Abstract
Toluene-degrading bacteria were isolated from hydrocarbon-contaminated soil by incubating liquid enrichment cultures and agar plate cultures in desiccators in which the vapor pressure of toluene was controlled by dilution with vacuum pump oil. Incubation in desiccators equilibrated with either 100, 10, or 1% (wt/wt) toluene in vacuum pump oil and testing for genomic cross-hybridization resulted in four genomically distinct strains (standards) capable of growth on toluene (strains Cstd1, Cstd2, Cstd5, and Cstd7). The optimal toluene concentrations for growth of these standards on plating media differed considerably. Cstd1 grew best in an atmosphere equilibrated with 0.1% (wt/wt) toluene, but Cstd5 failed to grow in this atmosphere. Conversely, Cstd5 grew well in the presence of 10% (wt/wt) toluene, which inhibited growth of Cstd1. 16S ribosomal DNA sequencing and cross-hybridization analysis indicated that both Cstd1 and Cstd5 are members of the genus Pseudomonas. An analysis of the microbial communities in soil samples that were incubated with 10% (wt/wt) toluene with reverse sample genome probing indicated that Pseudomonas strain Cstd5 was the dominant community member. However, incubation of soil samples with 0.1% (wt/wt) toluene resulted in a community that was dominated by Pseudomonas strain Q7, a toluene degrader that has been described previously (Y. Shen, L. G. Stehmeier, and G. Voordouw, Appl. Environ. Microbiol. 64:637–645, 1998). Q7 was not able to grow by itself in an atmosphere equilibrated with 0.1% (wt/wt) toluene but grew efficiently in coculture with Cstd1, suggesting that toluene or metabolic derivatives of toluene were transferred from Cstd1 to Q7.
Spain et al. (13) have stated that “bioremediation often involves complex mixtures of contaminants and undefined, mixed populations of microorganisms.” The bioremediation of C5+ by soil microbial communities is an example that aptly fits this statement. C5+ is obtained as a by-product of pyrolytic conversion of ethane into ethylene, which serves as a precursor for polymer production. The C5+ produced at a large polyethylene plant in Alberta, Canada is typically composed of 45% (wt/wt) benzene, 13% dicyclopentadiene (DCPD), 7% cyclopentadiene, 6% toluene, 3% styrene, and smaller quantities of other compounds. On-site spills of the C5+ product stream have created an interest in the rate and course of C5+ removal by soil microbial communities under both aerobic and anaerobic conditions. Aerobically, most of the components are rapidly removed; the only exceptions are cyclopentadiene, DCPD, and higher-molecular-weight components (14). We have shown that C5+-contaminated soil harbors a mixed community of microorganisms. In previous work 35 genomically distinct bacteria (standards 1 to 35) were isolated primarily by plating samples onto rich media (11). The genomic DNAs of these standards were spotted onto a master filter, which was used to analyze the change in community composition when slurries of soil and minimal salts medium were exposed in desiccators to either a saturated DCPD atmosphere or a saturated toluene atmosphere. After exposure to toluene Pseudomonas strain LQ20 (standard 11) was a dominant community component. LQ20 was subsequently shown to efficiently mineralize toluene. However, four other pseudomonads (standards 2, 8, 25, and 27; strains LQ5, LQ16, Q5, and Q7) were also shown to be capable of toluene mineralization. We wondered whether the contributions of these organisms to toluene mineralization can be defined in more detail. To study this, we isolated additional toluene degraders by using different toluene concentrations and determined the effect of toluene concentration on community composition by using a master filter containing 42 standards.
MATERIALS AND METHODS
Biochemical reagents.
[α-32P]dCTP (10 mCi/ml; 3,000 Ci/mmol) was purchased from ICN, while enzymes and bacteriophage λ DNA (0.5 mg/ml) were obtained from Pharmacia. Reagent grade chemicals were obtained from BDH, Fisher, or Sigma. Vacuum pump oil 19, a 100% paraffinnic oil with a density of 0.85 g cm−3, was obtained from VWR Scientific.
Culture media and regulation of toluene concentration.
Minimal salts (MS) medium and PTYG medium were prepared as described previously (11). Toluene was used as the carbon and energy source for growth in MS medium. Plates or liquid cultures were incubated at room temperature (22 to 26°C) in 15.3-liter glass desiccators, each of which contained a beaker or crystallizing dish with a mixture of toluene and vacuum pump oil (vpo) in the bottom. vpo was selected as the diluent because it has a negligible vapor pressure and is inexpensive. The toluene concentration in the gas or vapor phase (Vtol) was regulated by the fraction of toluene in the mixture (Ftol). The concentration of toluene which was dissolved in an aqueous solution (in medium or in an agar plate) (Ctol) in a desiccator was linearly dependent on Vtol. Ftol can be reported as either weight or volume percentages; these ratios are essentially the same since toluene and the vpo used in this study have similar densities (0.86 and 0.85 g cm−3, respectively). Preparations were incubated in desiccators with an Ftol of 0% (150 g of vpo), 0.01% (0.05 g of toluene, 500 g of vpo), 0.1% (0.5 g of toluene, 499.5 g of vpo), 1% (2.5 g of toluene, 247.5 g of vpo), 10% (10 g of toluene, 90 g of vpo), or 100% (15 g of toluene). Vtol and Ctol were estimated to be 0 and 0, 0.05 and 0.16, 0.5 and 1.6, 5 and 16, 45 and 143, and 170 and 540 mg/liter for Ftol values of 0, 0.01, 0.1, 1, 10, and 100%, respectively. These estimates were based on data provided by Evans et al. (3) for mixtures containing 1, 10, and 100% (vol/vol) toluene and hexadecane (the Vtol values were 5.3, 45, and 170 mg/liter, respectively) and on measurements obtained in our laboratory for mixtures of toluene and vpo in which the Ftol values were 33, 50, and 100% (the Vtol values were 96, 142, and 162 mg/liter, respectively). A value of 540 mg/liter for the solubility of toluene in water under our experimental conditions was used for these estimates. Opening of desiccators with equilibrated atmospheres was kept to a minimum. After opening, the toluene-vpo mixture was replaced or gravimetrically adjusted by adding toluene in order to maintain a constant Ftol. Experimentally set toluene concentrations are reported below in terms of the Ftol values of the toluene-vpo mixtures placed in the desiccators.
Isolation and characterization of toluene-degrading bacteria.
Soil samples that were obtained from either the northern end (N soil) or the southern end (S soil) of a C5+-contaminated soil pile (11, 14) were combined into two large samples of ca. 500 g each. Aliquots (1 g) were incubated with 10 ml of MS medium and an Ftol of either 1, 10, or 100%. Appropriate dilutions of these cultures were plated onto MS agar plates (MS medium supplemented with 15 g of agar per liter) and incubated under the same conditions. Single colonies were picked, grown in PTYG medium, and stored as PTYG-glycerol stock preparations at −70°C. Large-scale (100-ml) cultures of 18 isolates were used to prepare DNA by a modification of the method of Marmur (7, 17). Following cross-hybridization testing in which dot blots were used and identification by 16S rRNA sequencing, as described below, eight genomically distinct isolates (Table 1) were identified for reverse sample genome probing (RSGP) analysis, as described elsewhere (11, 16, 17, 19). Following heat denaturation, solutions containing 66 to 90 ng of DNAs of standards 36 to 42 (Table 1) were spotted onto a set of master filters together with defined amounts (20 to 100 ng) of bacteriophage λ DNA. These filters were incubated with labeled probes together with master filters containing standards 1 to 35 prepared in a previous study (11). There were two types of labeled probes. In order to determine the percentage of cross-hybridization and the ratio of hybridization constants (kλ/kx) (11, 16, 17), one filter from the set of filters containing standards 1 to 35 and one filter from the set of filters containing standards 36 to 42 were hybridized with a labeled mixture containing 97.5 ng of standard DNA and 2.5 ng of λ DNA. These hybridizations resulted in cross-hybridization plots, as shown in Fig. 1. In order to determine the community composition, one filter from each set of filters was hybridized with a labeled mixture containing 97.5 ng of community DNA and 2.5 ng of λ DNA. These hybridizations resulted in community profiles (expressed as the calculated fraction of each standard, uncorrected for cross-hybridization, versus standard number), as shown in Fig. 2. DNA was isolated from soil by using a modification of the technique described by Bakken (1), as described previously (11). DNA mixtures were labeled by extension of random hexamer primers by using Klenow polymerase and [α-32P]dCTP (11). Labeled probes were incubated with the master filter dot blots under very stringent conditions (18). Following washing and drying, the dot blots were exposed to BAS-III imaging plates. Hybridization intensities, which were determined with a Fuji model BAS1000 bioimaging analyzer, were used to calculate kλ/kx ratios and the fraction of each standard, as described previously (16). Standards were identified by partial 16S ribosomal DNA (rDNA) sequencing of PCR products obtained with primers f8 (10) and r1406 (4), as described elsewhere (11, 16). The best matching sequence in the Ribosomal Database Project (RDP) database was then identified with the program SIMILARITY_RANK (6). The advantages and problems associated with use of the RSGP method have been described recently (19).
TABLE 1.
Properties and identification of standards isolated in this study
| Standard no.a | Strain designation | Optimal toluene concn (%)b | kλ/kxc | Taxond | Nearest RDP homologe |
|---|---|---|---|---|---|
| 36 | Cstd1 | 0.1 | 56 | Pseudomonas sp. | Pseudomonas putida |
| 37 | Cstd2 | 1 | 255 | — | NDf |
| Cstd3 | — | — | Staphylococcus aureus | ||
| 38 | Cstd4 | — | 280 | Pseudomonas sp., Bordetella sp. | Rhodococcus sp. |
| 39 | Cstd5 | 10 | 226 | Pseudomonas sp. | Pseudomonas stutzeri |
| 40 | Cstd6 | — | 127 | — | Microbacterium lacticum |
| 41 | Cstd7 | 1-10 | 92 | Pseudomonas sp. | Pseudomonas fluorescens |
| 42 | Cstd8 | — | 120 | Pseudomonas sp. | Pseudomonas putida |
Assigned standard number. Standards 1 to 35 have been described by Shen et al. (11).
Percentage of toluene in the toluene-vpo mixture that is optimal for growth. —, no growth or poor growth under all of the conditions tested.
Ratio of hybridization constants, determined as described by Telang et al. (16).
Identity based on cross-hybridization (Fig. 3). —, no significant cross-hybridization occurred.
Nearest homolog based on a comparison of the 16S rRNA sequence determined with the 16S rRNA sequences in the RDP database.
ND, not determined (see text).
FIG. 1.
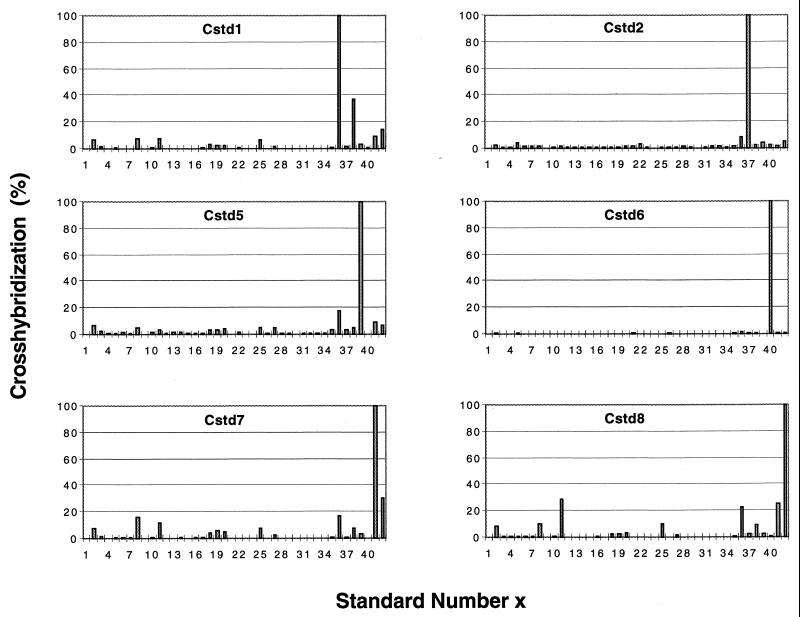
Hybridization of soil community master filters with genomic DNAs from six newly isolated standards. The net hybridization intensity relative to the hybridization intensity observed for the genome used as a probe (100%) is plotted versus the standard number (Table 1). Cstd1, Cstd5, Cstd7, and Cstd8 are all members of the genus Pseudomonas, as discussed in the text.
FIG. 2.
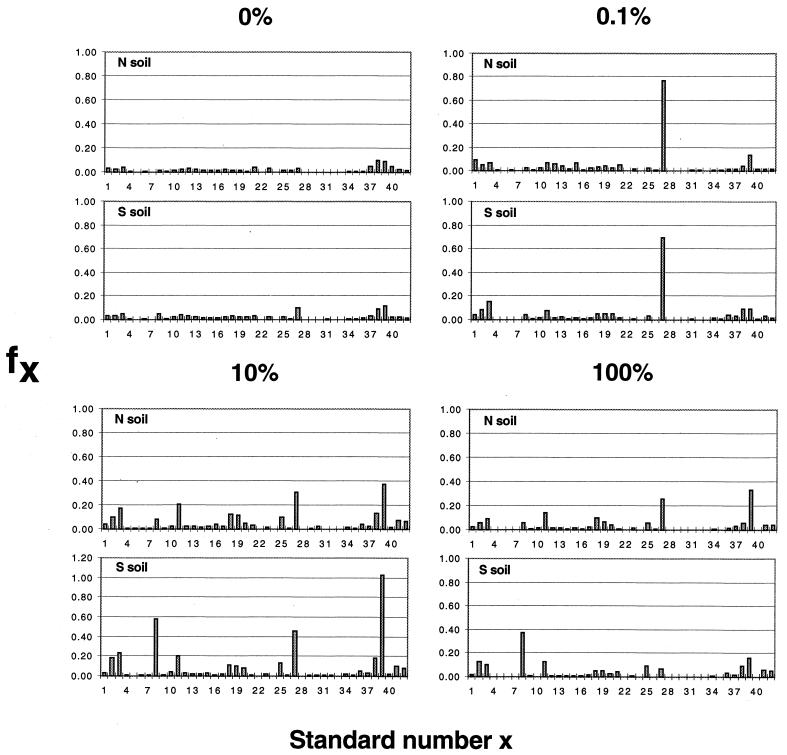
RSGP analysis of community DNAs from N soil and S soil from a contaminated soil pile. Soil aliquots (1 g) were incubated with MS medium in desiccators equilibrated with toluene-vpo mixtures containing 0, 0.1, 10, and 100% toluene, as indicated, for 2 weeks, after which community DNAs were extracted and used for RSGP analysis. The calculated fraction (fx) for each genome represented on the filters, uncorrected for cross-hybridization, is plotted against the standard number (Table 1). The results of two representative incubations are shown; a total of three, three, five, and two incubations were done for 0, 0.1, 10, and 100% toluene, respectively.
RESULTS
Molecular biological identification of bacterial standards.
Eighteen single-colony isolates were obtained from soil enrichment cultures plated onto MS agar and incubated with either 1, 10, or 100% toluene. Although these isolates were obtained from plates in which toluene (and perhaps agar) served as the sole carbon and energy source, several of them grew poorly in liquid MS medium in which toluene was the sole carbon and energy source. PTYG medium was, therefore, used to propagate these isolates and for large-scale (100-ml) cultures and DNA isolation. Following cross-hybridization testing eight standards (strains Cstd1 to Cstd8) (Table 1) were defined initially. The cross-hybridization patterns obtained for six of these standards are shown in Fig. 1. Four of the eight genomically distinct isolates grew well on toluene (Table 1 and Fig. 3). Cstd3, which was genomically unique, did not grow on toluene, and Staphylococcus aureus was the closest RDP homolog of this organism. This suggested that this strain did not originate from the soil samples, and Cstd3 was, therefore, not assigned a standard number (Table 1) and was not included on the master filter. The genome of Cstd4, which grew poorly with toluene as the sole carbon and energy source (Fig. 3), cross-hybridized with genomes of both Pseudomonas and Bordetella spp. (data not shown). The nearest RDP homolog of Cstd4 (Rhodococcus sp.) differed from both of these taxa, which suggested that the culture was not pure. The genomes of Cstd1, Cstd5, Cstd7, and Cstd8 (standards 36, 39, 41, and 42) (Table 1) exhibited limited cross-hybridization with each other and with the genomes of standards 2, 8, 11, 18 to 20, 25, and 27 (Fig. 1). The latter standards have all been shown to be members of the genus Pseudomonas by 16S rDNA sequencing (11), which means that Cstd1, Cstd5, Cstd7, and Cstd8 belong to the same genus. 16S rDNA sequencing confirmed that Cstd1, Cstd5, Cstd7, and Cstd8 are Pseudomonas spp. (Table 1). Cstd2 and Cstd6 had unique hybridization patterns (Fig. 1). So far, 16S rDNA sequencing of Cstd2 has not been successful, while 16S rDNA sequencing of Cstd6 identified Microbacterium lacticum as the closest RDP homolog.
FIG. 3.
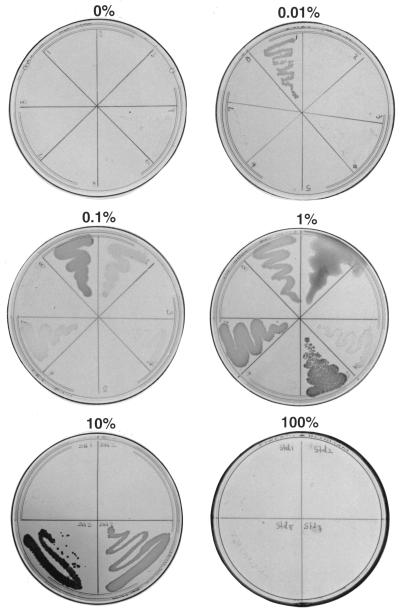
Growth of isolated standards on MS agar plates in desiccators equilibrated with toluene-vpo mixtures containing 0, 0.01, 0.1, 1, 10, and 100% toluene, as indicated. For 0, 0.01, 0.1, and 1% toluene, clockwise from the top left: Cstd1, Cstd2, Cstd3, Cstd4, Cstd5, Cstd6, Cstd7, and Cstd8. For 10 and 100% toluene, clockwise from the top left: Cstd1, Cstd2, Cstd7, and Cstd5. The plates were photographed after 2 weeks of incubation.
Growth at different toluene concentrations.
Figure 3 shows the growth of standards on agar plates in the presence of different toluene concentrations. Cstd3, Cstd4, Cstd6, and Cstd8 did not grow or grew poorly in the presence of 0.01, 0.1, and 1% toluene (Fig. 3), as well as 10 and 100% toluene (data not shown). Cstd8 grew well on benzene (data not shown). In the absence of toluene some growth of Cstd4 and Cstd7 occurred. On plates equilibrated with 0.01% toluene there was significant growth of only Cstd1, and this strain also grew best in the presence of 0.1% toluene. Incubation with 1% toluene resulted in significant growth of Cstd1, Cstd2, Cstd5, and Cstd7. Only the latter two strains grew well in desiccators equilibrated with 10% toluene, in which growth of Cstd1 and Cstd2 did not occur or was weak. Finally, only Cstd5 exhibited some growth in the presence of pure toluene (Fig. 3). Growth studies performed with liquid MS medium cultures confirmed these results. Growth was estimated after 2 weeks by determining the increase in absorbance at 600 nm (ΔA600) for 50-ml cultures in 250-ml Erlenmeyer flasks incubated in the desiccators. In the presence of 1, 10, and 100% toluene the ΔA600 values for Cstd1 were 0.5, 0.02, and 0.01, respectively, the ΔA600 values for Cstd2 were 0.19, 0.04, and 0.02, respectively, the ΔA600 values for Cstd5 were 0.26, 0.18, and 0.01, respectively, and the ΔA600 values for Cstd7 were 0.18, 0.16, and 0.00, respectively. Thus, Cstd1 and Cstd2 grew only in the presence of 1% toluene, whereas Cstd5 and Cstd7 grew in the presence of both 1 and 10% toluene. No significant growth occurred in the presence of 100% toluene. Although in principle measuring the ΔA600 provides a more quantitative comparison of growth, data collection was often complicated due to cell lysis or clumping and formation of pigments (Fig. 3). Comparing strains on plating media, as shown in Fig. 3 and 4, was therefore the preferred method for evaluating growth in the presence of different toluene concentrations.
FIG. 4.
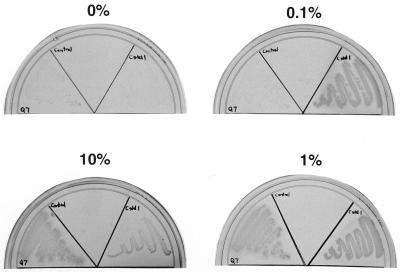
Comparison of growth of Pseudomonas strains Q7 and Cstd1 on MS agar plates in desiccators equilibrated with toluene-vpo mixtures containing 0, 0.1, 1, and 10% toluene, as indicated. Clockwise from the left: Q7, control (no bacteria), and Cstd1. The plates were photographed after 2 weeks of incubation.
Effect of toluene concentration on community composition.
The observation that toluene-utilizing standards Cstd1, Cstd2, Cstd5, and Cstd7 grow optimally in the presence of different toluene concentrations raises the possibility that the composition of microbial communities is affected by this parameter. This possibility was investigated by performing an RSGP analysis (19) of DNAs extracted from soils that were incubated in MS medium with different Ftol. Standard 39 (Pseudomonas strain Cstd5) was the dominant organism in all five incubations containing 10% toluene; the results obtained for two of these are shown in Fig. 2. Standards 3 (Azospirillum strain LQ6), 8 (Pseudomonas strain LQ16), 11 (Pseudomonas strain LQ20), and 27 (Pseudomonas strain Q7) were also significant community components under these conditions. The dominance of standard 39 (Pseudomonas strain Cstd5) is consistent with the ability of this organism to grow at a high Ftol (Fig. 3).
Following incubation at a low Ftol the community was totally dominated by standard 27 (Pseudomonas strain Q7). Standard 36 (Pseudomonas strain Cstd1) did not become a dominant community component, despite its ability to grow at a very low Ftol (Fig. 3). Pseudomonas strain Q7 was obtained in previous work by plating soil samples on rich media but was shown to be able to mineralize toluene (11). In order to determine whether Q7 can also grow in the presence of low toluene concentrations, plates were incubated in the presence of 0, 0.1, 1, and 10% toluene. Q7 grew best in the presence of 1 and 10% toluene (Fig. 4). Growth in the presence of 0.1% toluene was negligible (not stronger than growth in the absence of toluene) compared to the growth of Cstd1 (Fig. 4 and Table 2). The mechanism that allows Q7 to become the dominant community component during growth in the presence of 0.1% toluene is not clear; this dominance is not the result of an intrinsic high affinity for toluene, such as that demonstrated for Cstd1.
TABLE 2.
Growth of Pseudomonas strains Cstd1 and Q7 in the presence of low toluene concentrations
| Inoculuma | Ftol (%)b | A600c | % Q7d | % Cstd1d |
|---|---|---|---|---|
| Q7 | 0 | 0.012 | ||
| Cstd1 | 0 | 0.015 | ||
| Q7 + Cstd1 | 0 | 0.023 | ||
| Q7 | 0.1 | 0.045 | ||
| Cstd1 | 0.1 | 0.883 | ||
| Q7 + Cstd1 | 0.1 | 0.581 | 58 ± 7e | 42 ± 7e |
| Q7 | 1 | 0.260 | ||
| Cstd1 | 1 | 1.256 | ||
| Q7 + Cstd1 | 1 | 0.580 | 85 ± 3f | 15 ± 3f |
Bacteria (0.065 A600 unit) were inoculated into 5 ml of MS medium, as described in the text.
Fraction of toluene in the toluene-vpo mixture used for growth, expressed as a percentage.
Cell density after 2 weeks, determined as described in the text.
As determined by the RSGP method.
Average values based on four incubations.
Average values based on two incubations.
Competition of Q7 and Cstd1 at a low toluene concentration.
Q7 and Cstd1 were both grown in 10 ml of PTYG medium. Following centrifugation the cells were suspended in 10 ml of MS medium, recentrifuged, and suspended in 5 ml of MS medium. The cell densities, as measured by A600 were 0.653 and 0.774, respectively. Inocula representing equal cell densities (100 μl of Q7 and/or 84 μl of Cstd1) were inoculated into 5-ml portions of MS medium and incubated in desiccators equilibrated with 0, 0.1, or 1% toluene. The cell densities measured after 2 weeks are shown in Table 2. The data confirmed that as a monoculture, Q7 grew in the presence of 1% toluene but not in the presence of 0.1% toluene, while Cstd1 grew well (better than Q7) under both conditions. A mixture of the two strains grew to a lower cell density than Cstd1 alone grew. The composition of this mixture, as analyzed by RSGP, indicated that it was dominated by Q7, both in cultures grown in the presence of 0.1% toluene and in cultures grown in the presence of 1% toluene (Table 2). The dominance of Q7 was even greater (88% in the presence of 0.1% toluene and 96% in the presence of 1% toluene) if 1 g of sterilized soil, prepared as described by Shen et al. (11), was included in each mixed culture. These results are unusual, because when a mixture of the toluene-degrading bacteria isolated in this study (Cstd1, Cstd2, Cstd5, and Cstd7) was grown in MS medium in the presence of 0.1, 1, or 10% toluene, Cstd1 was the dominant organism in mixed cultures grown in the presence of 0.1% toluene and Cstd5 was the dominant organism in mixed cultures grown in the presence of 10% toluene (results not shown), as expected from the data in Fig. 3.
DISCUSSION
Spills of aromatic hydrocarbons at polyethylene plants often involve direct exposure of soil to an undiluted C5+ mixture (14). Following spreading by diffusion and transport, the aqueous concentration of each C5+ component varies from the maximum possible value (obtained when the undiluted mixture is equilibrated with water) to zero. In the case of toluene, which comprises 6% (wt/wt) of C5+, the aqueous concentration may vary from 0 to 20% of the value obtained when pure toluene is equilibrated with water (540 mg/liter) (note that the data in Materials and Methods indicate that a plot of Vtol or Ctol versus Ftol in the organic phase is nonlinear). Toluene-degrading soil bacteria, which are present at C5+-contaminated sites, can either be active over this entire range of Ctol values or specialize (e.g., they can remove toluene at either the low end or the high end of the range of concentrations). None of the toluene degraders isolated in the current study appeared to be able to act efficiently at the entire range of toluene concentrations (Fig. 3). Organisms that degrade toluene at low concentrations (e.g., Pseudomonas strain Cstd1) may have mechanisms, as yet unknown, for accumulating toluene at very low Ctol values. In a recent study of biofilm succession in fluidized bed reactors inoculated with toluene-degrading soil communities, the reactor effluent was shown to have toluene concentrations of 0.04 to 0.14 mg/liter (8). Cstd1 can grow at these concentrations; i.e., equilibration of an aqueous phase with a toluene-vpo mixture containing 0.01% toluene gives rise to a Ctol value of 0.16 mg/liter. Organisms that degrade toluene at the high end of the toluene concentration scale must be able to cope with the toxicity of toluene, which has been well-documented under these conditions. The coping mechanisms may involve an increase in the number of trans isomers of unsaturated fatty acids or removal of toluene from membranes via efflux pumps (5, 12). The latter mechanism, when operated in reverse, may allow active import at low toluene concentrations. We did not isolate degraders that grew well in the presence of 100% toluene (Fig. 3), perhaps because such a high toluene concentration has never occurred at the sampling site, at which toluene was spilled as part of the C5+ mixture.
A binary organic solvent system which provided a low but constant Vtol (and thus a constant Ctol) was used by Evans et al. (3) to isolate strain T1, a denitrifying, toluene-degrading bacterium. These workers used a binary mixture of toluene and hexadecane. Strain T1 grew best in cultures equilibrated with 1% (vol/vol) toluene in hexadecane. Raising the toluene concentration in the binary mixture to 10% (vol/vol) was found to be inhibitory. Hexadecane was estimated to have a gas phase concentration of 0.085 mg/liter, which was considered negligible (too low to support microbial growth). As pointed out by Evans et al. (3), this method is generally applicable for isolation of bacteria on a volatile substrate that is inhibitory at saturating concentrations. Using a chemically similar, nonvolatile diluent (e.g., hexadecane) results in a binary mixture that is thermodynamically ideal at a low substrate concentration. Thus, at a toluene concentration below 10% (vol/vol), Vtol decreases essentially linearly with the Ftol in the binary mixture. The method was considered to be particularly useful for isolating anaerobic hydrocarbon degraders in sealed containers (3). During aerobic isolation the toluene concentration is often less carefully controlled; e.g., a small amount of toluene is placed in the lid of an agar plate (3, 20), which results in a culture that experiences a range of toluene concentrations. Duetz et al. (2) maintained their aerobic cultures in an atmosphere equilibrated with 10% (vol/vol) toluene in hexadecane. Tay et al. (15) isolated two toluene-degrading Mycobacterium strains in desiccators containing a beaker of water equilibrated with toluene, which had to be replaced every 2 to 3 days.
Frequent replacement is not necessary with a binary organic solvent mixture. In a 15-liter desiccator containing 0.1% toluene in vpo (0.5 g of toluene, 499.5 g of vpo), Vtol is 0.5 mg/liter and only 1.5% of the toluene in the binary mixture evaporates into the gas phase. Thus, Vtol is efficiently buffered and can be accurately maintained by adding 7.5 mg of toluene to the toluene-vpo mixture or by replacing the toluene-vpo mixture altogether each time that the desiccator is opened. Adjustment or replacement is not necessary if the desiccator is opened infrequently. We believe that using vpo as a diluent is an improvement over using hexadecane. Although the vapor pressure of hexadecane is low, it is not negligible. For instance, Cstd1 can grow at a Vtol of 0.05 mg/liter (Ftol, 0.01%) (Fig. 3), which is less than the gaseous concentration of hexadecane (0.085 mg/liter). vpo is chemically similar to hexadecane but has a higher molecular weight and was selected because it has a very low vapor pressure (0.01 Torr at 110°C). It is also more economical to use. Although Vtol values can also be accurately adjusted by mixing gas streams, a method used in continuous-culture studies of toluene-degrading bacteria or toluene-degrading communities (2, 9), this method is not practical for plate incubation.
Using our improved method, we isolated four genomically distinct bacteria that degrade toluene at different concentrations. Cstd1 grows best at a low Ftol (0.1%), Cstd2 grows best at an intermediate Ftol (1%), Cstd7 grows best at high Ftol (1 to 10%), and Cstd5 grows best at a very high Ftol (10%) (Fig. 3). We also found that the composition of a toluene-degrading community obtained from C5+-contaminated soil is strongly dependent on the concentration of toluene (Fig. 2). When the concentration of toluene in the organic phase was 10%, Cstd5 (standard 39) was the dominant community member in both N soil and S soil, while in the presence of 100% toluene Cstd5 was the dominant community member in the N soil sample (Fig. 2). These results are consistent with the demonstrated ability of Cstd5 to grow in the presence of very high toluene concentrations (Fig. 3). Apparently, this Pseudomonas strain is well-equipped to cope with toluene toxicity at high toluene concentrations and may, therefore, be a primary catalyst for toluene degradation close to the origin of a spill. Standards 8, 11, and 27 (Pseudomonas strains LQ16, LQ20, and Q7) may have similar properties. Strain LQ20 was found to be the dominant community component in incubations with 100% toluene in a previous study (11). At the low end of the toluene activity scale we obtained an unexpected result. Although strain Q7 cannot grow by itself in the presence of 0.1% toluene (Fig. 4 and Table 2), it did nevertheless dominate the community under these conditions both when N soil and when S soil were incubated (Fig. 2). Apparently, Q7 can derive carbon and energy from strains such as Cstd1 that are able to grow in the presence of such low toluene concentrations (Fig. 3). The mechanism by which Q7 can benefit from growth of Cstd1 at low toluene concentrations is not known.
Other workers have also estimated the contributions of selected community components to toluene mineralization. Tay et al. (15) concluded that two Mycobacterium strains that were isolated from a contaminated stream contributed little to the overall toluene mineralization by the microbial community (based on a comparison of their estimated numbers and degradation activities and the degradation activity of the community). Duetz et al. (2) examined continuous cultures of mixtures of selected toluene-degrading Pseudomonas spp. and found that competitiveness under toluene-limiting conditions depends on the pathway used to oxidize toluene. The results obtained in the present study indicate that the role and importance of a given community component (such as Cstd1) cannot always be determined from its numerical abundance under a given set of conditions. Although we have not characterized the pathways used by the toluene-degrading soil bacteria isolated in this study, the possible presence of different pathways cannot explain the results shown in Fig. 4 and Table 2. The competitive interactions involving toluene in soil microbial communities can, apparently, be more complex than the interactions in the synthetic microcosms studied by Duetz et al. (2) and may perhaps include sharing of pathways by different microorganisms.
ACKNOWLEDGMENTS
This work was supported by a Strategic Grant from the Natural Science and Engineering Research Council of Canada to G.V. and by a financial contribution from NOVA Research & Technology Corporation.
REFERENCES
- 1.Bakken L R. Separation and purification of bacteria from soil. Appl Environ Microbiol. 1985;49:1482–1487. doi: 10.1128/aem.49.6.1482-1487.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duetz W A, de Jong C, Williams P A, van Andel J G. Competition in chemostat culture between Pseudomonas strains that use different pathways for the degradation of toluene. Appl Environ Microbiol. 1994;60:2858–2863. doi: 10.1128/aem.60.8.2858-2863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a dinitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks R, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isken S, de Bont J A M. Active efflux of toluene in a solvent-resistant bacterium. J Bacteriol. 1996;178:6056–6058. doi: 10.1128/jb.178.20.6056-6058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 8.Massol-Deya A, Weller R, Rios-Hernandez L, Zhou J-Z, Hickey R F, Tiedje J M. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol. 1997;63:270–276. doi: 10.1128/aem.63.1.270-276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matteau Y, Ramsay B. Active compost biofiltration of toluene. Biodegradation. 1997;8:135–141. doi: 10.1023/a:1008221805947. [DOI] [PubMed] [Google Scholar]
- 10.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Stehmeier L G, Voordouw G. Identification of hydrocarbon-degrading bacteria in soil by reverse sample genome probing. Appl Environ Microbiol. 1998;64:637–645. doi: 10.1128/aem.64.2.637-645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikkema J, de Bont J A M, Poolmans B. Mechanisms of membrane toxicity of hydrocarbons. Microb Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spain J C, Pettigrew C A, Haigler B E. Biodegradation of mixed solvents by a strain of Pseudomonas. In: Sayler G S, Fox R, Blackburn J W, editors. Environmental biotechnology for waste treatment. New York, N.Y: Environmental Science Research, Plenum Press; 1991. pp. 175–184. [Google Scholar]
- 14.Stehmeier L. Fate of dicyclopentadiene in the environment. Ph.D. thesis. Calgary, Alberta, Canada: The University of Calgary; 1997. [Google Scholar]
- 15.Tay S T-L, Hemond H F, Polz M F, Cavanaugh C M, Dejesus I, Krumholz L R. Two new Mycobacterium strains and their role in toluene degradation in a contaminated stream. Appl Environ Microbiol. 1998;64:1715–1720. doi: 10.1128/aem.64.5.1715-1720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telang A J, Ebert S, Foght J M, Westlake D W S, Jenneman G E, Gevertz D, Voordouw G. The effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl Environ Microbiol. 1997;63:1785–1793. doi: 10.1128/aem.63.5.1785-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voordouw G, Voordouw J K, Karkhoff-Schweizer R R, Fedorak P M, Westlake D W S. Reverse sample genome probing, a new technique for identification of bacteria in environmental samples by DNA hybridization and its application to the identification of sulfate-reducing bacteria in oil field samples. Appl Environ Microbiol. 1991;57:3070–3078. doi: 10.1128/aem.57.11.3070-3078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voordouw G, Strang J D, Wilson F R. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol. 1989;171:3881–3889. doi: 10.1128/jb.171.7.3881-3889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voordouw G. Reverse sample genome probing of microbial community dynamics. ASM News. 1998;64:627–633. [Google Scholar]
- 20.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla)) [sic] mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]