Abstract
Estuarine waters receive fecal pollution from a variety of sources, including humans and wildlife. Escherichia coli is a ubiquitous bacterium in the intestines of warm-blooded animals and is used as an indicator of fecal pollution. However, its presence does not specifically differentiate sources of pollution. A total of 238 E. coli isolates from human sources (HS) and nonhuman sources (NHS) were collected from the Apalachicola National Estuarine Research Reserve, from associated sewage treatment plants, and directly from animals and tested for ribotype (RT) profile. HS and NHS isolates showed 41 and 61 RT profiles, respectively. At a similarity index of ca. 50%, HS and NHS isolates demonstrated four clusters, with the majority of HS and NHS isolates located in clusters C and D; isolates obtained directly from human and animal feces also could be grouped within these clusters. Discriminant analysis (DA) of RT profiles showed that 97% of the NHS isolates and 100% of the animal fecal isolates were correctly classified. The average rate of correct classification for HS and NHS isolates was 82%. We conclude that DA of RT profiles may be a useful method for identifying HS and NHS fecal pollution and may potentially facilitate management practices.
Fecal pollution is a major concern for many estuaries, where it can originate from human sources (HS) and nonhuman sources (NHS). Its impact can degrade water quality and restrict its use for harvesting sea foods, as well as recreational activities. However, without knowing the precise source of fecal input, the human health risk cannot be accurately predicted.
The fecal coliform Escherichia coli has been used as an indicator of human enteric pathogens for many years (12). However, it is now well established that E. coli is not limited to humans but also exists in the intestines of many other warm-blooded animals (22). Consequently, its presence in water is not specific to human sources of pollution.
This is especially relevant when recognizing that human feces can carry various human enteric pathogens, such as Salmonella spp., Shigella spp., E. coli, hepatitis A virus, and Norwalk group viruses. In contrast, most of these pathogens do not colonize nonhuman species, potentially resulting in less risk posed by NHS fecal pollution (16, 17, 22, 32). Therefore, it is important to know whether fecal pollution originates from HS or NHS in order to properly assess risk, and research is needed to determine the characteristics of indicators, such as E. coli, that may be used to identify sources of pollution.
To meet this challenge, there have been various attempts to develop methods that differentiate the sources of fecal pollution. Initially, the ratio of fecal coliforms to fecal streptococci was proposed, where a ratio of ≥4.0 would indicate HS pollution, whereas a ratio of ≤0.7 would indicate NHS pollution (13). However, this approach was unreliable due to the variable survival rates of fecal streptococcus species (28).
Investigators have also reported that nonhuman and human feces contains different serotypes of RNA coliphages (11, 22), suggesting that phages could be used to predict the source of pollution. However, their usefulness is limited, because only a small percentage of human fecal samples contain phages (22).
More-traditional methods for discriminating bacteria have included biochemical tests (1, 21), phage susceptibility (38), outer membrane protein profiles (8), antibody reactivity (36), fimbriation (18), bacteriocin production and susceptibility, and other methods (14). However, these systems have serious disadvantages, including unstable phenotypes, low sensitivity at the intraspecies level, and limited specificity (33). Even with these limitations, we have recently reported that multiple antibiotic resistance (MAR) can be used to differentiate point and nonpoint sources of E. coli in an estuarine environment (25).
Some of the more recent techniques involve DNA analysis, such as plasmid profiles, pulsed-field gel electrophoresis (6, 23), and restriction fragment length polymorphism analysis at specific loci, such as fimbrial adhesion and rRNA (ribotyping) operons (15, 18). These methods are generally less dependent on the more unstable phenotypic traits.
Ribotyping has proved to be a useful epidemiological technique for various bacteria, including E. coli (33), Salmonella enterica (21), Vibrio cholerae O1 (27), and Vibrio vulnificus (7, 34). However, an examination of the literature shows that this method has not been used to discriminate HS and NHS E. coli strains.
Any new predictive test requires extensive statistical modeling of the data to correlate specific bacterial characteristics with their source. Discriminant analysis (DA) can classify individuals into groups on the basis of several classification variables (5).
To test the applicability of ribotyping to predict the source of E. coli pollution, we selected the Apalachicola National Estuarine Research Reserve (ANERR), a site where we have previously studied MAR in over 700 E. coli isolates (25). The ANERR consists of two barrier islands, the lower 20 miles of the Apalachicola River and its flood plain, adjoining uplands, and the Apalachicola Bay system (10, 25). The ANERR is also a significant harvest area for shellfish and a variety of finfish (10). We found that DA of E. coli ribotype (RT) profiles predicted the source of E. coli pollution.
MATERIALS AND METHODS
E. coli isolates.
E. coli isolates were selected from among 700 isolates previously described in an earlier study of MAR (25). For the present RT studies, 179 isolates were selected in proportion to the number of isolates in specific MAR clusters; an additional 30 and 29 isolates were directly isolated from human and wildlife feces, respectively (25). E. coli was isolated and identified by standard procedures (3, 4, 25).
RT analyses.
For RT analyses, E. coli ATCC 9637 was used as the control strain.
(i) DNA extraction.
DNA was extracted by the method of Sambrook et al. (29). Briefly, E. coli were grown in brain heart infusion broth (Difco Laboratories, Detroit, Mich.), and late-log-phase cultures were pelleted at 3,000 × g for 15 min and treated with lysozyme (10 mg/ml; Sigma), proteinase K (10 mg/ml; Fisher Biotech, Fair Lawn, N.J.), and RNase (150 μg/ml; Fisher) at 37°C for 30 min. DNA was extracted with chloroform-isoamyl alcohol (24:1), precipitated with 100% isopropanol, washed once in 70% ethanol, recovered, dried, and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0).
(ii) Determination of DNA concentration.
DNA concentration was determined by using a TKO 100 Mini-Fluorometer according to the manufacturer’s instructions (Hoefer Scientific Instruments, San Francisco, Calif.). Two microliters of a 100-μg/ml concentration of calf thymus DNA was used as a standard.
(iii) Restriction enzyme analysis.
Approximately 1 μg of DNA was digested with several restriction enzymes, including HindIII, EcoRI, SalI, and BglI (Gibco BRL, Gaithersburg, Md.), according to the manufacturer’s instructions. Digested DNA was separated in a 1.0% agarose gel at 30 V for 16 h in TBE buffer (0.09 M Tris-borate, 0.002 M EDTA), stained with ethidium bromide (5 μg/ml), and photographed under shortwave UV light. A HindIII digest of bacteriophage lambda (Promega Corp., Madison, Wis.) was used as a molecular weight marker.
(iv) Southern blot analysis.
After electrophoresis of restriction-digested DNA, agarose gels were denatured in 0.5 M NaOH–1.5 M NaCl for 35 min and neutralized in 0.5 M Tris-HCl (pH 7.2)–1.5 M NaCl–0.001 M disodium EDTA for 45 min. DNA was then blotted from gels onto nylon membranes (Bio-Rad Laboratories, Hercules, Calif.) by using a vacuum blotting system (VacuGene XL; Pharmacia Biotech, Piscataway, N.J.) and fixed with shortwave UV light for 5 min (31).
(v) Probe preparation.
E. coli 16S and 23S rRNA were reverse transcribed into cDNA with avian reverse transcriptase (Boehringer Mannheim Corp., Indianapolis, Ind.) and then labeled with digoxigenin-dUTP according to the manufacturer’s instructions (Boehringer Mannheim Corp.).
(vi) Hybridization and detection.
Membranes were prehybridized at 42°C for 2 h and then hybridized with a digoxigenin-labeled probe with 25 ng of DNA per 15- by 15-cm filter at 65°C for 16 h. After hybridization, membranes were washed twice for 5 min each time with 2× SSC (2× SSC is 0.3 M NaCl plus 30 mM sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at room temperature and twice for 15 min each time with 0.5× SSC–0.1% SDS at 65°C; membranes were then reacted with antibody and visualized by using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate for colorimetric detection according to the manufacturer’s instructions (Boehringer Mannheim Corp.).
Statistical analysis.
RT profiles were measured by using DNA Proscan software (Nashville, Tenn.). DNA fragments for each strain were translated into binary code. Similarity indices were determined by using Dice’s coincidence index (9), where restriction endonuclease digestion profile similarities (Sxy) for individuals x and y equals the numbers of common fragments in their DNA profiles (nxy) divided by the average number of fragments exhibited by both individuals (Sxy = 2nxy/nx + ny). Relationships among isolates were examined by using cluster analysis. Data management and cluster analysis were performed in SAS (SAS Institute, Inc., Cary, N.C.), and cluster dendrograms were plotted by using S-Plus software (Statistical Services, Inc., Seattle, Wash.). Binary codes were also analyzed by using statistical discrimination methodology (5, 19, 20), as implemented in SAS (SAS 1989 AIX, version 6.12). A test for homogeneity of within-group covariance matrices suggested an unpooled covariance analysis, resulting in the use of quadratic discriminant function. In addition, equal prior probabilities were assumed. Results of the discrimination model were summarized by use of the average rate of correct classification (ARCC) and the percentage of correctly and misclassified isolates from the classification table. The table is a source-by-source matrix in which the numbers and percentages of correctly classified isolates were found on the diagonal. The ARCC was computed by averaging the percentages along the diagonal of the table (the correctly classified isolates).
RESULTS
We analyzed RT profiles of 238 E. coli isolates (84 HS, 95 NHS, 30 human feces, and 29 animal feces). Preliminary experiments examined several restriction enzymes (HindIII, EcoRI, SalI, and BglI) with a panel of E. coli isolates to determine the appropriate enzyme for this study. Results showed that HindIII gave the most discriminating profile, consisting of 4 to 12 bands over a size range of 0.7 to 20.0 kb, and thus it was used for subsequent studies. Patterns were considered to be unique when differentiated by one or more bands.
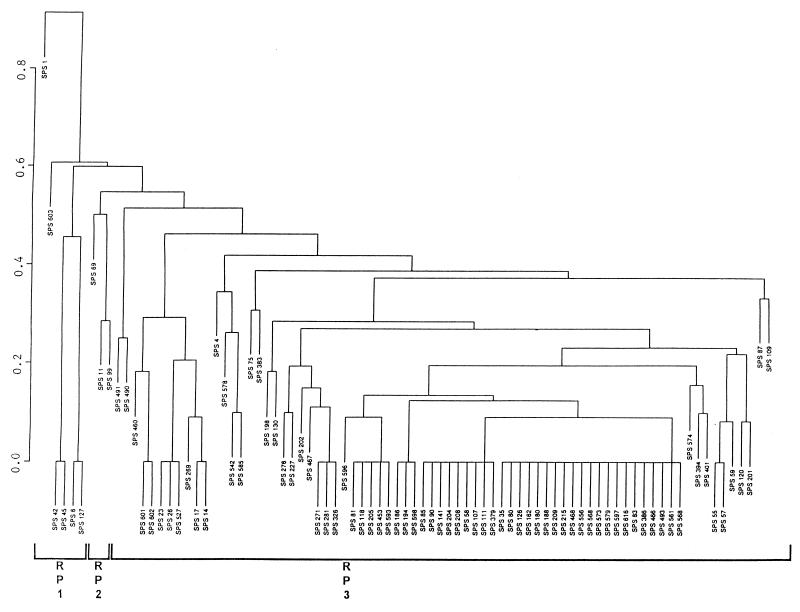
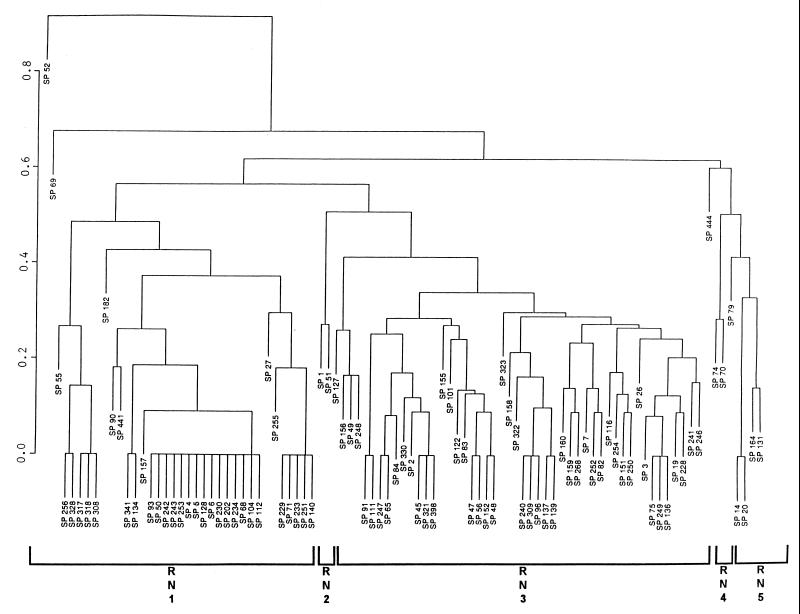
HS isolates showed 41 unique RT profiles, with more than 60% belonging to 11 RTs (Fig. 1). A binary metric average linkage plot of HS isolates at a similarity index of ca. 50% demonstrated three clusters, designated RP1, RP2, and RP3; two isolates (i.e., SPS 1 and SPS 603) were located outside of these clusters (Fig. 1). An even higher degree of homology was observed for HS isolates at a more stringent level of similarity (Fig. 1 and 2). In contrast, NHS isolates exhibited 61 different RT profiles, with 14 RTs accounting for 50% of the isolates (Fig. 2). A binary metric average linkage plot of NHS isolates at the 50% similarity index showed five clusters, designated RN1, RN2, RN3, RN4, and RN5; three isolates (i.e., SP 52, SP 69, and SP 444) were located outside of the clusters (Fig. 2).
FIG. 1.
Binary metric plot of average similarity linkage for RT profiles of HS isolates digested with HindIII. The prefix “SPS” indicates HS isolates. The similarity index is indicated on the left axis.
FIG. 2.
Binary metric plot of average similarity linkage for RT profiles of NHS isolates digested with HindIII. The prefix “SP” indicates NHS isolates. The similarity index is indicated on the left axis.
We combined individual databases for HS and NHS isolates, and at a similarity index of ca. 50% four clusters were obtained. The majority of the HS and NHS isolates were located in clusters C and D. There were 24 unique RT patterns that contained more than one isolate and included 64 and 50% of the HS and NHS isolates, respectively. Eight patterns contained only HS isolates, and another eight contained only NHS isolates; the remaining patterns contained both HS and NHS isolates (Table 1).
TABLE 1.
HS, NHS, human fecal, and animal fecal isolates at different similarity indices
| Cluster (n) | No. of HS/NHS isolates per cluster | Similarity index | Subclustersa (no. of HS/NHS isolates) |
|---|---|---|---|
| A (6) | 4/2 | 100 | A1 (2/0); A2 (2/0) |
| 90 | A1 (2/0); A2 (2/0) | ||
| 80 | A1 (2/0); A2 (2/0) | ||
| 70 | A1 (2/0); A2 (2/0); A3 (0/2) | ||
| 60 | A1 (2/0); A2 (2/2) | ||
| B (10) | 3/7 | 100 | B1 (0/2); B2 (1/1) |
| 90 | B1 (0/2); B2 (1/1); B3 (0/1) | ||
| 80 | B1 (0/2); B2 (1/2) | ||
| 70 | B1 (1/1); B2 (0/2); B3 (1/2); B4 (0/2) | ||
| 60 | B1 (2/5); B2 (1/2) | ||
| C (37) | 9/28 | 100 | C1 (2/0); C2 (1/2); C3 (3/5, 1AF); C4 (2/5) |
| 90 | C1 (3/0); C2 (1/2); C3 (3, 10HF/16, 1AF) | ||
| 80 | C1 (0/2); C2 (3/0); C3 (4/18); C4 (2/6) | ||
| 70 | C1 (7/20); C2 (2/7) | ||
| 60 | C1 (0/1); C2 (9/27) | ||
| D (122) | 66/56 | 100 | D1 (0/2); D2 (1/3); D3 (2/0); D4 (0/2); D5 (0/2); D6 (0/2); D7 (0/2); D8 (0/3); D9 (1/1, 6AF); D10 (0/2); D11 (1/3, 1AF); D12 (5/0); D13 (3, 15HF/0); D14 (9/0); D15 (21/3); D16 (3, 5HF/0) |
| 90 | D1 (0/2); D2 (1/3); D3 (1/0); D4 (1/0); D5 (0/1); D6 (1/1); D7 (0/1); D8 (2/1); D9 (0/1); D10 (0/1); D11 (0/4); D12 (0/2); D13 (1/0); D14 (1/0); D15 (0/3); D16 (0/1); D17 (1/0); D18 (0/4); D19 (0/1); D20 (1/0); D21 (0/1, 4AF); D22 (0/2); D23 (0/2, 9AF); D24 (1/2); D25 (1/0); D26 (0/1); D27 (0/2); D28 (1/0); D29 (1/0); D30 (1/0); D31 (1/14, 2AF); D32 (5/1); D33 (3/0); D34 (9/10); D35 (0/1, 5AF); D36 (21/3); D37 (0/1); D38 (0/1);D39 (1/0); D40 (1/0); D41 (3/0); D42 (0/1); D43 (1/1) | ||
| 80 | D1 (0/1); D2 (1/15); D3 (1/10); D4 (2/0); D5 (1/1); D6 (0/1); D7 (3/4); D8 (0/1); D9 (0/6); D10 (0/2); D11 (4/7); D12 (0/1); D13 (1/1); D14 (0/3); D15 (1/4); D16 (1/3); D17 (42/9); D18 (0/2); D19 (5/0); D20 (0/2); D21 (1/0); D22 (1/2) | ||
| 70 | D1 (1/6); D2 (1/0); D3 (3/0); D4 (2/1); D5 (3/12); D6 (5/11); D7 (49/21); D8 (2/4) | ||
| 60 | D1 (1/6); D2 (4/0); D3 (59/45); D4 (2/4) |
A1 to A3, B1 to B4, C1 to C4, and D1 to D43 indicate subclusters. HF, human fecal; AF, animal fecal. In subclusters D21, D23, and D35, AF isolates showed 100% similarity with NHS isolates.
Ten E. coli isolates were obtained from each of three separate human fecal samples and then tested for RT. Different individuals showed different RT profiles. However, all of these human isolates were grouped within previously defined HS RT profiles (Table 1).
In addition, there were approximately five E. coli isolates obtained from five separate raccoons, as well as five isolates from wildlife of unknown origin. The RT patterns of raccoon isolates varied from raccoon to raccoon but demonstrated high homology (i.e., 93%) within the previously defined NHS clusters (Table 1). When animal fecal isolates were grouped in a cluster that contained both HS and NHS isolates, in most instances animal fecal isolates (e.g., C3) showed 100% similarity with NHS isolates.
DA of RT profiles showed that the ARCC was 82%. A total of 97 and 67% of the NHS and HS isolates, respectively, were correctly classified by the RT method.
DA of E. coli isolates obtained directly from human and animal feces showed that 67 and 100% of human and animal fecal isolates, respectively, were correctly classified; the ARCC was 84%.
DISCUSSION
Techniques such as restriction endonuclease analysis (REA) of chromosomal DNA (i.e., DNA fingerprinting) and ribotyping (2, 30, 33) provide genotypic characteristics of bacteria that avoid the potential pitfalls associated with phenotypic methods. REA of chromosomal DNA is useful for the identification of many phenotypically indistinguishable bacteria (36). However, REA of E. coli produces many fragments, making visual comparisons of isolates difficult (data not shown). In contrast, RT yields patterns that allow much simpler comparisons among isolates (33). A major advantage of RT is that rRNA sequences are highly conserved and are present as multiple copies in bacterial genomes (15). Furthermore, one of the major benefits of RT is that 16S and 23S rRNA are commercially available.
In this study, ribotyping was used to discriminate HS and NHS E. coli isolates in a way similar to the way other investigators have used ribotyping to discriminate E. coli from different sources (24, 33, 35). From our study, we found that HS E. coli showed much less diversity than NHS isolates (Fig. 1 and 2). The latter situation may result from E. coli clones maintained in diverse reservoirs within the ANERR, such as raccoons, birds, deer, and other wildlife.
We also found that the HS and NHS clusters produced by the combined databases (Table 1) were in agreement with the RT profiles of isolates obtained directly from humans and wildlife. This finding strongly indicates that HS and NHS isolates may be derived from humans and raccoons and from unknown animal feces, respectively. Previously, we reported that MAR profiles of HS E. coli isolates showed greater resistance to antibiotics and higher MAR indices than those of NHS isolates. E. coli isolates obtained directly from human and animal feces could also be clustered by MAR among the HS and NHS isolates, respectively (25).
This study strongly indicates that DA of RT profiles can be used to differentiate HS and NHS of fecal pollution. Although some of the isolates were classified incorrectly, many more isolates were correctly classified than would be if the classification was random. This analysis also showed an ARCC well above that expected by chance (P < 0.05).
The advantage of DA is that it generates a classification rule based on all of the isolates; that rule can then be used to classify each individual isolate into one of many possible sources. Once a DA classification rule is developed by using RT profiles of E. coli isolates from known sources, DA can then be used to classify an unknown isolate to one of the known sources by using the unknown organism’s RT profiles.
Wiggins (37) demonstrated that DA of antibiotic resistance patterns of fecal streptococci is a useful tool for differentiating human and animal sources of fecal pollution in water. He found that 92% of HS isolates could be classified with an ARCC of 84%. We have reported that DA of MAR profiles of E. coli isolates from the ANERR classified 82 and 68% of HS and NHS isolates, respectively, and that the ARCC was 75% (26). However, this method has certain disadvantages: the antibiotic resistance patterns of bacteria are influenced by selective pressure and thus may be different in other geographical areas and may vary over time. In contrast, RT profiles are a genetic characteristic and are not as easily influenced by selective pressure. Therefore, on the basis of these data, DA of RT profiles provides a strong method for differentiating HS and NHS fecal pollution and may enhance efforts to improve the natural and commercial quality of estuarine ecosystems and to assess the importance of upstream activities, local storm water runoff, and marine activities.
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Commerce, National Oceanographic and Atmospheric Administration, Sanctuaries and Reserves Division (grant NA370R0166).
We are grateful to Leslie Miller, Tammi Stowers, and Mary McGinley for technical assistance. We thank Chip Bailey for assistance with sample collection.
Footnotes
Florida Agricultural Experiment Station journal series no. R-06879.
REFERENCES
- 1.Albritton W L, Penner S, Slaney L, Brunton J. Biochemical characteristics of Haemophilus influenzae in relationship to source of isolation and antibiotic resistance. J Clin Microbiol. 1978;7:519–523. doi: 10.1128/jcm.7.6.519-523.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allardet-Servent A, Bouziges N, Carles-Nurit M J, Bourg G, Gouby A, Ramuz M. Use of low-frequency-cleavage restriction endonucleases for DNA analysis in epidemiological investigations of nosocomial bacterial infections. J Clin Microbiol. 1989;27:2057–2061. doi: 10.1128/jcm.27.9.2057-2061.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. Compendium of methods for the microbiological examination of foods. 2nd ed. Washington, D.C: American Public Health Association; 1984. pp. 265–285. [Google Scholar]
- 4.American Public Health Association. Standard methods for the examination of water and wastewater. 17th ed. Washington, D.C: American Public Health Association; 1989. pp. 9.99–9.107. [Google Scholar]
- 5.Anderson T W. An introduction to multivariate statistical analysis. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1984. [Google Scholar]
- 6.Arbeit R D, Arthur M, Dunn R, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed-field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 7.Aznar R, Ludwig W, Schleifer K H. Ribotyping and randomly amplified polymorphic DNA analysis of V. vulnificus biotypes. Syst Appl Microbiol. 1993;16:303–309. [Google Scholar]
- 8.Barenkamp S J, Munson R S, Jr, Granoff D M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981;143:668–676. doi: 10.1093/infdis/143.5.668. [DOI] [PubMed] [Google Scholar]
- 9.Dice L R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 10.Edmiston H L, Tuck H A. Resource inventory of the Apalachicola River and Bay drainage basin. Tallahassee, Fla: Florida Game and Fresh-Water Fish Commission; 1987. [Google Scholar]
- 11.Furuse K, Ando A, Osawa S, Watanabe I. Distribution of ribonucleic acid coliphages in raw sewage from treatment plants in Japan. Appl Environ Microbiol. 1981;41:1139–1143. doi: 10.1128/aem.41.5.1139-1143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geldreich E E. Sanitary significance of fecal coliform in the environment. Water Pollution Control Research series, publication WP-20-3. Cincinnati, Ohio: Federal Water Pollution Control Administration, U.S. Department of the Interior; 1966. [Google Scholar]
- 13.Geldreich E E, Kenner B A. Concepts of fecal streptococci in stream pollution. J Water Pollut Control Fed. 1969;41:R336–R352. [PubMed] [Google Scholar]
- 14.Govan J R W, Harris G. Typing of Pseudomonas cepacia by bacteriocin susceptibility and production. J Clin Microbiol. 1985;22:490–494. doi: 10.1128/jcm.22.4.490-494.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimont F, Grimont P A D. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur/Microbiol (Paris) 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 16.Guzewich J J, Morse D L. Sources of shellfish in outbreaks of probable viral gastroenteritis: implications for control. J Food Prot. 1986;49:389–394. doi: 10.4315/0362-028X-49.5.389. [DOI] [PubMed] [Google Scholar]
- 17.Krumperman P H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latham R H, Stamm W E. Role of fimbriated Escherichia coli in urinary tract infections in adult women: correlation with localization studies. J Infect Dis. 1984;149:835–840. doi: 10.1093/infdis/149.6.835. [DOI] [PubMed] [Google Scholar]
- 19.McLachlan G J. Discriminant analysis and statistical pattern recognition. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 20.Morrison D F. Multivariate statistical methods. New York, N.Y: McGraw-Hill Book Co.; 1976. [Google Scholar]
- 21.Olsen J E, Brown D J, Baggesen D L, Bisgaard M. Biochemical and molecular characterization of Salmonella enterica serovar berta, and comparison of methods for typing. Epidemiol Infect. 1992;108:243–260. doi: 10.1017/s0950268800049724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orskov F, Orskov I. Enterobacteriaceae. In: Broude A I, editor. Medical microbiology and infectious diseases. Philadelphia, Pa: The W. B. Saunders Co.; 1981. pp. 340–352. [Google Scholar]
- 23.Ott M, Bender L, Blum G, Schmittroth M, Achtman M, Tschape H, Hacker J. Virulence patterns and long-range genetic mapping of extraintestinal Escherichia coli K1, K5, and K100 isolates: use of pulsed-field gel electrophoresis. Infect Immun. 1991;59:2664–2672. doi: 10.1128/iai.59.8.2664-2672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parveen S, Murphree R L, Tamplin M L. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Differentiating point and nonpoint sources of Escherichia coli in an estuarine environment by multiple antibiotic resistance and ribotype profile, abstr. Q-476; p. 469. [Google Scholar]
- 25.Parveen S, Murphree R L, Edmiston L, Kaspar C W, Portier K M, Tamplin M L. Association of multiple-antibiotic resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl Environ Microbiol. 1997;63:2607–2612. doi: 10.1128/aem.63.7.2607-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parveen S, Tamplin M L. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Methods for measuring levels of human and nonhuman sources of fecal pollution in water, abstr. Q-108; p. 439. [Google Scholar]
- 27.Popovic T, Bopp C, Olsvik O, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for V. cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pourcher A M, Devriese L A, Hernandez J F, Delattre J M. Enumeration by a miniaturized method of Escherichia coli, Streptococcus bovis and enterococci as indicators of the origin of fecal pollution of waters. J Appl Bacteriol. 1991;70:525–530. doi: 10.1111/j.1365-2672.1991.tb02752.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Smith C L, Cantor C R. Purification, specific fragmentation and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- 31.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 32.Stelma G N, Jr, McCabe L J. Nonpoint pollution from animal sources and shellfish sanitation. J Food Prot. 1992;55:649–656. doi: 10.4315/0362-028X-55.8.649. [DOI] [PubMed] [Google Scholar]
- 33.Stull T L, LiPuma J J, Edlind T D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988;157:280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- 34.Tamplin M L, Jackson J K, Buchrieser C, Murphree R L, Portier K M, Ganger V V, Miller L G, Kaspar C W. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. 1996;62:3572–3580. doi: 10.1128/aem.62.10.3572-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarkka E, Ahman H, Siitonen A. Ribotyping as an epidemiologic tool for Escherichia coli. Epidemiol Infect. 1994;112:263–274. doi: 10.1017/s0950268800057678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wachsmuth K. Molecular epidemiology of bacterial infections: example of methodology and investigations of outbreaks. Rev Infect Dis. 1986;8:682–692. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- 37.Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl Environ Microbiol. 1996;62:3997–4002. doi: 10.1128/aem.62.11.3997-4002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zierdt C H, Robertson E A, Williams R L, MacLowry J D. Computer analysis of Staphylococcus aureus phage typing data from 1957 to 1975, citing epidemiological trends and natural evolution within phage typing system. Appl Environ Microbiol. 1980;39:623–629. doi: 10.1128/aem.39.3.623-629.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]