gerber A.L., de C Guimarães A.P., Antunes A.K.E., et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296:198345. doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A., Andersson M., Otecko N., Wise E.L., Moore N., et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg082. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supporting data necessary for interpretation are provided herein. Complete genome sequences were deposited in GISAID and accession numbers are shown in Table 3. Amplicon sequence tracings are available upon request.