Abstract
The bacterial endosymbionts of the hydrothermal vent tubeworm Riftia pachyptila play a key role in providing their host with fixed carbon. Results of prior research suggest that the symbionts are selected from an environmental bacterial population, although a free-living form has been neither cultured from nor identified in the hydrothermal vent environment. To begin to assess the free-living potential of the symbiont, we cloned and characterized a flagellin gene from a symbiont fosmid library. The symbiont fliC gene has a high degree of homology with other bacterial flagellin genes in the amino- and carboxy-terminal regions, while the central region was found to be nonconserved. A sequence that was homologous to that of a consensus ς28 RNA polymerase recognition site lay upstream of the proposed translational start site. The symbiont protein was expressed in Escherichia coli, and flagella were observed by electron microscopy. A 30,000-Mr protein subunit was identified in whole-cell extracts by Western blot analysis. These results provide the first direct evidence of a motile free-living stage of a chemoautotrophic symbiont and support the hypothesis that the symbiont of R. pachyptila is acquired with each new host generation.
Symbioses between chemoautotrophic bacteria and marine invertebrates are found in a wide variety of marine environments including deep-sea hydrothermal vents, sewage outfalls, anoxic basins, seagrass beds, and coralline sands (reviewed in references 12, 15, and 33). The hydrothermal vent tubeworm Riftia pachyptila is one of the most conspicuous vent animals and is an example of an organism involved in a highly specialized symbiotic association. The adult tubeworm of this species lacks a mouth and digestive system (24) and is never found without symbionts. Early studies showed that chemoautotrophic bacteria are found within bacteriocyte cells localized in a specialized organ called the trophosome (6, 11). Because the symbionts of R. pachyptila have eluded cultivation, their classification has been based principally on rRNA sequence analysis (9, 28, 44). The symbionts of R. pachyptila, as with all of the thioautotrophic symbionts examined to date, unambiguously belong to the gamma subdivision of the proteobacteria.
Indirect evidence suggests that the development and survival of R. pachyptila is entirely dependent on the acquisition of symbionts from a free-living bacterial population via horizontal transmission. In situ probing studies have failed to detect symbionts in R. pachyptila gametes (5). While adult tubeworms lack a mouth and digestive system (23), young juveniles possess a transient mouth and a ciliated gut but lack symbiont-containing tissues (22, 25). Additionally, it appears that the chemoautotrophic symbionts have not coevolved with their vestimentiferan hosts (13, 29). Furthermore, our recent evidence suggests that the symbionts possess functional mechanisms for sensing and responding to their environment through two-component regulatory systems (21). Taken together, these results suggest the presence and importance of a free-living protosymbiont. However, such an organism has yet to be identified from the hydrothermal vent environment.
An obvious feature required to establish contact with and eventually invade a host cell is motility mediated by flagella. Motility and flagellum-associated structures are important colonization factors in a number of bacterial symbionts and pathogens of animals (16, 17, 32, 35, 37, 40, 41) and of plants (2, 4, 8). Motility is a complex phenotype, which in Escherichia coli requires the coordinated expression of more than 60 genes contained in at least 13 operons in order to synthesize and rotate the E. coli flagellar apparatus (30). Flagellin molecules, encoded by the fliC gene, are the subunits which polymerize to form filaments of the bacterial flagellum. Flagellin proteins from diverse bacterial species commonly share conserved amino acid residues, making it possible to identify flagellin genes by sequence similarity.
For the lack of cultivated symbionts and the failure to identify a free-living protosymbiont from the hydrothermal vent environment we used alternative methods to investigate the potential of the symbionts to colonize their host. Recent findings of functional two-component regulatory systems (21) suggest that the presence of motility genes are likely and support our approach. We report here the identification of a symbiont flagellin gene and its characterization by expression in E. coli.
MATERIALS AND METHODS
PCR amplification of a flagellin gene from R. pachyptila symbiont DNA.
The following degenerate primers were designed by aligning conserved sequences of known enteric FliC genes: 5′-ATGGCACAAGTCATTAATACmAAC-3′ and 5′-GCCTGCTGsAkAATCTGCGCTTT-3′. The primers align with the 5′ and 3′ terminal regions of known flagellin genes. PCR was performed with 1 ng of purified R. pachyptila symbiont genomic DNA per μl by using standard reagents and reaction conditions (30 cycles of 92°C for 90 s, 50°C for 90 s, and 72°C for 2 min) (42). Amplification products were cloned and sequenced to confirm their similarity to flagellin sequences.
Hybridization of the symbiont fosmid library.
The preparation of the symbiont fosmid library was previously reported (21). The 1,500-member fosmid library consists of clones that contain DNA inserts of 35 to 45 kbp. The hybridization of the library with labeled amplification products was performed by using standard methods (42). The hybridization was analyzed by autoradiography and confirmed by Southern hybridization of restriction-digested positive fosmid clones. A 3.8-kbp EcoRI-digested DNA fragment present in all 11 positive fosmids was subcloned into the plasmid pBluescript (Stratagene, La Jolla, Calif.) and sequenced (ABI Sequencer 480).
Expression of R. pachyptila symbiont flagellin in motility mutant E. coli.
DNA from fosmid 1O9 was digested with EcoRI, and a 3.8-kbp DNA fragment shown to contain the flagellar gene was cloned into pBluescript (Stratagene) resulting in pDH90. Two primers, fliC-for (TAGGAGAAAAGCTTTGGCACTCGT [FTF-F, 24-mer]) and fliC-rev (AGATCACCCGGATCCCGGTCGATG [FTF-R, 24-mer]), were used to PCR amplify the flagellin gene from symbiont genomic DNA. Both primers were designed such that restriction sites (underlined) were created in the PCR amplification. The resulting 874-bp amplification product was directionally cloned into pBluescript, resulting in pDH95. Plasmid pMS1 containing the Salmonella H2 gene was used as a positive control for complementation by a homologous gene (lab strain from M. Simon). The resulting constructs were then transferred into E. coli motility mutant strains JA11, CSH4, and RP4770. E. coli JA11 has the 5′ flagellin-encoding gene fliC of E. coli (26) deleted, and strain CSH4 contains an uncharacterized mutation of the fliC gene (43a).
Motility experiments.
The motility of the fliC mutant strains containing fosmid clones pDH90 and pDH95 was observed both by microscopy and by assessing motility on swarm agar plates. Motility was determined by measuring the movement of bacterial cells on swarm agar plates containing 1% tryptone, 0.5% NaCl, and 0.25% agar. The optical density at 600 nm was determined for cultures grown overnight and for cultures at the mid-exponential phase. Equivalent numbers of cells in 2-μl volumes were spotted in the centers of swarm agar plates, and movement away from the center was measured after 24 h at 30°C.
Purification of flagellar filaments.
E. coli JA11 carrying the plasmid pMS1 or pFOS1O9 was grown in 500-ml cultures to mid-log growth. The bacterial cells were pelleted (8,500 × g for 15 min at 4°C) and resuspended in phosphate-buffered saline (42). Flagella were sheared from the bacteria in a Waring blender set at high speed for 1 min, and the bacterial cells were pelleted (5,500 × g for 10 min at 4°C). The supernatant was collected, and the flagella were pelleted by ultracentrifugation (40,000 rpm in a VTi65 rotor [Beckman] for 2 h at 10°C). The flagella were resuspended overnight in water and denatured in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Electron microscopy.
Electron microscopy was performed with a JEOL(Tokyo, Japan) microscope, and negatively stained cells were prepared as described previously (31). Alternatively, grids were prepared by ultracentrifugation of cells directly onto copper grids (40,000 rpm in a SWTi65 rotor for 1 h at 10°C).
SDS-PAGE and Western immunoblotting.
SDS-PAGE was performed by using standard protocols (42). Transferred proteins were reacted with monoclonal antibody 15D8 (1:500 dilution) in 5% milk at 35°C for 2 h, to allow the antibody to bind to specific proteins. The antibody 15D8 recognizes a conserved epitope in flagellins expressed by E. coli and other members of the family Enterobacteriaceae (14). To detect the antigen-antibody complexes, horseradish peroxidase conjugated to goat anti-mouse immunoglobulin G was diluted 1:10,000 in 5% milk and incubated with the blots for 2 h at 35°C. The blots were treated for 5 min at room temperature with a chemiluminescent substrate following the manufacturer’s suggestions (Super Signal; Pierce). The developed blots were immediately exposed on autoradiographic film.
Nucleotide sequence accession number.
The nucleotide sequence for the R. pachyptila symbiont fliC gene encoding a protein similar to enteric bacterial flagellin proteins is available from the GenBank database under accession no. AF105060.
RESULTS
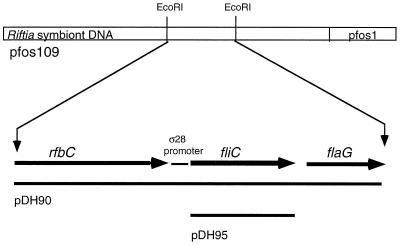
Probing the symbiont fosmid library resulted in the identification of 11 clones containing similar 40- to 45-kbp inserts with an overlapping 3.8-kbp EcoRI fragment that hybridized to the putative fliC amplification product (data not shown). Southern hybridization of R. pachyptila symbiont genomic DNA confirmed the origin of the amplification product to be the trophosome symbiont and showed hybridization of a single EcoRI DNA fragment, indicating that a single copy of the gene is present in the genome. Two fosmids, pFOS1O9 and pFOS2G9, and a subclone of the 3.8-kbp fragment (pDH90) were chosen for further work.
Analysis of the DNA sequence revealed three open reading frames (ORFs) (Fig. 1), one of which revealed high sequence similarity to previously characterized flagellin-encoding (fliC) genes. The second ORF consists of 846 nucleotides, starting with ATG and ending with the termination codon, TAA. A putative stem-loop structure (GCATCCGGGTGATCTAGCCCGGATGCAC) is found 13 nucleotide bases downstream of the termination codon, which could act as a transcriptional terminator (39). The inspection of the upstream region of this ORF revealed a 5′-AGGAG region which resides 8 bases upstream from the putative ATG translational start site and which resembles the AGGAGG E. coli Shine-Dalgarno ribosome-binding site consensus sequence. A comparison of the region upstream with corresponding regions in other bacteria showed that this region contains a putative promoter recognized by RpoF (ς28) containing RNA polymerases. This flagellar gene-specific RNA polymerase is responsible for the expression of flagellar genes in other bacteria (19). The putative symbiont fliC promoter (TAAA-N15-GCCGTTAC) contains a single mismatch compared to the Salmonella typhimurium H1 promoter (TAAA-N15-GCCGATAC) sequence and two mismatches compared to the E. coli consensus ς28 promoter (TAAA-N15-GCCGATAA) sequence.
FIG. 1.
Restriction map of the symbiont fliC gene region from the fosmid clone pFOS1O9. The size and transcriptional direction of the rbfC, fliC, and flaG genes are shown by arrows. The fliC fragments cloned for the expression study are indicated below the appropriate sequence.
The deduced amino acid sequence of the second ORF predicts a protein of 281 amino acids with a molecular mass of 29,422 Da. The sequence contains 82% similar and 65% identical amino acids throughout the amino-terminal (amino acids 1 to 106) and carboxy-terminal (amino acids 197 to 280) domains of the FliC protein from Legionella micdadei. L. micdadei was chosen for comparison since it belonged to the gamma subdivision of the proteobacteria and showed the highest sequence similarity to the symbiont protein. In addition, sequence information on more closely related marine bacteria is unavailable. Sequences throughout the entire length of the predicted symbiont protein are approximately 50% similar to the flagellin proteins of L. micdadei, E. coli, Pseudomonas aeruginosa, and Aeromonas salmonicida.
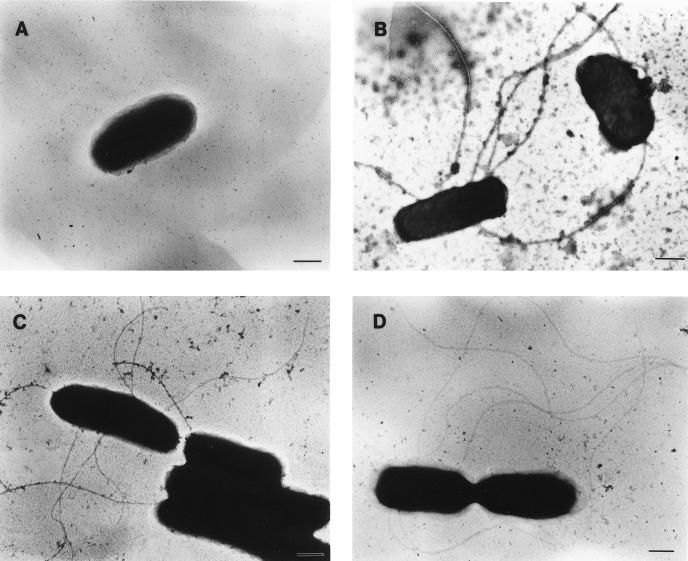
Plasmids containing the complete symbiont fliC gene under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lac promoter (pDH95) or its native promoter (pDH90) were expressed in motility mutant E. coli JA11 (ΔfliC). This strain was chosen because the mutation is stable and well characterized and the strain is impaired in recombination (26). A plasmid containing the S. typhimurium H2 gene, pMS1, was chosen as a positive control for complementation by a heterologous gene. On swarm plates, cells expressing the symbiont fliC gene did not show significant swarming ability; however, the cells appeared motile by light microscopy. Further inspection by electron microscopy revealed the presence of flagella. JA11 cells containing a vector alone (pBS or pBAC) never showed flagella (Fig. 2A), while JA11 cells containing the positive control pMS1 gene showed flagella (Fig. 2D).
FIG. 2.
Electron microscopy of motility mutant E. coli JA11 expressing the flagellin gene from the R. pachyptila symbiont. JA11 cells contained the following plasmids: pBluescript (vector control) (A); pDH90, containing the R. pachyptila symbiont fliC gene (FliCRS) (B); pDH95 (FliCRS) (C); and pMS1, containing the Salmonella H2 flagellin gene (FliCST) (D). Cells at the mid-exponential phase were prepared for electron microscopy by negative staining with 0.5% uranyl acetate. Bar, 500 nm.
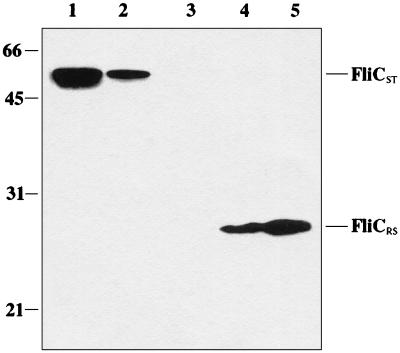
To support the observation of symbiont flagella by electron microscopy, we performed Western blot analysis. The Western blot shown in Fig. 3 demonstrates the recognition of the symbiont flagellin by a monoclonal antibody (15D8) that specifically recognizes a conserved epitope in flagellins expressed by E. coli and other members of the family Enterobacteriaceae (14). A 30,000-Mr protein in whole-cell purifications of JA11 cells containing plasmids with the symbiont flagellin gene (pDH90 or pDH95) reacted with the antibody (FliCRS) (Fig. 3, lanes 4 and 5, respectively). As expected, a 51,000-Mr protein corresponding to the E. coli flagellin subunit was not detected. The antibody did react with a 55,000-Mr species in both high-speed pellets and whole-cell preparations of JA11 containing the plasmid pMS1 encoding the S. typhimurium flagellin gene (FliCST) (Fig. 3, lanes 1 and 2, respectively). The vector control JA11 (pBS) showed no reactivity with the antibody (Fig. 3, lane 3).
FIG. 3.
Immunoblots showing reactivity of the anti-FliCE antibody (15D8) with purified Salmonella H2 flagellin (lane 1) and E. coli JA11 containing pMS1 (lane 2), pBluescript (lane 3), pDH90 (lane 4), or pDH95 (lane 5).
DISCUSSION
The chemoautotrophic bacterial symbionts of the hydrothermal vent tubeworm R. pachyptila are likely acquired from the environment and thus must be able to adapt outside the host prior to inoculation of the tubeworm gut. We have identified a flagellin gene from the symbiont and have begun to characterize the flagellin expressed in an E. coli host system. The alignment of the predicted symbiont amino acid sequence with those of other bacterial flagellin proteins shows that the structure of this protein is highly conserved within the amino- and carboxy-terminal regions. These conserved regions have been shown to be necessary for the export and polymerization of the flagellin subunits (20). Interestingly, the symbiont fliC gene product is lacking a large portion of the central domain, compared to the sequence of other flagellin proteins. It is unclear what effect if any the loss of the central domain would have on flagellar assembly or function; however, this region can be replaced with an unrelated sequence or can be removed without a loss of motility in other bacteria (27, 34).
To better understand the structure of the symbiont flagellum, we expressed the flagellin gene and attempted to complement motility in several nonmotile E. coli ΔfliC or fliC-negative strains. The expression of the symbiont fliC gene in cells of the E. coli motility mutant strain JA11 was shown by the presence of flagella observed by electron microscopy. However, the expression was at a low frequency, since approximately 5% of the recombinant cells appeared to be flagellated. It is possible that FliCRS production is toxic to E. coli, as supported by earlier findings in which the complete flagellin gene from L. micdadei and Treponema pallidum could not be expressed in E. coli (3, 36). Alternatively, the observed low-level expression may be due to the failure of the symbiont flagellin to be recognized for filament export or assembly, and thus the intracellular accumulation of flagellin could result in feedback control of operon expression. For this reason we investigated the expression of the symbiont protein by Western analysis with whole-cell extracts. The results indicated that a 30,000-Mr protein, of a size similar to that expected for the symbiont flagellin (Mr, 29,422) was identified in whole-cell extracts, suggesting the observed flagella is comprised of symbiont flagellin subunits.
Considering the high degree of similarity between the symbiont fliC gene product and the amino acid sequence of other bacterial flagellins, it seems reasonable to assume that the protein encoded by this gene serves a similar flagellar structural function in the symbiont. Additionally, the identification of a conserved regulatory sequence motif (ς28 promoter) upstream of the symbiont gene strongly suggests that the symbiont flagellin is regulated by a conserved mechanism (19). In other bacterial species, flagella expression is controlled by environmental signals (7, 10, 18, 38, 43). It is unclear what parameters would regulate the expression for a deep-sea chemoautotrophic symbiont, and such studies clearly must await the culturing of such organisms. In a number of nonpathogenic associations between animals and motile bacteria, flagella are required for invasion but are lost shortly after colonization of the host animal (12, 16, 41, 45). It is unknown what down-regulates flagellar expression for a symbiont or what regulates the expression of flagella once the bacteria are released from the host animal (41). In at least some cases this repression may be the result of a modulation of motility gene regulation such as that controlling both flagella synthesis and virulence determinants in Bordetella (1).
We hypothesize that motility is an important phenotype for the R. pachyptila symbiont by comparison to analogous types of associations in which motility is essential for symbiont colonization. Our assessment of the potential for motility of this symbiont and more recent evidence for the presence and function of chemotaxis genes (31a) lend direct support to the hypothesis that the symbionts are acquired by their host with each new generation. Studies such as this one will enable us to begin to assess the function of this uncultivated symbiont outside its host organism.
ACKNOWLEDGMENTS
We thank Peter Feng, Reid Johnson, and Sandy J. Parkinson for their generous gifts of antibody, plasmid, and strains, respectively.
This work was supported by National Science Foundation grant OCE93-14525 to H.F. and a Patricia Roberts Harris graduate fellowship to D.S.M.
REFERENCES
- 1.Ackerley B J, Monack D M, Falkow S, Miller J F. The bvg locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol. 1992;174:980–990. doi: 10.1128/jb.174.3.980-990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames P, Bergman K. Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J Bacteriol. 1981;148:728–729. doi: 10.1128/jb.148.2.728-729.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangsborg J M, Hindersson P, Shand G, Hoiby N. The Legionella micdadei flagellin: expression in Escherichia coli K12 and DNA sequence of the gene. APMIS. 1995;103:869–877. doi: 10.1111/j.1699-0463.1995.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 4.Caetano-Anolles G, Wall L G, De Micheli A T, Macchi E M, Bauer W D, Favelukes G. Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol. 1988;86:1228–1235. doi: 10.1104/pp.86.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary S C, Warren W, Anderson E, Giovannoni S J. Identification and localization of bacterial endosymbionts in specialized hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol Mar Biol Biotechnol. 1993;2:51–62. [PubMed] [Google Scholar]
- 6.Cavanaugh C M, Gardiner S L, Jones M L, Jannasch H W, Waterbury J B. Procaryotic cells in the hydrothermal vent tubeworm. Science. 1981;213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Helmann J D. The Bacillus subtilis ςD-dependent operon encoding the flagellar proteins FliD, FliS, and FliT. J Bacteriol. 1994;176:3093–3101. doi: 10.1128/jb.176.11.3093-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 9.Distel D L, Lane D J, Olsen G J, Giovannoni S J, Pace B, Stahl D A, Felbeck H. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol. 1988;170:2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelstein P H, Beer K B, DeBoynton E D. Influence of growth temperature on virulence of Legionella pneumophila. Infect Immun. 1987;55:2701–2705. doi: 10.1128/iai.55.11.2701-2705.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felbeck H. Chemoautotrophic potential of the hydrothermal vent tubeworm, Riftia pachyptila Jones (Vestimentifera) Science. 1981;213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 12.Felbeck H, Distel D L. Procaryotic symbionts of marine invertebrates. In: Ballows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The procaryotes. New York, N.Y: Springer Verlag; 1991. pp. 3891–3906. [Google Scholar]
- 13.Feldman R A, Black M B, Cary S C, Lutz R A, Vrijenhoek R C. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6:268–277. [PubMed] [Google Scholar]
- 14.Feng P, Sugasawara J R, Schantz A. Identification of a common enterobacterial flagellin epitope with a monoclonal antibody. J Gen Microbiol. 1990;136:337–342. doi: 10.1099/00221287-136-2-337. [DOI] [PubMed] [Google Scholar]
- 15.Fisher C R. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev Aquat Sci. 1990;2:399–436. [Google Scholar]
- 16.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant C C R, Konkel M E, Cieplak W, Jr, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzzo A, Diorio C, Dubow M S. Transcription of the Escherichia coli fliC gene is regulated by metal ions. Appl Environ Microbiol. 1991;57:2255–2259. doi: 10.1128/aem.57.8.2255-2259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmann J D, Masiarz F R, Chamberlin M J. Isolation and characterization of the Bacillus subtilis ς28 factor. J Bacteriol. 1988;170:1560–1567. doi: 10.1128/jb.170.4.1560-1567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homma M, Fuita H, Yamaguchi S, Lino T. Regions of Salmonella typhimurium flagellin essential for its polymerization and excretion. J Bacteriol. 1987;169:291–296. doi: 10.1128/jb.169.1.291-296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes D S, Felbeck H, Stein J L. A histidine protein kinase homolog from the endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. Appl Environ Microbiol. 1997;63:3494–3498. doi: 10.1128/aem.63.9.3494-3498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones M, Gardiner S. On the early development of the vestimentiferan tube worm Ridgeia sp. and observations on the nervous system and trophosome of Ridgeia sp. and Riftia pachyptila. Biol Bull (Woods Hole) 1989;177:254–276. [Google Scholar]
- 23.Jones M L. Riftia pachyptila Jones: observations on the vestimentiferan worm from the Galapagos rift. Science. 1981;213:333–336. doi: 10.1126/science.213.4505.333. [DOI] [PubMed] [Google Scholar]
- 24.Jones M L. The Vestimentifera, their biology, systematic and evolutionary patterns. Oceanol Acta. 1988;8:69–82. [Google Scholar]
- 25.Jones M L, Gardiner S L. Evidence for a transient digestive tract in Vestimentifera. Proc Biol Soc Wash. 1988;101:423–433. [Google Scholar]
- 26.Kawagishi I, Muller V, Williams A W, Irikura V M, Macnab R M. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J Gen Microbiol. 1992;138:1051–1065. doi: 10.1099/00221287-138-6-1051. [DOI] [PubMed] [Google Scholar]
- 27.Kuwajima G. Construction of a minimum-sized functional flagellin of Escherichia coli. J Bacteriol. 1988;170:3305–3309. doi: 10.1128/jb.170.7.3305-3309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane D J, Stahl D A, Olsen G J, Pace N R. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Biol Soc Wash Bull. 1985;6:389–400. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- 29.Laue B E, Nelson D C. Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tube worm hosts: an RFLP analysis. Mol Mar Biol Biotechnol. 1997;6:180–188. [PubMed] [Google Scholar]
- 30.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 31.McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 31a.Millikan, D. S., et al. Unpublished data.
- 32.Nachamkin I, Yang X-H, Stern N J. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. 1993;59:1269–1273. doi: 10.1128/aem.59.5.1269-1273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson D C, Fisher C R. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps. In: Karl D M, editor. The microbiology of deep-sea hydrothermal vents. Boca Raton, Fla: CRC Press, Inc.; 1995. pp. 125–167. [Google Scholar]
- 34.Newton S M C, Jacob C O, Stocker B A D. Immune response to cholera toxin epitope inserted in Salmonella flagellin. Science. 1989;244:70–72. doi: 10.1126/science.2468182. [DOI] [PubMed] [Google Scholar]
- 35.Norqvist A, Wolf-Watz H. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect Immun. 1993;61:2434–2444. doi: 10.1128/iai.61.6.2434-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris S J the Treponema pallidum Polypeptide Research Group. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Toole R, Milton D L, Wolf-Watz H. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol Microbiol. 1996;19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- 38.Ott M, Messner P, Hessemann J, Marre R, Hacker J. Temperature-dependent expression of flagella in Legionella. J Gen Microbiol. 1991;137:1955–1961. doi: 10.1099/00221287-137-8-1955. [DOI] [PubMed] [Google Scholar]
- 39.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 40.Richardson K. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Silverman M, Simon M. Characterization of Escherichia coli mutants that are insensitive to catabolite repression. J Bacteriol. 1974;120:1196–1203. doi: 10.1128/jb.120.3.1196-1203.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Silverman, M. Personal communication.
- 44.Stahl D A, Lane D J, Olsen G J, Pace N R. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984;224:409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- 45.Tebo B M, Linthicum D S, Nealson K H. Luminous bacteria and light-emitting fish: ultrastructure of the symbiosis. BioSystems. 1979;11:269–280. doi: 10.1016/0303-2647(79)90027-3. [DOI] [PubMed] [Google Scholar]