Abstract
During the ongoing investigation of bambusicolous ascomycetous fungi in Yunnan, China, 24 specimens belonging to the family Roussoellaceae were collected and identified based on morphological features and phylogenetic support. Maximum-likelihood (ML) analyses and Bayesian analyses were generated based on the combined data set of ITS, LSU, tef1, and rpb2 loci. The phylogenetic analyses revealed four novel lineages in Roussoella s. str.; thus, we introduced four new species viz., Roussoella multiloculate sp. nov., R. papillate sp. nov., R. sinensis sp. nov., and R. uniloculata sp. nov. Their morphological characters were compared with the known Roussoella taxa, which lack sequence data in the GenBank. Asexual morphs of R. kunmingensis and R. padinae were recorded from dead bamboo culms in China (from the natural substrates) for the first time. Neoroussoella bambusae, Roussoella japanensis, R. nitidula, R. padinae, R. scabrispora, and R. tuberculate were also reported as the first records from China. All new taxa are described and illustrated in detail. Plates are provided for new reports.
Keywords: bambusicolous ascomycetes, new records, new taxa, phylogeny, taxonomy
1. Introduction
The family Roussoellaceae Liu et al. [1] accommodates three genera, viz., Neoroussoella Liu et al., Roussoella Sacc. and Roussoellopsis Hino and Katum. Later, Jaklitsch and Voglmayr [2] synonymized Roussoellaceae under Thyridariaceae based on the multigene analysis of limited taxa. Tibpromma et al. [3] reinstated Roussoellaceae; treating Roussoellaceae and Thyridariaceae as distinct families within Pleosporales. Subsequent studies confirmed this separation using additional taxa and combining morphological and phylogenetic analyses [4,5,6,7,8]. Currently, twelve genera have been accepted (viz., Appendispora Hyde, Cytoplea Bizz. and Sacc., Elongatopedicellata Zhang et al., Immotthia Barr, Neoroussoella, Pararoussoella Wanas. et al., Pseudoneoconiothyrium Wanas et al., Pseudoroussoella Mapook and Hyde, Roussoella Sacc., Roussoellopsis Hino and Katum., Setoarthopyrenia Mapook and Hyde and Xenoroussoella Mapook and Hyde) in Roussoellaceae [9].
Roussoella (the type of genus of Roussoellaceae) was introduced by Saccardo and Paoletti [10] with R. nitidula Sacc. and Paol. as the type species recorded from bamboo in Malacca, Malaysia. Höhnel [11] proposed that Dothidea hysterioides Ces. is the former name for this taxon; thus, being transferred to the Roussoella genus, i.e., Roussoella hysterioides (Ces.) Höhn. However, Aptroot [12] and Müller and Arx [13] assigned Roussoella to Amphisphaeriaceae, which comprises cylindrical and unitunicate asci, immersed ascostromata, two-celled and brown ascospores. Aptroot [12] described Roussoella asci as unitunicate and moved the three species to this genus, while Aptroot [14] modified his concept of Roussoella asci and considered them as bitunicate. Roussoella is characterized by immersed, large, and loculate ascostromata, cylindrical and bitunicate asci, brown, fusiform to ellipsoidal, ornamented, and two-celled ascospores surrounded by a sheath [1,15,16]. Roussoella mainly occurs on gramineous plants, with most species found on bamboo and palms [1,16,17].
Neoroussoella is a monotypic genus, distinct from Roussoella, with uniloculate ascomata, absence of a clypeus, and an asexual morph forming hyaline to brown, oblong to ellipsoidal, and smooth-walled phoma-like conidia [1,18]. Further, Neoroussoella is characterized by a distinct asexual morph producing relatively smaller (3–4 × 1.5–2 μm), hyaline conidia with smooth wall [1]. Currently, this genus comprises eleven species [19,20].
The aims of this study are to introduce four new species, viz. R. multiloculate, R. papillate, R. sinensis, and R. uniloculata. In addition, the asexual morphs of R. kunmingensis and R. padinae isolated from dead bamboo culms (from a natural substrate) are also described for the first time. Finally, the species Neoroussoella bambusae, Roussoella japanensis, R. nitidula, R. padinae, R. scabrispora, and R. tuberculate are reported for the first time in China.
2. Materials and Methods
2.1. Fungal Sampling and Morphology
Bamboo culms were collected in Yunnan, China, and conserved in protection bags for two days until they arrived at the laboratory. Samples were examined, and single spore isolation was performed as previously described [17]. Morphological characters were examined using water slides and photographed (Olympus BX53 DIC compound microscope with an Olympus DP74 camera). Fruiting bodies, such as ascostromata and conidiomata, were also photographed (Leica S8AP0 stereomicroscope with an HDMI 200C camera). Measurements were registered (Tarosoft (R) Image Frame Work 80 software). Specimens and living cultures were deposited at the Herbarium of Guizhou Medical University (GMB) and Guizhou Medical University Culture Collection (GMBCC) in Guiyang, China. Duplicates of holotypes and ex-type cultures were also deposited at the herbarium of Research Institute of Resource Insects, Chinese Academy of Forestry (IFRD), and Research Institute of Resource Insects, Chinese Academy of Forestry Culture Collection (IFRDCC) in Kunming, China. Index Fungorum [20] numbers were provided for newly introduced taxa.
2.2. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and Phylogeny
Pure cultures were grown on PDA media, for 30–40 days, at 28 °C, in the dark. Fresh mycelium was scraped using a surgical knife and placed into a 1.5 mL centrifuge tube, and ground into powder using liquid nitrogen. Genomic DNA was extracted following the instruction book of the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®).
Information of primers used for the amplification of internal transcribed spacers (ITS), small subunit rDNA (SSU), large subunit rDNA (LSU), translation elongation factor 1-α gene region (tef1), and RNA polymerase II second largest subunit (rpb2) genes is presented in Table 1. ITS, SSU, LSU, tef1, and rpb2 loci amplifications were performed by polymerase chain reaction (Eppendorf Mastercycler nexus gradient) according to the conditions presented in Table 2 [17]. Both forward and reverse primers (Table 1) were used for sequencing and these primers were the same as those used for amplification. The PCR products were sequenced, and the sequences were deposited in GenBank, as shown in Table 3.
Table 1.
ITS, SSU, LSU, tef1, and rpb2 loci primers information.
| Genes | Primers and Base Pairs | References |
|---|---|---|
| Internal transcribed spacers (ITS) | Forward: ITS5 TCCTCCGCTTATTGATATGC Reverse: ITS4 GGAAGTAAAAGTCGTAACAAGG |
[21] |
| Large subunit rDNA (LSU) | Forward: LROR GTACCCGCTGAACTTAAGC Reverse: LR5 ATCCTGAGGGAAACTTC |
[22] |
| Small subunit rDNA (SSU) | Forward: NS1 GTAGTCATATGCTTGTCTC Reverse: NS4 CTTCCGTCAATTCCTTTAAG |
[21] |
| Translation elongation factor 1-α gene region (tef1) | Forward: EF1-983F GCYCCYGGHCAYCGTGAYTTYAT Reverse: EF1-2218R ATGACACCRACRGCRACRGTYTG |
[23] |
| RNA polymerase II second largest subunit (rpb2) | Forward: fRPB2-5f GAYGAYMGWGATCAYTTYGG Reverse: fRPB2-7cr CCCATRGCTTGTYYRCCCAT |
[24] |
Table 2.
ITS, SSU, LSU, tef1, and rpb2 loci PCR conditions.
| Genes | Initial Period | Cycles, Denaturation, Annealing and Elongation | Final Extension | References |
|---|---|---|---|---|
| ITS, LSU, SSU, tef1 | 94 °C for 3 min | 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min | 72 °C for 10 min | [17] |
| rpb2 | 95 °C for 5 min | 40 cycles of denaturation at 95 °C for 1 min, annealing at 52 °C for 2 min, elongation at 72 °C for 90 s | 72 °C for 10 min | [17] |
Table 3.
Isolates or specimens used in this study and their GenBank accession numbers. The newly generated sequences are marked with asterisk “★” and ex-type strains are in bold, “-” means sequences data are unavailable in the GenBank database.
| Taxa | Strain/Voucher No. | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | tef1 | rpb2 | ||
| Arthopyrenia sp. | UTHSC DI16-362 | LT796905 | LN907505 | LT797145 | LT797065 |
| Arthopyrenia sp. | UTHSC DI16-334 | LT796887 | LN907477 | LT797127 | - |
| Neoroussoella alishanense | FU31016 | MK503816 | MK503822 | MK336181 | MN037756 |
| Neoroussoella bambusae | MFLUCC 11-0124 | KJ474827 | KJ474839 | KJ474848 | KJ474856 |
| Neoroussoella bambusae ★ | GMBCC1116 | OM891810 | OM884022 | ON098358 | ON098377 |
| Neoroussoella bambusae ★ | GMBCC1118 | OM891812 | OM801294 | ON098359 | ON098376 |
| Neoroussoella entadae | MFLUCC 15-0098 | MH275075 | MH260309 | - | - |
| Neoroussoella heveae | MFLUCC 17-0338 | MH590693 | MH590689 | - | - |
| Neoroussoella heveae | MFLUCC 17-2069 | MT310634 | MT214589 | MT394647 | MT394703 |
| Neoroussoella lenispora | GZCC 16-0020 | - | KX791431 | - | - |
| Neoroussoella leucaenae | MFLUCC 18-1544 | MK347767 | MK347984 | MK360067 | MK434876 |
| Neoroussoella solani | CPC 26331 | KX228261 | KX228312 | - | - |
| Pararoussoella mangrovei | MFLUCC 16-0424 | MH025951 | MH023318 | MH028246 | MH028250 |
| Pararoussoella mukdahanensis | KUMCC 18-0121 | MH453489 | MH453485 | MH453478 | MH453482 |
| Pararoussoella mukdahanensis | MFLUCC 11-0201 | KU940129 | KU863118 | - | - |
| Pararoussoella rosarum | MFLUCC 17-0796 | NR_157529 | NG059872 | MG829224 | - |
| Parathyridaria percutanea | CBS 128203 | KF322117 | KF366448 | KF407988 | KF366453 |
| Parathyridaria percutanea | CBS 868.95 | KF322118 | KF366449 | KF407987 | KF366452 |
| Parathyridaria ramulicola | CBS 141479 | KX650565 | KX650565 | KX650536 | KX650584 |
| Parathyridaria ramulicola | MF4 | KX650564 | KX650564 | KX650535 | - |
| Parathyridaria robiniae | MFLUCC 14-1119 | KY511142 | KY511141 | KY549682 | - |
| Pseudoneoconiothyrium euonymi | CBS 143426 | MH107915 | MH107961 | - | MH108007 |
| Pseudoneoconiothyrium rosae | MFLUCC 15-0052 | NR_157523 | NG059868 | - | - |
| Pseudoroussoella chromolaenae | MFLUCC 17-1492 | MT214345 | MT214439 | MT235769 | - |
| Pseudoroussoella elaeicola | MFLUCC 17-1483 | MT214348 | MT214442 | MT235772 | MT235808 |
| Pseudoroussoella elaeicola | MFLUCC 15-0276b | MH742330 | MH742327 | - | - |
| Pseudoroussoella elaeicola | MFLUCC 15-0276a | MH742329 | MH742326 | - | - |
| Roussoella angusta | MFLUCC 15-0186 | - | KT281979 | - | - |
| Roussoella aquatica | MFLUCC 18-1040 | NR171975 | NG073797 | - | - |
| Roussoella arundinacea | CBS 146088 | MT223838 | MT223928 | MT223723 | - |
| Roussoella chiangraina | MFLUCC 10-0556 | KJ474828 | KJ474840 | KJ474849 | KJ474857 |
| Roussoella doimaesalongensis | MFLUCC 14-0584 | KY026584 | KY000659 | KY651249 | KY678394 |
| Roussoella elaeicola | MFLUCC 15-0276a | MH742329 | MH742326 | - | - |
| Roussoella elaeicola | MFLUCC 15-0276b | MH742330 | MH742327 | - | - |
| Roussoella guttulata | MFLUCC 20-0102 | NR_172428 | NG_075383 | MW022188 | MW022187 |
| Roussoella hysterioides | CBS 546.94 | KF443405 | KF443381 | KF443399 | KF443392 |
| Roussoella intermedia | NBRC 106245 | KJ474831 | AB524624 | - | - |
| Roussoella intermedia | CBS 170.96 | KF443407 | KF443382 | KF443398 | KF443394 |
| Roussoella japanensis | MAFF 239636 | KJ474829 | AB524621 | AB539114 | AB539101 |
| Roussoella japanensis ★ | GMBCC1067 | OM891802 | OM884018 | ON098344 | ON098381 |
| Roussoella japanensis ★ | GMBCC1117 | OM891811 | OM884023 | ON098345 | ON098382 |
| Roussoella kunmingensis | KUMCC 18-0128 | MH453491 | MH453487 | MH453480 | MH453484 |
| Roussoella kunmingensis ★ | GMBCC1055 | OM891797 | OM884013 | ON098353 | ON098385 |
| Roussoella kunmingensis ★ | GMBCC1057 | OM891798 | OM884014 | ON098354 | ON098362 |
| Roussoella kunmingensis ★ | GMBCC1086 | OM891804 | OM801287 | ON098355 | ON098363 |
| Roussoella magnatum | MFLUCC 15-0185 | - | KT281980 | - | - |
| Roussoella margidorensis | MUT 5329 | KU314944 | MN556322 | MN605897 | MN605917 |
| Roussoella mediterranea | MUT 5369 | KU314947 | MN556324 | MN605899 | MN605919 |
| Roussoella mexicana | CPC 25355 | KT950848 | KT950862 | - | - |
| Roussoella multiloculate ★ | GMB1219 | OM891801 | OM884017 | ON098341 | ON098366 |
| Roussoella multiloculate ★ | GMBCC1056 | OM891799 | OM884015 | ON098343 | ON098369 |
| Roussoella multiloculate ★ | GMBCC1065 | OM891800 | OM884016 | ON098338 | ON098364 |
| Roussoella multiloculate ★ | GMBCC1069 | OM891803 | OM884019 | ON098340 | ON098365 |
| Roussoella multiloculate ★ | GMBCC1071 | ON159383 | OM755586 | ON098342 | ON098368 |
| Roussoella multiloculate ★ | GMBCC1080 | ON159384 | OM755589 | ON098339 | ON098367 |
| Roussoella neopustulans | MFLUCC 11-0609 | KJ474833 | KJ474841 | KJ474850 | - |
| Roussoella neopustulans | MFLUCC 12-0003 | KU940130 | KU863119 | - | - |
| Roussoella nitidula | MFLUCC 11-0182 | KJ474835 | KJ474843 | KJ474852 | KJ474859 |
| Roussoella nitidula | MFLUCC 11-0634 | KJ474834 | KJ474842 | KJ474851 | KJ474858 |
| Roussoella nitidula ★ | GMBCC1097 | OM891805 | OM884020 | ON098351 | ON098384 |
| Roussoella padinae | MUT 5503 | KU158170 | MN556327 | MN605902 | MN605922 |
| Roussoella padinae ★ | GMBCC1126 | OM891816 | OM884025 | ON098356 | ON098383 |
| Roussoella papillate ★ | GMBCC1121 | OM891814 | OM755608 | ON098346 | ON098378 |
| Roussoella papillate ★ | IFRDCC 3103 | ON228188 | ON228184 | ON244452 | ON244450 |
| Roussoella pseudohysterioides | MFLUCC 13-0852 | KU940131 | KU863120 | KU940198 | - |
| Roussoella pseudohysterioides | KUMCC 18-0111 | MH453490 | MH453486 | MH453479 | MH453483 |
| Roussoella pustulans | MAFF 239637 | KJ474830 | AB524623 | AB539116 | AB539103 |
| Roussoella scabrispora | MFLUCC 11-0624 | KJ474836 | KJ474844 | KJ474853 | KJ474860 |
| Roussoella scabrispora | MFLUCC 14-0582 | KY026583 | KY000660 | - | - |
| Roussoella scabrispora ★ | GMBCC1101 | ON159385 | OM755615 | ON098347 | ON098371 |
| Roussoella scabrispora ★ | GMBCC1102 | OM891806 | OM884021 | ON098348 | ON098370 |
| Roussoella scabrispora ★ | GMBCC1104 | OM891807 | OM755616 | ON098349 | ON098373 |
| Roussoella scabrispora ★ | GMBCC1108 | OM891808 | OM755614 | ON098350 | ON098372 |
| Roussoella siamensis | MFLUCC 11-0149 | KJ474837 | KJ474845 | KJ474854 | KJ474861 |
| Roussoella sinensis ★ | GMBCC1119 | OM891813 | OM884024 | ON098357 | ON098379 |
| Roussoella sinensis ★ | IFRDCC 3101 | ON228187 | ON228183 | ON244453 | ON244451 |
| Roussoella thailandica | MFLUCC 11-0621 | KJ474838 | KJ474846 | - | - |
| Roussoella tuberculata | MFLUCC 13-0854 | KU940132 | KU863121 | KU940199 | - |
| Roussoella tuberculata ★ | GMBCC1123 | OM891815 | OM755613 | ON098352 | ON098380 |
| Roussoella uniloculata ★ | GMBCC1110 | OM891809 | OM801286 | ON098360 | ON098374 |
| Roussoella uniloculata ★ | DDQ01005-2 | OM891817 | OM884026 | ON098361 | ON098375 |
| Roussoella verrucispora | CBS 125434 | KJ474832 | AB524622 | AB539115 | AB539102 |
| Roussoella yunnanensis | KUMCC 18-0115 | MH453492 | MH453488 | MH453481 | - |
| Roussoellopsis macrospora | MFLUCC 12-0005 | - | KJ474847 | KJ474855 | KJ474862 |
| Roussoellopsis sp. | NBRC 106246 | - | AB524626 | - | - |
| Roussoellopsis tosaensis | KT 1659 | - | AB524625 | MG829199 | AB539104 |
| Setoarthopyrenia chromolaenae | MFLUCC 17-1444 | MT214344 | MT214438 | MT235768 | MT235805 |
| Thyridaria acaciae | CBS 138873 | KP004469 | KP004497 | - | - |
| Thyridaria broussonetiae | CBS 141481 | NR_147658 | KX650568 | KX650539 | KX650586 |
| Torula herbarum | CBS 111855 | KF443409 | KF443386 | KF443403 | KF443396 |
| Torula hollandica | CBS 220.69 | KF443406 | KF443384 | KF443401 | KF443393 |
| Xenoroussoella triseptata | MFLUCC 17-1438 | MT214343 | MT214437 | MT235767 | MT235804 |
Abbreviations: CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CPC: Culture collection of Pedro Crous, Netherlands; IFRDCC: Research Institute of Resource Insects, Chinese Academy of Forestry Culture Collection, Kunming, China; GMB: Herbarium of Guizhou Medical University, Guiyang, China; GMBCC: Guizhou Medical University Culture Collection, Guiyang, China; KUMCC: Kunming Institute of Botany Culture Collection, Kunming, China; MAFF: Ministry of Agriculture, Forestry and Fisheries, Japan; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUT: Mycotheca Universitatis Taurinensis, Department of Life Sciences and Systems Biology, University of Turin, Turin, Italy; NBRC: Biological Resource Center, National Institute of Technology and Evaluation, Chiba, Japan; UTHSC: The University of Tennessee Health Science Center, Memphis, USA; DDQ: Dong-Qin Dai; KT: K. Tanaka.
2.3. Phylogenetic Analyses
The quality of the sequences was verified (BioEdit v.7.0 [25]), and alignments from single genes were generated (MAFFT v.7.215 [26]) (http://mafft.cbrc.jp/alignment/server/index.html, accessed on 20 April 2021), being manually edited when needed (MEGA6 version 6.0 [27,28]). The combined alignment of multi-genes was carried out (MEGA6 [27]). Maximum-likelihood (ML) analyses were performed (software RAxMLGUI v.1.0 [29,30]) with a 1000 bootstrap. Multi-gene alignments were uploaded to the website (http://sing.ei.uvigo.es/ALTER/, accessed on 20 April 2022) to obtain the PHYLIP format file. The best nucleotide substitution model was determined using the online tool Findmodel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html, accessed on 20 April 2022) and was executed in RAxMLGUI to generate the best ML.
Bayesian analyses were performed using MrBayes v.3.0b4 [31]. MrModeltest v.2.2 selected the best evolution model [32]. Posterior probabilities (PP) [33,34] were performed by Markov Chain Monte Carlo sampling (MCMC) [35]. Six simultaneous Markov chains were run for 1,000,000 generations, and trees were sampled every 100th generation. The burn-in was set to 0.25, and the run was automatically stopped when the average standard deviation of split frequencies reached below 0.01 [36].
Trees were constructed (TreeView [37]) and formatted (Adobe Illustrator CS v.5). Maximum-likelihood bootstrap values (MLBP) equal to or greater than 50% and Bayesian posterior probabilities (BYPP) > 0.80 are given at the branches. The sequences used in this study are listed in Table 1. The alignment based on the combined loci and phylogenetic tree was submitted to TreeBASE under the code 29601 (https://www.treebase.org/, accessed on 20 April 2022).
3. Results
3.1. Phylogenetic Analyses
The sequence data set of combined ITS, LSU, tef1, and rpb2 loci was used to determine the phylogenetic position of the newly generated described strains. SSU sequences were not included in the alignment, as most Roussoella taxa lack SSU in the GenBank. The dataset comprised 90 strains, including two outgroup strains (Torula herbarum CBS 220.69 and CBS 111855, Table 3). The final alignment comprises 3381 characters used for the phylogenetic analyses, including gaps. The RAxML analysis of the combined dataset generated a best-scoring tree with a final ML optimization likelihood value of −30,685.326261 (Table 4). GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis.
Table 4.
Different parameters for ML analyses.
| Analyses | Parameters | Value |
|---|---|---|
| ML | Final ML Optimization Likelihood | −30,685.326261 |
| No of characters | 3372 | |
| Alignment patterns | 1513 | |
| Proportion of Undetermined characters or gaps | 29.97% | |
| Substitution model | GTR | |
| Tree length | 3.629102 | |
| Estimated base frequencies | A = 0.240311 | |
| C = 0.267630 | ||
| G = 0.270582 | ||
| T = 0.221477 | ||
| Substitution rates | AC = 1.886299 | |
| AG = 5.163488 | ||
| AT = 1.984078 | ||
| CG = 1.308792 | ||
| CT = 9.477974 | ||
| GT = 1.000000 | ||
| Gamma distribution shape parameter | α = 0.180205 |
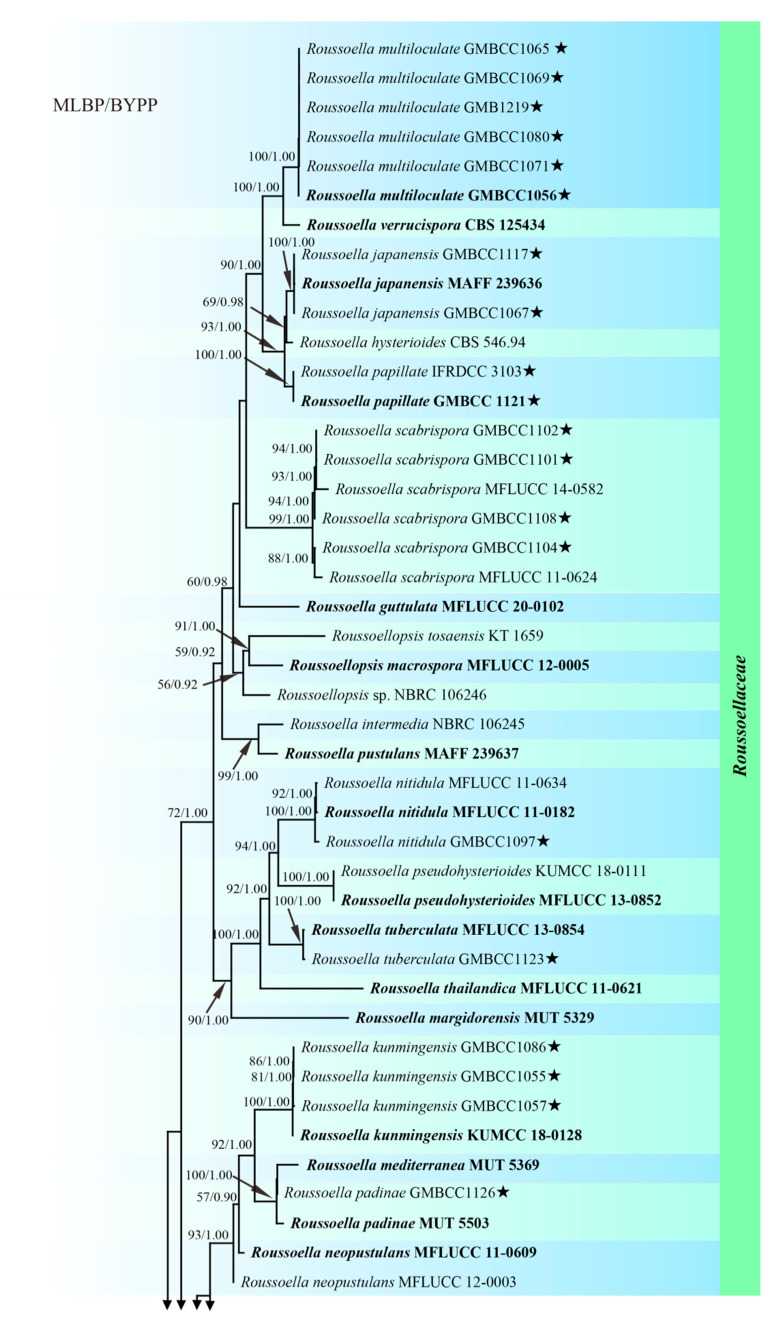
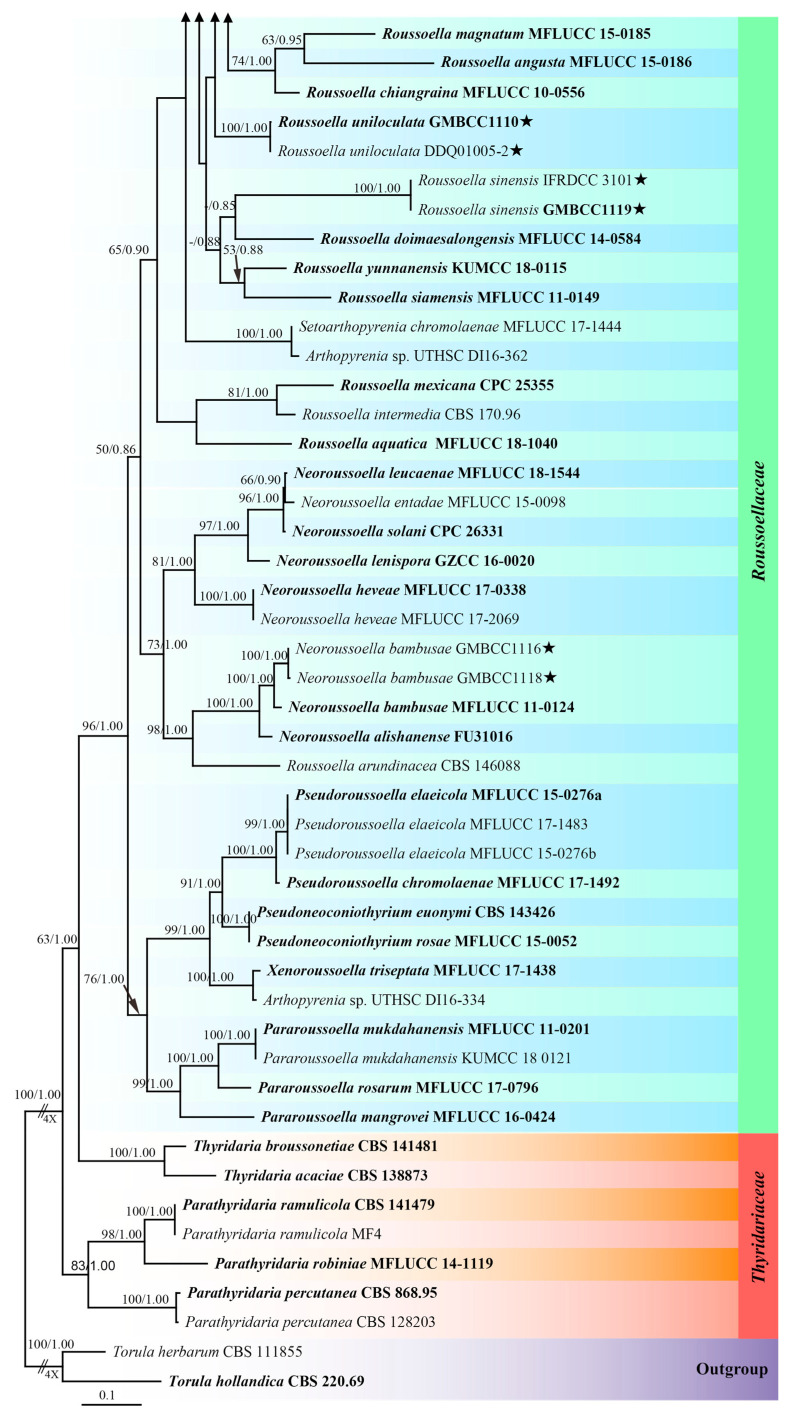
Based on the multi-gene phylogenetic analyses (Figure 1), 26 new isolates were grouped in the family Roussoellaceae (96% MLBP, 1.00 BYPP). Five isolates, GMBCC1056, GMBCC1065, GMBCC1069, GMBCC1071, and GMBCC1080, and one specimen, GMB1219, represented a novel species Roussoella multiloculate sp. nov. and formed a sister clade to R. verrucispora with high statistical support (100% MLBP, 1.00 BYPP). Roussoella papillate sp. nov. (GMBCC 1121 and IFRDCC 3103) grouped sister with R. japanensis and R. hysterioides with high bootstrap support (100% MLBP, 1.00 BYPP). The third new species, R. sinensis sp. nov., clustered together with R. doimaesalongensis, R. siamensis, and R. yunnanensis. The last new taxon, R. uniloculata, forms a distinct clade at the base of lineage, which contains R. angusta, R. chiangraina, R. kunmingensis, R. magnatum, R. mediterranea, R. neopustulans, and R. padinae. The R. sinensis and R. uniloculata clades are phylogenetically distant from the known species (Figure 1).
Figure 1.
Phylogenetic tree from the best scoring of the RAxML analysis based on combined ITS, LSU, rpb2 and tef1 loci is rooted to Torula herbarum (CBS 111855) and T. hollandica (CBS 220.69). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.80, respectively, are given at the branches. The newly generated sequences are marked with asterisk “★” and ex-type strains are indicated in bold. Bar = 0.1 expected number of nucleotide substitutions per site per branch.
3.2. Taxonomy
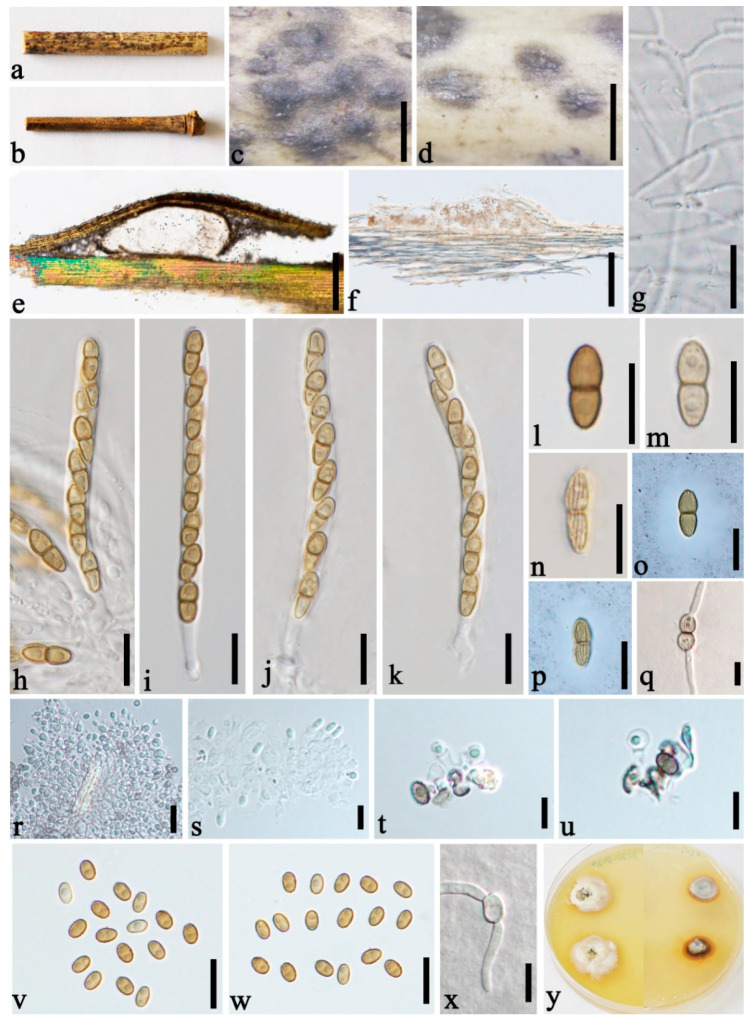
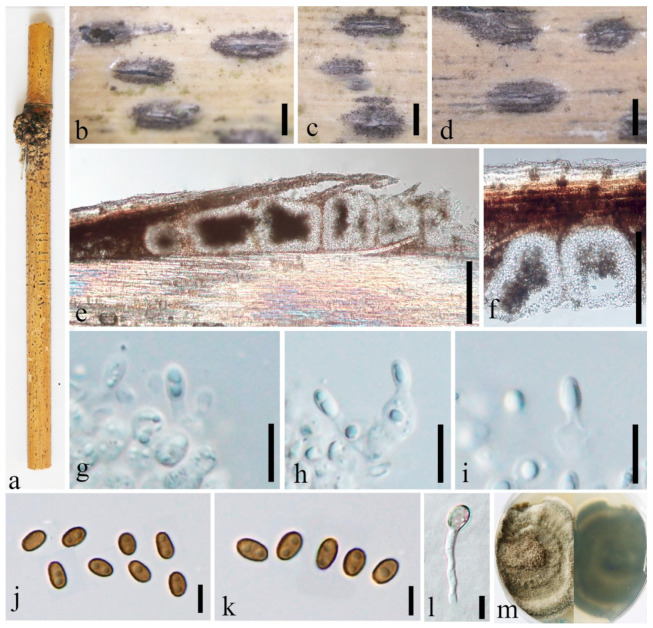
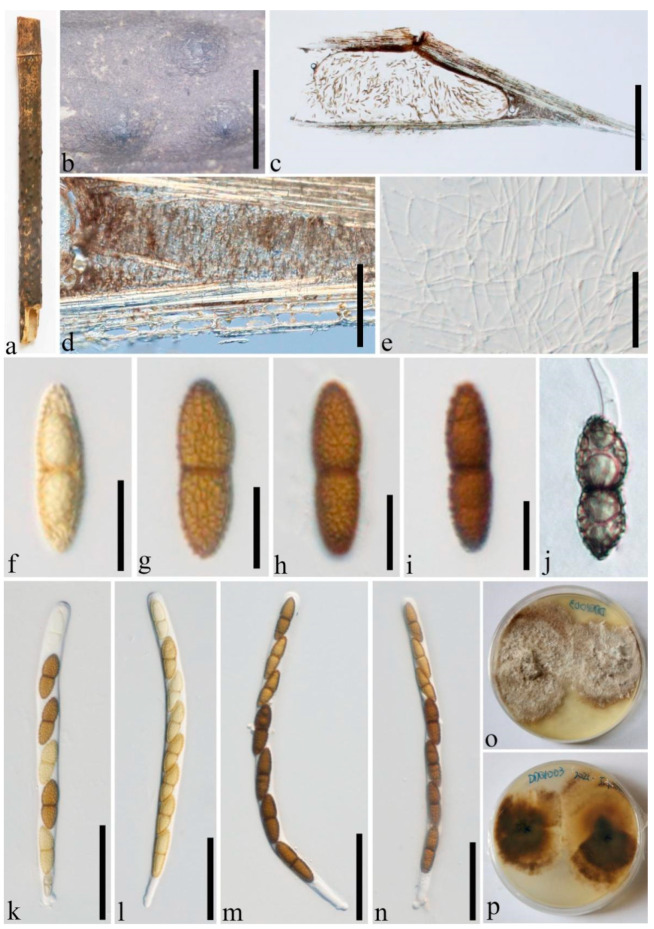
Neoroussoella bambusae Phook., Liu and Hyde, Phytotaxa 181(1):23 (2014) Figure 2.
Figure 2.
Neoroussoella bambusae (GMB1291, new country record). (a) Bamboo specimen; (b) Black ascostromata on host surface; (c) Vertical section of ascostroma; (d) Cells of locules walls; (e) Pseudoparaphyses; (f–i) Asci; (j–m) Ascospores; (n) Ascospore in India ink; (o) Germinating ascospore; (p,q) Cultures on PDA from above and below. Scale bars: (b) = 150 μm, (c) = 100 μm, (d) = 50 μm, (e,n) = 10 μm, (f–i) = 20 μm, (j–m) = 5 μm, (o) = 15 μm.
Index Fungorum number: IF 550669.
For descriptions of sexual and asexual morphs see Liu et al. [1].
Distributions: Thailand, China.
Material examined: China, Yunnan Province, Mengla, Jinghong, Xishuangbanna Topic Botany Garden, Bamboo Garden, (101°24′41.74″ N, 21°93′40.94″ E, 507.88 m), on dead culms of bamboo, 15 August 2020, Dong-Qin Dai, DDQ01025 (GMB1291); living culture, GMBCC1116, GenBank accession number SSU: OM891830; Ibid. DDQ01044 (GMB1295); living culture, GMBCC1118 (new country record), GenBank accession number SSU: OM891832.
Notes: In morphology, GMB1291 and GMB1295 are similar to each other and resemble N. bambusae. Furthermore, LSU, ITS, and rpb2 gene regions of GMBCC1116 and GMBCC1118 are identical to each other. Therefore, new collections GMBCC1116 and GMBCC1118 are identified as N. bambusae. Our new strains (GMBCC1116 and GMBCC1118) grouped in a clade comprising N. alishanense (FU31016, ex-type), N. bambusae (MFLUCC 11-0124, ex-type) and R. arundinacea (CBS 146088) with high statistical support (100% MLBP, 1.00 BYPP) (Figure 1). Neoroussoella bambusae differs from N. alishanense in having ascostromata that are visible as black dome-shaped or shield-shaped or flattened ovoid areas on the host surface while N. alishanense is saprobic on Pennisetum purpureum and having hemispherical to subconical ascomata and ascospores which lack mucilaginous sheath around the ascospores [18]. This is the first report of this species from China.
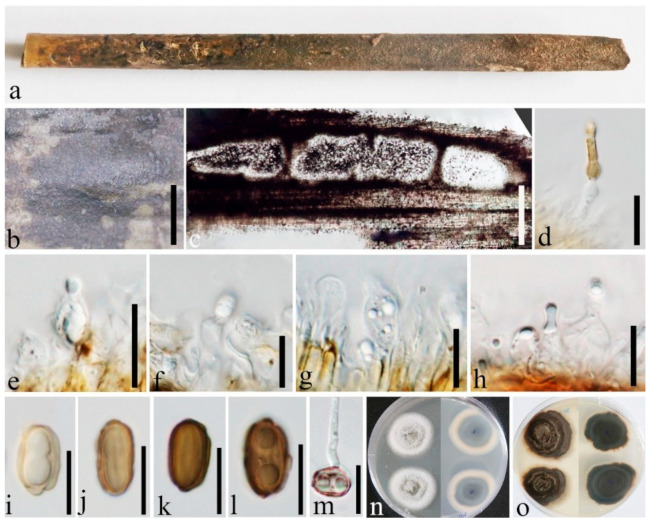
Roussoella japanensis Kaz. Tanaka, Liu and Hyde, Phytotaxa 181(1):14 (2014) Figure 3.
Figure 3.
Roussoella japanensis (GMB1292, new country record). (a) Bamboo specimen; (b) black ascostromata on host surface; (c) vertical section of ascoma; (d) cells between locules; (e) pseudoparaphyses; (f–j) ascospores; (k) ascospore in India ink; (l) germinating ascospore; (m) cultures on PDA from below and above; (n–s) asci. Scale bars: (b) = 2 mm, (c) = 200 μm, (d) = 50 μm, (e–j) = 10 μm, (k,l) = 20 μm, (n–s) = 30 μm.
Index Fungorum number: IF 550663.
For descriptions of sexual and asexual morphs see Liu et al. [1].
Distributions: Japan, China.
Material examined: China, Yunnan Province, Kunming Expo Park, on dead culms of bamboo, 24 July 2020, Dong-Qin Dai, DDQ00780 (GMB1220), living culture, GMBCC1067 (new country record), GenBank accession number SSU: OM891823; Luoping, Jiulong waterfall, on dead culms of bamboo, 29 August 2020, Dong-Qin Dai, DDQ01029 (GMB1292), living culture, GMBCC1117, GenBank accession number SSU: OM891831.
Notes: Our phylogenetic analyses showed that new strains GMBCC1067 and GMBCC1117 clustered together with R. japanensis (MAFF 239636, ex-type) with strong statistical supports (100% MLBS, 1.00 BYPP) (Figure 1). Furthermore, there are no base pair differences in ITS, tef1 and rpb2 loci of GMBCC1067, GMBCC1117, and MAFF 239636 which indicates that they are conspecific. Morphologically, they are identical to each other [1]. Hence, based on both morphology and phylogeny, we identified GMBCC1067 and GMBCC1117 as R. japanensis. Roussoella japanensis was introduced by Liu et al. [1] from twigs of Sasa veitchii var. veitchii in Japan. This is the first record of R. japanensis from China.
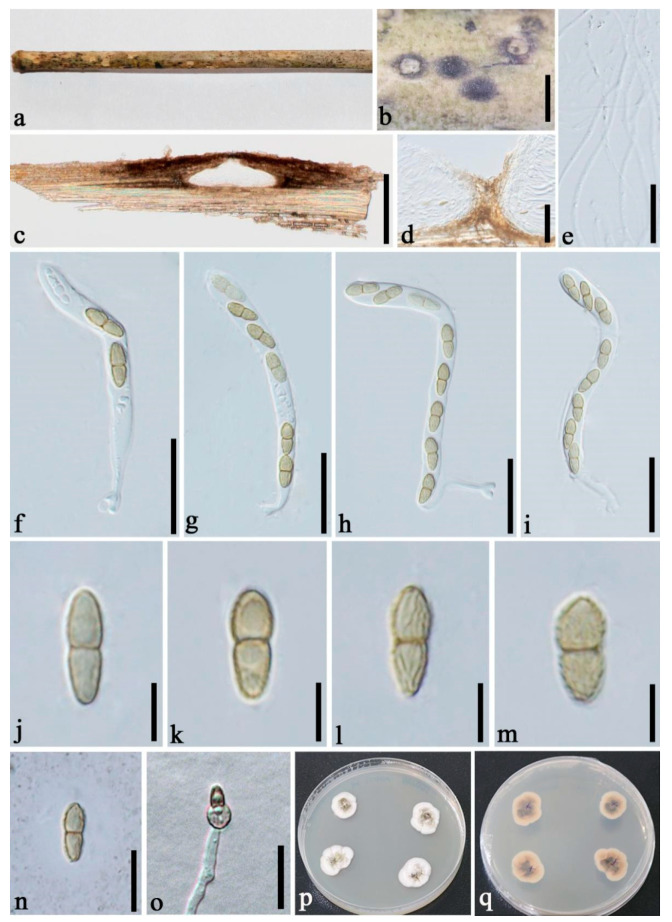
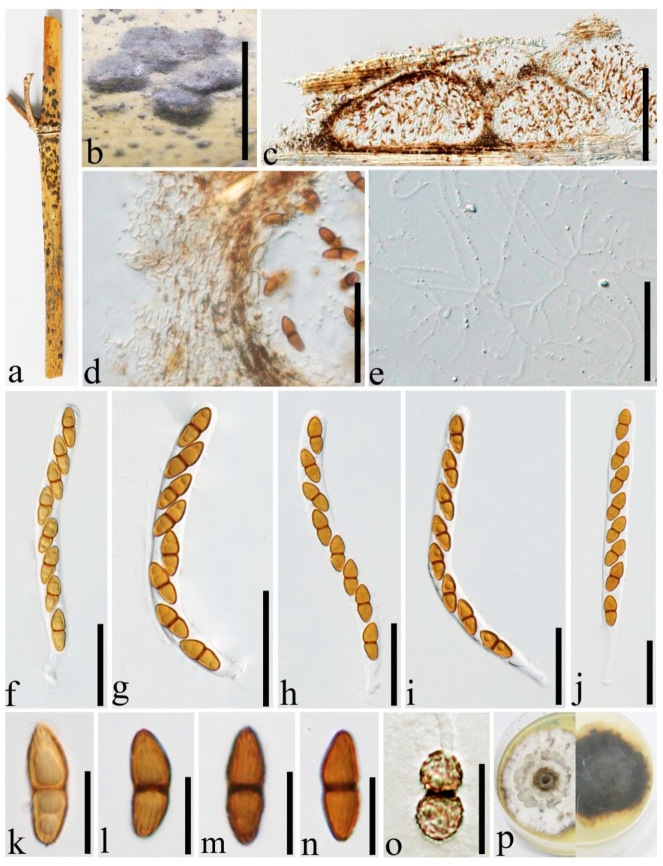
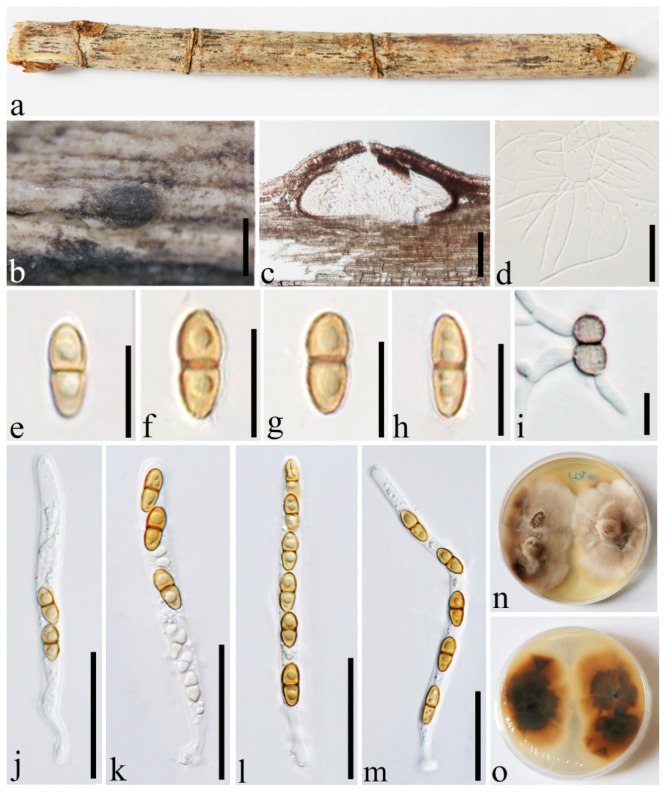
Roussoella kunmingensis H.B. Jiang, Phookamsak and Hyde, Mycol. Prog. 18(4):581 (2019) Figure 4.
Figure 4.
Roussoella kunmingensis (GMB1203, sexual morph, (a,c,e,g,h–q); GMB1259, first report of asexual morph, (b,d,f,r–y)). (a,b) Bamboo specimens; (c) black ascostromata on host surface; (d) black conidiomata on host surface; (e) vertical section of ascostromata; (f) vertical section of conidioma; (g) pseudoparaphyses; (h–k) asci; (l–n) ascospores; (o,p) ascospores in India ink; (q) germinating ascospore; (r–u) conidiogenous cells contacting with conidia; (v,w) conidia; (x) germinating conidium; (y) cultures on PDA from above and below. Scale bars: (c) = 500 μm, (d) = 300 μm, (e) = 200 μm, (f) = 100 μm, (g–x) = 10 μm.
Index Fungorum number: IF 827562.
Saprobic on decaying bamboo culms. Sexual morph: see Jiang et al. (2019). Asexual morph: Stromata forming under a light brown area, up to 250–300 µm long and 75–105 µm wide, and becoming raised at maturity, globose, ellipsoidal to irregular. Conidiomata 55–75 µm high, 200–300 µm diam., locules solitary immersed in the stromata, globose to subglobose, dark brown. Conidiomatal wall comprising two layers of cells of textura angularis, with dark to brown outer thin layer 6–8 µm thick, with a 12–21 µm thick, hyaline, conidiogenous inner layer. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3.5–6.5 × 3–5 µm ( = 4.7 × 3.9 µm, n = 5) enteroblastic, phialidic, indeterminate, discrete, ampulliform, hyaline, smooth-walled. Conidia 4–5 × 2.5–3.5 µm ( = 4.3 × 2.9 µm, n = 20), ellipsoidal, oblong, aseptate, straight, rounded at both ends, hyaline when immature, and becoming brown to dark brown when mature, smooth-walled, inside usually containing 1–2 small guttules.
Culture characters: Ascospores and conidia germinating on PDA within 24 h and germ tubes produced from both cells and both sides. Colonies slow-growing, 15 mm diam. after 20 days at 28 °C, under 24 h dark, circular, floccose at the centre, with even margin, white at margin, and light yellow at the centre.
Distributions: China.
Material examined: China, Yunnan Province, Kunming, Kunming Expo Park (25°07′77″ N, 102°76′23″ E, 1960 m), on dead culms of bamboo, 24 July 2020, Dong-Qin Dai, DDQ00742, GMB1203, living culture, GMBCC1055; Ibid., DDQ00745 (GMB1206), living culture GMBCC1057, GenBank accession number SSU: OM891820; Diqin, Shangri-La, Bigu Mountain, on dead culms of bamboo, 22 July 2020, 27°36′56.9″ N, 99°42′6.4″ E, 3460 m, Dong-Qin Dai, DDQ00905, GMB1259; living culture, GMBCC1086 (first report of the asexual morph), GenBank accession number SSU: OM891825.
Notes: Roussoella kunmingensis is characterized by having immersed, uniloculate ascomata, cylindrical to cylindric-clavate, bitunicate asci, and ellipsoidal to fusiform, light brown to brown, 2-celled ascospores with longitudinal ribs. Multi-gene phylogenetic analyses showed that GMBCC1055, GMBCC1057, and GMBCC1086 grouped with R. kunmingensis (KUMCC 18-0128, ex-type) with high statistic values (100% MLBP, 1.00 BYPP). Base pair arrangement of ITS, tef1, and rpb2 regions of strains GMBCC1055, GMBCC1057, and GMBCC1086 and KUMCC 18-0128 were identical. However, Jiang et al. [5] described this species with only sexual morph from bamboo in China. Here, one new collection with sexual morph and two new collections with asexual morph were examined. Hence, we reported the asexual morph of R. kunmingensis for the first time and provide a description of the asexual morph and an illustration of the holomorph.
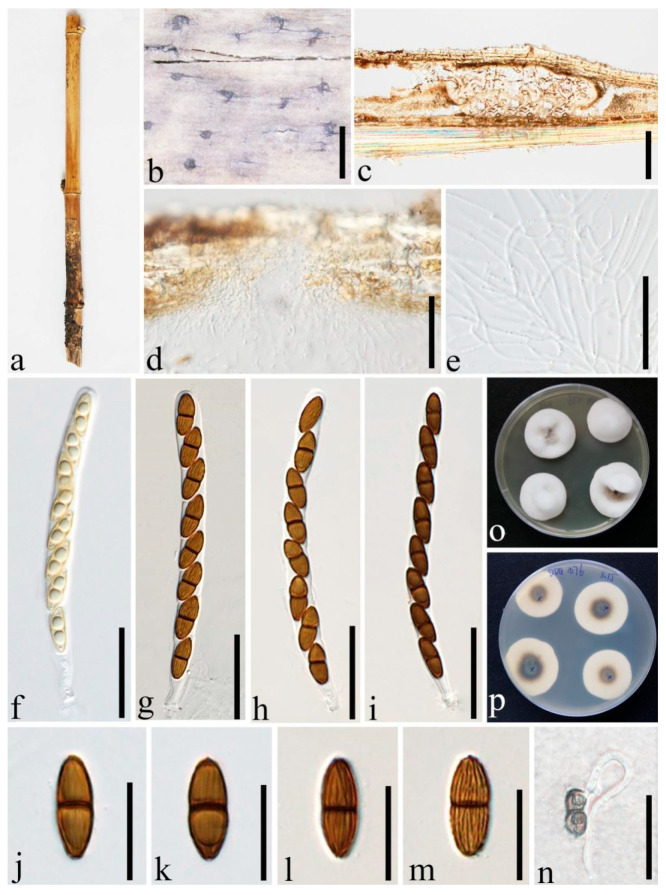
Roussoella multiloculate. Dai and Wijayaw., sp. nov. Figure 5.
Figure 5.
Roussoella multiloculate (GMB1207, holotype). (a) Bamboo specimen; (b–d) black conidiomata on host surface; (e,f) vertical sections of conidiomata; (g–i) conidia attached to conidiogenous cells; (j,k) conidia. (l) germinating conidium. (m) cultures on PDA from above and below. Scale bars: (b–d) = 500 µm, (e,f) = 100 µm, (g–l) = 5 µm.
Index Fungorum number: IF 556005.
Etymology: Reference to its multi-loculate conidiomata.
Holotype: GMB1207.
Saprobic on dead bamboo branches. Sexual morph: Undetermined. Asexual morph: Stromata forming under a blackened area, up to 0.7–1.5 mm long and 0.3–0.5 mm wide, and becoming raised at maturity, ellipsoidal to oblong, occasionally irregular. Conidiomata 30–110 µm wide, 50–1000 µm long, loculate, 3–10 locules gregarious immersed in the stromata, globose to subglobose, dark brown, with slit-like opening. Conidiomatal wall comprising several layers of cells of textura angularis, with dark to brown outer thin layer, 5–7 µm thick, with 10–15 µm thick, hyaline, conidiogenous inner layer. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 2.5–10 × 2–5 µm ( = 8.7 × 3.1 µm, n = 20) enteroblastic, phialidic, indeterminate, discrete, cylindrical to ampulliform, hyaline, smooth-walled. Conidia 4–5.5 × 2.5–3.7 µm ( = 5.1 × 3.1 µm, n = 20), ellipsoidal oblong, aseptate, straight, rounded at both ends, hyaline when immature and becoming brown to dark brown when mature, smooth-walled, inside usually containing 2 small guttules.
Culture characters: Conidia germinating on PDA within 24 h and germ tubes produced from one side. Colonies slow growing, 40 mm diam. after 20 days at 28 °C, under 24 h dark, circular, with even margin, floccose at the center, greenish brown, and white at margin, ring-like from below.
Distributions: China.
Material examined: China, Yunnan Province, Kunming, Kunming Expo Park (25°07′77″ N, 102°76′23″ E, 1960 m), on dead culms of bamboo, 24 July 2020, Dong-Qin Dai, DDQ00748 (GMB1207, holotype), ex-type GMBCC1056, GenBank accession number SSU: OM891821; Ibid. (IFRD500-21 isotype), ex-isotype IFRDCC 3100; Ibid. DDQ00774 (GMB1218), living culture, GMBCC1065, GenBank accession number SSU: ON124715; Ibid. DDQ00775 (GMB1219), GenBank accession number SSU: OM891822; Ibid. DDQ00783(GMB1221), living culture, GMBCC1069, GenBank accession number SSU: ON124716; Ibid. DDQ00792 (GMB1223), living culture, GMBCC1071, GenBank accession number SSU: ON124717; Ibid. DDQ00859 (GMB1248), living culture, GMBCC1080, GenBank accession number SSU: OM891824.
Notes: Newly generated strains, GMBCC1056, GMBCC1065, GMBCC1069, GMBCC1071 and GMBCC1080, and specimen GMB1219 grouped together as the sister clade to R. verrucispora (CBS 125434, ex-type) with high statistical supports (100% MLBP, 1.00 BYPP) (Figure 1). A comparison of nucleotide base pairs of ITS, tef1 and rpb2, shows that R. multiloculate differs from R. verrucispora. Considering ITS, tef1 and rpb2 loci, R. multiloculate differs from R. verrucispora in 24/520 (4.6%), 27/1045 (2.6%), and 20/924 bp (2%), respectively. However, the asexual morph of R. verrucispora is undetermined yet for the morphological comparison [1]. Roussoella multiloculate shares similar morphological features to R. chiangraina, R. neopustulans, R. pustulans, and R. siamensis [1], but phylogenetic reconstructions strongly supported that these species are phylogenetically distinct (Figure 1). Therefore, based on the phylogenetic analyses and guidelines provided by Jeewon and Hyde [38] for the delimitation of new species, we introduce R. multiloculate as a novel species of Roussoella.
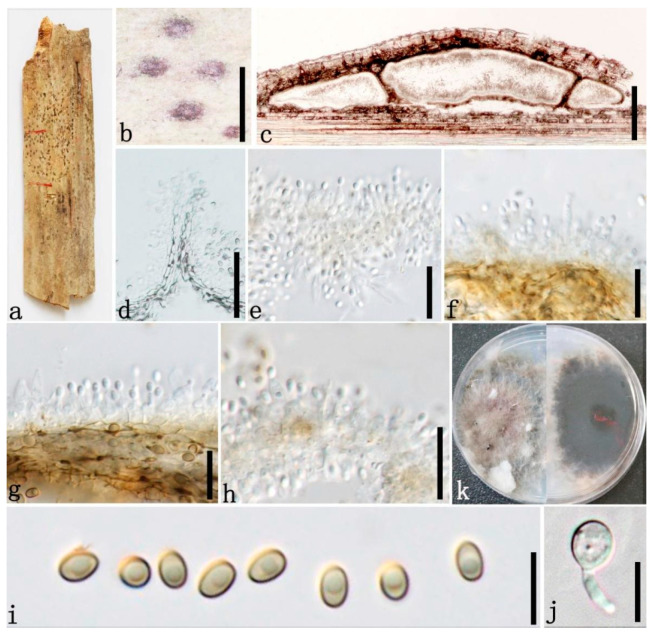
Roussoella nitidula Sacc. and Paol., Atti Inst. Veneto Sci. lett., ed Arti, Sér. 6 6:410 (1888) Figure 6.
Figure 6.
Roussoella nitidula (GMB1270, new country record). (a) Bamboo specimen; (b) black ascostromata on host surface; (c) vertical sections of ascomata; (d) cells of locule wall. (e) pseudoparaphyses; (f–j) asci; (k–n) ascospores; (o) Germinating ascospore; (p) Cultures on PDA from above and below. Scale bars: (b) = 1 mm, (c) = 500 μm, (d) = 50 μm, (e–j) = 30 μm, (k–n) = 10 μm, (o) = 15 μm.
Index Fungorum number: IF 177454.
Descriptions of sexual and asexual morphs see Liu et al. [1].
Distributions: China, Malaysia, Thailand.
Material examined: China, Yunnan Province, Dehong, Ruili, Yinlong Village (97°55′40.6″ N, 24°15′49.3″ E, 912 m), on dead culms of bamboo, 16 August 2020, Dong-Qin Dai, DDQ00957 (GMB1270); living culture, GMBCC1097 (new country record), GenBank accession number SSU: OM891826.
Notes: In our phylogenetic analyses, the new strain GMBCC1097 grouped with two strains of R. nitidula (MFLUCC 11-0182, ex-type) and MFLUCC 11-0634) (Figure 1) with high statistical supports (100% MLBS, 1.00 BYPP). Furthermore, GMBCC1097 morphologically resembles R. nitidula, having black dome-shaped ascostromata; hypha-like, septate, numerous, narrow pseudoparaphyses; cylindrical, relatively thin-walled asci and two-celled dark brown ornamented ascospores [1]. Our fresh collections are morphologically similar to the type of R. nitidula and have approximate sizes of asci and ascospore (asci 90–130 × 8–11 μm vs. 110–150 × 8–10(–11) μm; ascospores 14.5–17 × 5–6.5 μm vs. 16–18 × 6–7 μm). Therefore, we consider them to represent one species based on phylogeny and morphology. We herein report R. nitidula from China for the first time.
Roussoella papillate Dai and Wijayaw. sp. nov. Figure 7.
Figure 7.
Roussoella papillate (GMB129, holotype). (a) Bamboo specimen; (b) black ascostromata on host surface; (c) vertical section of ascoma; (d) cells of locule wall near the ostiole; (e) branching pseudoparaphyses; (f–i) asci; (j–m) ascospores; (n) germinating ascospore; (o,p) cultures on PDA from above and below. Scale bars: (b) = 500 µm, (c) = 150 µm, (d) = 50 µm, (e–i,n) = 30 µm, (j–m) = 15 µm.
Index Fungorum number: IF 556010.
Etymology: References to its papilate ascostromata.
Holptype: GMB1298.
Saprobic on decaying bamboo culms. Sexual morph: Ascostromata 250–350 μm high, 500–900 μm long, 500–700 μm wide, deeply immersed under a brown area, becoming slightly raised at maturity, ellipsoidal to irregular coriaceous, solitary to gregarious, elliptical, with a prominent, black papillate, uniloculate. Locules 200–300 μm high, 450–500 μm diam., solitary, subglobose, brown to dark brown, with a central ostiole. Wall of locules 9–20 μm wide, composed of 1–2 layers of textura angularis, thin-walled flattened at the base, light brown to brown. Hamathecium comprises 1–2 μm wide, numerous, anastomosing cellular pseudo paraphyses, branching at the apex, smooth-walled, often constrict at the septum, and embedded in a gelatinous matrix. Asci 108–125 × 7–10 μm ( = 114.1 × 8.3 μm, n = 20), 8-spored, bitunicate, cylindrical, short pedicellate, apically rounded with an ocular chamber (0.5–0.8 μm). Ascospores 15–17 × 5.5–7 μm ( = 16.4 × 6.2 μm, n = 20), uniseriate, ellipsoidal to broad fusiform, 2-celled, constricted at the septum, brown to dark brown, with longitudinal striations and surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Culture characters: Ascospores germinating on PDA within 24 h and germ tubes produced from one cell. Colonies slow-growing, 20 mm diam. after 20 days at 28 °C, under 24 h dark, rounded, with even margin, white, cottony from above, and white at the margin, yellowish-brown at the center, ring-like from below.
Distributions: China.
Material examined: China, Yunnan Province, Luoping, Jiulong fall waterfall, on dead culms of bamboo, 29 August 2020, Dong-Qin Dai, DDQ01076 (GMB1298, holotype); ex-type GMBCC1121; Ibid. (IFRD500-24 isotype), ex-isotype living culture IFRDCC 3103, GenBank accession number SSU: ON228186.
Notes: Roussoella papillate (GMBCC1121) formed a well distinct lineage (100% MLBP, 1.00 BYPP) basal to R. hysterioides (CBS 546.94) and R. japanensis (MAFF 239636, ex-type, GMBCC1067, and GMBCC1117) (Figure 1). Base pair differences in the tef1 gene region of R. papillate to R. hysterioides and R. japanensis are 4/841 (0.5%) and 18/923 bp (2%), respectively, while base pair differences of ITS locus of these three species are very less. Morphological differences between R. papillate and related species are listed in Table 5. Therefore, depending on morphological differences and slight base pair differences in tef1 region, we introduce this species as a new member of Roussoella.
Table 5.
Morphological comparison of R. papillate with R. hysterioides and R. japanensis.
| Characters | R. japanensis [1] |
R. papillate (in This Study) |
R. hysterioides [39] |
|---|---|---|---|
| Ascostromata | 500–2000 μm diam., immersed under a clypeus, raised, visible, black, dome-shape areas on host surface, uni-biloculate | 250–350 μm high, 500–900 μm long, 500–700 μm wide deeply immersed under a brown area, becoming raised at maturity, ellipsoidal to irregular coriaceous, solitary to gregarious, brown, with black papilla, uniloculate | 230–280 µm high, 2–2.5 mm wide, immersed, flattened at the base, multilocular |
| Locules | 190–210 μm high, 500–560 μm diam., depressed globose with a flattened base, single or 2–3 grouped, ostiolate | 200–300 μm high, 450–500 μm diam., solitary, subglobose, brown to dark brown, with a central ostiole | 75–150 × 35–50 µm, with the ostiole erumpent through the host epidermis |
| Peridium (Wall of locules) | 10–15 μm thick at sides, composed of 3–5 layers of polygonal flattened cells (3.5–12.5 × 1.5–2.5 μm), surrounded by wedge-shaped stromatic region (450–800 μm wide at sides) composed of rectangular to polygonal cells (3.5–15 × 4–10 μm) | 9–20 μm wide, composed of 1–2 layers of textura angularis, thin-walled flattened at the base, light brown to brown | |
| Asci | 107–132 × 8–9.5 μm, cylindrical, short pedicellate | 108–125 × 7–10 μm, cylindrical, short pedicellate | 105–120 × 4–6 µm, cylindrical, short- pedicellate |
| Ascospores | 16–22 × 5.5–7 μm, uniseriate, fusiform to ellipsoidal, with a median septum, 2-celled, brown, rough-walled more or less, covered with longitudinal striations and surrounded by an entire sheath of 0.5–4 μm wide | 15–17 × 5.5–7 μm, uniseriate, ellipsoidal to broad fusiform, 2-celled, constricted at the septum, brown to dark brown, with longitudinal striations, surrounded by a mucilaginous sheath | 13–20 × 4–6 µm, uniseriate, overlapping, fusiform, uniseptate, constricted at the septum, brown, slightly pointed at the ends, upper cell larger, with striate ornamentation on surface |
Roussoella padinae Prigione, Bovio, Poli, and Varese, Diversity 12(4, no. 144):14 (2020) Figure 8.
Figure 8.
Roussoella padinae (GMB1320, first report of asexual morph, first record from terrestrial habitat and first record from China). (a) Bamboo specimen; (b) reddish-brown conidiomata on host surface; (c) vertical sections of conidiomata; (d) conidioma wall; (e–h) conidiogenous cells; (i) conidia; (j) germinating conidium; (k) cultures on PDA from above and below. Scale bars: (b) = 1 mm, (c) = 150 µm, (d) = 30 µm, (e–h) =15 µm, (i) = 5 µm, (j) = 10 µm.
Index Fungorum number: IF 832843.
Saprobic on dead bamboo culms. Sexual morph: Undetermined. Asexual morph: Stromata forming under a light red area, up to 1–1.5 mm diam. and 100–200 µm high, and becoming raised when mature, ellipsoidal to globose. Conidiomata 85–175 µm high, 200–980 µm diam., loculate, 2–5 locules gregarious immersed in the stromata, fabiform, dark brown. Conidiomatal wall comprising two layers of cells of textura angularis, with dark to brown outer thin layer 3 µm thick, with 6.3–7.5 µm thick, hyaline, conidiogenous inner layer. Conidiophores were reduced to conidiogenous cells. Conidiogenous cells 1.5–4.5 × 1–2.5 µm ( = 2.7 × 1.6 µm, n = 20) enteroblastic, indeterminate, discrete, cylindrical to ampulliform, hyaline, smooth-walled. Conidia 3–4 × 2–3 µm ( = 3.3 ×2.4 µm, n = 20), ellipsoidal to globose, aseptate, straight, rounded at both ends, hyaline when young, and becoming brown at maturity, smooth-walled, inside usually containing one small guttule.
Culture characters: Conidia germinating on PDA within 24 h and germ tubes produced from one side. Colonies slow-growing, 40 mm diam. after 20 days at 28 °C, under 24 h dark, circular, with radialized margin, floccose at the center, brown from above and dark brown from below.
Distributions: China, Italy.
Material examined: China, Yunnan Province, Jinghong Menla, Manzhang (21°91′97.56″ N, 101°20′42.49″ E, 617.14 m), on dead culms of bamboo, 16 July 2020, Dong-Qin Dai, DDQ02019, GMB1320; living culture, GMBCC1126.
Notes: Phylogenetic analyses showed that the new strain GMBCC1126 grouped with R. padinae (MUT 5503, ex-type) which was introduced by Poli et al. [8] from brown alga Padina pavonica (i.e., from marine habitat). Base pair arrangement of ITS, LSU, and rpb2 loci are identical in strain GMBCC1126 and R. padinae. Therefore, GMB1320 (strain: GMBCC1126) and R. padinae are conspecific. However, sexual and asexual morphs of R. padinae are undetermined while Poli et al. [8] have described this species based only on the morphology of colonies and vegetative structures. In this study, we report the asexual morph of R. padinae for the first time from dead culms of bamboo. Further, this is the first record of this species from terrestrial habitats and the first record from China.
Roussoella scabrispora (Höhn.) Aptroot, Nova Hedwigia 60(3-4):368 (1995) Figure 9.
Figure 9.
Roussoella scabrispora (GMB1286, new country record). (a) Bamboo specimen; (b) ascomata on bamboo host; (c) vertical section of ascoma; (d) peridium; (e) pseudoparaphyses; (f–i) ascospores; (j) germinating ascospore; (k–n) asci; (o,p) cultures on PDA from above and below. Scale bars: (b) = 1 mm, (c) = 500 μm, (d) = 100 μm, (e) = 30 μm, (f–j) = 15 μm, (k–n) = 50 μm.
Index Fungorum number: IF 414110.
Description of sexual morph see Liu et al. [1], Asexual morph: Undetermined.
Distributions: China, Indonesia, Thailand.
Material examined: China, Yunnan Province, Ruili, Dehong Yinlong Village (97°55′40.6″ N, 24°15′49.3″ E, 912 m), on dead culms of bamboo, 16 August 2020, Dong-Qin Dai, DDQ00960 (GMB1274); living culture, GMBCC1101, GenBank accession number SSU: OM891827; Ibid. DDQ00961 (GMB1275), GenBank accession number SSU: ON124718; living culture, GMBCC1102; Ibid. DDQ01003 (GMB1286); living culture, GMBCC1108, GenBank accession number SSU: ON124719; Bamboo Garden, Xishuangbanna Topic Botany Garden, Mengla, Jinghong, Yunnan, China (101°24′41.74″ N, 21°93′40.94″ E, 507.88 m), on dead culms of bamboo, 15 August 2020, Dong-Qin Dai, DDQ00975 (GMB1279); living culture, GMBCC1104 (new country record), GenBank accession number SSU: OM891828.
Notes: Roussoella scabrispora comprises distinctive ascospores with reticulate wall ornamentation [1]. However, the ascospores of our new collection are slightly narrower (27–34 × 7–9.5 μm vs. (24−)25–29(−32) × (7−)9–10.5 μm) than in the protologue [39]. Nevertheless, based on phylogenetic analyses (Figure 1), we confirmed new collections (i.e., GMB1274, GMB1286 and GMB1279) are R. scabrispora. This is the first report of R. scabrispora from China.
Roussoella sinensis Dai and Wijayaw. sp. nov. Figure 10.
Figure 10.
Roussoella sinensis (GMB1296, holotype). (a) Bamboo specimen; (b) black ascostromata showing black ostioles with openings on host surface; (c,d) vertical section of ascostromata; (e) cells of locule wall; (f) pseudoparaphyses; (g–j) ascospores; (k) germinating ascospore; (l–o) different developmental stages of asci; (p,q) cultures on PDA from above and below. Scale bars: (c,d) = 200 μm, (e) = 50 μm, (f) = 10 μm, (g–k) = 15 μm, (l–o) = 30 μm.
Index Fungorum number: IF 556014.
Etymology: Reference to its first collection site China.
Holotype: GMB1296.
Saprobic on decaying bamboo culms. Sexual morph: Ascostromata 400–600 μm diam., forming under raised, visible, dark brown to black, globose areas near ostioles opening 100–200 diam. on host surface, deeply immersed, scattered to gregarious, uniloculate. Locules 160–200 μm high, 250–400 μm diam., subglobose, dark brown, with a short central ostiolate neck, 55–65 μm high, 40–45 μm diam. Wall of locules 10–20 μm thick, comprising host and fungal tissues, thin, 8–13 μm wide, composed of dark brown cells of textura angularis. Hamathecium comprises 1–2 μm wide, numerous, anastomosing branched pseudoparaphyses, rough-walled, and embedded in a gelatinous matrix. Asci 130–170 μm × 9–10.5 μm ( = 148.9 × 9.6 μm, n = 20), 8-spored, bitunicate, cylindrical, with a short knob-like pedicel, with an ocular apical chamber. Ascospores 16.5–20.5 × 6–7.5μm ( = 18.2 × 6.8 μm, n = 20), uniseriate, overlapping, ellipsoid to broad fusiform, 2-celled, upper cells bigger, occasionally curve, brown, constricted at the septum, narrowly at both ends, with longitudinal striations and surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Culture characters: Ascospores germinating on PDA within 24 h and germ tubes produced from upper cell. Colonies rapidly growing, 20 mm diam. after 20 days at 28 °C, under 24 h dark, ellipsoidal to rounded, with even, thallus-like margin, pale yellow, floccus at the margin, milk-white at the center and becoming light brown.
Distributions: China.
Material examined: China, Yunnan Province, Jinghong, Mengla, Bamboo Garden, Xishuangbanna Topic Botany Garden (101°24′41.74″ N, 21°93′40.94″ E, 507.88 m), on dead culms of bamboo, 15 August 2020, Dong-Qin Dai, DDQ01045 (GMB1296, holotype); ex-type GMBCC1119, GenBank accession number SSU: OM891833; Ibid. (IFRD500-22, isotype), ex-isotype IFRDCC 3101, GenBank accession number SSU: ON228185.
Notes: Roussoella sinensis (GMB1296, holotype) shows the typical morphological characters of the sexual morphs of Roussoella species [1,17], and is distinct by having locules with a short central ostiolate neck (Figure 10c), 55–65 μm high, 40–45 μm diam. Furthermore, Roussoella sinensis grouped as the sister species to R. doimaesalongensis (MFLUCC 14-0584, ex-type). ITS, tef1 and rpb2 base pair differences of R. sinensis (GMBCC1119) and R. doimaesalongensis are 10% (53/530), 21.30% (171/803), and 9.6% (70/728), respectively. Roussoella siamensis (MFLUCC 11-0149, ex-type) and R. yunnanensis (MFLUCC 18-0115, ex-type) as a separate lineage but with relatively less statistical support (Figure 1). ITS, tef1, and rpb2 base pair differences of R. sinensis (GMBCC1119) and R. siamensis (MFLUCC 11-0149) are 15.20% (62/515), 18.53% (192/1036), and 6.60% (60/909) respectively. Base pair differences of ITS and rpb2 genes locus of Roussoella sinensis (GMBCC1119) and R. yunnanensis (MFLUCC 18-0115) are 15.20% (71/467) and 5.51% (50/907), respectively. However, tef1 sequences from R. yunnanensis (MFLUCC 18-0115) are not available in the GenBank database. Roussoella doimaesalongensis was introduced by Thambugala et al. [40] based on a specimen collected on dead bamboo in Thailand. However, only the asexual morph is known for R. doimaesalongensis [40]. According to the recommendations suggested by Jeewon and Hyde [38] regarding base pair differences, Roussoella sinensis is a phylogenetically different species from R. doimaesalongensis, R. siamensis, and R. yunnanensis; hence, we introduced Roussoella sinensis as a new member to Roussoella.
Roussoella tuberculata Dai and Hyde, Fungal Diversity 82:37 (2016) Figure 11.
Figure 11.
Roussoella tuberculata (GMB1317, new country record). (a) Bamboo specimen; (b) black conidioma on host surface; (c) vertical sections of conidiomata; (d–h) conidiogenous cells and developing conidia; (i–l) conidia; m: germinating conidium; (m) germinating conidium; (n) cultures on PDA from above and below after two weeks; (o) cultures on PDA from above and below after four weeks. Scale bars: (b) = 500 µm, (c) = 200 µm, (d–f,m) = 15 µm, (h–l) = 10 µm.
Index Fungorum number: IF 552027.
Sexual morph: Undetermined. Description of asexual morph see Dai et al. [17].
Distributions: China, Thailand.
Material examined: China, Yunnan Province, Jinghong, Mengla, Bamboo Garden, Xishuangbanna Topic Botany Garden (101°24′41.74″ N, 21°93′40.94″ E, 507.88 m), on dead culms of bamboo, 15 August 2020, Dong-Qin Dai, DDQ02000 (GMB1317); living culture, GMBCC1123 (new country record), GenBank accession number SSU: OM891834.
Notes: Our fresh collection (GMB1317) shares similar morphologies of R. tuberculata which was introduced by Dai et al. [17] from Thailand. Roussoella tuberculata is characterized by large, immersed, eustromatic conidiomata, which are rather flattened, phialidic, annellidic conidiogenous cells and conidia covered by small tubercules [17]. Usually, the conidia of Roussoella species comprise verrucose wall ornamentation [1,39], while R. tuberculata has conidia with small and roughened tubercules. The above morphological identification is confirmed by our phylogenetic analyses and base-pair comparisons as well (100% MLBS, 1.00 BYPP, Figure 1). This is the first record of this species from China.
Roussoella uniloculata Dai and Wijayaw., sp. nov. Figure 12.
Figure 12.
Roussoella uniloculata (GMB1288, holotype). (a) Bamboo specimen; (b) black ascostroma on host surface; (c) vertical section of ascomata; (d) pseudoparaphyses; (e–h) ascospores; (i) germinating ascospore; (j–m) asci; (n,o) cultures on PDA from above and below. Scale bars: (b) = 300 µm, (c) = 50 µm, (d,j–m) = 30 µm, (e–i) = 10 µm.
Index Fungorum number: IF 556016.
Etymology: Uniloculata means single locule.
Holotype: GMB1288.
Saprobic on decaying bamboo culms. Sexual morph: Ascostromata 250–350 μm diam., forming under raised, visible, black, round areas on host surface and becoming prominent when maturity, containing a single locule. Locules 120–145 μm high, 270–380 μm diam., solitary, subglobose to ellipsoidal, brown, with an inconspicuous central ostiole. Wall of locules comprising host and fungal tissues, thin, 7–10 μm wide, composed of dark brown cells of textura angularis. Hamathecium comprises 1–2.5 μm wide, numerous, anastomosing branched cellular pseudoparaphyses, rough-walled, and embedded in a gelatinous matrix. Asci 61.5–102.5 × 4–6.5 μm ( = 78.8 × 5.2 μm, n = 20), 6–8-spored, bitunicate, cylindrical, with a short knob-like pedicel, with an ocular apical chamber. Ascospores 8.5–12 × 3.5–4.5(−4.8) μm ( = 10.3 × 4.1 μm, n = 20), uniseriate, ellipsoid to broad fusiform, 2-celled, upper cells bigger, occasionally curved, brown, constricted at the septum, with longitudinal striations and surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Culture characters: Ascospores germinating on PDA within 24 h and germ tubes produced from both cells. Colonies slow-growing, 30 mm diam. after 20 days at 28 °C, under 24 h dark, elliptic to round, with irregular margin, milk-white at margin, and light brown to dark brown at the central.
Distributions: China.
Material examined: China, Yunnan Province, Ruili, Dehong Yinlong Village (97°55′40.6″ N, 24°15′49.3″ E, 912 m), on dead culms of bamboo, 16 August 2020, Dong-Qin Dai, DDQ01005 (holotype GMB1288), ex-type GMBCC1110, GenBank accession number SSU: OM891829; Ibid. (IFRD500-23 isotype), ex-isotype IFRDCC 3102; Ibid. DDQ01005-2, GenBank accession number SSU: OM891835.
Notes: Roussoella uniloculata (GMB1288) formed a basal lineage in a clade comprising R. angusta (MFLUCC 15-0186, ex-type), R. chiangraina (MFLUCC 10-0556), R. kunmingensis (KUMCC 18-0128), R. magnatum (MFLUCC 15-0185), R. mediterranea (MUT 5369), R. neopustulans (MFLUCC 11-0609), and R. padinae (MUT 5503) with high statistical supports (100% MLBP, 1.00 BYPP) (Figure 1). Base pair differences of ITS and rpb2 gene loci of R. uniloculata and other related species showed that they are phylogenetically distinct species [38] (Table 6).
Table 6.
Base pair differences of ITS and RPB2 gene loci of R. uniloculata and other related species.
| Species | ITS | rpb2 |
|---|---|---|
| R. chiangraina | 7.22% (33/457) | 3.68% (34/925) |
| R. kunmingensis | 5.29% (25/472) | 4.61% (39/845) |
| R. mediterranea | 4.26% (20/469) | 3.84% (25/651) |
| R. neopustulans | 5.56% (27/469) | 3.80% (35/922) |
| R. padinae | 4.71% (22/467) | 3.78% (35/925) |
Roussoella uniloculata is similar to R. yunnanensis and R. siamensis in having ellipsoidal to fusiform and 2-celled ascospores with longitudinal striations. However, R. uniloculata can be distinguished from R. yunnanensis in having smaller ascostromata (250–350 μm diam. vs. 1–1.3 mm diam.) with single locule vs. with multiple locules, and asci with a short knob-like pedicel vs. asci with a slightly furcate short pedicel [5]. Roussoella uniloculata can be distinguished from R. siamensis in having smaller ascostromata (250–350 μm diam. vs. 620–750 μm diam. [17], higher locules (120–145 μm high vs. 70–120 μm high; [17]. Roussoella uniloculata can be compared with R. pustulans, in having small ascospores with bigger upper cells (8.5–12 × 3.5–4.5 μm vs. 10–16 × 4–5 μm) [1]. However, R. uniloculata differs by smaller ascostromata (165–300 μm diam. vs. 1 mm diam.), and in the phylogenetic tree, they form separate lineages (Figure 1). Moreover, morphology, host, and distribution of the new species were compared with the known species which are lacking sequence data (Table 7). Hence, based on molecular phylogenetic and morphological analyses, we introduced R. uniloculata as a novel species of Roussoella.
Table 7.
Ascospores, host, and distribution comparison of eleven sequence lacking known Roussoella taxa with three new species in this study. “-”: not available in the protologue.
| Taxa | Ascospores | Host | Known Distribution | References |
|---|---|---|---|---|
| Roussoella aequatoriensis Hyde | 26–33 × 9–11 µm, fusiform-ellipsoidal, 1-septate, constricted at the septum, brown, with oblique wall, striations running the entire length of the ascospore and with yellow coloured mucilaginous, pad-like appendages at each end | Palm | Ecuador, Puerto Rico | [41] |
| R. alveolata Ju, Rogers, and Huhndorf | 34–42 × 11–13 µm, with ridges between the longitudinal striations. | Bamboo | Indonesia (Java) | [41] |
| R. angustispora Zhou, Cai, and Hyde | 24–28 × 6–8 µm, ellipsoid-fusiform, 1-septate, constricted at the septum, brown, with reticulate wall ornamentations | Bamboo (Bambusa changii) | China (Hong Kong) | [42] |
| R. bambusae (Pat.) Monod | 23 × 5 µm, elliptical elongated, often acute at both ends, not constricted at the septum, colorless and surrounded by a fleeting hyaline sheath | - | - | [43] |
| R. calamicola Fröhl., Hyde, and Aptroot | 20–27(–29.5) × 7–8.5 μm, ellipsoidal, 1-septatae, brown, verrucose, surrounded by a mucilaginous sheath | Calamus | Australian (Queensland) | [44] |
| R. chilensis (Speg.) Ju, Rogers, and Huhndorf | asci contain only four 20–25(–28) × 6–8 µm, ascospores with longitudinal wall striations This fungus is unique amongst Roussoella in having four ascospores per ascus | Bamboo (Chusquea) | Chile | [41] |
| R. donacicola (Speg.) Ju, Rogers, and Huhndorf | ascospores are (6–)6.5–8(–8.5) × 3–3.5 µm, with longitudinal striations | Bamboo (Arundo, Phyllostachys) | Argentina, France | [41] |
| R. palmicola Fröhl., Hyde, and Aptroot | 12.5–24 × 2.5–4 μm, fusiform, 1-septate, brown, striate, with small pads of mucilage at both ends | Rattan (Calamus flabellatus) | Brunei | [44] |
| R. saltuensis Hyde | 25–30 × 8–11 µm, overlapping ellipsoidal, 1-septate, constricted at the central septum, dark-brown, covered with irregular longitudinal striations and surrounded by a mucilaginous sheath which spreads in water | Palm (indet.) | Ecuador | [41] |
| R. serrulata (Ellis and Martin) Hyde and Aptroot | 18–20 × 5–6 µm, characterized by the often deeply (up to 1 mm) immersed ascomata | Pam (Serenoa serrulata) | USA, Florida | [12] |
| R. verruculosa Cand. and Katum | 7–8 × 5 µm, fusoid, septate, slightly constricted at the septum, rounded at the ends, verruculose | Bamboo (Phyllostachys mitis) | France | [45] |
| R. papillate Dai and Wijayaw | 15–17 × 5.5–7 μm, brown to dark brown, rough-walled, with longitudinal striations | Bamboo | China (Yunnan) | In this study |
| R. sinensis Dai and Wijayaw | 16.5–20.5 × 6–7.5 μm, ellipsoid to broad fusiform, upper cells bigger, constricted at the septum, narrowly at both ends, with longitudinal striations | Bamboo | China (Yunnan) | In this study |
| R. uniloculata Dai and Wijayaw | 8.5–12× 3.5–4.5, ellipsoid to broad fusiform, 2-celled, upper cells bigger, occasionally curve, brown, constricted at the septum, with longitudinal striations | Bamboo | China (Yunnan) | In this study |
4. Discussion
The family Roussoellaceae (in Pleosporales), comprises the genera Neoroussoella, Roussoella and Roussoellopsis [1], which are saprobes in different hosts, especially bamboo and palms (terrestrial and aquatic environments) or human pathogens [46]. Recently, three new species have been isolated from marine environments [8]. Currently, the family comprises 12 genera reported as sexual, asexual, or holomorph [1,8,9,17,46,47]. This study introduced four new species of Roussoella: three reported as sexual morphs (i.e., R. papillate, R. sinensis, and R. uniloculata), and another as an asexual morph (i.e., R. multiloculate). All species have been reported as saprobes of bamboo plants.
However, asexual morphs of R. kunmingensis (described initially as a sexual taxon [8]) and R. padinae (described initially as without sexual or asexual characteristics fide [8]) were reported for the first time in the present study. Sexual and asexual links were established based on DNA sequence analyses (Figure 1). The sexual morph of R. kunmingensis was also reported from Kunming, Yunnan Province. Nevertheless, Poli et al. [8] introduced R. padinae from brown alga Padina pavonica (Italy), and thus from the marine environment. This finding indicates that Roussoella species present a broad range of habitats and distribution. Further, the same species could be reported from different environments but as its alternative morph.
Neoroussoella bambusae (Thailand [1]), R. japanensis (Japan [1]), R. nitidula (Malaysia [1]), R. scabrispora (Indonesia [41]), and R. tuberculata (Thailand) species were reported from China for the first time. These records confirmed that the members of Roussoellaceae have a broad range of geographical distribution in Southeast Asia and Central Asia. We predict that novel species could occur in other tropical Asian countries, such as India, Laos, Myanmar, Pakistan, and Sri Lanka.
The genus Roussoella comprises 51 epithets [47], but only 46 species are listed in Species Fungorum [47]. Eight species were transferred to Dothideaceae, Thyridariaceae, Phyllachoraceae, and Diaporthales [2,7,28,48]. Further, two species were transferred to Neoroussoella and Pseudoroussoella, which are also nested in Roussoellaceae [49]. Moreover, R. hysterioides var. minuta (Hino and Katum) Hino and Katum were listed as synonyms of R. hysterioides in Index Fungorum [47], and two species R. phyllostachydis and R. minutella were synonymized as R. pustulans by Hyde [41]. Thus, a total of 33 taxa were accepted in Roussoellaceae. Hongsanan et al. [46] mentioned that DNA sequence data are available only for 22 species. Hence, it is essential to recollect known species lacking DNA sequence data and designate epitypes. In this study, we compared the morphological characters of the sequence lacking Roussoella taxa [12,41,42,43,44,45,50] prior to introducing the new taxa (Table 7).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. NSFC 31860620, 31760013, 31950410558), High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program and “High-End Foreign Experts” Program) and partially supported by Chiang Mai University, Thailand. Authors would like to thank Aptroot for helping with references.
Author Contributions
Conceptualization, D.-Q.D. and N.N.W.; methodology, D.-Q.D.; software, M.C.D. and D.-Q.D.; validation, H.-H.C., D.-Q.D., T.-T.Z. and X.Z.; formal analysis, T.-T.Z. and G.-Q.Z.; investigation, N.N.W.; resources, L.-S.H. and D.-Q.D.; writing—original draft preparation, D.-Q.D., M.C.D. and N.N.W.; writing—review and editing, D.-Q.D., N.N.W. and J.K.; supervision, D.-Q.D. and N.N.W.; project administration, D.-Q.D.; funding acquisition, D.-Q.D., N.N.W. and H.-H.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The research was supported by the National Natural Science Foundation of China (No. NSFC 31860620, 31760013, 31950410558), the High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program and “High-End Foreign Experts” Program), CAS President’s International and partially supported by Chiang Mai University, Thailand.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu J.K., Phookamsak R., Dai D.Q., Tanaka K., Jones E.G., Xu J.C., Chukeatirote E., Hyde K.D. Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa. 2014;181:1–33. doi: 10.11646/phytotaxa.181.1.1. [DOI] [Google Scholar]
- 2.Jaklitsch W.M., Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016;85:35–64. doi: 10.1016/j.simyco.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibpromma S., Hyde K.D., Jeewon R., Maharachchikumbura S.S., Liu J.K., Bhat D.J., Jones E.B., McKenzie E.H., Camporesi E., Bulgakov T.S., et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–261. doi: 10.1007/s13225-017-0378-0. [DOI] [Google Scholar]
- 4.Hyde K.D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K.W.T., Dayarathne M.C., De Silva N.I., Dissanayake A.J., Ekanayaka A.H., et al. Mycosphere notes 169–224. Mycosphere. 2018;9:271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- 5.Jiang H.B., Hyde K.D., Jayawardena R.S., Doilom M., Xu J., Phookamsak R. Taxonomic and phylogenetic characterizations reveal two new species and two new records of Roussoella (Roussoellaceae, Pleosporales) from Yunnan, China. Mycol. Prog. 2019;18:577–591. doi: 10.1007/s11557-019-01471-9. [DOI] [Google Scholar]
- 6.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B., Maharachchikumbura S.S., Raspé O., Karunarathna S.C., Wanasinghe D.N., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 7.Mapook A., Hyde K.D., McKenzie E.H., Jones E.B., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 8.Poli A., Bovio E., Ranieri L., Varese G.C., Prigione V. News from the sea: A new genus and seven new species in the Pleosporalean families Roussoellaceae and Thyridariaceae. Diversity. 2020;12:144. doi: 10.3390/d12040144. [DOI] [Google Scholar]
- 9.Wijayawardene N.N., Hyde K.D., Dai D.Q., Sánchez-García M., Goto B.T., Saxena R.K., Erdoğdu M., Selçuk F., Rajeshkumar K.C., Aptroot A., et al. Outline of Fungi and fungus-like taxa-2021. Mycosphere. 2022;13:53–453. doi: 10.5943/mycosphere/13/1/2. [DOI] [Google Scholar]
- 10.Saccardo P.A., Paoletti G. Mycetes Malacenses. Funghi della penisola di Malacca raccolti nel 1885 dell’ Ab. Benedetto Scortechini. Atti Accad. Sci. Veneto-Trentino-Istriana. 1885;6:387–428. [Google Scholar]
- 11.Höhnel F. Fragmente zur Mykologie XXIII. Sitzungsber Akad Wiss Wien Math-Naturwiss KI. 1919;128:535–625. [Google Scholar]
- 12.Aptroot A. Redisposition of some species excluded from Didymosphaeria (Ascomycotina) Nova Hedwig. 1995;60:325–379. [Google Scholar]
- 13.Müller E., von Arx J.A. Die Gattungen der Didymosporen Pyrenomyceten. Kommissionsverlag Buchdruckerei Büchler; Wabern, Switzerland: 1962. [Google Scholar]
- 14.Aptroot A. A monograph of Didymosphaeria. Stud. Mycol. 1995;37:1–160. [Google Scholar]
- 15.Tanaka K., Hirayama K., Yonezawa H., Hatakeyama S., Harada Y., Sano T., Shirouzu T., Hosoya T. Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 2009;64:175–209. doi: 10.3114/sim.2009.64.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 17.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 18.Karunarathna A., Phookamsak R., Jayawardena R.S., Cheewangkoon R., Hyde K.D., Kuo C.H. The holomorph of Neoroussoella alishanense sp. nov. (Roussoellaceae, Pleosporales) on Pennisetum purpureum (Poaceae) Phytotaxa. 2019;406:218–236. doi: 10.11646/phytotaxa.406.4.1. [DOI] [Google Scholar]
- 19.Phukhamsakda C., McKenzie E.H., Phillips A.J., Gareth Jones E.B., Jayarama Bhat D., Stadler M., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 20.Index Fungorum. [(accessed on 20 April 2022)]. Available online: http://www.indexfungorum.org/names/Names.asp.
- 21.White T.J., Bruns T., Lee S.J., Taylor J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 22.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehner S. Primers for Elongation Factor 1-α (EF1-α) [(accessed on 30 March 2022)]. Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf/
- 24.Liu Y., Whelen S., Hall B. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 25.Hall T. BioEdit. Ibis Therapeutics. 2004. [(accessed on 30 March 2022)]. Available online: http://www.mbio.ncsu.edu/BioEdit/bioedit.html/
- 26.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crous P.W., Schumacher R.K., Akulov A., Thangavel R., Hernández-Restrepo M., Carnegie A.J., Cheewangkoon R., Wingfield M.J., Summerell B.A., Quaedvlieg W., et al. New and interesting fungi. 2. Fungal Syst. Evol. 2019;3:57–134. doi: 10.3114/fuse.2019.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 30.Silvestro D., Michalak I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2011;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 31.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 32.Nylander J.A.A. MrModeltest 2.0. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. Program Distributed by the Author. [Google Scholar]
- 33.Rannala B., Yang Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 34.Zhaxybayeva O., Gogarten J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002;3:4. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 36.Maharachchikumbura S.S., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Huang S.K., Abdel-Wahab M.A., Daranagama D.A., Dayarathne M., D’souza M.J., Goonasekara I.D., et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015;72:199–301. doi: 10.1007/s13225-015-0331-z. [DOI] [Google Scholar]
- 37.Page R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 38.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 39.Hyde K.D., Eriksson O.E., Yue J.Z. Roussoëlla, an ascomycete genus of uncertain relationships with a Cytoplea anamorph. Mycol. Res. 1996;100:1522–1528. doi: 10.1016/S0953-7562(96)80089-X. [DOI] [Google Scholar]
- 40.Thambugala K.M., Wanasinghe D.N., Phillips A.J.L., Camporesi E., Bulgakov T.S., Phukhamsakda C., Ariyawansa H.A., Goonasekara I.D., Phookamsak R., Dissanayake A., et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere. 2017;8:697–796. doi: 10.5943/mycosphere/8/4/13. [DOI] [Google Scholar]
- 41.Hyde K.D. The genus Roussoella, including two new species from palms in Cuyabeno, Ecuador. Mycol. Res. 1997;101:609–616. doi: 10.1017/S0953756296003061. [DOI] [Google Scholar]
- 42.Zhou D.Q., Cai L., Hyde K.D. Astrosphaeriella and Roussoella species on bamboo from Hong Kong and Yunnan, China including a new species of Roussoella. Cryptogam. Mycol. 2003;24:191–197. [Google Scholar]
- 43.Monod M. Monographie taxonomique des Gnomoniaceae (Ascomycètes de l’ordre des Diaporthales) I. Beihefte Sydowia. 1983;9:1–314. doi: 10.1016/S0007-1536(85)80245-X. [DOI] [Google Scholar]
- 44.Hyde K.D., Aptroot A., Fröhlich J., Taylor J.E. Fungi from palms. XLII. Didymosphaeria and similar ascomycetes from palms. Nova Hedwig. 1999;69:449–471. doi: 10.1127/nova.hedwigia/69/1999/449. [DOI] [Google Scholar]
- 45.Candoussau F., Katumoto K., Sherwood-Pike M.A. Bambusicolous fungi from the southwest of France I. Two new species of Pyrenomycetes and a new genus of the Phacidiaceae. Sydowia. 1985;38:28–34. [Google Scholar]
- 46.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Lücking R., Boonmee S., Bhat J.D., Liu N., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 47.Species Fungorum. [(accessed on 20 April 2022)]. Available online: http://www.speciesfungorum.org/Names/Names.asp.
- 48.Theissen F., Sydow H. Die Dothideales. Kritisch-systematische Originaluntersuchungen. Ann. Mycol. 1915;13:147–746. [Google Scholar]
- 49.Jayasiri S.C., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Jeewon R., Phillips A.J.L., Bhat D.J., Wanasinghe D.N., Liu J.K., Lu Y.Z., et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere. 2019;10:1–186. doi: 10.5943/mycosphere/10/1/1. [DOI] [Google Scholar]
- 50.Ju Y.M., Rogers J.D., Huhndorf S.M. Valsaria and notes on Endoxylina, Pseudothyridaria, Pseudovalsaria, and Roussoella. Mycotaxon. 1996;58:419–481. doi: 10.1002/(SICI)1097-0061(199605)12:63.0.CO;2-B. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.