Abstract
We purified a secreted fungal laccase from filtrates of Gaeumannomyces graminis var. tritici cultures induced with copper and xylidine. The active protein had an apparent molecular mass of 190 kDa and yielded subunits with molecular masses of 60 kDa when denatured and deglycosylated. This laccase had a pI of 5.6 and an optimal pH of 4.5 with 2,6-dimethoxyphenol as its substrate. Like other, previously purified laccases, this one contained several copper atoms in each subunit, as determined by inductively coupled plasma spectroscopy. The active enzyme catalyzed the oxidation of 2,6-dimethoxyphenol (Km = 2.6 × 10−5 ± 7 × 10−6 M), catechol (Km = 2.5 × 10−4 ± 1 × 10−5 M), pyrogallol (Km = 3.1 × 10−4 ± 4 × 10−5 M), and guaiacol (Km = 5.1 × 10−4 ± 2 × 10−5 M). In addition, the laccase catalyzed the polymerization of 1,8-dihydroxynaphthalene, a natural fungal melanin precursor, into a high-molecular-weight melanin and catalyzed the oxidation, or decolorization, of the dye poly B-411, a lignin-like polymer. These findings indicate that this laccase may be involved in melanin polymerization in this phytopathogen’s hyphae and/or in lignin depolymerization in its infected plant host.
Laccase (p-diphenol:oxygen oxidoreductase; EC 1.10.3.2) is a copper-containing enzyme that catalyzes the oxidation of a phenolic substrate by coupling it to the reduction of oxygen to water. Fungal laccases display a wide substrate range, are known to catalyze the polymerization, depolymerization, and methylation and/or demethylation of phenolic compounds (11, 12), and may play a role in plant pathogenicity (23, 24) or lignin degradation (8).
Many fungi produce dihydroxynaphthalene (DHN) melanin, and even though phenol oxidase(s), including laccases, is hypothesized to catalyze the oxidation of DHN into polymeric melanin, the in vivo mechanism of polymerization is unknown (1, 21). While cell-free homogenates of Cochliobolus heterostrophus catalyzed the oxidation of DHN, purified laccases that efficiently catalyze the polymerization of DHN in vitro have not yet been described (21).
The fungus Gaeumannomyces graminis var. tritici is a phytopathogenic ascomycete that causes take-all, a severe root disease of wheat and barley in temperate regions worldwide (9, 25). G. graminis var. tritici produces DHN melanin and initiates root infection with melanized, ectotrophic “runner” hyphae; these produce hyaline infection hyphae that penetrate the root cortex and endodermis. Albino mutants of G. graminis are nonpathogenic, perhaps because runner hyphae must be melanized for anchoring or for producing the invasive infection hyphae (10). The host forms lignin-impregnated papillae, or lignitubers, around invading hyphal tips (20); these are physical barriers that impede hyphal tip advancement. The means by which G. graminis var. tritici penetrates the lignitubers is unknown at present, but the use of lignin-degrading enzymes, such as laccase, is a plausible mechanism. Hence, G. graminis laccase(s) may be involved in melanin biosynthesis and/or lignin degradation. Our objective in this study was to purify and characterize the secreted laccase of G. graminis var. tritici. This laccase is of interest because it polymerizes DHN and oxidizes a lignin-like dye in vitro. These activities suggest that this secreted laccase facilitates fungal infection of host plants.
MATERIALS AND METHODS
Strains, media, and culture conditions.
G. graminis var. tritici was isolated (by D. Mathre, strain DM528) from Montana wheat (Triticum aestivum cv. Pondera) and purified by single-ascospore isolation. The fungal strain was maintained on Luria-Bertani medium (19) plus 1% (wt/vol) agar at 24 to 26°C. Long-term stocks were maintained on potato dextrose agar (Difco, Detroit, Mich.) slants stored at 4°C.
Cultures were grown in modified Fahraeus minimal medium (7). Sucrose was substituted for glucose. Biotin and CuSO4 were added to final concentrations of 1.02 mM and 400 μM, respectively.
For laccase purification, 2-liter flasks containing 500 ml of minimal medium were inoculated with 60 plugs of hyphae transferred with Pasteur pipettes (1 mm in diameter) from the growing margin of a G. graminis var. tritici colony. Flasks were shaken (200 rpm) at 23 to 25°C. After 2 days, 10% (vol/vol) xylidine in ethanol (95%) was added to a final concentration of 40 μM xylidine to stimulate laccase production. Cultures were harvested at maximum laccase activity, generally 5 to 8 days after the addition of xylidine.
Laccase assay.
Laccase activity was determined spectrophotometrically by monitoring the conversion of 2 mM 2,6-dimethoxyphenol (DMOP) to 3,5,3′,5′-tetramethoxydiphenoquinone in pH 4.5 citrate-phosphate (15 mM) buffer at 468 nm (ɛ = 49.6 mM−1 cm−1) (5). All spectrophotometric assays were performed with a Gilford (Oberlin, Ohio) model 2600 spectrophotometer. A unit of laccase activity is defined as the amount of enzyme needed to oxidize 1 μmol of DMOP per min.
Protein concentration.
Protein concentrations were estimated spectrophotometrically at 280 nm (ɛ = 10%−1 cm−1) (17) with the absorbance corrected for DNA contamination, determined by the A280/A260 ratio (17).
Laccase purification.
Cultures were filtered through Whatman no. 1 filter paper, and culture filtrates were combined and concentrated to 50 ml with an Amicon (Beverly, Mass.) Stirred Cell protein concentrator with a 50,000-molecular-weight (MW) cutoff membrane. The protein was concentrated to 1 ml with a Millipore (Bedford, Mass.) Ultrafree 15 centrifuge cell with a 50,000-MW cutoff membrane. The concentrated protein solution was cooled on ice for 20 min and centrifuged at 15,000 × g for 10 min. The concentrated protein solution was decanted from the precipitate, and 100 μl of glycerol was added.
The enzyme was electrophoresed with a Bio-Rad (Hercules, Calif.) Prep Cell by using a 28-mm-inner-diameter gel tube at 2 W constant power, a running buffer of pH 6.6 His-MOPS [25 mM histidine–30 mM 3-(N-morpholino)propanesulfonic acid], and a 15-ml, 5.5% polyacrylamide nondenaturing gel. The elution buffer was pH 5.6 MES [10 mM 3-(N-morpholino)-ethanesulfonic acid], and laccase was eluted after 7 to 8 h as determined by DMOP assays. Collected fractions with laccase activity were concentrated to 0.5 ml with a Millipore Ultrafree 15 filter (50,000-MW cutoff) and quick-frozen in liquid nitrogen for storage. Samples (2 μl) of laccase in 2× loading buffer [75 mM sodium dodecyl sulfate (SDS), 20% (vol/vol) glycerol, 40 μM bromophenol blue, 125 mM tris(hydroxymethyl)aminomethane (Tris), and 5% (vol/vol) β-mercaptoethanol adjusted to pH 6.8 with HCl] were electrophoresed on a 7.5% (wt/vol) SDS-polyacrylamide gel by using a Bio-Rad Fast Gel System. Samples were visualized after electrophoresis by activity staining with DMOP or protein staining with silver stain (2) or Pierce (Rockford, Ill.) Gelcode Blue Stain Reagent.
Laccase characterization.
To detect monomer subunits, 4-μl samples of pure laccase were electrophoresed on a 7.5% (wt/vol) SDS-polyacrylamide gel by using a Bio-Rad Fast Gel System. The first sample was loaded with pH 6.8 loading buffer. The second sample was incubated in pH 8.8 loading buffer for 2 min at 23°C before loading. The third sample was incubated in pH 8.8 loading buffer for 10 min at 23°C before loading. The gel was silver stained for protein detection after electrophoresis.
Pure laccase (∼20 μg) was denatured at 100°C in a solution of 0.5% (wt/vol) SDS and 1% (vol/vol) β-mercaptoethanol. Denatured subunits were deglycosylated according to the manufacturer’s instructions with 2,000 U of New England Biolabs (Beverly, Mass.) PNGase F added to the denatured protein solution and incubated at 37°C for 1 h. The laccase monomer was detected by electrophoresis with a 7.5% (wt/vol) SDS-polyacrylamide gel.
The isoelectric point was determined by isoelectric focusing using a Bio-Rad Fast Gel System. A pH 3 to 9 wide-range isoelectric focusing gel with Bio-Rad wide-range ampholytes (pH 3 to 9) as standards and a low-range (pH 4.5 to 6) isoelectric focusing gel with Bio-Rad low-range ampholytes (pH 5.3 to 6.4) as standards were stained with Pierce Gelcode Blue Stain Reagent.
Kinetic measurements were made at 23°C, with initial velocity measurements performed in 3-ml glass cuvettes with 1-cm path lengths. Reactions were initiated by the addition of laccase, and initial rates were obtained from the linear portion of the progress curve. The velocities of laccase-catalyzed reactions were measured at 468 nm for DMOP, 450 nm for catechol, 450 nm for pyrogallol, 436 nm for guaiacol, and 275 nm for l-tyrosine. Kinetic data were fitted to the appropriate equation with the programs of Cleland (6) to obtain the desired kinetic parameters.
Inhibitor studies were performed by using the DMOP assay with separate pH 4.5 citrate-phosphate buffer solutions containing either 0.5 mM sodium azide, 0.5 mM EDTA, 0.5 mM potassium cyanide, 0.5 mM hydrogen peroxide, or 200 U of catalase/ml (all from Sigma, St. Louis, Mo.).
The pH rate profile for laccase was determined by using 2 mM DMOP in 15 mM citrate-phosphate buffer (buffering range from pH 3 to 8). The activity was monitored spectrophotometrically at 486 nm.
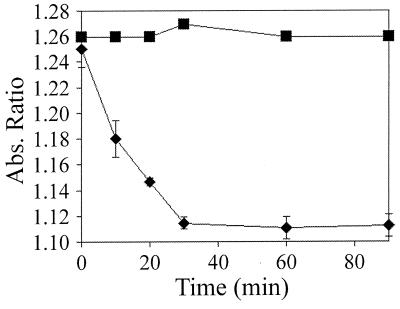
Lignin degradation potential was estimated by diluting 20 μl of a 0.2% (wt/vol) poly B-411 dye solution (in H2O) to 4 ml with pH 4.5 citrate-phosphate buffer. Laccase (400 U) was added, and decolorization was assayed after 0, 10, 20, 30, 60, and 90 min at 23°C. Decolorization was monitored by diluting 0.5 ml of the dye solutions to 2 ml with a 10 mM sodium azide solution in 10 mM pH 4.5 citrate-phosphate buffer. The ratio of absorbance (A593/A483) was measured for the azide-dye solution. Decolorization assays were performed in triplicate, and absorbances were compared with that of a control without added laccase.
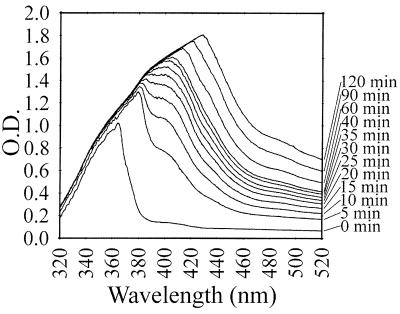
Polymerization of 1,8-DHN was measured by the addition of 15 U of laccase to a 2-ml solution of 1 mM DHN in 5 mM pH 4.5 citrate-phosphate buffer in 50% ethanol. The polymerization solution was incubated at 23°C and monitored spectrophotometrically by scanning the solution from 320 to 520 nm at 0 to 120 min. An identical control solution without the enzyme that auto-oxidized for 16 h was also scanned. Partially solubilized polymer (in 50% ethanol) and DHN monomer allowed to auto-oxidize overnight were dialyzed with 60,000-MW-cutoff dialysis tubing to determine if the oxidized products were larger than 60 kDa.
RESULTS
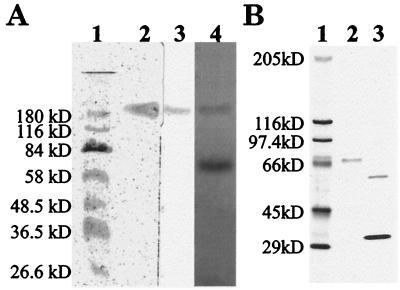
We concentrated laccase from culture filtrates of G. graminis var. tritici and purified the enzyme by preparative gel electrophoresis (Table 1). We did not detect laccase in minimal medium without copper, and only low levels of laccase production were detected in the absence of xylidine (data not shown). We used pH 6.6 buffer in the Prep Cell because higher pH running buffers irreversibly denatured the protein. We purified laccase, as identified by activity staining (Fig. 1A, lane 3), to homogeneity, as visualized by a single, silver-stained protein band on acrylamide gels (Fig. 1A, lane 2). Because protein assay methods, such as the Bradford method (3), produced inconsistent results, we estimated protein concentrations spectrophotometrically.
TABLE 1.
Purification of secreted laccase
| Purification step | Vol (ml) | Total activity (U) | Total amt of protein (mg) | Sp act (U/mg) | Yield (%) | Fold purification |
|---|---|---|---|---|---|---|
| 1. Culture filtrate | 1,000 | 100,000 | 2,900 | 34 | 100 | |
| 2. Stirred Cell | 50 | 76,000 | 1,900 | 40 | 76 | 1.2 |
| 3. Ultrafree 15 | 1 | 63,000 | 1,100 | 57 | 63 | 1.7 |
| 4. Prep Cell | 8 | 4,600 | 1.1 | 4,200 | 4.6 | 120 |
FIG. 1.
Determination of molecular masses of purified G. graminis var. tritici laccase and laccase subunits. (A) Lane 1, molecular size standards (silver stain); lane 2, native laccase (silver stain); lane 3, native laccase (activity stain with DMOP); lane 4, native and denatured laccase (silver stain). (B) Lane 1, molecular size standards (silver stain); lane 2, denatured laccase (silver stain); lane 3, denatured, deglycosylated laccase (60 kDa) and PNGase F deglycosylase (36 kDa; silver stain).
The exact molecular mass of the active, glycosylated laccase complex is unknown, as glycosylation slows protein migration in polyacrylamide gels in an unpredictable manner (15) and the protein did not denature on SDS-polyacrylamide gels (Fig. 1A, lane 3), making the protein’s migration partially dependent on the enzyme shape (27). However, purified undenatured laccase had an apparent molecular mass of approximately 190 kDa (Fig. 1A, lane 2). During long-term storage, approximately 2 months at 4°C, the enzyme degraded to a band at 70 kDa that lacked activity (Fig. 1A, lane 4). The process of subunit dissociation accelerated at pH values of 7 and higher (data not shown). After deglycosylation, the molecular mass of the protein subunits was 60 kDa (Fig. 1B, lane 3). Based on the observed molecular mass of the glycosylated monomer (70 kDa), the estimated molecular mass of a glycosylated homotrimer would be ∼210 kDa and that of a homodimer would be ∼140 kDa. Since the active complex migrated at ∼190 kDa, the active complex was likely a homodimer (15, 28).
The wide substrate specificity of laccase sometimes makes it difficult to distinguish from tyrosinase (EC 1.14.18.1) and peroxidase (EC 1.11.1.7). The enzyme oxidized common laccase substrates, such as DMOP (Km = 2.6 × 10−5 ± 7 × 10−6 M), catechol (Km = 2.5 × 10−4 ± 1 × 10−5 M), pyrogallol (Km = 3.1 × 10−4 ± 4 × 10−5 M), and guaiacol (Km = 5.1 × 10−4 ± 2 × 10−5 M). The enzyme did not oxidize tyrosine and was reversibly inhibited by hydrogen peroxide but not by catalase, and thus it was not a tyrosinase or peroxidase by the classical definitions (16, 22).
Irreversible inhibition of the enzyme by metal chelators (sodium azide, potassium cyanide, and EDTA) suggested that there was at least one metal center in the enzyme required for activity. Laccases commonly contain 4 Cu atoms in three different types of copper binding sites (a type 1, a type 2, and a type 3 binuclear site). Spectral studies did not reveal the characteristic peak near 600 nm expected for the type 1 copper site found in most laccases (data not shown). However, inductively coupled plasma spectroscopy conducted by Little Bear Laboratories (Denver, Colo.) detected copper in the purified G. graminis laccase enzyme complex. Recently, laccases that lack the type 1 copper site were reported (14). However, sequencing of five cloned putative laccase genes of G. graminis demonstrated that the consensus sequences for all three copper sites typical of laccases are encoded in all five genes (13). Therefore, the absence of a peak in the blue spectral region may be due to a low extinction coefficient for the type 1 copper site in this protein.
We determined by isoelectric focusing that the pI of the protein was 5.6 (data not shown), which was slightly higher than the pH at which the maximal activity for DMOP occurred (pH 4.5, as determined by the pH rate profile). Low-range isoelectric focusing also indicated that there were no laccase isoforms with similar molecular weights but slightly different pI’s (data not shown).
The ability to decolorize poly B-411 dye by ring-opening oxidation is utilized to predict the lignocellulose degradation abilities of fungi (4). Although a direct comparison to previously tested lignin degraders cannot be made due to the difference in reaction conditions, the laccase described here did oxidize poly B-411 (Fig. 2), an activity that suggested a possible role in lignin degradation (4). The enzyme also polymerized DHN into a high-molecular-mass melanin (Fig. 3). No polymerization occurred in control reactions without added laccase; the absorbance maximum remained at 360 nm with an increase of less than 0.1 optical density unit (data not shown). While the melanin polymer was not soluble in many solvents tested, it did remain soluble in the initial 50% ethanol solution for a few hours after the polymerization reaction was initiated. This partially solubilized melanin did not dialyze through 60,000-MW-cutoff dialysis tubing, but the DHN monomer, allowed to auto-oxidize, did. Thus, the polymer synthesized by laccase had a molecular mass greater than 60 kDa.
FIG. 2.
Decolorization of poly B-411 dye by laccase. Decolorization was monitored at different times by observing the absorbance ratios at 593 and 483 nm. Squares, control reaction (no laccase added); diamonds, reactions where laccase was added. Results shown are averages.
FIG. 3.
Polymerization of 1,8-DHN by laccase, monitored by scanning changes in absorbance from 320 to 520 nm.
DISCUSSION
The pathway for DHN melanin biosynthesis in fungi is almost completely characterized (1). However, the enzyme that catalyzes the final oxidation step in the pathway, the conversion of DHN to polymerized melanin, is unknown at present (1). The laccase characterized in this paper is the first purified laccase shown to catalyze the polymerization of DHN and may polymerize DHN in vivo.
It is not known if this laccase is secreted when G. graminis var. tritici infects wheat; however, reverse transcription-PCR has shown that the mRNA for this laccase is present in infected rice and wheat plants (13). Wheat plants are known to contain enough copper to allow the laccase to be active if it is expressed (20). Melanin synthesis catalyzed by laccase is required for the pathogenicity of Cryptococcus neoformans; melanin protects this fungal pathogen from animal host oxidative immune responses (18, 26). Similarly, G. graminis var. tritici laccase may function to confer protection from plant host oxidative defense responses.
In conclusion, these studies provide evidence that the extracellular laccase produced by G. graminis var. tritici is a glycosylated homodimer that may be involved in lignin degradation and/or melanin synthesis. Future studies will focus on environmental factors that affect laccase expression and the role(s) of laccase in melanization, delignification, and/or protection from antimicrobial compounds produced by the infected plant host.
ACKNOWLEDGMENTS
The work presented in this paper was funded by the U.S. Army Research Office (grant DAAH04-96-1-0194).
We thank Jeffrey Dean and Michele McGuirl for valuable discussions and help with equipment, and we thank L. J. Cookson for the gift of poly B-411.
REFERENCES
- 1.Bell A, Wheeler M. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986;24:411–451. [Google Scholar]
- 2.Blum H, Beier H, Gross H. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chet I, Trojanowski J, Huttermann A. Decolourisation of the dye poly B-411 and its correlation with lignin degradation by fungi. Microbios Lett. 1985;29:37–43. [Google Scholar]
- 5.Claus H, Filip Z. The evidence of a laccase-like enzyme activity in a Bacillus sphaericus strain. Microbiol Res. 1997;152:209–216. [Google Scholar]
- 6.Cleland W. Kinetics. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 7.Fahraeus G, Reinhammer B. Large scale production and purification of laccase from cultures of the fungus Polyporus versicolor and some properties of laccase. Acta Chem Scand. 1967;21:2367–2378. doi: 10.3891/acta.chem.scand.21-2367. [DOI] [PubMed] [Google Scholar]
- 8.Hatakka A. Lignin-modifying enzymes from select white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1998;13:125–135. [Google Scholar]
- 9.Huber D, McCay-Buis T. A multiple component analysis of the take-all disease of cereals. Plant Dis. 1993;77:437–447. [Google Scholar]
- 10.Kelly C, Osbourn A, Caten C. The genetics of Gaeumannomyces graminis with particular reference to pigment production and pathogenicity. Fungal Genet Newsl. 1997;44A:108. [Google Scholar]
- 11.Leonowicz A, Szklarz G, Wojtas-Wasilewska M. The effect of fungal laccase on fractionated lignosylphonates (Peritan Na) Phytochemistry. 1985;24:393–396. [Google Scholar]
- 12.Leonowicz A, Trojanowski J, Barbara O. Basidiomycetes: apparent activity of the inducible and constitutive forms of laccase with phenolic substrates. Acta Biochim Pol. 1979;25:369–378. [PubMed] [Google Scholar]
- 13.Litvintseva, A., and J. Henson. 1999. Personal communication.
- 14.Palmier G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G. A novel white laccase from Pleurotus ostreatus. J Biol Chem. 1997;272:31301–31307. doi: 10.1074/jbc.272.50.31301. [DOI] [PubMed] [Google Scholar]
- 15.Perry C, Matcham S, Wood D, Thurston C. The structure of laccase protein and its synthesis by the commercial mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:171–178. doi: 10.1099/00221287-139-1-171. [DOI] [PubMed] [Google Scholar]
- 16.Robb D. Tyrosinase. In: Lontie R, editor. Copper proteins and copper enzymes. III. Boca Raton, Fla: CRC Press; 1992. pp. 207–240. [Google Scholar]
- 17.Robyt J, White B. Biochemical techniques: theory and practice. Prospect Heights, Ill: Waveland Press, Inc.; 1987. pp. 232–234. [Google Scholar]
- 18.Salas S, Bennett J, Kwon-Chung K, Perfect J, Williamson P. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. p. A1. [Google Scholar]
- 20.Soon Y, Clayton G, Clarke P. Content and uptake of phosphorus and copper by spring wheat: effect of environment, genotype, and management. J Plant Nutr. 1997;20:925–937. [Google Scholar]
- 21.Tanaka C, Tajima S, Furusawa I, Tsuda M. The Prg1 mutant of Cochliobolus heterostrophus lacks a ρ-diphenol oxidase involved in naphthalenediol melanin synthesis. Mycol Res. 1992;96:959–964. [Google Scholar]
- 22.Tijssen P. Practice and theory of enzyme immunoassays. In: Burden R, VanKnippenberg P, editors. Laboratory techniques in biochemistry and molecular biology. Amsterdam, The Netherlands: Elsevier; 1985. pp. 173–189. [Google Scholar]
- 23.VanEtten H, Matthews D, Matthews P. Phytoalexin detoxification: importance for pathogenicity and practical implications. Annu Rev Phytopathol. 1989;27:143–164. doi: 10.1146/annurev.py.27.090189.001043. [DOI] [PubMed] [Google Scholar]
- 24.VanEtten H, Sandrock R, Wasmann C, Soby S, McCluskey K, Wang P. Detoxification of phytoanticipins and phytoalexins by phytopathogenic fungi. Can J Bot. 1995;73:S518–S525. [Google Scholar]
- 25.Walker J. Taxonomy of take-all fungi and related genera and species. In: Asher M, Shipton P, editors. Biology and control of take-all. London, United Kingdom: Academic Press Inc.; 1981. pp. 15–74. [Google Scholar]
- 26.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 28.Wood D. Production, purification and properties of extracellular laccase of Agaricus bisporus. J Gen Microbiol. 1980;117:327–338. [Google Scholar]