Abstract
Tannins in forages complex with protein and reduce the availability of nitrogen to ruminants. Ruminal bacteria that ferment protein or peptides in the presence of tannins may benefit digestion of these diets. Bacteria from the rumina of sheep and goats fed Calliandra calothyrsus (3.6% N and 6% condensed tannin) were isolated on proteinaceous agar medium overlaid with either condensed (calliandra tannin) or hydrolyzable (tannic acid) tannin. Fifteen genotypes were identified, based on 16S ribosomal DNA-restriction fragment length polymorphism analysis, and all were proteolytic and fermented peptides to ammonia. Ten of the isolates grew to high optical density (OD) on carbohydrates (glucose, cellobiose, xylose, xylan, starch, and maltose), while the other isolates did not utilize or had low growth on these substrates. In pure culture, representative isolates were unable to ferment protein that was present in calliandra or had been complexed with tannin. One isolate, Lp1284, had high protease activity (80 U), a high specific growth rate (0.28), and a high rate of ammonia production (734 nmol/min/ml/OD unit) on Casamino Acids and Trypticase Peptone. Phylogenetic analysis of the 16S ribosomal DNA sequence showed that Lp1284 was related (97.6%) to Clostridium botulinum NCTC 7273. Purified plant protein and casein also supported growth of Lp1284 and were fermented to ammonia. This is the first report of a proteolytic, ammonia-hyperproducing bacterium from the rumen. In conclusion, a diverse group of proteolytic and peptidolytic bacteria were present in the rumen, but the isolates could not digest protein that was complexed with condensed tannin.
Ruminant production in dry tropical regions is commonly limited by adequate protein supply to the rumen. Supplementary protein can be made available via shrub and tree legumes, such as Calliandra calothyrsus, that are high in protein, produce large amounts of leaf material, and can be readily introduced into tropical environments (1). However, these plants often contain condensed tannins (polyphenolics) which complex with protein, thus reducing nitrogen availability to rumen microorganisms. Polyphenolics also inhibit growth of predominant rumen bacteria (Fibrobacter succinogenes, Butyrivibrio fibrisolvens, Ruminobacter amylophilus, and Streptococcus bovis) (4, 22), but tolerance to tannins has been demonstrated in some rumen microorganisms (35). Strains of bacteria capable of degrading protein may therefore proliferate in response to tannin-rich diets such as calliandra. We examined whether proteolytic bacteria, which are present as significant populations in the rumina of animals fed calliandra, are able to ferment amino acids in the presence of tannins or hydrolyze protein that is complexed with tannin.
Nutrient medium containing precipitated tannin-protein complexes has been used previously to isolate enteric bacteria capable of degrading these complexes (37). Brooker et al. (6) used this technique to isolate a ruminal bacterium, Streptococcus caprinus (gallolyticus) (42), which produced clearing zones in this medium and was tolerant of tannins. Streptococcus gallolyticus appears to be widely distributed to ruminants fed tannin-rich diets, but their ecological role in digestion of these forages is yet to be defined (32). In this study we used similar media (37) in an attempt to identify other bacteria that may be beneficial in the digestion of protein in tanniniferous diets.
MATERIALS AND METHODS
The anaerobic techniques of Hungate (19) as modified by Bryant (7) were used for the growth of organisms and preparation of media. The media were gassed with CO2, and 10-ml aliquots were dispensed into 25-ml Balch tubes (18 by 250 mm), which were stoppered and autoclaved for 15 min at 100 kPa. B vitamins (28) were added to each tube of medium just prior to inoculation, and incubations were at 39°C.
Isolation procedures.
Rumen digesta (100 g) was taken from four sheep and four goats (2-year-old male castrates, rumen fistulated) held in metabolism crates and fed a 100% fresh-calliandra diet (3.6% N) ad libitum. Calliandra fed to animals included leaf and stem material from young regrowth material, which was cut from trees and offered to the animals within an hour of harvest. The animals were sampled 18 h after feed was offered, and digesta was transferred immediately to an anaerobic chamber containing a gas phase of CO2-H2 (95:5). A 5-g subsample was diluted (1:10) with cold anaerobic diluent (29), homogenized for 1 min (Bamix, Mettlen, Switzerland), and serially diluted in diluent to a 10−9 dilution. Tannin overlay agar plates were prepared in an anaerobic chamber as follows (i) a 2% calliandra condensed tannin extract or tannic acid (Ajax Chemicals, Sydney, Australia) which was filter sterilized (22-μm pore size) was poured onto brain heart infusion (BHI) medium or rumen habitat-simulating (RHS) medium (30) agar plates (Table 1), (ii) the tannin solution was allowed to precipitate with protein on the plate surface for 20 min, (iii) the tannin solution was then aspirated off, and (iv) the plate was rinsed with diluent (37). Droplets (20 μl) were pipetted onto dried plates in an anaerobic chamber from the 10−5, 10−6, and 10−7 dilutions (29). More colonies grew on BHI medium, and therefore isolations were made only from these plates. Individual colonies that produced clearing zones or had distinct morphologies but did not produce zones were picked and inoculated into liquid BHI medium without tannin and restreaked onto BHI agar plates. Cultures were considered to be pure after three successive isolations from agar plates showed a single colony and cell morphological types. Cells were treated with Gram stain and examined by light microscopy. The number of proteolytic bacteria in sheep was estimated on RHS medium (30), and proteolytic colonies were counted as those with zones of hydrolysis after the plates were flooded with 1 M HCl.
TABLE 1.
Compositions of media used to select bacteria and for growth and metabolism experiments under various conditions
| Ingredient | Amt (ml or g/100 ml of indicated medium)a
|
|||
|---|---|---|---|---|
| BHI medium | RHS medium | PM | PMC | |
| Clarified rumen fluid | 30 | 15 | 15 | |
| Mineral solution Ib | 15 | 15 | ||
| Mineral solution IIb | 15 | 15 | ||
| Pfennings’ trace elements | 0.1 | 0.1 | ||
| Macro mineral solutionc | 20 | 20 | ||
| Buffer solutionc | 20 | 20 | ||
| Micro mineral solutionc | 0.01 | 0.01 | ||
| Volatile fatty acidsb | 1.0 | 1.0 | 1.0 | |
| Sodium hydrogen carbonate | 0.6 | 0.6 | 0.78 | 0.78 |
| Hemin | 0.0001 | 0.0001 | 0.0001 | |
| dl-Lactic acid | 0.2 | |||
| BHI (Difco) | 3.7 | |||
| Yeast extract | 0.5 | 0.25 | 0.25 | |
| Casamino Acids | 0.5 | 0.5 | ||
| Trypticase | 0.05 | 0.5 | 0.5 | |
| Cellobiose | 0.05 | 0.2 | ||
| Xylan (birchwood) | 0.05 | 0.1 | ||
| Starch | 0.05 | 0.1 | ||
| Maltose | 0.05 | 0.1 | ||
| Casein (soluble) | 1.0 | |||
| Xylose | 0.05 | 0.1 | ||
| Glucose | 0.25 | |||
l-Cysteine hydrochloride, resazurin, and agar for culture plates were added to media at 0.1, 0.001, and 2.0 g/100 ml, respectively, and deionized water was used to make up to volume.
Final concentration and composition were identical to those of the solution described by Caldwell and Bryant (8).
Final concentration and composition were identical to those of the solution described by Menke et al. (31).
Growth and nitrogen digestion studies in the absence of tannin.
Growth, ammonia production, and protease activity were determined for individual isolates grown on (i) peptide medium (PM), which contained primarily Casamino Acids (Difco) and Trypticase Peptone (Becton Dickinson) as substrates for growth, and (ii) PM with carbohydrates (PMC), which contained cellobiose, birchwood xylan, xylose, starch, and maltose (Table 1). Bacteria that showed little growth response to carbohydrates were grown on PMC containing 0.5% glucose to simulate rumen conditions on a concentrate diet. Isolate Lp1284 was also grown in PM that lacked yeast extract but contained 1.5% Trypticase Peptone and 1.5% Casamino Acids, or the peptides were replaced with either 1.5% casein (BDH Laboratory Supplies) or 3% fraction 1 leaf protein so that specific rates of ammonia production from peptides and growth on protein could be compared with published data (3). Fraction 1 leaf protein, which is ribulose-1,5-bisphosphate carboxylase-oxygenase (Rubisco), was isolated and partially purified from the leguminous plant lucerne (Medicago sativa) by gel filtration (23, 24). PM which lacked both yeast extract and rumen fluid was used to definitively confirm that Lp1284 could grow on peptides without carbohydrate.
Growth was measured as change in optical density (OD) at 600 nm (Spectronic spectrophotometer; Milton Roy Co.). Aliquots of culture fluid were taken at regular intervals during growth for measurements of ammonia production. The indophenol method for the determination of ammonia as described by Chaney and Marbach (9) was used to estimate the rate and amount of ammonia production in cultures.
Protease activity was measured during exponential growth in PM and PMC. Cells for protease assays were separated from culture fluid by centrifugation (7,000 × g for 20 min at 4°C), washed, and suspended in 0.1 M Bis-Tris [bis(2-hydroxyethyl)imino-tris(hydroxymethyl)methane (pH 7)] and recentrifuged as described above. The cell pellet was resuspended in 0.01 of the original volume and disrupted twice by ultrasonication with a Sanophon ultrasonic disintegrator (Ultrasonic Industries Pty. Ltd., Sydney, Australia) at 60 W for 10 min at a time. Centrifuged culture fluid was also assayed for protease activity.
Proteolytic activities in cell-associated and extracellular fractions of cultures were determined spectrophotometrically with azocasein as the substrate (5). Control experiments were performed by incubating enzyme samples and the azocasein substrate separately and by combining these solutions at the time of acid addition as described by Cotta and Hespell (13). One proteolytic enzyme unit equalled 1 μg of azocasein digested per h at 39°C. Extracellular proteolytic activity was expressed per milliliter of culture fluid per the OD of the culture prior to centrifugation. Cell-associated proteolytic activity per milliliter of culture fluid was also expressed per the OD of the culture. Assays were performed on centrifuged cell pellets that had been resuspended in buffer to the volume of the original culture. Enzyme activities were not expressed per milligram of microbial protein since the media contained large concentrations of the substrate nitrogen, some of which appeared to precipitate and could interfere with analysis of pelleted cell protein. The proteolytic ruminal bacterium Prevotella ruminicola B14 was used for comparison of protease activities, with azocasein as the substrate.
Volatile fatty acids, ethanol, formate, lactate, succinate, and ethanol in the PMC culture fluid were analyzed by high-performance liquid chromatography with a Waters System equipped with an Aminex HPX-87 cation-exchange column (300 by 7.8 mm) for organic acids and a microguard column (Bio-Rad Laboratories, Richmond, Calif.) with a column heater (Waters model 1122/WTC-120). Organic acids and ethanol were eluted with a mobile phase of 2.5% acetonitrile in 0.2% (vol/vol) phosphoric acid at a flow rate of 0.7 ml/min and a column temperature of 60°C, with UV detection at 210 nm. All assays were performed at least in duplicate.
Protein digestion in the presence of tannins.
Bacterial isolates were also screened for the ability to degrade protein that was complexed with tannin. These isolates were grown with medium in which substrate protein was included only as a tannin-protein complex, and evolution of ammonia was used as an indicator of fermentation of protein as described previously.
Tannin-protein complexes (50 mg) were added to PMC (5 ml), but Casamino Acids, Trypticase peptone, and yeast extract were not included so that the only protein available was in a complexed form. The complexes were weighed into sterile Balch tubes, and autoclaved medium was added before each tube was closed with a sterile stopper. Tannin-protein complexes were mainly insoluble and could not be sterilized by heat, UV light, or ethylene oxide gas, because each of these processes causes changes to the tannin molecule, or by filtration. Duplicate cultures were each inoculated with 0.1 ml of individual isolates which had been grown to late log phase on PMC. Uninoculated controls were routinely included in all assays to account for any microbial activity introduced by the tannin-protein complexes, all of which were not sterilized.
The ability to ferment protein in calliandra was also examined with selected isolates alone or in combination with a fiber-degrading microbe, Ruminococcus flavefaciens AR67, and results were compared with results with a mixed rumen fluid inoculum. Cocultures with AR67 were performed because this strain degrades fiber in calliandra, and it is expected that the availability of plant protein is enhanced by the activities of polysaccharide-hydrolyzing bacteria (16). Finely milled lyophilized calliandra (50 mg, 3.64% N) was added to the basal medium (10 ml) used for fermentation experiments of tannin-protein complexes, but NH4Cl was also included at a final concentration of 3 mM as a nitrogen source for the AR67. Polyethylene glycol 4000 (PEG 4000; 1 mg/10 mg of calliandra) was also included in some tubes to counteract the effect of tannin on protein (see reference 23). PEG 4000 at this concentration does not affect the rate of ammonia production in cultures where tannin is not present (30a). Ammonia production was monitored during growth for 72 to 96 h. Procedures for inoculation of cultures were essentially the same as those described for experiments with tannin-protein complexes, except triplicate assays and uninoculated control experiments were performed. The rumen fluid inoculum (0.1 ml) was obtained from a steer fed a diet comprising (70%) rhodes grass (Chloris gayana) and (30%) lucerne. Rumen digesta was strained through muslin cloth and incubated anaerobically at 39°C for 30 min so that the larger particulate matter would float to the surface, and aliquots for inoculation were taken from the fluid phase beneath this layer.
Tannin purification and complexes.
Total condensed tannin in calliandra was 6% as determined by the butanol-HCl method (46). Condensed tannin in calliandra used in these studies was obtained by extraction with 7:3 (vol/vol) acetone-water from fresh calliandra leaves that had been lyophilized and ground through a 1-mm-pore-size screen (46). The acetone was removed by rotary evaporation, and the aqueous solution was washed with diethyl ether to remove nontanniniferous material before chromatographic purification on Sephadex LH-20 as described by Terrill et al. (47).
Tannin-protein complexes were made as follows: calliandra tannin which had been purified as described previously was dissolved (3% [wt/vol]) in 0.2 M sodium acetate buffer (pH 5) and then added slowly to a solution of BHI medium (2% [wt/vol] BHI, 0.2 M sodium acetate buffer [pH 5]), allowing tannin-protein complexes to form. The complexes were precipitated by centrifugation (2,000 × g, 10 min), washed in the same buffer, and then centrifuged again and washed in culture medium to remove any soluble complexes before a final centrifugation and lyophilization of the pellet.
Bacterial genotyping and extraction of genomic DNA.
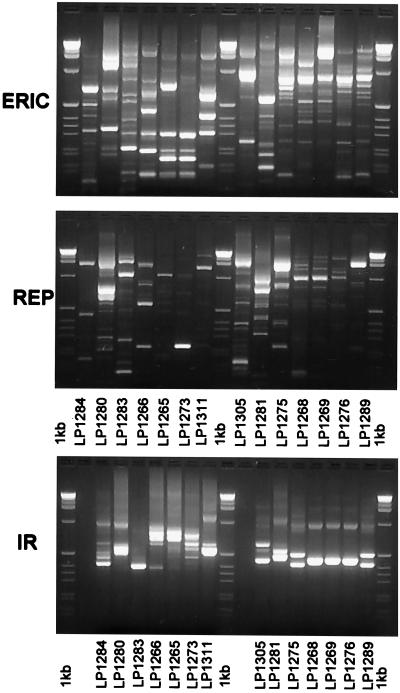
The genotypic diversity and phylogeny of the isolates were determined with restriction fragment length polymorphisms (RFLP) of 16S ribosomal DNA (rDNA) amplified by PCR which had been digested with restriction endonucleases (33). In addition, repetitive extragenic palindromic sequences (REP), enterobacterial repetitive intergenic consensus sequences (ERIC), and amplified rDNA intergenic spacer regions between 16 and 23S rDNAs (IR) were also used for genotyping (15, 21, 49). The REP, ERIC, and IR techniques are able to differentiate bacteria at the subspecies level and can be used to rapidly confirm the identity and purity of strains.
Bacterial isolates were grown on PMC, and cells were centrifuged at high speed (10,000 × g, 10 min). The pellet was resuspended and washed in 500 μl of TE (10 mM Tris-Cl, 1 mM Na2-EDTA), and DNA was extracted according to the method of Stahl et al. (44) as modified by Krause et al. (27). DNA concentration was measured at 260 nm and adjusted to a final concentration of 10 ng/μl.
ERIC, REP, and IR PCR protocols.
Each 50-μl PCR mixture included a 1/100 dilution of bacterial cells, 5 μl of 10× reaction buffer (Bresatec, Adelaide, Australia), 2.0 mM MgCl2 (ERIC and REP) or 3 mM MgCl2 (IR), 0.2 mM each deoxynucleotide triphosphate, 10 pmol of each primer, and 2.5 U of Taq polymerase (Promega, Sydney, Australia). For ERIC- and REP-PCR, cycling conditions were denaturation for 1 min at 94°C, annealing for 1 min at 47°C (ERIC) or 40°C (REP), and a final extension at 72°C for 2 min. This cycle was repeated 30 times. PCR cycling conditions for the 16-to-23S spacer were denaturation at 94°C for 5 min for the first cycle only and, 1 min thereafter, annealing at 50°C for 1 min, extension at 72°C for 1.5 min for 30 cycles, and a final extension at 72°C for 7 min. ERIC and REP primers were as previously described (15). The 16-to-23S rDNA spacer was amplified with a conserved 16S forward primer (5′-AAG TCG TAA CAA GGT AG/AC CGT A-3′), and a conserved 23S reverse primer (5′-GGG TTT/G/C CCC CAT TCG G-3′).
16S rDNA RFLP analysis.
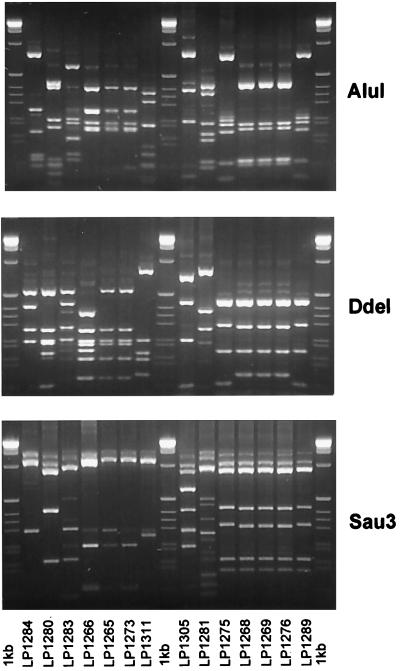
The 16S rDNA was amplified from a 1/100 dilution of overnight culture with universal primers (27f and 1492r). PCR mixtures contained (per 20 μl) 5 μl of 10× PCR buffer, 0.5 μl of MgCl2 (250 mM), 1 μl of deoxynucleoside triphosphates (10 mM), 10 pmol of each primer, 1 U of Taq polymerase (Promega), and 0.5 μl of a 1/100 dilution of culture. Cycling conditions were one cycle of 94°C for 5 min, 60°C for 1 min, and 72°C for 90 s, and then 31 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 90 s. The final PCR cycle was 94°C for 1 min, 60°C for 1 min, and 72°C for 8 min. Approximately 100 ng of the 16S rDNA PCR product was digested with tetrameric restriction enzymes (AluI, DdeI, and Sau3a) for at least 2 h according to the manufacturer’s instructions. Five microliters of the restricted product was run on a 1% (0.5× Tris-borate-EDTA) agarose gel.
Multivariate cluster analysis.
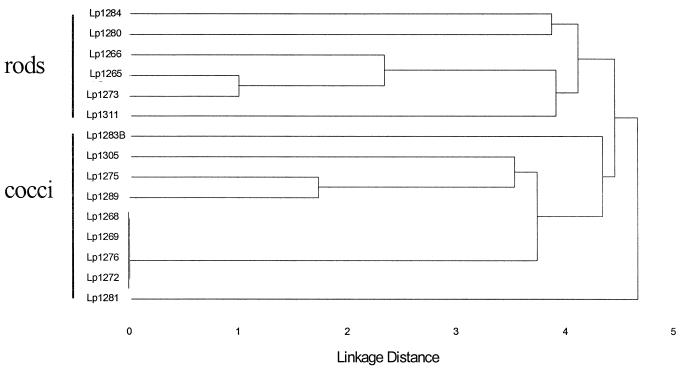
The isolates were grouped on similarity of patterns of DNA fragments in agarose gels by the 16S rDNA RFLP typing method described above. Individual isolates were scored visually for the presence or absence of DNA fragments (range, 70 to 1.4 kb) generated by this typing method. Cluster analysis of similarity matrices was performed by the unweighted-pair-group method with arithmetic averages (43).
Amplification and sequencing of 16S rDNA.
Bacterial biomass cultured in PMC for 2 days was centrifuged, washed once in sterile distilled water, and frozen at −20°C. Methods detailed in references 26 and 27 were employed for isolation and storage of DNA and PCR amplification of the 16S rDNA. Automated sequencing (27) was employed to obtain the nearly complete sequences (1,450 bp) of the 16S rDNAs from strains Lp1265, Lp1275, Lp1276, and Lp1284. Primers 27f and 1492r were employed for PCR, while primers 343r, 519r, 787r, 907r, 1100r, 1241r, 1385r, and 1492r were employed for sequencing.
Comparative sequence analysis.
Sequence data was aligned with CLUSTAL W (48), and the alignments were manually adjusted to take account of the conserved nature of helices. Phylogenetic analysis was by the distance methods of Jukes and Cantor (25), and tree topology was inferred by using the neighbor-joining algorithm (34) with Treecon. All sequence-based trees were analyzed by bootstrapping of 1,000 trees.
Statistical analysis.
Statistical analysis of the effects of carbohydrates on growth, ammonia production, and protease activity was by analysis of variance, with differences being determined by the method of least significant difference at the 5% level (P < 0.05).
RESULTS
Isolation, morphology, and genotype.
Proteolytic bacteria were present in the rumina of calliandra-fed sheep at 1.5 × 108 cells/g of digesta. Fifteen (data for Lp1272 are not shown) different proteolytic isolates were identified based on polymorphisms of IR, ERIC, and REP sequences (Fig. 1; Table 2). The bacteria were grouped on the basis of similarity according to results of cluster analysis of 16S rDNA RFLP patterns (Fig. 2 and 3). Several strains (Lp1268, Lp1269, Lp1272, Lp1276, Lp1275, and Lp1289) were closely related phylogenetically based on 16S rDNA RFLP patterns, and all produced lactate, formate, and acetate as end products of fermentation (Table 2). Strain Lp1284 produced both isobutyrate and isovalerate, while Lp1311 was the only isolate that formed succinate. Both strains Lp1265 and Lp1266 produced butyrate as a major end products. Two bacteria (Lp1265 and Lp1284) produced clearing zones on both tannic acid and calliandra tannin plates, while the other isolates produced zones with only one tannin type or no zones at all (Table 2).
FIG. 1.
DNA fragments amplified by primers specific for ERIC, REP, and 16-to-23S IR sequences for proteolytic isolates.
TABLE 2.
Characteristics of bacterial strains isolated on agar plates of BHI medium overlaid with tannic acid (hydrolyzable tannin) and calliandra tannin (condensed tannin)
| Level of carbohydrate fermentation | Bacterial strain | Origin | Gram stain reaction | Production of clearing zone on tannin overlay platesa
|
Production of indicated fermentation end product (amt [mM])b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA | CT | Lactate | Formate | Acetate | Propionate | Butyrate | Isovalerate | Valerate | ||||
| Low | Lp1284 | Goat | + | + | + | − | − | + | + (5.7) | − | + (8.9) | + (6.9) |
| Lp1281 | Sheep | + | − | + | − | − | + | − | + | + | + | |
| Lp1305 | Goat | − | − | − | − | + | + | − | + | + (1.8) | − | |
| Lp1280 | Sheep | − | − | NG | − | + (35.3) | + (12.7) | − | − | − | − | |
| Lp1283B | Goat | + | − | − | + (6.3) | − | − | + | + | − | + (2.9) | |
| High | Lp1268 | Goat | + | + | − | + (23.3) | + (27.6) | + (3.7) | + | + | − | − |
| Lp1269 | Goat | + | + | − | + (47.4) | + (8.7) | + | − | + | − | − | |
| Lp1275 | Sheep | + | − | + | + (23.6) | + (5.7) | + (7.4) | + | − | − | − | |
| Lp1276 | Sheep | + | − | − | + (39.1) | + | + (3.7) | − | + | − | − | |
| Lp1289 | Goat | + | − | NG | + (29.6) | + (10.0) | + | + (6.6) | − | − | − | |
| Lp1265 | Goat | − | + | + | + (11.2) | − | − | − | + (15.5) | − | − | |
| Lp1266 | Sheep | − | + | − | + | + | − | − | + (25.4) | − | − | |
| Lp1273 | Goat | − | + | NG | − | − | + | − | + | − | − | |
| Lp1272 | Goat | + | + | − | + (40.6) | + (8.7) | + | − | + | − | − | |
| Lp1311 | Sheep | + | − | − | + (3.6) | + (5.4) | − | − | − | − | − | |
TA, tannic acid; CT, calliandra tannin; +, clearing zones around colonies; −, no clearing zone; NG, no growth.
A + with no value in parentheses indicates that only a trace amount (<1 mM) of the end product was produced; a − indicates that no product was produced. Succinate (23.6 mM) and a trace amount of isobutyrate were produced by Lp1311 and Lp1284, respectively.
FIG. 2.
Restriction fragment length patterns of 16S rDNAs digested with AluI, DdeI, and Sau3 for proteolytic isolates.
FIG. 3.
Cluster analysis of restriction patterns of the amplified 16S rDNAs shown in Fig. 2 for proteolytic isolates.
Carbohydrate fermentation and deaminase and protease activities.
The isolates were placed into two groups (high and low fermentation of carbohydrate) based on a significantly higher (P < 0.05) growth response to inclusion of carbohydrates other than glucose in the medium (Table 3). Inclusion of glucose of PMC increased the maximum OD by only 0.2 to 0.4 for two of the bacteria with low levels of carbohydrate fermentation (Lp1280 and Lp1283B), which was less than the growth response to sugars in the group with high levels of carbohydrate fermentation. Deletion of yeast extract and rumen fluid from PM resulted in a decrease in growth of Lp1284 from 0.70 to 0.42 OD units.
TABLE 3.
Growth, ammonia production, and protease activities of bacteria grown on peptide medium with and without carbohydratea
| Level of carbohydrate fermentation | Bacteria | OD (600 nm)
|
Ammonia produced (mM)
|
Rate of ammonia production (μmol/h/OD unit)
|
Protease activity (U)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell associated
|
Extracellular
|
||||||||||
| With sugar | Without sugar | With sugar | Without sugar | With sugar | Without sugar | With sugar | Without sugar | With sugar | Without sugar | ||
| Low | Lp1284 | 0.73 | 0.78 | 16.30 | 24.41* | 933.0 | 1,316.0 | 5.7 | 1.9 | 37.3 | 80.1 |
| Lp1281 | 0.14 | 0.19 | 4.17 | 4.08 | 1,238.5 | 893.0 | 0.5 | 0.0 | 85.2 | 10.6* | |
| Lp1305 | 0.28 | 0.32 | 6.73 | 7.39 | 1,006.0 | 968.50 | 0.0 | 0.0 | 30.4 | 5.4 | |
| Lp1280 | 0.08 | 0.02 | 0.70 | 0.71 | 231.0 | 1,479.0 | 1.6 | 0.0 | 163.7 | 94.5* | |
| Lp1283B | 0.04 | 0.02 | 1.41 | 2.40 | 1,921.0 | 8,334.0* | 84.9 | 19.5* | 87.5 | 162.7** | |
| High | Lp1268 | 1.00 | 0.09* | 1.34 | 2.10 | 55.5 | 926.0 | 11.3 | 3.0 | 2.8 | 12.7 |
| Lp1269 | 1.13 | 0.11* | 1.00 | 2.53 | 36.5 | 987.0 | 84.1 | 0.0* | 3.0 | 11.8 | |
| Lp1275 | 0.97 | 0.30* | 3.95 | 5.80* | 227.5 | 1,096.0 | 0.8 | 0.9 | 0.2 | 2.4 | |
| Lp1276 | 1.06 | 0.15* | 3.14 | 8.47* | 123.5 | 3,178.0* | 4.9 | 1.6 | 2.8 | 7.9 | |
| Lp1289 | 0.97 | 0.20* | 3.23 | 3.67 | 139.5 | 748.5 | 1.4 | 0.0 | 1.5 | 6.0 | |
| Lp1265 | 1.14 | 0.07* | 0.28 | 4.73* | 10.3 | 3,912.5* | 4.9 | 3.4 | 1.9 | 14.8 | |
| Lp1266 | 1.01 | 0.04* | 0.75 | 2.32** | 30.6 | 2,645.0* | 11.9 | 0.0 | 4.1 | 39.3 | |
| Lp1273 | 1.10 | 0.06* | 0.63 | 2.55* | 23.8 | 2,271.5* | 8.9 | 3.8 | 3.6 | 18.4 | |
| Lp1272 | 1.05 | 0.06* | 1.10 | 2.37 | 43.5 | 1,732.5 | 7.8 | 0.0 | 4.9 | 26.7 | |
| Lp1311 | 0.57 | 0.07* | 1.05 | 4.60 | 104.8 | 3,877.0* | 25.9 | 4.7 | 0.2 | 10.7 | |
Values marked with single asterisks are significantly different (P < 0.05) from values in corresponding columns (values for medium with sugar). **, P < 0.1.
One proteolytic enzyme unit equalled 1 μg of azocasein digested per h per OD unit.
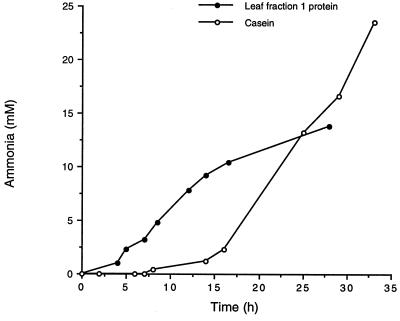
All isolates showed deaminase activity when they were grown on PM, but strain Lp1284 grew to the highest OD (P < 0.05) and produced the greatest amount of ammonia (Table 3) (P < 0.05). Specific growth rate, OD, linear rate of ammonia production, and total ammonia production for Lp1284 in PM that contained 1.5% Trypticase peptone and 1.5% Casamino Acids were 0.28, 1.45, 734 μmol/min/ml/OD unit and 62.3 mM, respectively. Strain Lp1284 also produced ammonia from growth on casein (OD, 0.65) and fraction 1 leaf protein (Fig. 4). Approximately 18.2 and 31.5% of N from Trypticase-Casamino Acids and casein, respectively, were fermented to ammonia N by Lp1284. Proteolysis of azocasein was detected for all isolates and varied from <10 to >160 U compared with 105 U of activity for P. ruminicola B14 (Table 3). Protease activity in bacteria with low carbohydrate fermentation was predominantly extracellular, whereas carbohydrate fermenters produced both cell-associated and extracellular activities. The protease activity of carbohydrate-fermenting bacteria (except for Lp1269) was not significantly (P < 0.05) affected by the presence or absence of carbohydrate in PM, although this activity tended to be lower in the absence of sugar in the bacteria that fermented carbohydrate at low levels (Table 3).
FIG. 4.
Ammonia production from growth of Lp1284 on casein (1.5%) and leaf fraction 1 protein (3%).
Protein digestion in the presence of tannins.
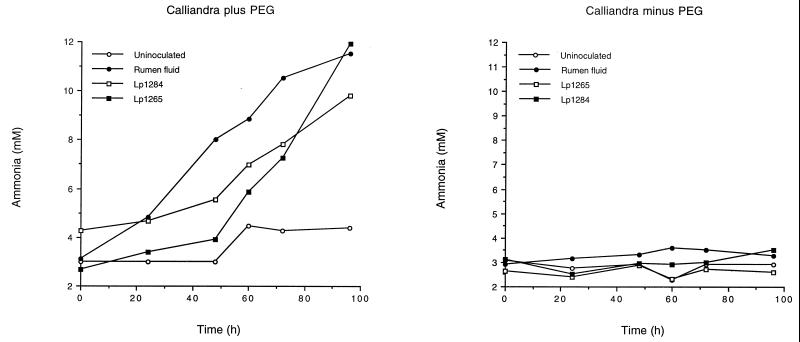
Initial screens showed that two isolates (Lp1284 and Lp1265) produce ammonia (>1 mM) from calliandra, and these isolates were thus compared with mixed rumen fluid. Both isolates and mixed rumen fluid fermented protein in calliandra to ammonia (5 to 10 mM) when PEG 4000 was included in the medium, but the cultures accumulated significantly less ammonia (1.5 mM) when PEG 4000 was absent (Fig. 5). The percentages of calliandra protein N fermented to ammonia N by Lp1284, Lp1265, and rumen fluid in the presence of PEG 4000 were 52.2, 71.4, and 64.8% respectively. Coculturing of either Lp1284 or Lp1265 with the fibrolytic strain R. flavefaciens AR67 did not significantly affect the rate or extent of protein fermentation (data not shown).
FIG. 5.
Ammonia production by Lp1284 and Lp1265 and mixed rumen microorganisms grown on calliandra with and without PEG 4000 (1 mg/10 mg of calliandra).
Ammonia production from growth on tannin-protein complexes (containing 22% protein) was not greater than that of the control cultures, which lacked the complexes (data not shown).
Nucleic acids and 16S rDNA comparisons.
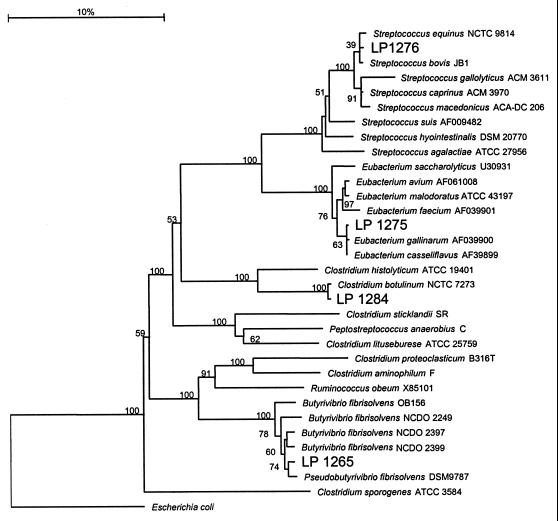
The GenBank accession numbers for the nucleotide sequences used are AF105402 (Lp1284) and AF105403 (Lp1265). Lp1284 was closely related to Clostridium botulinum NCTC 7273 (97.6%). The sequence of the closest relative to strain Lp1265 was Pseudobutyrivibrio ruminis DSM 9798 (96.3%). Strains Lp1275 (accession no. AF135452) and Lp1276 (accession no. AF135453) grouped with Eubacterium spp. and Streptococcus spp., respectively, and the closest relatives were Eubacterium gallinarium (accession no. AF03990) (98.3%) and Streptococcus bovis JB1 (99.1%). Phylogenetic trees based on 16S rDNA sequences of these bacteria and closely related organisms are shown in Fig. 6.
FIG. 6.
Phylogenetic tree of Lp1265, Lp1275, Lp1276, and Lp1284 and related organisms based on 16S rDNA sequences. Bootstrap values are given as percentages of 1,000 random trees. The scale bar represents 0.1 mutation per site and is expressed as a percentage.
DISCUSSION
This study demonstrates that proteolytic bacteria which ferment peptides to ammonia occur in relatively high numbers (108/g of digesta) in ruminants fed a highly tanniniferous diet and that the isolated organisms fall into two classes: those with high and low levels of carbohydrate fermentation. However, it cannot be definitively stated that the presence of these bacteria was due to the tannin-containing calliandra since animals not fed a tannin-enriched diet may also harbor these microorganisms. Therefore, the characterization of these proteolytic bacteria is discussed (i) within the broader context of rumen microbial ecology and (ii) in relation to their potential role in digesting tannin-rich forage.
Hungate (18) stated that he had “encountered” rumen bacteria “able to digest casein and requiring no carbohydrate,” but he did not isolate these bacteria. This is the first report of a proteolytic ruminal bacterium that grows rapidly on peptides and amino acids and does not require carbohydrates for growth. Previously, only obligate amino-acid-fermenting bacteria that were not proteolytic had been isolated (14). Strain Lp1284 is an ammonia hyperproducer, grows rapidly on peptides and amino acids, and is highly proteolytic. This organism also ferments peptides to branched-chain fatty acids and propionate, which has also been observed with the obligate amino-acid-fermenting rumen bacterium Clostridium sticklandii SR (11), which is not proteolytic. The rate and amount of ammonia production of Lp1284 grown on peptide medium were similar to those of the obligate amino-acid-fermenting rumen bacteria Peptostreptococcus sp. strain D1 and strain D4 (3). The high specific rates of ammonia production by Lp1284 are also comparable with those of Peptostreptococcus anaerobius (type C), C. sticklandii (type SR) and Clostridium aminophilum (type F) (10, 11, 26). Strain Lp1284 apparently converted approximately 18.3% of peptide N from Trypticase and Casamino Acids to ammonia and grew to an OD similar to that of Peptostreptococcus anaerobius (type C), which utilized 23 to 31% of the N from these nitrogen sources (10). A highly proteolytic clostridium (C. proteoclasticum) has also been isolated from the bovine rumen, but this organism grows on carbohydrate and does not produce ammonia (2). Most of these ammonia-hyperproducing bacteria fall within the genus Clostridium, as described by Collins and coworkers (12) (Fig. 6).
Phylogenetic analysis based on 16S rDNA sequence indicates that Lp1284 is related to C. botulinum group 1. This group of bacteria contain proteolytic C. botulinum types A, B, and F (20). They are also closely related to Clostridium sporogenes and fall within cluster 1 of the polysaccharolytic clostridia (40). Phylogenetic analysis and phenotypic characteristics demonstrate that strain Lp1265 belongs to cluster XIVa of the Clostridium subphylum (12, 50) and that it is closely related to several strains of B. fibrisolvens (17) but that Lp1268, Lp1269, Lp1272, and Lp1276 are Streptococcus spp.
This is the first report of an attempt to isolate rumen bacteria with an ability to digest protein in the presence of condensed tannins. Although the bacteria isolated were proteolytic, we were unable to demonstrate significant degradation of calliandra-protein complexes or fermentation of in situ complexed calliandra protein. However, when the interaction between tannin and protein in calliandra was counteracted with PEG 4000, then proteolytic bacteria were able to readily degrade protein. This result indicates that a substantial amount of calliandra protein is readily available for fermentation, provided that it is not totally complexed with tannin, which occurs in fresh plant material.
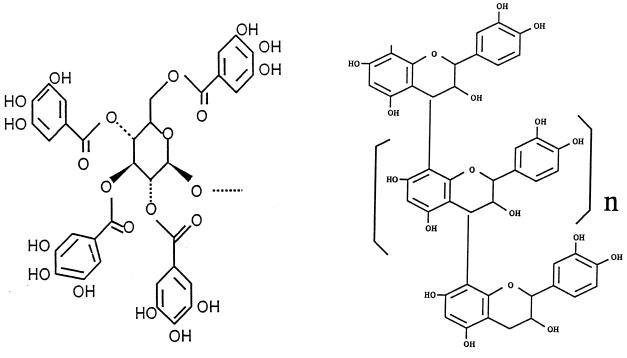
Anaerobic bacteria that degrade hydrolyzable tannins or hydrolyzable tannin-protein complexes have been isolated from the digesta of many nonruminant species, including koala (Phascolarctos cinereus), by using tannic acid as the model tannin in the selection medium (36–39). A reason why bacteria that degrade calliandra tannin-protein complex were not isolated is probably due to the difference in chemical structures of these two classes of tannins. Hydrolyzable tannins (e.g., tannic acid) are polymerized units of glucose esterified to gallic and hexahydroxydiphenic acid, whereas condensed tannins (e.g., calliandra tannin) are polymers of flavan-3-ols or flavon-3,4-diols (Fig. 7). Both tannins complex with protein by forming hydrogen bonds between the phenolic subunits of the polymer and aliphatic and aromatic side chains (carbonyl groups of peptides) of the protein. The most likely explanation for degradation of hydrolyzable tannin-protein complexes is that enzymes (tannin acylhydrolases and esterases) depolymerize the tannin polymer by cleaving the ester linkages between glucose and the phenolic subunits (41). A mechanism for the anaerobic degradation of the condensed tannin molecule has not yet been described. However, nonenzymatic cleavage of both types of tannin-protein may occur under acidic conditions in the rumen.
FIG. 7.
Diagram of the generic chemical structures of hydrolyzable (left panel) and condensed (right panel) tannins. n, repeating subunit of the polymer.
Further studies should be undertaken to determine whether some ruminal populations are better adapted to (tolerant of) tannin-containing forages and thus more efficient at digesting protein under those circumstances. Tolerance of the isolates to tannins was not measured, but the isolation procedures have previously yielded tolerant bacteria (6, 35). However, the predominate use of hydrolyzable tannin in these experiments rather than condensed tannin should be reevaluated, considering their relative levels of importance in the nutrition of herbivores. Condensed tannins are more widely distributed in plants than the hydrolyzable type and are thus regarded as a more significant nutritional problem (45). Hydrolyzable tannins are absent from nonvascular plants, and even in vascular plants, they are restricted to only dicotyledons and some classes of flowering plants in monocotyledons.
ACKNOWLEDGMENTS
This work was partly supported by the Australian Centre for International Agricultural Research.
We thank Christina Fraser for technical assistance.
REFERENCES
- 1.Ahn J H, Robertson B M, Elliott R, Gutteridge R C, Ford C W. Quality assessment of tropical browse legumes: tannin content and protein degradation. Anim Feed Sci Technol. 1989;27:147–156. [Google Scholar]
- 2.Attwood G T, Reilly K, Patel B K C. Clostridium proteoclasticum sp. nov., a novel proteolytic bacterium from the bovine rumen. Int J Syst Bacteriol. 1996;46:753–758. doi: 10.1099/00207713-46-3-753. [DOI] [PubMed] [Google Scholar]
- 3.Attwood G T, Klieve A V, Ouwerkerk D, Patel B K C. Ammonia-hyperproducing bacteria from New Zealand ruminants. Appl Environ Microbiol. 1998;64:1796–1804. doi: 10.1128/aem.64.5.1796-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae H D, McAllister T K, Yanke L J, Cheng K-J, Muir A. Effect of condensed tannins on endoglucanase activity and filter paper digestion by Fibrobacter succinogenes S85. Appl Environ Microbiol. 1993;59:2132–2138. doi: 10.1128/aem.59.7.2132-2138.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock F M, Forsberg C W, Buchanan-Smith J G. Proteolytic activity of rumen microorganisms and effects of proteinase inhibitors. Appl Environ Microbiol. 1982;44:561–569. doi: 10.1128/aem.44.3.561-569.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooker J D, O’Donovan L A, Skene I, Clarke K, Blackall L, Muslera P. Streptococcus caprinus sp. nov., a tannin-resistant ruminal bacterium from feral goats. Lett Appl Microbiol. 1994;18:313–318. [Google Scholar]
- 7.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell D R, Bryant M P. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966;14:794–801. doi: 10.1128/am.14.5.794-801.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaney A L, Marbach E P. Modified reagents for determination of urea and ammonia. Clin Chem. 1962;8:130–132. [PubMed] [Google Scholar]
- 10.Chen G, Russell J B. Fermentation of peptides and amino acids by a monensin-sensitive ruminal peptostreptococcus. Appl Environ Microbiol. 1988;54:2742–2749. doi: 10.1128/aem.54.11.2742-2749.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Russell J B. More monensin-sensitive, ammonia-producing bacteria from the rumen. Appl Environ Microbiol. 1989;55:1052–1057. doi: 10.1128/aem.55.5.1052-1057.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 13.Cotta M A, Hespell R B. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1986;52:51–58. doi: 10.1128/aem.52.1.51-58.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotta M A, Russell J B. Digestion of nitrogen in the rumen: a model for metabolism of nitrogen compounds in gastrointestinal environments. In: Mackie R I, White B S A, editors. Gastrointestinal microbiology. Vol. 1. New York, N.Y: Chapman and Hall; 1996. pp. 380–423. [Google Scholar]
- 15.de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolfing J, Gottschal J C. Microbe-microbe interactions. In: Mackie R I, White B S A, editors. Gastrointestinal microbiology. Vol. 2. New York, N.Y: Chapman and Hall; 1997. pp. 373–433. [Google Scholar]
- 17.Forster R J, Teather R M, Gong J, Deng S-J. 16S rDNA analysis of Butyrivibrio fibrisolvens: phylogenetic position and relation to butyrate-producing anaerobic bacteria from the rumen of white-tailed deer. Lett Appl Microbiol. 1996;23:218–222. doi: 10.1111/j.1472-765x.1996.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 18.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press; 1966. p. 297. [Google Scholar]
- 19.Hungate R E. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 1969;3:117–132. [Google Scholar]
- 20.Hutson R A, Thompson D E, Lawson P A, Schocken-Itturino R P, Bottger E C, Collins M D. Genetic interrelationships of proteolytic Clostridium botulinum types A, B, and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie Leeuwenhoek. 1993;64:273–283. doi: 10.1007/BF00873087. [DOI] [PubMed] [Google Scholar]
- 21.Jensen M A, Hubner R J. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in identification of Salmonella serovars. Appl Environ Microbiol. 1996;62:2741–2746. doi: 10.1128/aem.62.8.2741-2746.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones G A, McAllister T A, Cheng K-J, Muir A D. Effect of sainfoin (Onobrychis viciifolia Scop) on growth and proteolysis by four strains of rumen bacteria: resistance of Prevotella (Bacteroides) ruminicola B14. Appl Environ Microbiol. 1994;60:1374–1378. doi: 10.1128/aem.60.4.1374-1378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones W T, Mangan J L. Complexes of the condensed tannins of sainfoin (Onobrychis viciifolia Scop.) with fraction 1 leaf protein and with submaxillary-mucoprotein, and their reversal by polyethylene glycol and pH. J Sci Food Agric. 1977;28:126–136. [Google Scholar]
- 24.Jones W T, Lyttleton J W. Bloat in cattle 29. The foaming properties of clover proteins. N Z J Agric Res. 1969;12:31–46. [Google Scholar]
- 25.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 26.Krause D O, Russell J B. An rRNA approach for assessing the role of obligate amino acid-fermenting bacteria in ruminal amino acid deamination. Appl Environ Microbiol. 1996;62:815–821. doi: 10.1128/aem.62.3.815-821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause D O, White B A, Mackie R I. Ribotyping of adherent Lactobacillus from weaning pigs: a basis for probiotic selection based on diet and gut compartment. Anaerobe. 1997;3:317–325. doi: 10.1006/anae.1997.0118. [DOI] [PubMed] [Google Scholar]
- 28.Lowe S E, Theodorou M K, Trinci A P J, Hespell R B. Growth of anaerobic rumen fungi on defined and semi-defined media lacking rumen fluid. J Gen Microbiol. 1985;131:2225–2229. [Google Scholar]
- 29.Mackie R I, Gilchrist F M C, Robberts A M, Hannah P E, Schwartz H M. Microbiological and chemical changes in the rumen during the stepwise adaption of sheep to high concentrate diets. J Agric Sci. 1978;90:241–254. [Google Scholar]
- 30.Mackie R I, Wilkins C A. Enumeration of anaerobic bacterial microflora of the equine gastrointestinal tract. Appl Environ Microbiol. 1988;54:2155–2160. doi: 10.1128/aem.54.9.2155-2160.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.McSweeney, C. S. Unpublished data.
- 31.Menke K H, Raab L, Salewski A, Steingass H, Fritz D, Schneider W. The estimation of the digestibility and metabolisable energy content of ruminant feeding stuffs from the gas production when they are incubated with rumen liquor in vitro. J Agric Sci. 1979;93:217–222. [Google Scholar]
- 32.Miller S M, Brooker J D, Phillips A, Blackall L L. Streptococcus caprinus is ineffective as a rumen inoculum to improve digestion of mulga (Acacia aneura) by sheep. Aust J Agric Res. 1996;47:1323–1331. [Google Scholar]
- 33.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson K E, Thonney M L, Woolston T K, Zinder S H, Pell A N. Phenotypic and phylogenetic characterization of ruminal tannin-tolerant bacteria. Appl Environ Microbiol. 1998;64:3824–3830. doi: 10.1128/aem.64.10.3824-3830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemoto K, Osawa R, Hirota K, Ono T, Miyake Y. An investigation of gram-negative tannin-protein degrading bacteria in fecal flora of various mammals. J Vet Med Sci. 1995;57:921–926. doi: 10.1292/jvms.57.921. [DOI] [PubMed] [Google Scholar]
- 37.Osawa R O. Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from faeces of koalas. Appl Environ Microbiol. 1990;56:829–831. doi: 10.1128/aem.56.3.829-831.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osawa R O. Tannin-protein complex-degrading enterobacteria isolated from the alimentary tracts of koalas and a selective medium for their enumeration. Appl Environ Microbiol. 1992;58:1754–1759. doi: 10.1128/aem.58.5.1754-1759.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa R O, Walsh T P, Cork S J. Metabolism of tannin-protein complex by facultatively anaerobic bacteria isolated from koala feces. Biodegradation. 1993;4:91–99. [Google Scholar]
- 40.Rainey F A, Stackebrandt E. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 41.Skene I, Brooker J D. Characterisation of the tannin acylhydrolase in the ruminal bacterium Selenomonas ruminantium. Anaerobe. 1995;1:321–327. doi: 10.1006/anae.1995.1034. [DOI] [PubMed] [Google Scholar]
- 42.Sly I, Cahill M M, Osawa R O, Fujisawa T. The tannin-degrading species Streptococcus gallolyticus and Streptococcus caprinus are subjective synonyms. Int J Syst Bacteriol. 1997;47:893–894. doi: 10.1099/00207713-47-3-893. [DOI] [PubMed] [Google Scholar]
- 43.Sokal R R, Michener C D. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 44.Stahl M, Molin G, Persson A, Ahrne S, Stahl S. Restriction endonuclease patterns and multivariate analysis as a classification tool for Lactobacillus spp. Int J Syst Bacteriol. 1990;40:189–193. [Google Scholar]
- 45.Swain T. Tannin and lignins. In: Rosenthal A, Janzen J H, editors. Herbivores: their interaction with secondary plant metabolites. New York, N.Y: Academic Press; 1979. pp. 657–682. [Google Scholar]
- 46.Terrill T H, Rowan A M, Douglas G B, Barry T N. Determination of extractable and bound condensed tannin concentrations in forage plants, protein concentrate meals and cereal grains. J Sci Food Agric. 1992;58:321–329. [Google Scholar]
- 47.Terrill T H, Windham W R, Evans J J, Hoveland C S. Condensed tannin concentration in Sericea lespedeza as influenced by preservation method. Crop Sci. 1990;30:219–224. [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willems A, Amatmarco M, Collins M D. Phylogenetic analysis of Butyrivibrio strains reveals three distinct groups of species within the Clostridium subphylum of the gram-positive bacteria. Int J Syst Bacteriol. 1996;46:195–199. doi: 10.1099/00207713-46-1-195. [DOI] [PubMed] [Google Scholar]