Abstract
We obtained nine bacterial isolates from root or collar nodules of the non-stem-nodulated Aeschynomene species A. elaphroxylon, A. uniflora, or A. schimperi and 69 root or stem nodule isolates from the stem-nodulated Aeschynomene species A. afraspera, A. ciliata, A. indica, A. nilotica, A. sensitiva, and A. tambacoundensis from various places in Senegal. These isolates, together with 45 previous isolates from various Aeschynomene species, were studied for host-specific nodulation within the genus Aeschynomene, also revisiting cross-inoculation groups described previously by D. Alazard (Appl. Environ. Microbiol. 50:732–734, 1985). The whole collection of Aeschynomene nodule isolates was screened for synthesis of photosynthetic pigments by spectrometry, high-pressure liquid chromatography, and thin-layer chromatography analyses. The presence of puf genes in photosynthetic Aeschynomene isolates was evidenced both by Southern hybridization with a Rhodobacter capsulatus photosynthetic gene probe and by DNA amplification with primers defined from photosynthetic genes. In addition, amplified 16S ribosomal DNA restriction analysis was performed on 45 Aeschynomene isolates, including strain BTAi1, and 19 reference strains from Bradyrhizobium japonicum, Bradyrhizobium elkanii, and other Bradyrhizobium sp. strains of uncertain taxonomic positions. The 16S rRNA gene sequence of the photosynthetic strain ORS278 (LMG 12187) was determined and compared to sequences from databases. Our main conclusion is that photosynthetic Aeschynomene nodule isolates share the ability to nodulate particular stem-nodulated species and form a separate subbranch on the Bradyrhizobium rRNA lineage, distinct from B. japonicum and B. elkanii.
Rhizobia symbiotically interact with leguminous plants to form nitrogen-fixing nodules most often exclusively occurring on the roots. A few legumes, however, including several Aeschynomene species, form nodules also on stem-located sites. The genus Aeschynomene includes 22 stem-nodulated species that readily nodulate all along the stem and many other species, considered non-stem nodulated, since their nodulation is restricted to the lower (collar) and submerged part of the stem (6). A number of stem isolates of Aeschynomene spp. are of special interest because of their unusual ability to produce photosynthetic pigments, including both bacteriochlorophyll a (Bchl a) and carotenoids (15, 16, 34, 46). The well-studied strain BTAi1, isolated from stem nodules of Aeschynomene indica, was the first bacteriochlorophyll-synthesizing rhizobial strain described (14, 15, 17). When grown aerobically and heterotrophically under a light-dark cycle, strain BTAi1 synthesizes photosynthetic pigments and forms photosynthetic reaction centers like those of the purple nonsulfur photosynthetic bacteria (17, 42). Light-induced CO2 and light-decreased O2 uptakes gave evidence of the photosynthetic activity of this strain (17, 26, 27). Because of its functional photosynthetic apparatus, strain BTAi1 can be considered photosynthetic, and by extension, so can all the rhizobia producing photosynthetic pigments.
Rhizobia are taxonomically very diverse. By polyphasic taxonomy, 20 species have been identified and assigned to six genera, Rhizobium, Sinorhizobium, Mesorhizobium, Bradyrhizobium and Azorhizobium (for a review, see reference 58), and Allorhizobium (10). The unusual presence of a photosynthetic system in strain BTAi1 led to the tentative name “Photorhizobium thompsonum” (16) or “Photorhizobium thompsonianum” (15) for this strain. However, 16S rRNA gene sequence analysis showed that strain BTAi1 was very closely related to both Bradyrhizobium japonicum and Rhodopseudomonas palustris, suggesting that BTAi1 could be appropriately named Bradyrhizobium sp. (A. indica) (59). This was later confirmed by additional 16S rRNA gene sequencing of other photosynthetic rhizobia (47, 55) and by fatty acid analysis (47). Additional data came from numerical taxonomy (150 phenotypic characteristics) indicating that the photosynthetic rhizobia constitute a unique phenon that could be considered distinct from Bradyrhizobium (33). The precise taxonomic status of Aeschynomene photosynthetic rhizobia thus remained unclear.
Each rhizobium can nodulate only a limited number of legumes, referred to as its host range. Depending on the extent of their host range, rhizobia can be considered specific or nonspecific. Aeschynomene symbionts comprise both nonspecific rhizobia of the cowpea group and rhizobia specific to the stem-nodulated species (1). No correlation between symbiotic properties and photosynthetic pigment synthesis could be established (48).
Since the different reports on Aeschynomene bradyrhizobia generally studied different rhizobium collections and focused on either nodulation (1), phylogeny (55), or photosynthesis (48), the data appeared fragmentary and did not allow a comprehensive view of the diversity and evolution of Bradyrhizobium isolates from Aeschynomene species. Our objective was thus to examine possible links among the presence of photosynthetic pigments, nodulation capacity, and 16S rRNA gene-based phylogeny among bradyrhizobia from Aeschynomene species. These three topics were studied with a large collection of isolates. We first enlarged our collection of 45 Aeschynomene strains (1–3a) with 78 new bacterial isolates from nodules of diverse native stem- and non-stem-nodulated Aeschynomene species from different places in Senegal. We screened the isolates for photosynthetic pigment production (Bchl a and carotenoids), for DNA hybridization with a Rhodobacter capsulatus photosynthetic gene probe, and for DNA amplification with photosynthetic gene primers. We characterized their host range among Aeschynomene species and performed amplified 16S ribosomal DNA (rDNA) restriction analysis (ARDRA) including B. japonicum, Bradyrhizobium elkanii, and other Bradyrhizobium reference strains (1–3, 13, 37). We also determined the 16S rRNA gene sequence of a photosynthetic strain and compared it to sequences of reference strains, including strain BTAi1.
MATERIALS AND METHODS
Bacterial strains and culture growth conditions.
The strains used are listed in Tables 1 and 2. By use of the isolation procedure described by Alazard (1), 78 new isolates from Aeschynomene species were obtained from naturally occurring root or stem nodules collected in different regions of Senegal. Yeast extract-mannitol medium (54) was the routine medium used for isolation, purification, and maintenance of the rhizobia. Strains were grown at 30°C for 4 to 7 days under aerobic conditions. Type or representative strains of B. japonicum, B. elkanii, and the various clusters of Bradyrhizobium described by Moreira et al. (37) and by Dupuy et al. (13) were included in this study (Table 1).
TABLE 1.
Reference strains used in this studya
| Sp. and strain | LMG no. | Original host plant | Reference | ARDRA group |
|---|---|---|---|---|
| Bradyrhizobium japonicum | ||||
| NZP5533 | 6136 | Glycine max | B | |
| NZP5549T | 6138T | Glycine max | B | |
| USDA135 | 8321 | Glycine max | B | |
| Bradyrhizobium elkanii | ||||
| NZP5531T | 6134T | Glycine max | C | |
| NZP5532 | 6135 | Glycine max | C | |
| Bradyrhizobium sp. | ||||
| ORS348 | 12200 | Aeschynomene sp. | 3 | D |
| ORS103 | 10665 | Faidherbia albida | 13 | B |
| ORS110 | 10666 | Faidherbia albida | 13 | B |
| ORS121 | 10677 | Faidherbia albida | 13 | C |
| ORS133 | 10689 | Faidherbia albida | 13 | C |
| ORS162 | 10705 | Faidherbia albida | 13 | C |
| ORS169 | 10712 | Faidherbia albida | 13 | B |
| ORS174 | 10717 | Faidherbia albida | 13 | C |
| ORS175 | 10718 | Faidherbia albida | 13 | C |
| ORS180 | 10719 | Faidherbia albida | 13 | Sep. |
| ORS187 | 10726 | Faidherbia albida | 13 | B |
| BR29 | 9520 | Unknown | C | |
| BR3621 | 9966 | Acacia mangium | 37 | C |
| BR4406 | 9980 | Enterolobium ellipticum | 37 | C |
| INPA9A | 10029 | Derris sp. | 37 | B |
Abbreviations and designations: ORS, I.R.D. Collection, Institut de Recherche pour le Développement, Montpellier, France; LMG, Collection of Bacteria of the Laboratorium voor Microbiologie, University of Ghent, Ghent, Belgium; BR, strain from the CNPBS-EMBRAPA, Centro Nacional de Pesquisa cm Biologia do Solo, Seropedica, and Emprasa Brasiliera de Pesquisa Agropequaria, Rio de Janeiro, Brazil; INPA, National Institute of Amazonia Research, Manaus, Brazil; NZP, Culture Collection of the Department for Scientific and Industrial Research, Biochemistry Division, Palmerston North, New Zealand; USDA, U.S. Department of Agriculture, Beltsville, Md.; Sep., separate.
TABLE 2.
Nodulation specificity, Bchl a and carotenoid (Crt) content, and ARDRA grouping of bradyrhizobia from Aeschynomene
| Cross-inoculation group and host plant | Bacterial straina | LMG no. | Reference or source | Nodulation on groupd:
|
Crt type spectrume | ARDRA groupf | |||
|---|---|---|---|---|---|---|---|---|---|
| I (A. elaphroxylon) | II (A. afraspera) | III
|
|||||||
| A. indica | A. sensitiva | ||||||||
| Group I | |||||||||
| A. americana | ORS301 (R)b | 8290 | 1 | E | e | 0 | 0 | W | B |
| A. elaphroxylon | ORS377 (C) | 15420 | This study | E | i | 0 | 0 | W | |
| ORS378 (C) | 15421 | This study | E | i | 0 | 0 | W | ||
| ORS379 (C) | 15422 | This study | E | i | 0 | 0 | W | ||
| ORS381 (C) | 15423 | This study | E | i | 0 | 0 | W | ||
| ORS304 (C)c | 8069 | 1 | E | e | i | 0 | W | Sep. | |
| A. pfundii | ORS302 (C)b | 10296 | 1 | E | E | 0 | 0 | W | |
| A. schimperi | ORS271 (R) | This study | nt | E | 0 | 0 | W | ||
| ORS272 (R) | 15408 | This study | nt | E | 0 | 0 | W | ||
| ORS275 (R) | 15410 | This study | nt | E | 0 | 0 | W | ||
| ORS273 (R) | This study | nt | e | 0 | 0 | W | |||
| ORS274 (R) | 15409 | This study | nt | e | 0 | 0 | W | ||
| ORS305 (R)b | 8291 | 2 | E | e | 0 | 0 | W | Sep. | |
| ORS343 (R)b | 11801 | 3 | nt | e | 0 | 0 | W | ||
| ORS360 (R) | D. Alazard | nt | 0 | 0 | 0 | W | |||
| A. uniflora | ORS309 (C) | 7838, 8070 | 2 | E | e | 0 | 0 | W | C |
| ORS355 (C)b | 10301 | 3 | nt | e | 0 | 0 | W | ||
| Group II | |||||||||
| A. ciliata | ORS332 (U) | 8296 | D. Alazard | 0 | e | 0 | 0 | nt | |
| ORS333 (U) | 8307, 10289 | D. Alazard | 0 | e | i | i | nt | ||
| A. nilotica | ORS358 (U) | 10303 | D. Alazard | nt | e | 0 | 0 | W | C |
| ORS364 (S) | 11802 | 3 | nt | e | E | i | LP | ||
| A. afraspera | ORS313 (R) | 15416 | D. Alazard | i | e | 0 | 0 | W | |
| ORS336 (S)b | 8298, 11800 | 3 | i | e | 0 | 0 | W | C | |
| ORS284 (S) | This study | nt | e | 0 | 0 | W | |||
| ORS317 (R) | 15417 | D. Alazard | nt | e | 0 | 0 | W | ||
| ORS349 (U) | 10299 | D. Alazard | nt | e | 0 | 0 | W | ||
| ORS354 (S) | 15424 | D. Alazard | nt | e | 0 | 0 | W | ||
| ORS347 (S) | 10298 | 3 | nt | E | 0 | 0 | W | ||
| ORS323 (S) | 15380 | D. Alazard | nt | i | 0 | 0 | LP | ||
| ORS325 (S) | 8304 | D. Alazard | nt | i | e | E | LP | ||
| ORS288 (S) | 12189, 15412 | This study | nt | e | e | i | LP | ||
| ORS289 (S) | 15413 | This study | nt | e | e | e | LP | ||
| ORS290 (S) | 15414 | This study | nt | e | e | e | LP | ||
| ORS363 (S) | 15418 | This study | nt | e | e | e | W | ||
| ORS365 (S) | 15419 | This study | nt | e | e | e | LP | ||
| ORS291 (S) | 15415 | This study | nt | e | e | E | LP | ||
| ORS308 (R)c | 8293, 15401 | 2 | nt | E | e | E | LP | A | |
| ORS303 (S) | 8292 | 1 | 0 | E | e | i | LP | ||
| ORS322 (S)c | 8073, 15403 | 1 | 0 | E | e | i | LP | A | |
| ORS312 (R)c | 8294 | 2 | nt | E | e | i | LP | A | |
| ORS324 (R)c | 8295 | 3 | nt | E | e | i | LP | A | |
| ORS337 (R)c | 8299 t2 | 2 | nt | E | e | i | LP | A | |
| ORS374 (S) | 11960 | This study | nt | E | E | i | O | ||
| ORS286 (S) | 15377 | This study | nt | E | E | e | LP | ||
| ORS351 (S) | 10300 | 3 | nt | E | E | e | LP | ||
| ORS285 (S) | 15376 | This study | 0 | E | E | E | LP | ||
| ORS287 (S) | 15378 t1 | This study | nt | E | E | E | LP | A | |
| ORS335 (U) | 8297 | D. Alazard | nt | E | E | E | LP | ||
| ORS352 (S) | 15384 | D. Alazard | nt | E | E | E | LP | ||
| ORS353 (S) | 15385 | D. Alazard | nt | E | E | E | LP | ||
| ORS357 (U) | 10302 | D. Alazard | nt | E | E | E | LP | ||
| ORS362 (S) | 10305 | 3 | nt | E | E | E | LP | ||
| ORS356 (S) | 15386 | This study | nt | e | E | E | LP | ||
| Group III | |||||||||
| A. tambacoundensis | ORS266 (S) | 15442 | This study | 0 | 0 | e | e | LP | A |
| ORS268 (S) | This study | 0 | e | e | E | LP | |||
| ORS331 (S)c | 11799 | 3 | 0 | 0 | E | E | LP | A | |
| ORS334 (S)c | 8308 | 1 | 0 | 0 | E | E | LP | ||
| A. indica | ORS280 (S) | 15411 | This study | 0 | 0 | e | e | LP | |
| ORS306 (S)c | 8300, 11797, 10286 | 1 | 0 | 0 | e | e | LP | A | |
| ORS307 (S) | 15379 | This study | 0 | 0 | e | e | LP | ||
| ORS310 (S)c | 8071 t1 | 1 | 0 | 0 | e | e | LP | ||
| ORS311 (S) | This study | 0 | 0 | e | e | LP | |||
| ORS318 (S)c | 8301 t1 | 2 | 0 | 0 | e | e | LP | ||
| ORS319 (S)c | 8302 t1 | 2 | 0 | 0 | e | e | LP | ||
| ORS339 (S) | This study | 0 | 0 | e | e | LP | |||
| ORS375 (S) | 15388 | This study | 0 | 0 | e | e | LP | ||
| ORS397 (S) | This study | 0 | 0 | e | e | LP | |||
| ORS320 (S)c | 8303 | 2 | 0 | E | e | e | LP | A | |
| ORS321 (S) | This study | 0 | 0 | e | e | LP | |||
| ORS328 (S)c | 1 | 0 | 0 | e | E | LP | |||
| ORS338 (S) | 3 | 0 | 0 | e | E | LP | |||
| ORS376 (S) | This study | 0 | 0 | e | E | LP | |||
| ORS383 (S) | This study | 0 | 0 | e | E | LP | |||
| ORS386 (S) | This study | 0 | 0 | e | E | LP | A | ||
| ORS400 (S) | This study | 0 | 0 | E | E | LP | |||
| BTAi1 (S) | 14 | 0 | 0 | E | e | LP | A | ||
| ORS340 (S) | This study | 0 | 0 | E | e | LP | |||
| ORS341 (S) | This study | 0 | 0 | E | e | LP | |||
| ORS372 (S) | 11805 | This study | 0 | 0 | E | e | LP | ||
| ORS388 (S) | 11814 | This study | 0 | 0 | E | e | LP | ||
| ORS389 (S) | 15393 | This study | 0 | 0 | E | e | LP | ||
| ORS390 (S) | 11813 | This study | 0 | 0 | E | e | LP | A | |
| ORS391 (S) | 15394 | This study | 0 | 0 | E | e | LP | ||
| ORS392 (S) | 11812, 12205 | This study | 0 | 0 | E | e | O | A | |
| ORS393 (S) | 11811, 15407 | This study | 0 | 0 | E | e | LP | A | |
| ORS394 (S) | 15395 | This study | 0 | 0 | E | e | LP | ||
| ORS269 (S) | This study | 0 | 0 | E | E | LP | |||
| ORS270 (S) | This study | 0 | 0 | E | E | LP | |||
| ORS282 (S) | This study | 0 | 0 | E | E | O | |||
| ORS342 (S) | 15383 | This study | 0 | 0 | E | E | LP | ||
| ORS344 (S) | 12198 | This study | 0 | 0 | E | E | DP | ||
| ORS346 (S) | 12199 | This study | 0 | 0 | E | E | LP | ||
| ORS371 (S) | 11804, 12203 | This study | 0 | 0 | E | E | DP | A | |
| ORS373 (S) | 11806 | This study | 0 | 0 | E | E | LP | ||
| ORS382 (S) | 11816, 15406 | This study | 0 | 0 | E | E | LP | A | |
| ORS384 (S) | 15390 | This study | 0 | 0 | E | E | LP | A | |
| ORS385 (S) | 15391 | This study | 0 | 0 | E | E | LP | ||
| ORS387 (S) | 15392 | This study | 0 | 0 | E | E | LP | ||
| ORS395 (S) | 15396 | This study | 0 | 0 | E | E | LP | ||
| ORS396 (S) | 11808 | This study | 0 | 0 | E | E | LP | ||
| ORS399 (S) | 12206 | This study | 0 | 0 | E | E | LP | ||
| ORS327 (S) | 15381 | This study | 0 | 0 | i | e | LP | ||
| ORS368 (S) | 15387 | This study | 0 | 0 | e | i | LP | A | |
| ORS398 (S) | 11809 | This study | 0 | i | E | i | O | ||
| ORS380 (S) | 11817 | This study | 0 | E | E | i | LP | ||
| A. sensitiva | ORS297 (S) | 12195 | This study | 0 | 0 | e | e | LP | A |
| ORS279 (S) | 12188 | This study | 0 | 0 | e | E | O | A | |
| ORS292 (S) | 12205 | This study | 0 | 0 | e | E | LP | ||
| ORS294 (S) | 12192 | This study | 0 | 0 | e | E | LP | A | |
| ORS330 (S)c | 8306, 15404 | 1 | 0 | 0 | e | E | LP | ||
| ORS293 (S) | 12191 | This study | 0 | 0 | E | e | O | ||
| ORS300 (S) | 12197, 15400 | This study | 0 | E | E | e | LP | A | |
| ORS295 (S) | This study | 0 | 0 | E | e | LP | |||
| ORS296 (S) | 12194 | This study | 0 | 0 | E | e | DP | D | |
| ORS298 (S) | 12196 | This study | 0 | 0 | E | e | LP | A | |
| ORS299 (S) | 15399 | This study | 0 | 0 | E | e | LP | Sep. | |
| ORS278 (S) | 12187 | This study | 0 | 0 | E | E | O | A | |
| ORS276 (S) | 12185 | This study | 0 | 0 | E | E | LP | ||
| ORS277 (S) | 12186 | This study | 0 | 0 | E | E | O | A | |
| ORS359 (S) | 12201 | This study | 0 | 0 | E | E | LP | A | |
| ORS361 (S) | 12202 | This study | 0 | 0 | E | E | LP | A | |
Photosynthetic strains are in boldface. R, strain isolated from root nodules; C, strain from nodule located on the stem collar (nodule on the lower and submerged part of the stem); S, strain from stem nodule; U, unknown; ORS, I.R.D. Collection, Institut de Recherche pour le Développement, Montpellier, France; LMG, Collection of Bacteria of the Laboratorium voor Microbiologie, University of Ghent, Ghent, Belgium.
nt, not tested; 0, no nodulation; i, ineffective root nodulation; e, partially effective root nodulation; E, effective root nodulation.
Crt, carotenoid. The spectrum is described in Fig. 1. W, white (strain lacking Bchl a and carotenoid). DP, dark pink; O, orange; LP, light pink.
Sep., separate.
Nodulation tests.
Seeds and plants of Aeschynomene species were prepared for root nodulation trials according to previously described procedures (1). Plants were grown under continuous light (20 W/m2) at 28°C. Four to six plants were tested for each strain. Plants were observed for nodule formation over 6 to 8 weeks, and effectiveness was estimated from visual observation of plant vigor and foliage color.
Photosynthetic pigment determination.
Cultures were grown at 30°C for 7 days under aerobic conditions on a 15-h–9-h light-dark cycle. Bchl was extracted under dim light with cold acetone-methanol (7:2 [vol/vol]) at 4°C for 30 min (35). The supernatant was analyzed with a Beckman DU40 spectrophotometer. Absorption spectra were generated by scanning over a wavelength range from 350 to 800 nm. Carotenoids were further purified and analyzed by high-pressure liquid chromatography (HPLC) or thin-layer chromatography (TLC) as previously described (35).
Southern hybridization.
Genomic DNA was extracted as previously described (36). Total DNA was digested by EcoRI, PstI, or HindIII as specified by the manufacturer (Boehringer Mannheim or Pharmacia). Restricted DNA was run in a 0.8% agarose gel and transferred to a nylon membrane under alkaline conditions by the Southern blot standard procedure (43). Hybridization was carried out with the digoxigenin labeling and detection kit from Boehringer Mannheim. The probe was labeled by randomly primed incorporation of digoxigenin-linked dUTP, (DIG-dUTP) and hybridization was performed overnight at 37°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50% (vol/vol) deionized formamide, 2% (wt/vol) blocking reagent in maleic acid buffer (100 mM maleic acid, 150 mM NaCl, pH 7.5), 0.1% (wt/vol) N-lauroylsarcosine (Sarkosyl), and 0.02% (wt/vol) sodium dodecyl sulfate. After stringency washes, hybrids were revealed by a chemiluminescence reaction, and detection was performed on X-ray film. The hybridization probe was a 3.3-kb EcoRI-HindIII fragment (pufBALMX) from the plasmid pUC13::pufBALMX (5, 8) provided by A. Lilburn (University of British Columbia, Vancouver, Canada). This fragment is part of the 46-kb region of the R. capsulatus chromosome which encodes the photosynthetic apparatus.
PCR amplification of puf genes.
Part of the pufLM genes was amplified from genomic DNA with the nondegenerated primers pufL278f (5′-CACCCATCTCGATTGGGTGTCG-3′) and pufM278r (5′-CTCCAGCTGCCCATGAAGATCG-3′), specifically defined from the pufLM sequence of Bradyrhizobium sp. strain ORS278 and amplifying a 926-bp fragment from the 3′ end of pufL and the 5′ end of pufM in the ORS278 sequence (20a). Amplification reactions were performed in a 50-μl final volume containing 0.1 μg of DNA, each deoxyribonucleoside triphosphate at a concentration of 0.2 mM, 0.8 μM (each) primer, 1.5 mM MgCl2, 1.25 U of Taq DNA polymerase (Gibco BRL), and the buffer supplied with the enzyme. The amplification conditions were 5 min at 94°C, 30 cycles consisting of 30 s at 94°C and 30 s at 60°C, and 1 min at 72°C.
ARDRA.
DNA was extracted according to the procedure of Pitcher et al. (41) with slight modifications. A loopful of cells was washed with 500 μl of 150 mM NaCl–10 mM EDTA, pH 8.0. DNA was extracted by a procedure involving lysis with Sarkosyl-guanidinium thiocyanate (Sigma), phenol-isoamylic alcohol treatment, and isopropanol precipitation. DNA was further purified by treatment for 1 h at 37°C with RNase at a final concentration of 250 μg/ml. Milli-Q water (Millipore) was used for all enzymatic reactions and amplification procedures.
ARDRA was performed as described by Vaneechoutte et al. (49). 16S rDNA was amplified with a forward primer (5′-AGAGTTTGATCATGGCTCAG-3′) and a reverse primer (5′-TACCTTGTTACGACTTCACCCCA-3′) supplied by Pharmacia. PCR was carried out in a 50-μl reaction volume by mixing 250 ng of template DNA with the polymerase reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl, pH 8.4), 10 nmol of each deoxynucleoside triphosphate (Pharmacia), 20 pmol of each primer (Pharmacia), and 1.25 U of EuroTaq polymerase (Eurogentec, Seraing, Belgium). Amplification was achieved in a Perkin-Elmer PCR GeneAmp 9600 thermocycler (Perkin-Elmer Cetus) with the following temperature profile: an initial denaturation at 95°C for 7 min; 50 cycles of denaturation (45 s at 95°C), annealing (30 s at 57°C), and extension (2 min at 72°C); and a final extension at 72°C for 10 min. The PCR products were purified with the PCR Clean Up kit (Boehringer) according to the manufacturer’s recommendations.
Restriction was carried out as specified by the manufacturers in 20-μl volumes of commercially supplied incubation buffer containing 10 μl of PCR product and 5 U each of restriction endonucleases HinfI, DdeI, and MwoI (New England Biolabs, Leusden, The Netherlands) or AluI and HhaI (Pharmacia). Restriction fragment length polymorphism patterns were analyzed by horizontal gel electrophoresis of each restriction mixture at 90 V for 130 min in 2% (wt/vol) Metaphor agarose (FMC Bioproducts, Rockland, Maine) in TBE buffer (Tris-HCl, 89 mM; boric acid, 89 mM; EDTA, 2 mM; pH 8.0) containing 0.5 μg of ethidium bromide per ml. Gels were viewed under UV illumination (312 nm) and photographed with a charge-coupled device camera (768 by 494 pixels). Images were scanned for normalization of restriction patterns with the Gelcompar 3.1 software package (53) with AluI-digested pBR322 as molecular weight markers. For each strain, the five normalized restriction patterns were assembled into a combined profile and analyzed with the Dice similarity coefficient (SD) expressed as a percentage and the unweighted pair group method with average linkage (UPGMA) clustering algorithm.
Analysis of the 16S rRNA genes.
A few colonies of strain ORS278 (LMG 12187) were suspended in 50 μl of water, and a small amount of sterile glass beads was added. The suspension was mixed for 1 min, boiled for 5 min, and again mixed for 1 min, and finally, the cell debris was spun down. Five microliters of the supernatant was used in a PCR to amplify the nearly complete 16S rRNA gene (positions 28 to 1521 of the Escherichia coli 16S rRNA gene). The PCR product was purified with a Prep-A-Gene kit (Bio-Rad Laboratories, Hercules, Calif.) and sequenced with primers to universally conserved fragments, a Taq dye-deoxy terminator cycle sequencing kit (Perkin-Elmer Corp., Foster City, Calif.), and an automatic DNA sequencer (model 377; Perkin-Elmer Corp.). The obtained sequence fragments were aligned, and a consensus sequence was constructed with the program AutoAssembler (Perkin-Elmer). For further phylogenetic analysis, the Genetics Computer Group (GCG) package (12) and the phylogeny inference (PHYLIP) package (18), available on the Belgian EMBnet Node of the Brussels Free University Computing Centre, were used. The new sequence was aligned, together with reference sequences obtained from the EMBL data library, with the program PILEUP of the GCG package. In total, a continuous stretch of 1,401 base positions (including gaps) was used for further analysis. Distances, modified according to the Kimura-2 model, were calculated by using the DNADIST program of the PHYLIP package, and the program NEIGHBOR of the same package was used to produce an unrooted phylogenetic tree. The stability of the groupings was verified by bootstrap analysis (500 replications) with the PHYLIP programs DNABOOT, DNADIST, NEIGHBOR, and CONSENSE.
Nucleotide sequence accession number.
The EMBL accession number for the 16S rRNA gene sequence of strain ORS278 (LMG 12187) is AJ133779.
RESULTS
Isolation of rhizobia from Aeschynomene spp.
We obtained nine isolates from root or collar nodules of non-stem-nodulated Aeschynomene species (A. elaphroxylon, A. uniflora, or A. schimperi) and 69 root or stem nodule isolates from the stem-nodulated A. afraspera, A. ciliata, A. indica, A. nilotica, A. sensitiva, and A. tambacoundensis (Table 2) from various places in Senegal. Included in this study were 45 isolates from various Aeschynomene species previously isolated by Alazard (1–3a). Growth on yeast-mannitol agar produced small typical Bradyrhizobium-like colonies after incubation at 30°C for 5 to 7 days. Colonies formed by numerous isolates turned either light pink (LP), dark pink (DP), or orange (O; see below and Table 2), especially after light exposure, suggesting photosynthetic pigments (15, 35).
Host-specific nodulation within the genus Aeschynomene.
On the basis of nodulation tests performed with 15 rhizobial strains and 20 different Aeschynomene species, Alazard (1) identified three cross-inoculation groups among Aeschynomene species. Group I contained only non-stem-nodulated Aeschynomene species, while true stem-nodulated Aeschynomene spp. belonged to groups II and III. Strains isolated from Aeschynomene spp. of groups I and II were able to nodulate plants from other cross-inoculation groups, whereas rhizobia isolated from group III plants nodulated only plants of the homologous cross-inoculation group.
To confirm and extend these results, all the new Aeschynomene isolates (except ORS326 and ORS348) were tested for root nodulation on plants representative of nodulation group I (A. elaphroxylon), group II (A. afraspera), and group III (A. indica and A. sensitiva) (Table 2).
Isolates from A. americana, A. elaphroxylon, A. pfundii, A. schimperi, and A. uniflora nodulated A. elaphroxylon (group I) and A. afraspera (group II) but rarely plants of group III. According to the work of Alazard (1), we thus classified A. schimperi and A. uniflora in cross-inoculation group I together with A. americana, A. elaphroxylon, and A. pfundii. Isolates from A. ciliata nodulated A. afraspera (group II) and generally representative plants of group III (A. indica and A. sensitiva). A. ciliata, previously classified in cross-inoculation group III by Alazard (1), thus rather belongs to group II, comprising A. nilotica and A. afraspera. Isolates from plants of group III (A. tambacoundensis, A. indica, and A. sensitiva) all nodulated group III representatives but never nodulated A. elaphroxylon from group I and hardly ever nodulated A. afraspera from group II. Their nodulation ability thus appeared to be restricted to group III plants.
Bchl a synthesis is a specific characteristic of group III stem-nodulated Aeschynomene symbionts.
To evaluate the extent of photosynthesis among Aeschynomene isolates and to determine whether there is a relationship between the photosynthetic nature of the microsymbiont and the host plant or the host plant cross-inoculation group, we screened our collection for Bchl a and carotenoid content by spectrometry, HPLC, and TLC analyses.
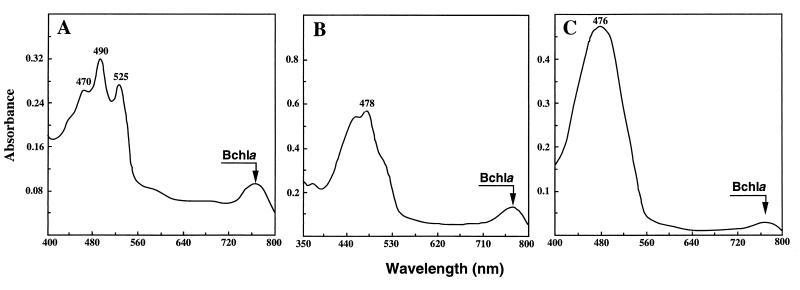
Absorption spectra from Aeschynomene isolates were obtained by using acetone-methanol extracts of bacterial cells. All strains originating from Aeschynomene species belonging to group III (A. tambacoundensis, A. indica, and A. sensitiva) produced an absorbance peak at 770 nm, characteristic of Bchl a, and peaks around 400 to 500 nm, corresponding to carotenoids. These peaks were absent in all strains originating from the group I plants. The content of isolates originating from group II plants appeared variable: 70% of the strains synthesized Bchl a and carotenoids. Three different absorption spectra, corresponding to three pigmentation groups, were obtained for Bchl-synthesizing strains. LP, DP, and O strains (Table 2) exhibited spectra A, B, and C, respectively (Fig. 1). The determination of the carotenoid composition of several representative strains of each group by TLC and HPLC analysis confirmed our previous results (35). Rf (TLC) and retention time (HPLC) values determined for each pigment were found to be identical to those already found. Bchl a and carotenoid contents were also determined by spectrophotometry and HPLC analysis and were found to have values similar to those of our previous report (35). LP strains produced only spirilloxanthin, whereas DP and O strains synthesized both spirilloxanthin and canthaxanthin, together with several other minor carotenoids. The difference in pigmentation between DP and O strains is due to the different ratios of canthaxanthin to spirilloxanthin in these strains. This ratio was found to be 88 to 93% for O strains and 70 to 77% for DP strains, thus confirming our previous observations (35).
FIG. 1.
Absorption spectra of acetone-methanol extracts from stem-nodulating photosynthetic rhizobia. (A) Extract from LP strain ORS266; (B) extract from DP strain ORS397; (C) extract from O strain ORS277. Each extract was obtained from a 50-ml culture.
Up to now, Bchl a has been found in only photosynthetic organisms (20, 24, 39, 40, 45). Consequently, Bchl-containing Aeschynomene strains will be referred to as photosynthetic strains in the following sections.
Among isolates originating from group II plants, the photosynthetic rhizobia are those nodulating A. indica and A. sensitiva, representatives of group III plants (with two exceptions being ORS323 and ORS333). A high correlation was thus found between the ability to synthesize Bchl a and the ability to nodulate A. indica and A. sensitiva, suggesting a relationship between the rhizobial photosynthetic nature and nodulation ability. It should also be noticed that the Bchl a-synthesizing strains from A. afraspera were more effective than the nonphotosynthetic strains.
Genetic evidence for the presence of bacterial photosynthetic genes in Aeschynomene stem-nodulating bradyrhizobia.
The photosynthetic apparatus of purple nonsulfur bacteria (belonging to the alpha subclass of the Proteobacteria together with Bradyrhizobium) is mainly composed of pigment-protein complexes, namely, reaction center and light-harvesting complexes (see reference 50 for a review). To evaluate the occurrence of genes encoding photosynthetic proteins in Bchl-synthesizing rhizobia, we screened 16 selected Aeschynomene strains for the presence of DNA sequences hybridizing to probes consisting of the pufBALMX genes from R. capsulatus (5). The pufBALMX genes have been shown to encode the α- and β-polypeptides of the light-harvesting complex B875 (genes pufBA), the reaction center polypeptides (genes pufLM), and an open reading frame (pufX) (4, 5, 60). Genomic DNA from photosynthetic Aeschynomene strains ORS266, ORS277, ORS278, ORS294, ORS306, ORS322, ORS364, ORS371, and BTAi1 hybridized with the pufBALMX probe. Conversely, genomic DNA from nonphotosynthetic strains ORS301, ORS304, ORS305, ORS309, ORS347, ORS358, and ORS377 did not show any detectable hybridization with this probe (results not shown).
The presence of puf genes among Aeschynomene isolates was also evaluated by pufLM partial amplification with primers defined from the pufLM sequence of Bradyrhizobium sp. strain ORS278. All the photosynthetic strains studied (ORS266, ORS268, ORS277, ORS278, ORS282, ORS285, ORS287, ORS294, ORS296, ORS300, ORS306, ORS320, ORS322, ORS324, ORS330, ORS335, ORS344, ORS352, ORS353, ORS357, ORS362, ORS363, ORS364, ORS368, ORS371, and ORS380) gave a fragment of the expected 926-bp size, while the nonphotosynthetic strains studied (ORS292, ORS301, ORS302, ORS304, ORS305, ORS309, ORS336, ORS347, and ORS358) gave no amplification band.
ARDRA.
Nearly full-length 16S rDNAs from 46 Aeschynomene nodule isolates (including BTAi1) and from 19 reference strains of B. japonicum, B. elkanii, and other Bradyrhizobium spp. previously characterized (13, 37) were amplified, yielding an expected single band of about 1,500 bp (data not shown). The amplified 16S rDNA of all strains was restricted with the enzymes HinfI, DdeI, MwoI, AluI, and HhaI. The combined restriction patterns were used to construct a dendrogram based on the UPGMA algorithm (Fig. 2).
FIG. 2.
ARDRA results presented as a dendrogram based on SD values, calculated by UPGMA. Pseudomonas fluorescens LMG 1799, present in our database, was included as an outgroup organism.
At or above a mean Dice similarity coefficient (SD) value of ± 88%, four main clusters were delineated. Except for ORS296 and ORS299, all photosynthetic Aeschynomene strains belong to the large cluster A. B. japonicum constituted cluster B together with two Aeschynomene strains, ORS301 (nonphotosynthetic) and ORS326 (photosynthetic status not determined); one strain isolated from Derris sp. (LMG 10029); and four strains from Faidherbia albida (ORS103, ORS110, ORS169, and ORS187). Cluster C contained B. elkanii, three nonphotosynthetic strains of Aeschynomene spp. (ORS309, ORS336, and ORS358), strain LMG 9520, and strains isolated from diverse other hosts including Enterolobium ellipticum (LMG 9980), Acacia mangium (LMG 9966), and F. albida (ORS121, ORS133, ORS162, ORS174, and ORS175). The strains ORS296 (photosynthetic) and ORS348 (photosynthetic status not determined) formed cluster D, and the photosynthetic strain ORS299 is the closest relative of this cluster. No evident relationship between the original host plant and the ARDRA clustering could be found.
16S rRNA gene sequence analysis.
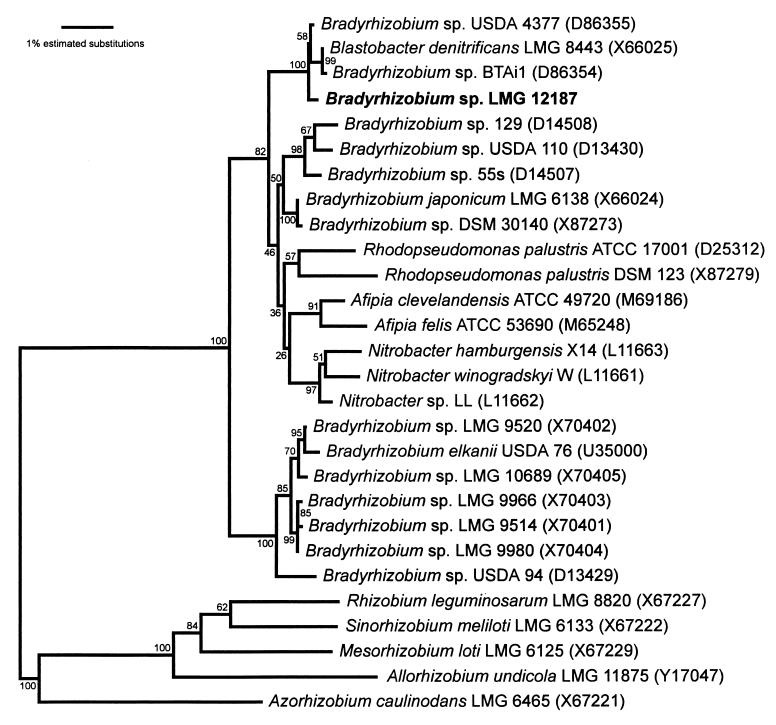
Strain ORS278 (LMG 12187) was chosen as a representative of the photosynthetic strains, and its 16S rRNA gene sequence was determined. It consisted of 1,441 nucleotides and was very similar to the sequence of the photosynthetic strains BTAi1 (11 differences) and USDA 4377 (5 differences). A phylogenetic tree was constructed to determine the position of this strain among other bradyrhizobia (Fig. 3). Strain ORS278 (LMG 12187) formed a separate cluster together with Bradyrhizobium strains BTAi1 and USDA 4377 and Blastobacter denitrificans LMG 8443. This grouping was supported by a bootstrap value of 100% and was distinct from B. japonicum and B. elkanii.
FIG. 3.
Neighbor-joining dendrogram showing the position of photosynthetic strain ORS278 (LMG 12187) among bradyrhizobia and closely related taxa. Bootstrap values, expressed as percentages of 500 replications, are given at the branching points. Numbers in parentheses are the accession numbers of the sequences used. The bar represents one estimated substitution per 100 nucleotide positions.
DISCUSSION
Since the isolation of the strain BTAi1, which displays heterotrophic photosynthesis (15, 17, 28), several bradyrhizobia from various Aeschynomene species have been reported to be photosynthetic, which is a rare property among rhizobia (for a review, see reference 19). Phylogenetic investigations established that the photosynthetic rhizobia belonged to the B. japonicum-R. palustris lineage (55). Previously, nodulation investigations showed that a group of Aeschynomene bradyrhizobia specifically nodulated stem-nodulated species (1). However, no correlation among photosynthetic ability, phylogenetic position, and host specificity could be established, mainly because the different results were established with different bradyrhizobial collections.
In this study, by characterizing a collection of isolates from the genus Aeschynomene, specifically by determining their Bchl content, nodulation abilities, and 16S rRNA gene-based phylogeny, we demonstrate that photosynthetic rhizobia are mainly monophyletic and share the ability to nodulate particular stem-nodulated Aeschynomene species.
To obtain more photosynthetic isolates, we extended the Senegalese collection of Aeschynomene rhizobia (1–3a), mainly by isolating bacteria from naturally occurring stem nodules, since photosynthetic rhizobia are generally isolated from stem-nodulated Aeschynomene spp. When grown under a light-dark cycle, nearly all the stem isolates examined (Table 2) were found to produce Bchl a, a photosynthetic pigment found in only photosynthetic organisms (20, 24, 39, 40, 45), confirming previous reports suggesting that photosynthesis is widespread among stem-nodulating strains (34). The photosynthetic nature of the Bchl-synthesizing bradyrhizobia was confirmed by both Southern hybridization and gene amplification studies. Indeed, the presence of DNA sequences homologous to reaction center and light-harvesting genes from R. capsulatus was detected in all Bchl-synthesizing strains examined, while the presence of pufLM genes in Bchl-synthesizing strains was evidenced by DNA amplification with pufLM primers designed from the pufLM sequence of Bradyrhizobium sp. strain ORS278 (20a). Although the primers used were not designed from a conserved motif in the puf genes, they were found suitable to amplify a puf fragment from all the photosynthetic bradyrhizobia tested in this study. All Bchl-synthesizing strains produced the carotenoid spirilloxanthin, which is known to be bound to the light-harvesting protein-associated complex in purple nonsulfur bacteria and members of the family Chromatiaceae (9, 21–23). A few of them also synthesized other carotenoids, including canthaxanthin (35). The role of the carotenoid canthaxanthin in photosynthesis is unknown, but this pigment has great biotechnological value (38).
Within the past 15 years, the taxonomy of the rhizobia has greatly changed with the discovery of several new species and genera (58). Quite a number of diverse nodule isolates have been characterized and described in the literature as belonging to the large group of bradyrhizobia (13, 37, 52), but only a few studies brought sufficient taxonomic data for clear taxonomic conclusions and nomenclatural decisions (30–32, 56). Several authors have reported the difficulties encountered in studying bradyrhizobia and contradictory results from phenotypic and genotypic studies (13, 33). Here we add further taxonomic data on a collection of 123 isolates from Aeschynomene species, either stem nodulated or non-stem nodulated, together with 19 Bradyrhizobium reference strains, including B. japonicum (30), B. elkanii (32), and Bradyrhizobium sp. strains partially characterized in the literature (13, 37, 52).
Alazard (3) showed that the free-living nitrogen-fixing Aeschynomene symbionts form a single phenon within the Bradyrhizobium genus. Moreover, two representative strains of this phenon, ORS310 and ORS322, were able to grow in the free-living state at the expense of N2 (2), like Azorhizobium caulinodans, which is highly specialized in the stem nodulation of Sesbania rostrata (6). Our results demonstrate that these two free-living nitrogen-fixing Aeschynomene bradyrhizobia also synthesize Bchl a (Table 2). These observations corroborate the work of Ladha and So (33), who found that 52 photosynthetic Aeschynomene nodule isolates belonging to a separate phenon had the ability to grow and fix N2 in the absence of combined nitrogen. Therefore, diazotrophy is probably a general property of photosynthetic isolates, both properties together probably conferring a great selective saprophytic advantage on these bacteria.
We observed a strong correlation between photosynthetic and nodulation abilities. Indeed, all the photosynthetic strains were isolated from stem-nodulated Aeschynomene species belonging to cross-inoculation groups II and III (1, 6). Moreover, among isolates originating from stem-nodulated Aeschynomene spp. of group II, the photosynthetic strains corresponded to those which are also able to nodulate plants of group III (A. sensitiva and A. indica). In contrast to the photosynthetic strains, the nonphotosynthetic rhizobia isolated from plants of groups I and II were able to nodulate F. albida and thus belong to the cowpea group (data not shown). This study thus confirms the occurrence of nonspecific and specific bradyrhizobia among Aeschynomene symbionts with the photosynthetic strains being highly specific. In rhizobium-legume interactions, host specificity is mainly controlled by extracellular bacterial signal molecules, which are called Nod factors (see references 11 and 51 for reviews). All Bradyrhizobium Nod factors examined so far bear a substituted or a nonsubstituted methyl fucose group on their reducing ends (7, 18a, 44). Specific Aeschynomene photosynthetic symbionts thus represent an interesting model to determine which structural features of Nod factors account for host specificity.
From our 16S rDNA-based phylogenetic analysis (Fig. 3), it is apparent that the photosynthetic strains, represented by strains ORS278 (LMG 12187), BTAi1, and USDA 4377, form a separate cluster together with B. denitrificans, an unpigmented budding organism from lake water (25). This small group is supported by a bootstrap value of 100%. In a separate analysis, in which a shorter stretch of approximately 1,000 positions was used (data not shown), we included the shorter sequences for the photosynthetic strains MKAa2 and IRBG 230 (55) in the analysis and found both strains belonging to the same small group. It is clear that this photosynthetic cluster, including B. denitrificans, is distinct from B. japonicum, B. elkanii, and other Bradyrhizobium sp. strains related to both these species. The photosynthetic cluster would seem about equally distant from B. japonicum, the photosynthetic species R. palustris, the nitrifying genus Nitrobacter, and the pathogenic genus Afipia and slightly more distant still from B. elkanii (Fig. 3).
The ARDRA technique confirmed that all Aeschynomene isolates clustered on the Bradyrhizobium phylogenetic branch (Fig. 2) and showed that the majority of the photosynthetic strains formed a distinct sublineage (sublineage A), related to B. japonicum (sublineage B) at a correlation coefficient of 86%; both clusters are related to the B. elkanii cluster (sublineage C) at a correlation coefficient of 80%.
rRNA-based phylogenetic investigations have shown that the genus Bradyrhizobium is closely related to R. palustris, a photosynthetic bacterium able to grow photoautotrophically under anaerobic conditions (29, 57). This would suggest that Bradyrhizobium may have evolved from photosynthetic free-living bacteria by the acquisition of symbiotic functions. Most bradyrhizobia are root symbionts living in a soil-root environment where they are not exposed to significant levels of light. As a consequence of low selection pressure, photosynthetic function may have been lost during evolution from a free-living existence to a symbiotic one. In the particular case of stem nodule symbionts, however, the ancestral trait of photosynthesis may have been retained since remaining genetic information for heterotrophic photosynthesis could still be a selective advantage in both free-living and symbiotic states. The natural habitat of stem-nodulated legumes is restricted to tropical waterlogged or very humid, nitrogen- and carbon-deficient soils. In waterlogged soil or on the plant surface, bacterial photosynthesis may sustain better growth and survival of bacteria and give a competitive advantage for stem nodulation. In symbiosis, bacterial photosynthesis may allow more efficient interaction by reducing the need of the microsymbiont for carbon. Our simultaneous observation in the same Bradyrhizobium phylogenetic group of photosynthetic characteristics and specific nodulation abilities supports the hypothesis that a branch of ancestral photosynthetic bacteria has adapted to the particular stem-nodulated Aeschynomene environment through acquisition of specific symbiotic functions and conservation of photosynthetic characteristics. However this remains speculative and, alternatively, the possibility that symbiotic bradyrhizobia acquired photosynthetic genes by lateral transfer cannot be excluded. Phylogenetic studies of nodulation and photosynthetic genes may elucidate the origin of these genes. Further investigation is needed to evaluate the role of bacterial photosynthesis in the symbiotic interaction and to evaluate whether preservation of photosynthetic functions reflects an adaptation to the stem-nodulated Aeschynomene environment.
ACKNOWLEDGMENTS
We thank D. Alazard for kindly providing Aeschynomene strains.
This work was supported in part by the Commission of the European Communities (STD3 programme, contract TS2 0169-F; BRIDGE programme, contracts BIOT-CT91-0263 and BIOT-CT91-0294), in part by the French and Belgian Embassies through Programme d’Actions Intégrées franco-belge Tournesol, in part by the Bureau des Ressources Génétiques (France), and in part by CNRS (Dynamique de la Biodiversité et Environnement). F.M. is indebted to I. R. D. (ex ORSTOM) for a doctoral grant. M.G. is indebted to the Fund for Scientific Research-Flanders (Belgium), for research and personnel grants. A.W. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral research fellow.
REFERENCES
- 1.Alazard D. Stem and root nodulation in Aeschynomene spp. Appl Environ Microbiol. 1985;50:732–734. doi: 10.1128/aem.50.3.732-734.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alazard D. Nitrogen fixation in pure culture by rhizobia isolated from stem nodules of tropical Aeschynomene species. FEMS Microbiol Lett. 1990;68:177–182. [Google Scholar]
- 3.Alazard D. La nodulation caulinaire dans le genre Aeschynomene. Ph.D. thesis. Lyon, France: University Claude Bernard-Lyon I; 1991. [Google Scholar]
- 3a.Alazard, D. Unpublished data.
- 4.Bauer C E, Young D A, Marrs B L. Analysis of the Rhodopseudomonas capsulata puf operon. Location of the oxygen-regulated promoter region and identification of an additional puf-encoded gene. J Biol Chem. 1988;263:4820–4827. [PubMed] [Google Scholar]
- 5.Belasco J G, Beatty J T, Adams C W, von Gabian A, Cohen S N. Differential expression of photosynthetic genes in Rhodopseudomonas capsulata results from segmental differences in stability within a polycistronic transcript. Cell. 1985;40:171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- 6.Boivin C, N’doye I, Molouba F, de Lajudie P, Dupuy N, Dreyfus B. Stem nodulation in legumes: diversity, mechanisms and unusual characters. Crit Rev Plant Sci. 1997;16:1–30. [Google Scholar]
- 7.Carlson R W, Juan S J, Bhat U R, Glushka J, Spaink H P, Wijfjes A H M, van Brussel A N N, Stokkermans T J W, Peters N K, Stacey G. The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type-1 and type-2 strains of Bradyrhizobium japonicum. J Biol Chem. 1993;268:18372–18381. [PubMed] [Google Scholar]
- 8.Chen C Y A, Beatty J T, Cohen S, Belasco J G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988;52:609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- 9.Cogdell R J, Thornber J P. The preparation and characterization of different types of light-harvesting complexes from some purple bacteria. Ciba Found Symp. 1979;61:61–79. doi: 10.1002/9780470720431.ch4. [DOI] [PubMed] [Google Scholar]
- 10.de Lajudie P, Fulele-Laurent E, Willems A, Tork U, Coopman R, Collins M D, Kersters K, Dreyfus B L, Gillis M. Description of Allorhizobium undicola gen. nov. sp. nov. for nitrogen-fixing bacteria efficiently nodulating Neptunia natans in Senegal. Int J Syst Bacteriol. 1998;48:1277–1290. doi: 10.1099/00207713-48-4-1277. [DOI] [PubMed] [Google Scholar]
- 11.Dénarié J, Debellé F, Promé J C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupuy N, Willems A, Pot B, Dewettinck D, Vandenbruaene I, Maestrojuan G, Dreyfus B, Kersters K, Collins M D, Gillis M. Phenotypic and genotypic characterization of bradyrhizobia nodulating the leguminous tree Acacia albida. Int J Syst Bacteriol. 1994;44:461–473. doi: 10.1099/00207713-44-3-461. [DOI] [PubMed] [Google Scholar]
- 14.Eaglesham A R J, Szalay A A. Aerial stem nodules on Aeschynomene spp. Plant Sci Lett. 1983;29:265–272. [Google Scholar]
- 15.Eaglesham A R J, Ellis J M, Evans W R, Fleischman D E, Hungria M, Hardy R W F. The first photosynthetic N2-fixing Rhizobium: characteristics. In: Gresshoff P M, Roth L E, Stacey G, Newton W L, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. pp. 805–811. [Google Scholar]
- 16.Ellis J M, Eardly B D, Hungria M, Hardy R W F, Rizzo N W, Eaglesham A R J. 12th North American Symbiotic Nitrogen Fixation Conference proceedings, Ames, Iowa. 1989. Photosynthetic N2-fixing Rhizobium; p. 101. [Google Scholar]
- 17.Evans W R, Fleischman D E, Calvert H E, Pyati R V, Alter G M, Subba Rao N S. Bacteriochlorophyll and photosynthetic reaction centers in Rhizobium strain BTAi1. Appl Environ Microbiol. 1990;56:3445–3449. doi: 10.1128/aem.56.11.3445-3449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein J. Numerical methods for inferring evolutionary trees. Q Rev Biol. 1982;57:379–404. [Google Scholar]
- 18a.Ferro, M., et al. Personal communication.
- 19.Fleischman D, Kramer D. Photosynthetic rhizobia. Biochim Biophys Acta. 1998;1364:17–36. doi: 10.1016/s0005-2728(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst J A, Hawkins J A, Holmes A, Sly L I, Moore C J, Stackebrandt E. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from fresh water. Int J Syst Bacteriol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 20a.Giraud, E. Unpublished data.
- 21.Goodwin T W. The carotenoids of photosynthetic bacteria. II. The carotenoids of a number of non-sulphur bacteria (Athiorhodaceae) Arch Mikrobiol. 1956;24:313–322. doi: 10.1007/BF00693102. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin T W. Plants. 2nd ed. Vol. 1. London, United Kingdom: Chapman & Hall, Ltd.; 1980. The biochemistry of the carotenoids; pp. 257–345. [Google Scholar]
- 23.Goodwin T W, Osman H G. Studies on carotenogenesis: spirilloxanthin synthesis by washed cells of Rhodospirillum rubrum. Biochem J. 1954;56:222–227. doi: 10.1042/bj0560222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harashima K, Shiba T, Totsuka T, Simidu U, Taga N. Occurrence of bacteriochlorophyll a in a strain of an aerobic heterotrophic bacterium. Agric Biol Chem. 1978;42:1627–1628. [Google Scholar]
- 25.Hirsch P, Müller M. Blastobacter aggregatus sp. nov., Blastobacter capsulatus sp. nov., and Blastobacter denitrificans sp. nov., new budding bacteria from freshwater habitats. Syst Appl Microbiol. 1985;6:281–286. [Google Scholar]
- 26.Hungria M, Eaglesham A R J, Hardy R W F. Physiological comparisons of root and stem nodules of Aeschynomene scabra and Sesbania rostrata. Plant Soil. 1992;139:7–13. [Google Scholar]
- 27.Hungria M, Ellis J M, Eaglesham A R J, Hardy R W F. Light-driven 14CO2 fixation, light-decreased O2 uptake, and acetylene reduction activity by free-living Rhizobium strain BTAi1. In: Gresshoff P M, Roth L E, Stacey G, Newton W E, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. p. 351. [Google Scholar]
- 28.Hungria M, Ellis J M, Hardy R W F, Eaglesham A R J. Light-stimulated 14CO2 uptake and acetylene reduction by bacteriocholorophyll containing stem nodule isolate BTAi1. Biol Fertil Soils. 1993;15:208–214. [Google Scholar]
- 29.Jarvis B D W, Gillis M, De Ley J. Intra- and intergeneric similarities between the ribosomal ribonucleic acid cistrons of Rhizobium and Bradyrhizobium species and some related bacteria. Int J Syst Bacteriol. 1986;36:129–138. [Google Scholar]
- 30.Jordan D C. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing root nodule bacteria from leguminous plants. Int J Syst Bacteriol. 1982;32:136–139. [Google Scholar]
- 31.Jordan D C. Rhizobiaceae Conn 1938, 321AL. In: Krieg N R, Holt J C, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams and Wilkins Co.; 1984. pp. 234–236. [Google Scholar]
- 32.Kuykendall L M, Saxena B, Devine T E, Udell S E. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol. 1992;38:501–503. [Google Scholar]
- 33.Ladha J K, So R B. Numerical taxonomy of photosynthetic rhizobia nodulating Aeschynomene species. Int J Syst Bacteriol. 1994;44:62–73. [Google Scholar]
- 34.Ladha J K, Pareek R P, Becker M. Stem-nodule symbiosis and its unusual properties. In: Gresshoff P M, Roth L E, Stacey G, Newton W L, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. pp. 633–640. [Google Scholar]
- 35.Lorquin J, Molouba F, Dreyfus B L. Identification of the carotenoid canthaxanthin from photosynthetic Bradyrhizobium strains. Appl Environ Microbiol. 1997;63:1151–1154. doi: 10.1128/aem.63.3.1151-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lortet G, Mear N, Lorquin J, Dreyfus B, De Lajudie P, Rosenberg C, Boivin C. Nod factor thin-layer chromatography profiling as a tool to characterize symbiotic specificity of rhizobial strains: application to Sinorhizobium saheli, S. teranga and Rhizobium sp. strains isolated from Acacia and Sesbania. Mol Plant-Microbe Interact. 1996;9:736–747. [Google Scholar]
- 37.Moreira F, Gillis M, Pot B, Kersters K, Franco A A. Characterization of rhizobia from different divergence groups of tropical Leguminosae by comparative polyacrylamide gel electrophoresis of their total proteins. Syst Appl Microbiol. 1993;16:135–146. [Google Scholar]
- 38.Nelis H J, De Leenheer A P. Microbial sources of carotenoid pigments used in foods and feeds. J Appl Bacteriol. 1991;70:181–191. [Google Scholar]
- 39.Nishimura Y, Shimadzu M, Iizuka H. Bacteriocholorophyll formation in radiation-resistant Pseudomonas radiora. J Gen Appl Microbiol. 1981;27:427–430. [Google Scholar]
- 40.Pfennig N. General physiology and ecology of photosynthetic bacteria. In: Clayton R K, Sistrom W R, editors. The photosynthetic bacteria. New York, N.Y: Plenum Press; 1978. pp. 3–18. [Google Scholar]
- 41.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 42.Remsen C C. Comparative subcellular architecture of photosynthetic bacteria. In: Clayton R K, Sistrom W R, editors. The photosynthetic bacteria. New York, N.Y: Plenum Press; 1978. pp. 31–60. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sanjuan J, Carlson R W, Spaink H P, Baht U R, Barbour W M, Glushka J, Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato K. Bacteriochlorophyll formation by facultative methylotrophs, Protaminobacter ruber and Pseudomonas AM 1. FEBS Lett. 1978;85:207–210. doi: 10.1016/0014-5793(78)80456-6. [DOI] [PubMed] [Google Scholar]
- 46.Shanmugasundaram S, Suguna S, Fleischman D. 12th North American Symbiotic Nitrogen Fixation Conference proceedings, Ames, Iowa. 1989. Genetic evidence for the presence of bacterial photosynthetic gene(s) in Rhizobium sp. isolated from Aeschynomene indica; p. 56. [Google Scholar]
- 47.So R B, Ladha J K, Young J P W. Photosynthetic symbionts of Aeschynomene spp. form a cluster with Bradyrhizobia on the basis of fatty acid and rRNA analyses. Int J Syst Bacteriol. 1994;44:392–403. doi: 10.1099/00207713-44-3-392. [DOI] [PubMed] [Google Scholar]
- 48.Van Berkum P, Tully R E, Keister D L. Nonpigmented and bacteriochlorophyll-containing Bradyrhizobia isolated from Aeschynomene indica. Appl Environ Microbiol. 1995;61:623–629. doi: 10.1128/aem.61.2.623-629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaneechoutte M, de Beenhouwer H, Clayes G, Verschraegen G, de Rouck A, Paepa N, Elaichouni A, Portaels F. Identification of Mycobacterium by using amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1993;31:2061–2065. doi: 10.1128/jcm.31.8.2061-2065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Grondelle R, Dekker J P, Gillbro T, Sundström V. Energy transfer and trapping in photosynthesis. Biochim Biophys Acta. 1994;1187:1–65. [Google Scholar]
- 51.Van Rhijn P J S, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Rossum D, Schuurmans F P, Gillis M, Muyotcha A, Van Verseveld H W, Stouthamer A H, Boogerd F C. Genetic and phenotypic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl Environ Microbiol. 1995;61:1599–1609. doi: 10.1128/aem.61.4.1599-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 54.Vincent J M. A manual for the practical study of root nodule bacteria. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1970. [Google Scholar]
- 55.Wong F Y K, Stackebrandt E, Ladha J K, Fleischman D E, Date A R, Fuerst J A. Phylogenetic analysis of Bradyrhizobium japonicum and photosynthetic stem-nodulating bacteria from Aeschynomene species grown in separated geographical regions. Appl Environ Microbiol. 1994;60:940–946. doi: 10.1128/aem.60.3.940-946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu L M, Ge C, Cui Z, Li J, Fan H. Bradyrhizobium liaoningensis sp. nov. isolated from the root nodules of soybean. Int J Syst Bacteriol. 1995;45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 57.Yanagi M, Yamasato K. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett. 1993;107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 58.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 59.Young J P W, Downer H L, Eardly B D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Youvan D C, Bylina E J, Alberti M, Begusch H, Hearst J E. Nucleotide and deduced polypeptide sequence of the photosynthetic reaction center, B870 antenna, and flanking polypeptides from Rhodopseudomonas capsulata. Cell. 1984;37:949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]