Abstract
The catabolic or biodegradative threonine dehydratase (E.C. 4.2.1.16) of Escherichia coli is an isoleucine feedback-resistant enzyme that catalyzes the degradation of threonine to α-ketobutyrate, the first reaction of the isoleucine pathway. We cloned and expressed this enzyme in Corynebacterium glutamicum. We found that while the native threonine dehydratase of C. glutamicum was totally inhibited by 15 mM isoleucine, the heterologous catabolic threonine dehydratase expressed in the same strain was much less sensitive to isoleucine; i.e., it retained 60% of its original activity even in the presence of 200 mM isoleucine. To determine whether expressing the catabolic threonine dehydratase (encoded by the tdcB gene) provided any benefit for isoleucine production compared to the native enzyme (encoded by the ilvA gene), fermentations were performed with the wild-type strain, an ilvA-overexpressing strain, and a tdcB-expressing strain. By expressing the heterologous catabolic threonine dehydratase in C. glutamicum, we were able to increase the production of isoleucine 50-fold, whereas overexpression of the native threonine dehydratase resulted in only a fourfold increase in isoleucine production. Carbon balance data showed that when just one enzyme, the catabolic threonine dehydratase, was overexpressed, 70% of the carbon available for the lysine pathway was redirected into the isoleucine pathway.
Corynebacteria have a long history of use in the industrial production of amino acids, which are used as food additives (most notably lysine and other essential amino acids) and as flavor enhancers (monosodium glutamate). The overall global market for amino acids as animal feed additives is estimated to be worth more than $2 billion and totals about 700,000 metric tons of material. Lysine and methionine account for an overwhelming majority of the market, which also includes lower-volume products, such as threonine (14). The current level of production of isoleucine is less than 400 metric tons per year. This amino acid is currently used as a constituent of infusions and special dietary products. Like the demand for other amino acids, the demand for isoleucine is increasing, and industrial production of isoleucine is expected to open up additional animal feed additive markets. Due to the tight control of isoleucine biosynthesis in bacteria, some isoleucine is still produced commercially by direct extraction from protein hydrolysates.
The gram-positive organism Corynebacterium glutamicum is currently used in industry to produce more than 100 g of lysine per liter of culture. The flux of carbon through metabolic pathways can be diverted from the production of lysine to the production of related amino acids by the processes and methods of metabolic engineering. Traditional metabolic manipulation involves random mutagenesis and screening for desired changes in physiology, but in the last 10 years workers have developed transformation and genetic manipulation tools which allow more direct engineering of specific pathway elements in Corynebacterium spp. (11, 14).
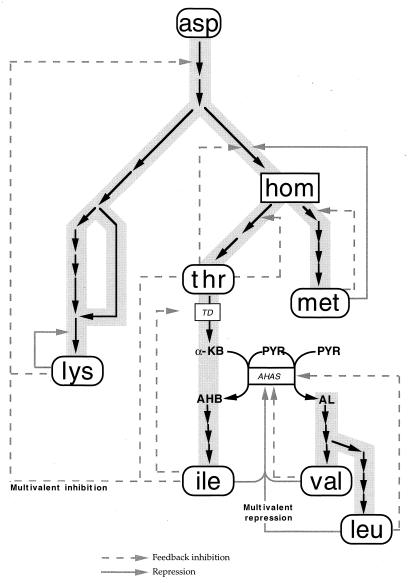
l-Isoleucine belongs to the aspartate-derived family of amino acids, as do lysine, homoserine, threonine, and methionine. The enzymes that synthesize this family of amino acids have been well-characterized in Corynebacterium spp., as has the regulation of these enzymes (Fig. 1). The first important regulatory point during production of isoleucine by C. glutamicum is end product inhibition of the first committed enzyme, threonine dehydratase (E.C. 4.2.1.16), which is encoded by the ilvA gene . Isoleucine has been overproduced by introducing excess threonine dehydratase (encoded by ilvA) into threonine-producing strains (3). Threonine dehydratase is normally feedback inhibited by isoleucine. Mutant derivatives of threonine dehydratase with reduced sensitivity to isoleucine have been an additional dividend in this isoleucine production system (9, 16). Despite these gains, it appears that amino acid export has seriously limited the effectiveness of amino acid production (13).
FIG. 1.
Aspartate-derived amino acid pathway. Abbreviations: asp, aspartic acid; hom, homoserine; thr, threonine; met, methionine; lys, lysine; ile, isoleucine; val, valine; leu, leucine; α-KB, alpha-ketobutyrate; AHB, acetohydroxybutyric acid; PYR, pyruvate; AL, acetolactate; AHAS, acetohydroxyacid synthase; TD, threonine dehydratase.
Work on artificial enzyme evolution has shown that it is difficult to subtly alter a task for which an enzyme specifically evolved, while it is easier to coopt an enzyme for a completely new task (1). This may be the case for an enzyme like threonine dehydratase, which specifically evolved sensitivity to isoleucine in order to regulate its biosynthetic function. It would be much more effective to select an enzyme that is not subject to the same mechanism of inhibition and to employ this enzyme in the isoleucine synthesis pathway than to try to change the role of an enzyme already in the pathway.
One such alternative enzyme might be the catabolic enzyme threonine dehydratase, also called biodegradative threonine dehydratase (E.C. 4.2.1.16), of Escherichia coli, which is encoded by the gene tdcB. This threonine dehydratase is produced in E. coli cells when the organism is grown anaerobically in a medium containing high concentrations of amino acids and no glucose (20). In contrast to the threonine dehydratase encoded by ilvA, the enzyme encoded by tdcB is not sensitive to inhibition by l-isoleucine and is activated by AMP (20). The tdcB gene of E. coli has been cloned and sequenced previously (8).
In order to assess the feasibility of increasing isoleucine production by using an E. coli catabolic threonine dehydratase in C. glutamicum, we cloned and overexpressed the tdcB gene in a lysine-producing strain. In this work, we demonstrated the utility of this approach. We found that a strain of C. glutamicum expressing the E. coli tdcB gene produced significantly more isoleucine than an isogenic strain overexpressing the native threonine dehydratase (encoded by ilvA) produced.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium or 2xYT medium (18) was used as the standard medium, and a medium containing 40 g of brain heart infusion per liter, 20 g of sorbitol per liter, and 10 g of sucrose per liter was used as the recovery broth for electroporated cells. The minimal medium used for E. coli was M9 medium (18). E. coli AB1255 (17) was obtained from the E. coli Genetic Stock Center, courtesy of Barbara Bachman, and the minimal medium used for this strain was supplemented with 100 mg of histidine per liter, 100 mg of arginine per liter, and 100 mg of methionine per liter.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or descriptiona | Source or referenceb |
|---|---|---|
| C. glutamicum AS019-E12 | Restriction-deficient derivative of C. glutamicum AS019 | 7 |
| C. lactofermentum ATCC 21799 | Lysine-producing strain, Leu− AECr | ATCC |
| E. coli strains | ||
| BW310 | λ−ung-1 relA1 spoT1 thi-1 | 4 |
| AB1255 | ilvA201 metB1 hisG1 argH1 | 17 |
| Plasmids | ||
| pGC77 | Kmr Apr, lacIqhomdrtac::thrB ilvA | 3 |
| pCRscript | Apr | Stratagene |
| tdcB-PCRscript | Apr, tdcB (no promoter) | This study |
| pTrc99a | Apr, lacIqtrc | Pharmacia |
| pEP2 | Kmr, NG2 rep | 21 |
| pAPE5 | Apr, lacIqtrc:tdcB | This study |
| pAPE7 | Kmr, lacIqtrc:tdcB | This study |
| pAPE12 | Kmr, lacIqtrc | This study |
| pAPE13 | Kmr, lacIqtrc:ilvA | This study |
| pAPE16 | Apr, trc:ilvA | This study |
| pAPE17 | Apr, lacIqtrc:ilvA | This study |
AECr, resistance to aminoethylcysteine.
ATCC, American Type Culture Collection, Rockville, Md.
The defined medium used for C. glutamicum contained (per liter) 35 g of glucose, 2 g of NaCl, 3 g of citrate (trisodium salt, dihydrate), 0.1 g of CaCl2 · 2H2O, 0.5 g of MgSO4 · 7H2O, 75 mg of Na2EDTA · 2H2O, 50 mg of FeSO4 · 7H2O, 20 ml of a 100× salt solution, 4 g of K2HPO4, 2 g of KH2PO4, 7.5 g of (NH4)2SO4, 3.75 g of urea, 0.85 g of leucine, 0.45 mg of thiamine, 0.45 mg of biotin, and 4.5 mg of pantothenic acid; the pH was 7.0. The salt solution contained (per liter) 200 mg of MnSO4, 20 mg of Na2B4O7 · 10H2O, 10 mg of (NH4)6Mo7O24 · 4H2O, 200 mg of FeCl3 · 6H2O, 50 mg of ZnSO4 · 7H2O, and 20 mg of CuCl2 · 2H2O, and the pH was 2.0. When appropriate, kanamycin (150 mg/liter) and isopropyl-β-d-thiogalactopyranoside (IPTG) (150 mg/liter) were included.
For the growth study performed with amino acid supplements, the defined medium was supplemented with Bacto Casamino Acids (Difco Laboratories, Detroit, Mich.) at a concentration of 2 g liter−1 or with an amino acid (alanine, glycine, methionine, or valine) at a concentration of 0.5 g liter−1.
Cloning.
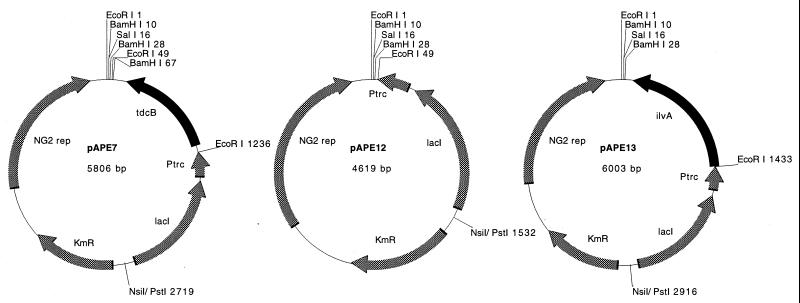
tdcB and ilvA coding regions were amplified by the PCR (Table 2) from BW310 genomic DNA and from pGC77 (Table 1), respectively. The PCR products were cloned into the pCRScript system (Stratagene, La Jolla, Calif.), which resulted in tdcB-pCRscript and pAPE16, respectively. The EcoRI-BamHI fragments of these pCRscript derivatives were cloned into pTrc99a (Pharmacia, Uppsala, Sweden) to create pAPE5 and pAPE17, respectively. Subsequently, the NsiI-SalI fragments of pAPE5, pAPE17, and pTrc99a (including the lacIq and Ptrc elements) were subcloned into pEP2 which had been cut with PstI and with SalI; this resulted in pAPE7, pAPE13, and pAPE12, all of which could replicate both in Corynebacterium spp. and in E. coli and expressed the appropriate gene product (or empty control) under control of the trc promoter. Plasmid maps are shown in Fig. 2.
TABLE 2.
PCR products and primers
| PCR product | Template | Description | Primer 1 | Primer 2 |
|---|---|---|---|---|
| tdcB | BW310 genomic DNA | Coding region for catabolic threonine dehydratase from E. coli | 5′ GGGAATTCGGTGTCGGTTACGGTTACCT 3′ | 5′ CCGGTACCCCAAACAAGCCTAACGTCCA 3′ |
| ilvA | pGC77 | Coding region for anabolic threonine dehydratase from C. glutamicum | 5′ GGAATTCATGAGTGAAACATACGTGTCTGA 3′ | 5′ CCACGCGTGGGGCTTTGCGATCCT 3′ |
FIG. 2.
Plasmid maps. Abbreviations: NG2 rep, open reading frame from the NG2 replicon permitting plasmid replication in both E. coli and C. glutamicum; Ptrc, trc promoter from pTrc99a; lacI, open reading frame encoding the lac repressor from pTrc99a; KmR, kanamycin resistance gene from pEP2. NsiI/PstI 2719, NsiI/PstI 1532, and NsiI/PstI 2916 indicate the positions of hybrid NsiI and PstI sites resulting from ligation.
Enzyme assays.
Enzyme assays were performed with cell-free crude extracts prepared by the following method. Cells were harvested by centrifugation for 10 min at 5,000 × g at 4°C and were washed with 10 ml of the enzyme assay buffer (100 mM Tris-HCl [pH 7.5] containing 20 mM KCl, 5 mM MnSO4, 0.1 mM EDTA, and 2 mM dithiothreitol) (12). Each cell pellet was resuspended in the same buffer containing a protease inhibitor cocktail (Boerhinger, Mannheim, Germany) so that the final concentration was 20 g (dry cell weight [DCW]) of cells · liter−1. The resuspended cells were disrupted with a glass bead mixer (model 5100 Mixer-Mill; SPEX, Metuchen, N.J.). A 2.5-ml portion of each bacterial suspension was poured into a frozen steel vial containing 5 g of cold 106-μm-diameter glass beads (Sigma) and one stainless steel ball bearing. The vials containing bacterial suspensions and glass beads were vigorously shaken at 4°C by using the mixer and 10 30-s shaking cycles separated by 1-min rest cycles. Cell debris was removed by centrifugation for 20 min at 47,000 × g at 4°C. Each supernatant (crude extract) was then tested for enzyme activity. Protein concentrations were determined by the method of Bradford (2) with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.); bovine serum albumin was used as the standard.
To determine threonine dehydratase activity, the 1-ml (final volume) assay mixtures contained 40 mM threonine, 1 mM pyridoxal phosphate, crude extract, and 100 mM potassium phosphate buffer (pH 8.0). Each reaction was started by adding threonine and was terminated by adding 1 ml of a solution containing 1% semicarbazide and 0.9% sodium acetate. After 15 min of incubation at room temperature, the amounts of α-ketobutyrate formed at various times were determined spectrophotometrically by monitoring the semicarbazone derivative at 254 nm (ɛ = 0.52 mmol · cm−1). Relevant standards and controls were treated in the same manner. To determine catabolic threonine dehydratase activity, the assay mixtures contained 20 mM isoleucine in order to inhibit the anabolic threonine dehydratase.
In Fig. 3 through 5 the relative threonine dehydratase activities are reported as the ratios of the specific activities of the enzymes from the recombinant strains to the specific activity of enzyme from the wild-type strain. Each assay was replicated five times, and the results were reproducible within 15%.
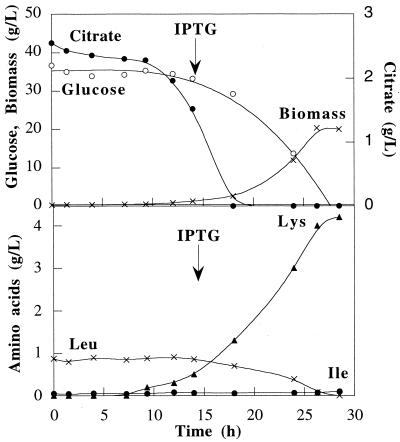
FIG. 3.
C. glutamicum ATCC 21799 culture grown in a batch reactor containing defined medium: kinetics of growth, substrate consumption, amino acid production, and production of threonine dehydratase. Arrows indicate the time at which IPTG was added to the cultures.
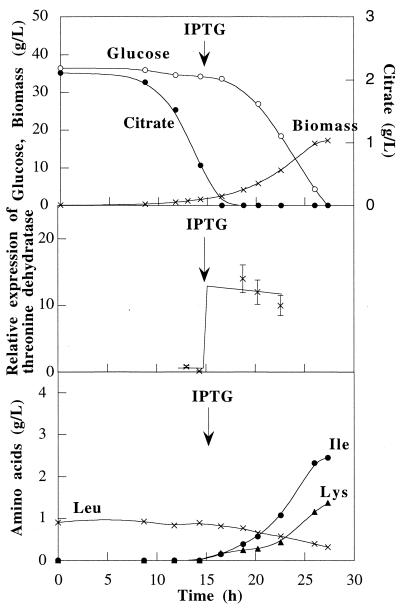
FIG. 5.
C. glutamicum ATCC 21799(pAPE7) culture grown in a batch reactor containing defined medium: kinetics of growth, substrate consumption, amino acid production, and production of catabolic threonine dehydratase.
Fermentations.
Starter cultures of C. glutamicum were prepared by transferring single colonies from LB agar plates to 5 ml of LB medium. The resulting cultures were incubated for 2 days at 30°C and 200 rpm. Then 500-ml Erlenmeyer flasks containing 50 ml of defined medium were inoculated with 1-ml portions of the starter cultures. The resulting cultures were incubated at 30°C and 200 rpm for 30 h. These flask cultures were used as 5% (vol/vol) inocula for 2-liter portions of defined medium in a 4-liter Chemap CMF100 reactor (Alfa-Laval Chemap, Dietlikon, Switzerland). Each culture was grown at 30°C with an aeration rate of 0.45 vol/vol/min (vvm) and with agitation at 1,500 rpm. The pH was maintained at 7.5 with ammonium hydroxide and hydrochloric acid solutions. Fermentations were carried out twice for each strain.
Determination of biomass, sugars, organic acids, and amino acids.
During fermentation, samples were collected and centrifuged at 10,000 × g and 4°C for 10 min. For biomass determinations, cell dry weights were determined gravimetrically. To determine glucose, organic acid, and amino acid contents, samples were collected and filtered through 0.2-μm-pore-size Acrodisc filters (Gelman Sciences, Ann Arbor, Mich.). Sugar and organic acid concentrations were determined by high-pressure liquid chromatography by using a model 1050 system (Hewlett-Packard, Waldbronn, Germany) and an Aminex HPX-87H column (Bio-Rad). Samples were analyzed at 40°C by using 5 mM sulfuric acid as the mobile phase at a flow rate of 0.6 ml · min−1. Sugars were detected with a refractive index detector (Hewlett-Packard model 1047A), and organic acids were detected with a UV detector (Hewlett-Packard model 1050). The results of replicate measurements of glucose contents were reproducible within 5%.
Amino acids were analyzed as ortho-phthaldialdehyde derivatives by reversed-phase chromatography by using a C18 AminoQuant column and a Hewlett-Packard model 1050 high-pressure liquid chromatography system. The results of amino acid determinations were reproducible within 5% in replicate assays.
RESULTS
Overexpression of tdcB in E. coli and in C. glutamicum.
Threonine dehydratase activity assays were performed with crude extracts obtained from cultures of E. coli AB1255, which cannot produce anabolic threonine dehydratase (encoded by the ilvA gene), and the same strain carrying plasmid pAPE5 containing the tdcB gene. Even in the presence of very high concentrations of isoleucine (up to 47 mM), the AB1255(pAPE5) extracts exhibited consistent activity of about 80 μmol of product · min−1 · mg of protein−1, while AB1255(pTrc99a) control extracts exhibited no measurable activity at any isoleucine concentration. These results indicate that the tdcB gene product can be overexpressed under aerobic conditions in E. coli and still retain its function.
In order to test expression of the catabolic threonine dehydratase in Corynebacterium sp., the trc:tdcB fusion was subcloned from pAPE5 into the expression vector pAPE12, and the resulting plasmid (pAPE7) was expressed in C. glutamicum AS019-E12. Whereas the control strain, C. glutamicum AS019-E12 carrying pAPE12, exhibited no detectable catabolic threonine dehydratase activity, the strain carrying pAPE7 produced 9 μmol of product · min−1 · mg of protein−1. Although the level of activity in crude extracts of this strain was about 10-fold lower than the level of activity in E. coli extracts, the levels of activity were actually proportional to the plasmid copy numbers in these two bacteria, since the copy number of pAPE7 in Corynebacterium sp. should be 10-fold lower than the copy number of pAPE5 in E. coli. These results showed that tdcB was expressed in the heterologous species.
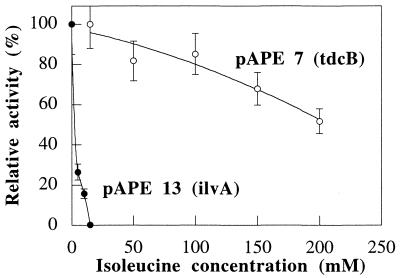
Isoleucine sensitivity of the threonine dehydratases.
In order to confirm that the catabolic dehydratase expressed in a lysine-producing strain of C. glutamicum was not sensitive to isoleucine, threonine dehydratase activities were measured in crude extracts of strains ATCC 21799 and ATCC 21799 carrying pAPE7 in the presence of different concentrations of isoleucine (Fig. 6). The anabolic threonine dehydratase of wild-type strain ATCC 21799 was completely inhibited by an isoleucine concentration of 15 mM. On the other hand, the catabolic threonine dehydratase expressed in strain ATCC 21799(pAPE7) was much less sensitive to isoleucine; it retained 60% of its original activity even at an isoleucine concentration of 200 mM.
FIG. 6.
Sensitivities of threonine dehydratases expressed in C. glutamicum ATCC 21799 to different isoleucine concentrations.
Fermentation results.
In order to determine whether there was an advantage to using the catabolic threonine dehydratase for production of isoleucine, fermentations were performed with C. glutamicum ATCC 21799 as a control and two derivatives of this strain carrying either plasmid pAPE13 containing the trc:ilvA fusion or plasmid pAPE7 containing the trc:tdcB fusion.
(i) Fermentation with the wild-type strain.
C. glutamicum ATCC 21799 was grown twice in defined medium in a 4-liter reactor. IPTG (150 mg/liter) was added when the biomass reached 1.5 g DCW · liter−1. The growth kinetics, substrate consumption, threonine dehydratase specific activity, and amino acid production data are shown Fig. 3. This strain grew exponentially with a specific growth rate of 0.3 h−1 and a glucose-to-biomass conversion yield of 0.53 g DCW · g of glucose−1. The basal level of threonine dehydratase remained constant around 1 μmol of α-ketobutyrate · min−1 · mg of protein−1 during fermentation. The strain produced mainly lysine at final concentrations of 3.9 and 4.2 g · liter−1 in the two fermentations. An isoleucine concentration of 50 mg · liter−1 was detected at the end of fermentation for both cultures. Oxygenation of the cultures was sufficient, and thus neither lactate nor acetate was detected during fermentation.
(ii) Fermentation with the ilvA-overexpressing strain.
C. glutamicum ATCC 21799 harboring pAPE13 was grown twice in defined medium in a 4-liter reactor. As described above, IPTG (150 mg/liter) was added when the biomass reached 1.5 g DCW · liter−1 (Fig. 4). The specific growth rate and the glucose-to-biomass conversion yield of this strain were identical to those of the wild-type strain. Addition of IPTG resulted in a 20-fold increase in the level of threonine dehydratase activity. ATCC 21799(pAPE13) still produced mainly lysine; the final concentrations of lysine were 3.2 and 2.9 g · liter−1 in the two fermentations. The concentration of isoleucine was 0.2 g · liter−1 at the end of both fermentations.
FIG. 4.
C. glutamicum ATCC 21799(pAPE13) culture grown in a batch reactor containing defined medium: kinetics of growth, substrate consumption, amino acid production, and production of threonine dehydratase.
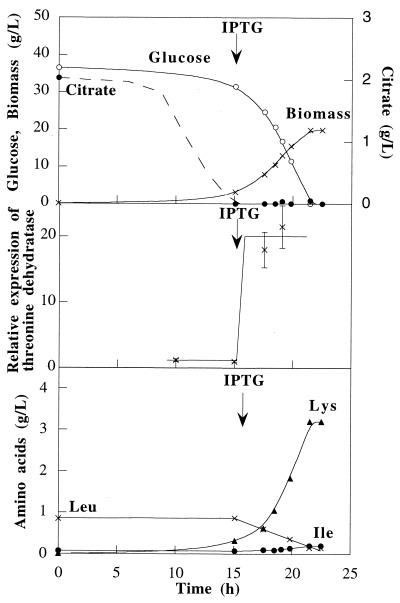
(iii) Fermentation with the tdcB-overexpressing strain.
C. glutamicum ATCC 21799 harboring pAPE7 was grown twice in defined medium in a 4-liter reactor under the agitation, aeration, and IPTG addition conditions described above for strains ATCC 21799 and ATCC 21799(pAPE13) (Fig. 5). The specific growth rate (0.22 h−1) and the glucose-to-biomass conversion yield (0.46 g · g−1) of this strain were lower than the values obtained for the wild-type strain and the ilvA-overexpressing strain. Addition of IPTG resulted in the synthesis of catabolic threonine dehydratase, and the concentration of this enzyme was 15-fold higher than the concentration of the original enzyme. The level of catabolic threonine dehydratase activity remained high during the fermentation. As a result, ATCC 21799(pAPE7) produced 2.5 g of isoleucine · liter−1 and 1.3 g of lysine · liter−1 during the first fermentation and 2.3 g of isoleucine · liter−1 and 1.3 g of lysine · liter−1 during the second fermentation.
(iv) Growth study performed with amino acid supplements.
To investigate the slow growth of the tdcB-expressing strain, ATCC 21799 containing pAPE7 and ATCC 21799 containing pAPE13 were cultured in conical flasks containing defined medium supplemented with amino acids (Table 3). When the tdcB-expressing strain was cultured on defined medium and defined medium supplemented with a mixture of amino acids obtained from a casein hydrolysate (Casamino Acids), the optimal growth rates were 0.17 h−1 without Casamino Acids and 0.29 h−1 with Casamino Acids. The latter growth rate was comparable to the growth rates of the wild-type and ilvA-overexpressing strains (0.30 h−1). In order to determine which specific amino acid was limiting and caused the low growth rate in the tdcB-expressing strain, we supplemented the defined medium with specific amino acids based on their potential interactions with the isoleucine pathway. We tested valine and methionine due to their direct connections with the isoleucine pathway and alanine and glycine due to their indirect connections through the use of pyruvate, which is also a substrate in the isoleucine pathway. Our results showed that addition of valine and addition of methionine resulted in increases in the specific growth rate (to 0.24 and 0.26 h−1, respectively). Addition of both valine and methionine to the defined medium resulted in growth rates comparable to the growth rate obtained when Casamino Acids were added to the medium (0.29 h−1). Addition of alanine or glycine alone did not increase the growth rate of the tdcB-expressing strain (0.17 h−1).
TABLE 3.
Growth of ilvA- and tdcB-expressing C. glutamicum strains on defined medium supplemented with amino acids and 150 mg of IPTG per liter
| Supplement(s) added to defined medium | Specific growth rate (h−1) of:
|
|
|---|---|---|
| ATCC 21799(pAPE13) (ilvA) | ATCC 21799(pAPE7) (tdcB) | |
| None | 0.30 | 0.17 |
| Casamino Acids | 0.30 | 0.29 |
| Valine | NDa | 0.24 |
| Methionine | ND | 0.26 |
| Alanine | ND | 0.17 |
| Glycine | ND | 0.17 |
| Valine plus methionine | ND | 0.29 |
ND, not determined.
DISCUSSION
The first two enzymes in the threonine-to-isoleucine pathway (threonine dehydratase and acetohydroxy acid synthase [AHAS]) have been found to be important in isoleucine biosynthesis. Threonine dehydratase is sensitive to feedback inhibition by isoleucine, and therefore this enzyme can be a limiting factor for improvement of isoleucine biosynthesis. AHAS is also feedback inhibited by isoleucine, but this enzyme has been shown to be highly inducible in the presence of its substrate, α-ketobutyrate (3, 5). Therefore, threonine dehydratase has been the preferred target for metabolic engineering studies performed to increase carbon flux into the isoleucine pathway. In this study, we examined using the E. coli isoleucine-insensitive catabolic threonine dehydratase in C. glutamicum for production of isoleucine. To succeed, we had to deal with the following potential problems: (i) the efficiency of expression of an E. coli gene in C. glutamicum, (ii) the efficiency of expression in aerobic C. glutamicum cultures of an enzyme that is usually expressed under anaerobic conditions in E. coli, and (iii) conservation of the enzyme’s insensitivity to isoleucine in Corynebacterium sp.
In this study, the tdcB gene of E. coli encoding catabolic threonine dehydratase was cloned and inserted into an expression vector for C. glutamicum. We were able to express the tdcB gene in two different strains of C. glutamicum, AS019-E12 and ATCC 21799. In vitro enzymatic assays showed that the catabolic threonine dehydratase expressed in C. glutamicum was not sensitive to isoleucine. An isoleucine concentration of 200 mM resulted in only 40% inhibition of the catabolic threonine dehydratase, whereas 15 mM isoleucine completely inhibited the native threonine dehydratase in strain ATCC 21799. Morbach et al. (16) observed complete inhibition of the endogenous threonine dehydratase (encoded by ilvA) with an isoleucine concentration of 5 mM in C. glutamicum MH20-22B. Some authors have obtained deregulated threonine dehydratase by generating mutations in the ilvA gene (9, 15), and their mutated threonine dehydratase, V323A, exhibited 22% activity in the presence of 50 mM isoleucine (16), whereas the catabolic threonine dehydratase of E. coli retained 70 to 80% of its activity at the same isoleucine concentration. These results show the potential for using the catabolic threonine dehydratase of E. coli in Corynebacterium spp.
To determine whether expression of the catabolic threonine dehydratase (encoded by the tdcB gene) had any greater benefit in isoleucine production than overexpression of the native enzyme (encoded by ilvA) had, we constructed two overexpression vectors, one carrying the ilvA gene and the other carrying the tdcB gene. These plasmids were introduced into a lysine-producing strain, ATCC 21799, whose feedback-resistant aspartokinase permits a high level of lysine production (10). We showed that 20-fold overexpression of the ilvA gene product, threonine dehydratase, led to a fourfold increase in isoleucine production (0.2 g · liter−1). This low yield can be explained by a reduction in the activity of the threonine dehydratase due to feedback inhibition by isoleucine and/or by the inhibiting effect of leucine added to the medium. ATCC 21799 is a leucine auxotroph that requires leucine in the medium. However, excess leucine may decrease the activity of AHAS, the second enzyme of the isoleucine pathway, via feedback inhibition. AHAS has been found to be inhibited by leucine and valine, and its expression is repressed multivalently by all three branched-chain amino acids (5, 19). In comparison, 15-fold overexpression of the catabolic threonine dehydratase led to a 50-fold increase in isoleucine production (2.5 g · liter−1) at the expense of lysine production (1.4 g · liter−1). This observation eliminates the possibility that leucine inhibition of AHAS was responsible for the low isoleucine yield in the ilvA-overexpressing strain, since excess leucine was included in the tdcB-expressing cultures as well.
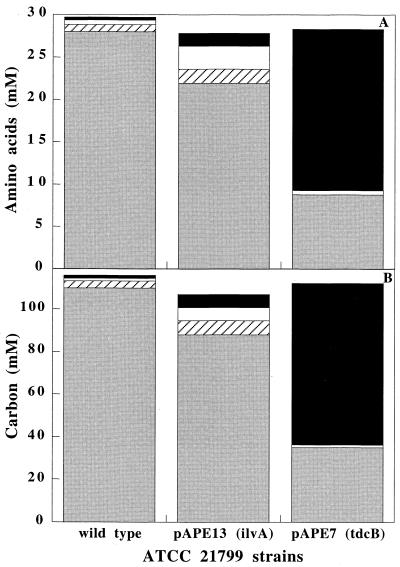
In order to determine the distribution of carbon throughout the aspartate-derived amino acid pathways in the different strains, the amino acid and carbon balances in the amino acid pathways of the strains were compared (Fig. 7). The carbon balances were corrected to account for incorporation of 1 mol of pyruvate into the lysine and isoleucine pathways and for economy of 1 mol of CO2 during synthesis of alanine and thus represent only derivatives of the carbon skeleton of aspartate. The three strains produced comparable amounts of amino acids. In the ilvA-overexpressing strain, only 5% of the carbon available for the aspartate-derived amino acid pathway was directed to isoleucine and 75% was directed to lysine. Higher concentrations of homoserine and alanine were produced in this strain. This finding can be explained by the fact that an increase in the homoserine concentration at the expense of lysine (as a result of ilvA overexpression) leads to greater availability of pyruvate that can be converted into alanine.
FIG. 7.
Amino acid concentrations (A) and carbon balances (B) for the three strains calculated from final fermentation titers. The lysine and isoleucine carbon values were multiplied by 0.67 to account for the pyruvate contributions in the branches. The alanine carbon value was multiplied by 0.75 to account for the economy of 1 mol of CO2 compared to the amino acids of the aspartate-derived pathway. Shaded bars, lysine; open bars, alanine; cross-hatched bars, homoserine; solid black bars, isoleucine.
The balances show that in the strain carrying the plasmid containing the tdcB gene the carbon flux was redirected from the lysine pathway to the isoleucine pathway. In this strain, 70% of the carbon available for the aspartate-derived amino acid pathway was converted into isoleucine. For comparison, Colon et al. (3) found that 80% of the carbon available for the aspartate-derived amino acid pathway was converted to isoleucine in ilvA-overexpressing strain ATCC 21799, but these authors used a threonine-overproducing strain as the host for ilvA overexpression. This host strain also overexpressed a deregulated homoserine dehydrogenase (encoded by homdr) and the homoserine kinase (encoded by thrB). Our study demonstrated that overexpression of tdcB alone in a lysine-producing strain is sufficient to increase isoleucine production to a level comparable to the level observed with a three-gene system (homdr thrB ilvA).
Growth kinetics showed that the strain carrying the plasmid containing the tdcB gene grew more slowly than the wild-type or ilvA-overexpressing strain. This could be explained by depletion of one or more amino acids, a consequence of the redirection of the carbon flux after expression of the tdcB gene. The inhibitory effect of α-ketobutyrate on the growth of C. glutamicum has been described previously (5). We found that addition of a mixture of amino acids obtained from a casein hydrolysate to the medium reestablished optimal growth of the strain expressing the tdcB gene. Further investigation showed that, specifically, addition of valine and addition of methionine led to partial recovery of the growth of this strain (80 and 86% recovery, respectively). Addition of these two amino acids together resulted in total growth recovery. Thus, overexpression of the feedback-resistant threonine dehydratase draws carbon away from the valine pathway. The isoleucine and valine pathways compete for pyruvate (Fig. 1) as a substrate. The enzyme AHAS catalyzes the second reaction of the isoleucine pathway by condensing α-ketobutyrate and pyruvate and also catalyzes the first reaction of the valine pathway by condensing two molecules of pyruvate. In contrast to E. coli, it has been shown that in C. glutamicum no isoenzymes of AHAS exist (5). The same authors showed that this enzyme has a higher Vmax and a threefold-higher affinity for α-ketobutyrate than for pyruvate. Moreover, the inhibitory effect of α-ketobutyrate on the growth of C. glutamicum has been described previously (5). Similarly, this inhibitory effect can be overcome by adding valine plus leucine. Thus, an increase in the catabolic threonine dehydratase activity leads to an increase in the amount of available α-ketobutyrate. This α-ketobutyrate outcompetes pyruvate, which leads to more acetohydroxybutyrate synthesis than acetolactate synthesis. The end result is that the valine supply falls short (excess leucine is supplied in the medium). Similarly, the fact that inhibition of the growth of the strain containing pAPE7 was overcome by adding methionine to the medium could be explained by the finding that overexpression of the catabolic threonine dehydratase directs carbon flux preferentially from homoserine to threonine at the expense of the methionine pathway, which also results in a reduced supply of methionine precursors.
We were able to direct expression of the tdcB gene under aerobic conditions by introducing a heterologous promoter. However, we made no attempt to alter the regulatory properties of the enzyme. The catabolic threonine dehydratase of E. coli is normally inhibited by high concentrations of pyruvate and some other α-keto acids, but this inhibition can be completely overcome by increased levels of AMP (6). While these effectors may operate in Corynebacterium spp., it is clear that there is sufficient threonine dehydratase activity in the tdcB-carrying strain to promote isoleucine production. As a result, we were able to increase the production of isoleucine by a factor 50, and 70% of the carbon available for the lysine pathway was directed into the isoleucine pathway. Further investigations are under way to study the regulation of this enzyme in Corynebacterium spp. and to determine whether expressing this enzyme in strains with different genetic backgrounds (e.g., a threonine-overproducing strain) provides any additional benefit in terms of isoleucine production.
ACKNOWLEDGMENTS
This work was funded by a grant from the Archer Daniels Midland Corporation. A. Rodal was supported by the MIT Undergraduate Research Opportunities Program.
REFERENCES
- 1.Benner S A. Enzyme kinetics and molecular evolution. Chem Rev. 1989;89:789–806. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Colón G E, Nguyen T T, Jetten M S, Sinskey A J, Stephanopoulos G. Production of isoleucine by overexpression of ilvA in a Corynebacterium lactofermentum threonine producer. Appl Microbiol Biotechnol. 1995;43:482–488. doi: 10.1007/BF00218453. [DOI] [PubMed] [Google Scholar]
- 4.Duncan B K, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982;151:750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggeling I, Cordes C, Eggeling L, Sahm H. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl Microbiol Biotechnol. 1987;25:346–351. [Google Scholar]
- 6.Feldman D A, Datta P. Catabolite inactivation of bidegradative threonine dehydratase of Escherichia coli. Biochemistry. 1975;14:1760–1767. doi: 10.1021/bi00679a031. [DOI] [PubMed] [Google Scholar]
- 7.Follettie M T, Shin H K, Sinskey A J. Organization and regulation of the C. glutamicum hom-thrB and thrC loci. Mol Microbiol. 1988;2:53–62. doi: 10.1111/j.1365-2958.1988.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 8.Goss T J, Datta P. Molecular cloning and expression of the biodegradative threonine dehydratase gene (tdc) of Escherichia coli K12. Mol Gen Genet. 1985;201:308–314. doi: 10.1007/BF00425676. [DOI] [PubMed] [Google Scholar]
- 9.Hashiguchi K, Kojima H, Sato K, Sano K. Effects of an Escherichia coli ilvA mutant gene encoding feedback-resistant threonine deaminase on l-isoleucine production by Brevibacterium flavum. Biosci Biotechnol Biochem. 1997;61:105–108. doi: 10.1271/bbb.61.105. [DOI] [PubMed] [Google Scholar]
- 10.Jetten M S M, Follettie M T, Sinskey A J. Effect of different levels of aspartokinase on the lysine production by Corynebacterium lactofermentum. Appl Microbiol Biotechnol. 1995;43:76–82. doi: 10.1007/BF00170626. [DOI] [PubMed] [Google Scholar]
- 11.Jetten M S M, Gubler M E, McCormick M M, Colón G E, Follettie M T, Sinskey A J. Molecular organization and regulation of the biosynthetic pathway for aspartate-derived amino acids in Corynebacterium glutamicum. In: Baltz R H, Hegeman G, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 97–104. [Google Scholar]
- 12.Jetten M S M, Pitoc G A, Follettie M T, Sinskey A J. Regulation of phospho(enol)-pyruvate- and oxaloacetate-converting enzymes in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1994;41:47–52. [Google Scholar]
- 13.Kelle R, Hermann T, Weuster-Botw D, Eggeling L, Kramer R, Wandrey C. Glucose-controlled l-isoleucine fed-batch production with recombinant strains of Corynebacterium glutamicum. J Biotechnol. 1996;50:123–136. [Google Scholar]
- 14.Lessard P A, Guillouet S, Willis L B, Sinskey A J. Corynebacteria, brevibacteria. In: Flickinger M C, Drew S W, editors. The encyclopedia of bioprocess technology: fermentation, biocatalysis and bioseparation. Vol. 2. New York, N.Y: John Wiley & Sons; 1999. pp. 729–740. [Google Scholar]
- 15.Mockel B, Eggeling L, Sahm H. Threonine dehydratases of Corynebacterium glutamicum with altered allosteric control: their generation and biochemical and structural analysis. Mol Microbiol. 1994;13:833–842. doi: 10.1111/j.1365-2958.1994.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 16.Morbach S, Sahm H, Eggeling L. Use of feedback-resistant threonine dehydratases of Corynebacterium glutamicum to increase carbon flux towards l-isoleucine. Appl Environ Microbiol. 1995;61:4315–4320. doi: 10.1128/aem.61.12.4315-4320.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pittard J, Adelberg E A. Gene transfer by F′ strains of E. coli K-12. II. Interaction between F-merogenote and chromosome during transfer. J Bacteriol. 1963;85:1402–1408. doi: 10.1128/jb.85.6.1402-1408.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Tsuchida T, Momose H. Genetic changes of regulatory mechanisms occurred in leucine and valine producing mutants derived from Brevibacterium lactofermentum 2256. Agric Biol Chem. 1975;39:2193–2198. [Google Scholar]
- 20.Umbarger H E, Brown B. Threonine deamination in Escherichia coli. II. Evidence for the two l-threonine deaminases. J Bacteriol. 1957;73:105–112. doi: 10.1128/jb.73.1.105-112.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Praszkier J, Hodgson A, Pittard A J. Molecular analysis and characterization of a broad-host-range plasmid, pEP2. J Bacteriol. 1994;176:5718–5728. doi: 10.1128/jb.176.18.5718-5728.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]