Abstract
A diverse series of imine linked 1,2,3-triazole hybrids has been synthesized via Cu(I)-promoted click reaction, with an aim to develop some new antimicrobial molecules. The structural characterization of the synthesized triazole hybrids was accomplished using various spectral techniques like 1H NMR, 13C NMR, FTIR and HRMS. Among the synthesized hybrids, 7d exhibited highest antimicrobial efficacy against R. oryzae and S. aureus with MIC of 0.0123 µmol/mL and 0.0061 µmol/mL, respectively. Docking studies of the terminal alkynes (4a, 4b) and triazole hybrids (6a, 6d, 7c, 7g) showing high potency towards E. coli DNA gyrase and C. albicans sterol 14-α demethylase were also undertaken to understand the binding behaviour.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11164-022-04737-2.
Keywords: Imine; Click chemistry; 1,2,3-triazoles; Antimicrobial activity; Docking
Introduction
Despite the availability of various antimicrobial drugs in the market, microbial infections are a foremost cause of deaths worldwide [1]. Just like Covid-19, microbial resistance is a threat to the health security, it may become next global health emergency, if necessary actions are not taken to prevent the microbial resistance [2, 3]. According to the annual report of GARDP, 1.2 million people die every year due to the infections caused by drug resistant microbes. Europe and US are facing a financial loss of 13.5 billion dollars every year due to the hospital associated infections and also by 2050, approximate 28.3 million people could be pushed into the extreme poverty (https://gardp.org/?gclid=Cj0KCQjwnoqLBhD4ARIsAL5JedK4wcgihTB_tiMNA5mWXcrz6y0nxXmDl-dJSHJWejvguu6L3cIG8EQaAmFbEALw_wcB (accessed on 31.01.2022)). The situation is worsened by the inappropriate use of the drugs available in the market and it results into the diminishing drug’s efficacy [4]. The main cause of drug resistant development in case of antibiotics is the formation of biofilms by bacteria [5]. To overcome this worsened situation, the development of microbial drugs with novel mode of action is highly desired. Scientists are using various approaches to develop antimicrobial drugs for saving human life from the health emergencies of future and molecular hybridization approach is evolved as a most promising method to achieve this target [6, 7].
A large number of natural and synthetic pharmacophores have been reported as antimicrobial agents [8]. In the last two decades, 1,2,3-triazoles have emerged as the one of the most promising pharmacophores exhibiting important biological activities like antibacterial [9, 10], antifungal [11], anti-Alzheimer [12], anti-cancer [13], anti-human immunodeficiency virus (anti-HIV) [14], antidiabetic [15], antimalarial [16], antitubercular [17], anti-inflammatory activity [18]. The hybridization of 1,2,3-triazole unit with several other pharmacophores like Schiff bases [19], chalcones [20], indole [21], pyrazolines [22], semicarbazone [23], urea [24], oxazolones [25] has empowered to develop various molecular hybrid displaying excellent inhibition against a variety of microorganisms. The high stability of 1,2,3-triazoles against acidic and basic hydrolysis and inertness towards reducing and oxidizing agents motivated scientists to exploit 1,2,3-triazole hybrids for bioactivity evaluation [26].
Imine unit also known as azomethine is also an important class of pharmacophores because of their ease of synthesis from carbonyl and amino derivatives as well as their abilities to exhibit various interactions with different biological targets. Compounds containing imine unit display a wide range of pharmacological properties including antibacterial [27], antifungal [28], anti-HIV [29], anti-cancer [30], anti-inflammatory [31], antimalarial [32], antitubercular [33]. Imines can easily be obtained by nucleophilic addition of carbonyl compounds and amino compounds [34, 35]. As reported in the literature, Gurjaspreet et al. synthesized schiff base linked triazole silatranes and evaluated their antimicrobial potential [36, 37]. Yuqin et al. derived paeonol schiff base containing 1,2,3-triazole and assessed their antilung adenocarcinoma potential [38]. Therefore, inspired from bioactivity profile of imine, triazole and amide units it was envisioned to synthesize some derivatives containing these units for the antimicrobial screening (Fig. 1). Herein, we have outlined the synthesis of various imine linked 1,2,3-triazoles as antimicrobial compounds. The docking studies of potent triazole hybrids against C. albicans sterol 14-α demethylase and E. coli DNA gyrase was carried out and obtained results are in support with the in vitro antimicrobial screening results.
Fig. 1.

Designed imine linked 1,2,3-triazole hybrids
Materials and methods
General
Starting materials employed for the synthesis of triazole hybrids were obtained from the commercial sellers, open capillary tube method was employed to record the melting points of the compounds and are reported as such. TLC plates were used to visualize the progression of the reaction. IR spectral data was recorded on Shimazdu IR Affinity FTIR spectrophotometer. Bruker Avance III 400 nano bay spectrometer operating at 400 and 100 MHz and Bruker Avance NMR spectrometer operating at 500 and 125 MHz in deuterated (DMSO-d6) using tetramethyl silane (TMS) as internal standard (chemical shift in ppm) was used to record 1H and 13C NMR.
General procedure for the synthesis of imine linked alkynes (4a, 4b)
To synthesize imines, an equimolar amount of m/p-hydroxy benzaldehydes (8.2 mmol) and p-fluoro aniline (8.2 mmol) was dissolved in 20 mL of dry ethanol and refluxed for 6 h at 80 °C in the presence of catalytic amount of HCl. When reaction was accomplished, obtained solid was washed with ethanol and then recrystallized from ethanol. Further, these synthesized imines (7.0 mmol) were propargylated with propargyl bromide (80% in toluene) (8.4 mmol) on refluxing in acetone in presence of K2CO3 (21 mmol). Propargylation was completed after continue refluxing for 7 h. Then ice-cold water was poured in the reaction mixture, precipitates were obtained in case of 4b alkyne, which was further washed with ice-cold water and recrystallized in ethanol. But in case of 4a alkyne on addition of water in reaction mixture, no precipitates obtained and it was extracted with ethyl acetate and 4a was obtained from organic layer after evaporating solvent under vacuum.
General procedure for the synthesis of imine linked 1,2,3-triazoles (6a–6g, 7a–7g)
To synthesize imine linked 1,2,3-triazole hybrids, N-phenyl acetamide bromide derivatives 5a–5g (1.0 mmol), imine linked alkynes 4a, 4b (1.0 mmol), sodium azide (3.0 mmol) in DMF: water (8:2) were stirred for 6 h at 50 °C, after the addition of copper sulphate pentahydrate (10 mol%) and sodium ascorbate (20 mol%). Reaction was monitored by using TLC under the UV lamp, on completion reaction mixture was cooled and diluted with ice-cold ammonium chloride (aq.): ammonia solution (9: 1 v/v). Solid thus obtained was filtered and washed with cold water to furnish imine linked 1,2,3-triazoles (6a–6g, 7a–7g).
Pharmacology
The preliminary in vitro pharmacological potential of the synthesized imine linked 1,2,3-triazole hybrids (6a–6g, 7a–7g) as well as their precursor terminal alkynes (4a, 4b) were studied against several microbial strains using serial dilution method [39].
Docking details
The three-dimensional structures of molecules were drawn and developed with Marvin Sketch [40] and docking simulations were carried out using Autodock Vina software [41] in to protein (DNA gyrase of E. coli and sterol 14-α-demethylase of C. albicans) with PDB ID: 5TZ1 downloaded from protein data bank. The protein preparation was carried out in UCSF Chimera [42]. The protocols followed while carrying out docking simulations were same as described in literature [43]. The visualization of the docking results was done using Discovery Studio Visualizer [44] and Chimera X [45].
Results and discussion
Chemistry
In continuation to our work on synthesis and bio-evaluation of 1,2,3-triazoles, a library of new imine linked 1,2,3-triazoles were synthesized. Synthetic protocol used to achieve imine appended 1,2,3-triazoles involved three steps. In first step m/p- hydroxy benzaldehydes (1a–1b) were condensed with p-fluoro aniline in dry ethanol with few drops of HCl as catalyst under refluxing to obtain imine derivatives (3a–3b). The propargylation of imine derivatives (3a–3b) was performed in acetone with propargyl bromide (80% in toluene) in presence of K2CO3 on refluxing. Finally, convenient click chemistry approach was used for one pot synthesis of 1,2,3-triazoles, where synthesized imines with a terminal alkyne (4a–4b) were reacted with N-bromoacetamide derivatives (5a–5g) using sodium azide, copper sulphate pentahydrate-sodium ascorbate catalytic system in DMF: water (8:2) as shown in scheme 1. Imine linked 1,2,3-triazole compounds were furnished in high yield i.e. 76–89% as shown in Table 1.
Scheme 1.
Synthesis of imine linked 1, 2, 3-triazole hybrids (6a–6g, 7a–7g)
Table 1.
Synthesis of imine linked 1, 2, 3-triazole hybrids (6a–6g, 7a–7g) via click chemistry
| Entry | Compounds | R | Yield (%)[a] | M. P. (°C) |
|---|---|---|---|---|
| 1 | 4a | – | 85 | 35–37 |
| 2 | 4b | – | 88 | 56–59 |
| 3 | 6a | H | 82 | 160–163 |
| 4 | 6b | F | 79 | 144–146 |
| 5 | 6c | Cl | 86 | 162–165 |
| 6 | 6d | Br | 82 | 174–176 |
| 7 | 6e | NO2 | 89 | 220–223 |
| 8 | 6f | OCH3 | 82 | 190–193 |
| 9 | 6g | CH3 | 77 | 187–190 |
| 10 | 7a | H | 81 | 228–231 |
| 11 | 7b | F | 77 | 219–221 |
| 12 | 7c | Cl | 82 | 223–224 |
| 13 | 7d | Br | 79 | 231–234 |
| 14 | 7e | NO2 | 82 | 220–222 |
| 15 | 7f | OCH3 | 76 | 226–229 |
| 16 | 7g | CH3 | 88 | 243–245 |
[a] Yield of synthesized imine linked 1,2,3-triazole hybrids
All the newly synthesized imine-based 1,2,3-triazole hybrids (6a–6g, 7a–7g) were characterized by various spectral techniques like FTIR, 1H, 13C NMR and HRMS data. The characteristics band/ peaks of two representative compounds 6a and 7a is discussed below:
FTIR analysis
In the IR spectra of synthesized hybrids 6a and 7a bands were observed at 3257 cm−1 and 3280 cm−1 respectively confirmed the presence of –NH group of amide functionality [46]. Formation of triazole ring was confirmed by the appearance of a band at 3142 cm−1 in both 6a and 7a because of C–H stretching vibration of triazole unit [47]. In compounds 6a and 7a, a band of strong intensity because of the amidic C=O stretching was observed at 1669 cm−1 and 1681 cm−1, respectively. While band due to the C=N stretching was detected at 1600 cm−1 for 6a and at 1607 cm−1 for 7a [48, 49]. For the compound 6a out of plane bending vibrations of benzene appeared at 696 cm−1 and 787 cm−1, whereas in compound 7a out of plane bending vibration of benzene appeared at 837 cm−1 [50]. In plane bending vibrations of benzene ring were recognized in the region of 1300–1000 cm−1 and are overlapped with the other stretching vibrations. In compounds 6a and 7a C–N stretching vibrations of azole unit was spotted at 1313 cm−1 and 1312 cm−1 respectively, whereas N=N stretching was detected at 1412 cm−1 in 6a, whereas in 7a this stretching vibration is observed at 1415 cm−1 [51].
1H NMR analysis
In the 1H NMR spectrum of synthesized triazole hybrid 6a, peak observed at δ 10.49 ppm is due to the –NH proton of amide functionality. Singlets observed at δ 5.37 and δ 5.27 ppm were due to the methylene protons present adjacent to the amide functionality and oxygen atom. A singlet at δ 8.62 ppm assigned to the –C = NH– proton [37]. Whereas, a confirmatory singlet was observed at δ 8.29 ppm is due to the triazolyl proton [52]. While, compound 7a showed a peak due to –NH proton of amide at δ 10.50 ppm, two singlets due to methylene protons were observed at δ 5.38 and δ 5.29 ppm. Singlets due to –C = NH– and triazolyl proton were observed at δ 8.55 ppm and δ 8.31 ppm, respectively.
13C NMR analysis
From the analysis of 13C NMR spectrum of synthesized triazole hybrids it was found that in compound 6a, two peaks at δ 61.64 and δ 52.68 ppm confirmed the existence of two methylenes, whereas in compound 7a these two peaks observed at δ 61.67 and δ 52.70 ppm. C4 carbon peak of triazole ring in 6a appeared at δ 142.81 ppm, while in 7a it is appeared at δ 142.58 ppm. The presence of all these peaks in synthesized compounds 13C NMR spectra confirms the formation of triazoles [52].
Mass analysis
HRMS data of synthesized triazole hybrid 6a was in good agreement with the calculated value. The molecular ion peak [M + H]+ of 6a C24H20FN5O2 was obtained at 430.1746 in HRMS which was in complete agreement with the calculated mass: 430.1674. When molecule get fragmented then a peak corresponding to C18H18N5O2 was obtained at 337.1354 which was in agreement with the calculated value 337.1533. Fragmentation of amide linkage produced molecular ion C18H14FN4O2 which gives a [M + H]+ peak at 338.1385 which is closely resembles with the calculated value 338.1174.
Antimicrobial activity
The preliminary antimicrobial potency of the newly prepared imine linked 1,2,3-triazole hybrid compounds (6a–6g, 7a–7g) as well as their precursor terminal alkynes (4a, 4b) were examined against some bacterial and fungal strains using standard serial dilution method [39]. All samples were examined at five different concentrations of 50, 25, 12.5, 6.25, 3.125 µg/mL against Bacillus subtilis (MTCC 441), Staphylococcus aureus (MTCC 3160), Pseudomonas aeruginosa (MTCC 424), Escherichia coli (MTCC 16,521), Salmonella enterica, Candida albicans (MTCC 183), and Rhizopus oryzae microbial species.
The in vitro activity results evinced that most of the synthesized imine clubbed 1,2,3-triazoles (6a–6g, 7a–7g) were more potent than their precursor terminal alkynes (4a, 4b) against the tested microbial strains. It shows that the introduction of triazole scaffold augmented the antimicrobial potential of the synthesized hybrid molecules. Activity of all the synthesized hybrid molecules was recorded in terms of minimum inhibitory concentration and compared with their precursor terminal alkynes as well as with the ciprofloxacin (antibacterial) and fluconazole (antifungal) as shown in Table 2 and Table 3, respectively. From the antimicrobial assay results, it was observed that compound 6d (MIC = 0.0123 µmol/mL) was found to be most potent against Gram-positive bacteria B. subtilis. Whereas, compound 7d (MIC = 0.0061 µmol/mL) showed excellent and better efficacy than the ciprofloxacin (MIC = 0.0094 µmol/mL) against S. aureus. In case of Gram-negative bacteria, compound 6d and 7d showed most potent activity among synthesized compounds against two bacterial strains S. enterica and P. aeruginosa, whereas against bacterial strain E. coli, compounds 7c (MIC = 0.0135 µmol/mL) and 7f (MIC = 0.0136 µmol/mL) were reported as most active.
Table 2.
Antibacterial screening data of the synthesized derivatives (4a, 4b, 6a-6g, 7a-7g)
| Minimum inhibitory concentration (MIC, μmol/mL) | |||||
|---|---|---|---|---|---|
| Entry | S. enterica | B. subtilis | S. aureus | P. aeruginosa | E. coli |
| 4a | 0.0987 | 0.0987 | 0.0493 | 0.0987 | 0.0987 |
| 4b | 0.0493 | 0.0987 | 0.0493 | 0.0987 | 0.0987 |
| 6a | 0.0291 | 0.0145 | 0.0073 | 0.0291 | 0.0145 |
| 6b | 0.0559 | 0.0279 | 0.0140 | 0.0279 | 0.0279 |
| 6c | 0.0539 | 0.0135 | 0.0269 | 0.0135 | 0.0269 |
| 6d | 0.0246 | 0.0123 | 0.0246 | 0.0123 | 0.0246 |
| 6e | 0.0263 | 0.0132 | 0.0263 | 0.0132 | 0.0263 |
| 6f | 0.0272 | 0.0136 | 0.0136 | 0.0272 | 0.0272 |
| 6g | 0.0282 | 0.0141 | 0.0282 | 0.0141 | 0.0282 |
| 7a | 0.0291 | 0.0145 | 0.0073 | 0.0145 | 0.0582 |
| 7b | 0.0279 | 0.0279 | 0.0070 | 0.0140 | 0.0279 |
| 7c | 0.0539 | 0.0539 | 0.0067 | 0.0135 | 0.0135 |
| 7d | 0.0246 | 0.0246 | 0.0061 | 0.0123 | 0.0492 |
| 7e | 0.0263 | 0.0132 | 0.0066 | 0.0132 | 0.0263 |
| 7f | 0.0544 | 0.0136 | 0.0068 | 0.0136 | 0.0136 |
| 7g | 0.0282 | 0.0564 | 0.0141 | 0.0282 | 0.0282 |
| Ciprofloxacin | 0.0094 | 0.0094 | 0.0094 | 0.0094 | 0.0094 |
Table 3.
Antifungal screening results of the synthesized hybrids (4a, 4b, 6a–6g, 7a–7g)
| Minimum inhibitory concentration (MIC, µmol/mL) | ||
|---|---|---|
| Compounds | C. albicans | R. oryzae |
| 4a | 0.0493 | 0.0247 |
| 4b | 0.0987 | 0.0987 |
| 6a | 0.0582 | 0.0291 |
| 6b | 0.0559 | 0.0279 |
| 6c | 0.0539 | 0.0135 |
| 6d | 0.0246 | 0.0492 |
| 6e | 0.0527 | 0.0132 |
| 6f | 0.0272 | 0.0544 |
| 6g | 0.0564 | 0.0282 |
| 7a | 0.0291 | 0.0291 |
| 7b | 0.0279 | 0.0140 |
| 7c | 0.0539 | 0.0270 |
| 7d | 0.0246 | 0.0123 |
| 7e | 0.0527 | 0.0263 |
| 7f | 0.0272 | 0.0272 |
| 7g | 0.0141 | 0.0282 |
| Fluconazole | 0.0408 | 0.0408 |
The in vitro antifungal assay of the synthesized compounds displayed that most of the imine based 1,2,3-triazole hybrids were more efficient than the fluconazole (MIC = 0.0408 µmol/mL). 1,2,3-Triazole hybrids prepared from 4b displayed more potency than the alkyne against both the tested fungal strains. Compound 7g displayed excellent activity against C. albicans having MIC (0.0141 µmol/mL), whereas compound 6d (MIC = 0.0246 µmol/mL) and 7d (MIC = 0.0246 µmol/mL) also displayed better activity than the fluconazole (MIC = 0.0408 µmol/mL) against C. albicans. All newly synthesized triazoles except 6d, 6f exhibited better potency than the fluconazole (MIC = 0.0408 µmol/mL) against R. oryzae.
Structure-activity relationship
Following SAR was established after analysing the results obtained from the antimicrobial assay of the synthesized imine linked 1,2,3-triazole hybrids:
-
(i)
As most of the synthesized imine-based 1,2,3-triazole hybrids displayed more efficacy than their corresponding terminal alkynes which led to the concept that molecular hybridization enhanced the activity.
-
(ii)
Triazoles bearing bromine atom on the phenyl ring found to be more potent against S. enterica and P. aeruginosa and there is no general trend of activity was observed for other compounds.
-
(iii)
Most of the triazole hybrids synthesized from meta substituted alkyne 4a were found to be equally or more potent than the triazoles synthesized from para substituted alkyne 4b against B. subtilis. Compound 6d bearing bromine atom and 7e bearing nitro group on the phenyl ring were found to be most active.
-
(iv)
Compound 7d with bromine atom on the phenyl ring was found to be the most active against S. aureus. Most of the imine linked 1,2,3-triazole hybrids synthesized from alkyne 4b were found to be more potent than the corresponding imine linked 1,2,3-triazole hybrids obtained from the 4a alkyne against S. aureus.
-
(v)
Triazole hybrids 7c and 7f bearing chlorine and methoxy substituent, respectively, on phenyl ring exhibited excellent activity against E. coli.
-
(vi)
Compound 7g bearing methyl group on phenyl ring found to be most active against C. albicans.
-
(vii)
Compounds 6d and 7d having bromine substituent on phenyl ring displayed good to better antimicrobial efficacy against all the microbial strains.
Docking studies
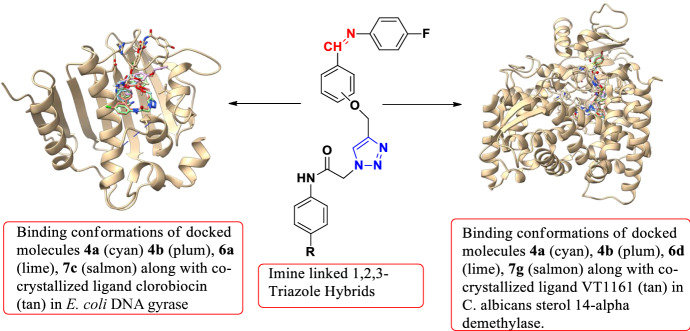
In order to theorize the plausible mechanism responsible for the antimicrobial activity and to understand the effect of triazole ring insertion into the molecular structure. In silico studies were carried out. As reported in the literature, organic molecules having 1,2,3-triazole scaffold act as a good inhibitor of E. coli DNA gyrase, C. albicans 14-α sterol demethylase, α-glucosidase, etc. [15, 22]. Therefore, compounds 4a, 4b, 6a and 7c having good inhibition potential against E. coli were docked in the active site of E. coli DNA Gyrase. While, C. albicans enzyme sterol 14-alpha demethylase was chosen as a target for docking of compounds 4a, 4b, 6d and 7g as the latter molecules are better inhibitor of C. albicans growth.
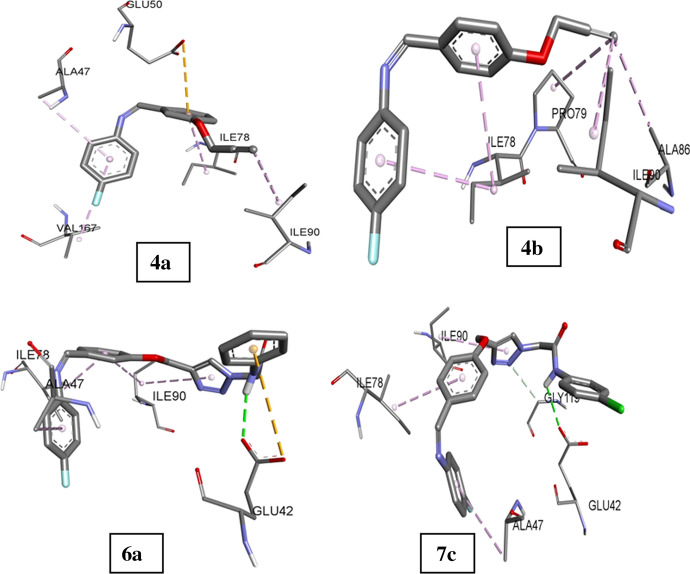
Terminal alkynes 4a and 4b mainly interacted through hydrophobic interactions with in the active sites of DNA gyrase. Middle phenyl ring of 4a showed one electrostatic interaction (π-anion) with GLU50. Residues ALA47, ILE78, PRO79, ALA86, ILE90 and VAL167 were involved in interactions. Triazole hybrids 6a and 7c made hydrogen bond with GLU42 via amide hydrogen. Nitrogen atom of triazole ring of compound 7c also created an additional carbon-hydrogen bond with GLY119. These hydrogen bonds might be the cause of higher potency of triazole hybrids against E. coli as compared to their terminal alkyne precursors as the other interactions shown by these triazole hybrids are hydrophobic and with the same residues as observed in terminal alkyne interactions. The docking score of 4a, 4b, 6a and 7c was − 7.0, − 6.4, − 8.0 and − 8.1 kcal/mol, respectively. The value of correlation coefficient between the observed activity and the calculated binding energy was 0.95 kcal/mol which shows very good relationship between these two studies. The diagrammatic representation of docked conformation along with interacting residues is shown in Fig. 2. Also, the cartoon representation of compounds 4a, 4b, 6a and 7c with E. coli DNA gyrase is highlighted in Fig. 3.
Fig. 2.
Binding interactions of compound 4a, 4b, 6a and 7c in active site of E. coli DNA gyrase
Fig. 3.

Cartoon diagram showing binding conformations of docked molecules 4a (cyan) 4b (plum), 6a (lime), 7c (salmon) along with co-crystallized ligand clorobiocin (tan) in E. coli DNA gyrase
Further, in sterol alpha demethylase, oxygen atom of terminal alkyne 4a made carbon-hydrogen bond with GLY307 and middle phenyl ring was tangled in π-π stacked interactions with HEME molecule and an electrostatic π-cation interaction with HEME iron. ILE131, ILE304 and LEU376 helped through hydrophobic interactions. 4b interacted with HEME molecule via carbon hydrogen bond, halogen bond, π-π stacked interactions and π-cation interactions. Phenoxy ring displayed π-π stacked interactions with TYR118. Triazole derivatives 6d and 7g created hydrogen bonds with TYR64 through amide hydrogen atom. Phenoxy ring was situated against TYR118 through T shaped π-π stacked interactions. Fluorophenyl ring was stacked near HEME via π-π stacked interactions. These interactions along with other hydrophobic interactions involved in anchorage of these molecules in the active site are pictured in Fig. 4. The binding affinity of 4a, 4b, 6d and 7g in 5TZ1 was − 10.1, − 9.4, − 11.6 and − 11.8 kcal/mol. The correlation coefficient between observed antifungal activity against C. albicans and calculated binding affinity was 0.95 which shows good agreement between experimental and theoretical results. The docked conformations of these molecules along with co-crystalized ligand are shown in Fig. 4. Cartoon representation of compounds 4a, 4b, 6d and 7g with C. albicans sterol 14-alpha demethylase is highlighted in Fig. 5.
Fig. 4.
Binding interactions of compound 4a, 4b, 6d and 7g in active site of C. albicans sterol 14-α demethylase
Fig. 5.

Cartoon diagram showing binding conformations of docked molecules 4a (cyan), 4b (plum), 6d (lime), 7g (salmon) along with co-crystallized ligand VT1161 (tan) in C. albicans sterol 14-alpha demethylase
ADME analysis
The bioactivity score of the synthesized triazole hybrids and terminal alkynes was predicted by using an online software Molinspiration (https://www.molinspiration.com/ (Accessed on 24.01.2022)). As per Lipinski’s rule of five, an orally active drug must have to follow at least four criteria out of the five which is as follows; (i) Molecular weight ≤ 500 (ii) hydrogen bond acceptors ≤ 10 (iii) hydrogen bond donors ≤ 5, (iv) miLogP ≤ 5 (v) rotatable bonds ≤ 10. All synthesized compounds except 6d and 7d showed drug likeness as predicted by the bioactivity score. Presence of the rotatable bonds in synthesized hybrids makes them flexible. The higher values of TPSA for synthesized triazole hybrids as compared to their precursor terminal alkynes clearly showed that they have better transport properties and hydrogen bond formation capacity with the targeted enzymes. Percentage absorption was obtained by following the formula % ABS = 109(0.345xTPSA) [53].
| Entry | Compounds | %ABS | MW | miLogP | TPSA | Natoms | nON | nOHNH | Nrotb | Volume |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4a | 101.55 | 253.28 | 3.83 | 21.60 | 19 | 2 | 0 | 4 | 231.69 |
| 2 | 4b | 101.55 | 253.28 | 3.85 | 21.60 | 19 | 2 | 0 | 4 | 231.69 |
| 3 | 6a | 80.91 | 429.45 | 4.22 | 81.41 | 32 | 7 | 1 | 8 | 377.49 |
| 4 | 6b | 80.91 | 447.44 | 4.38 | 81.41 | 33 | 7 | 1 | 8 | 382.42 |
| 5 | 6c | 80.91 | 463.90 | 4.90 | 81.41 | 33 | 7 | 1 | 8 | 391.02 |
| 6 | 6d | 80.91 | 508.35 | 5.03 | 81.41 | 33 | 7 | 1 | 8 | 395.38 |
| 7 | 6e | 65.10 | 474.45 | 4.18 | 127.24 | 35 | 10 | 1 | 9 | 400.82 |
| 8 | 6f | 77.72 | 459.48 | 4.28 | 90.65 | 34 | 8 | 1 | 9 | 403.04 |
| 9 | 6g | 80.91 | 443.48 | 4.67 | 81.41 | 33 | 7 | 1 | 8 | 394.05 |
| 10 | 7a | 80.91 | 429.45 | 4.24 | 81.41 | 32 | 7 | 1 | 8 | 377.49 |
| 11 | 7b | 80.91 | 447.44 | 4.41 | 81.41 | 33 | 7 | 1 | 8 | 382.42 |
| 12 | 7c | 80.91 | 463.90 | 4.92 | 81.41 | 33 | 7 | 1 | 8 | 391.02 |
| 13 | 7d | 80.91 | 508.35 | 5.05 | 81.41 | 33 | 7 | 1 | 8 | 395.38 |
| 14 | 7e | 65.10 | 474.45 | 4.20 | 127.24 | 35 | 10 | 1 | 9 | 400.82 |
| 15 | 7f | 77.72 | 459.48 | 4.30 | 90.65 | 34 | 8 | 1 | 9 | 403.04 |
| 16 | 7g | 80.91 | 443.48 | 4.69 | 81.41 | 33 | 7 | 1 | 8 | 394.05 |
Conclusion
In summary, various new imine-based 1,2,3-triazole derivatives was synthesized via Cu(I)-catalyzed 1,3-dipolar cycloaddition reaction and their in vitro antimicrobial efficacy was also evaluated against various microbial strains. The antimicrobial screening results indicated that, the presence of triazole scaffold with imine led to augmentation in inhibition potential of the synthesized compounds. Triazole hybrid 7d displayed most potent antibacterial potency against S. aureus with MIC of 0.0061 µmol/mL, also compound 7d displayed effective antifungal efficacy against R. oryzae with MIC of 0.0123 µmol/mL. The docking simulations of selected triazole hybrids with E. coli DNA Gyrase and C. albicans sterol 14-α demethylase were also in agreement with the observed antimicrobial screening data. Compound 7c binds within E. coli DNA gyrase active site with binding energy of − 8.1 kcal/mol, whereas compound 7g displayed binding affinity of − 11.8 kcal/mol for the sterol 14-α demethylase binding sites. Further, the value of correlation coefficient between the observed activity and the calculated binding energy was 0.95 kcal/mol which shows very good relationship between these two studies. We expect that imine linked 1,2,3-triazole hybrids may open new opportunities for the medicinal chemists as antimicrobial agents.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors acknowledge the assistance from PURSE program No. SR/PURSE 2/40(G) from DST, New Delhi. AK thanks University Grants Commission for providing financial assistance as SRF. The NMR and MS spectral facility at A P J Abdul Kalam Central instrumentation laboratory, Guru Jambheshwar University of Science & Technology, Hisar, India are highly acknowledged.
Declarations
Conflict of interest
No conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Asadi A, Razavi S, Talebi M, Gholami M. Infection. 2019;47:13. doi: 10.1007/s15010-018-1222-5. [DOI] [PubMed] [Google Scholar]
- 2.dos Santos Ramos MA, dos Santos KC, da Silva PB, de Toledo LG, Marena GD, Rodero CF, de Camargo BAF, Fortunato GC, Bauab TM, Chorilli M. Int. J. Pharm. 2020;589:119780. doi: 10.1016/j.ijpharm.2020.119780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz MP, Santos E, Cervantes MV, Juarez ML. Rev. Clin. Esp. 2021;221:55. doi: 10.1016/j.rce.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusini A, Rampini SK, Bansal V, Ledergerber B, Kuster SP, Ruef C, Weber R. PLoS ONE. 2010;5:e14011. doi: 10.1371/journal.pone.0014011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi L, Li H, Zhang C, Liang B, Li J, Wang L, Du X, Liu X, Qiu S, Song H. Front. Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viegas-Junior C, Danuello A, da Silva Bolzani V, Barreiro EJ, Fraga CAM. Curr. Med. Chem. 2007;14:1829. doi: 10.2174/092986707781058805. [DOI] [PubMed] [Google Scholar]
- 7.Sunil R, Pal S, Jayashree A. Med. Chem. 2019;9:93. [Google Scholar]
- 8.O’donnell F, Smyth TJP, Ramachandran VN, Smyth WF. Int. J. Antimicrob. Agent. 2010;35:30. doi: 10.1016/j.ijantimicag.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Kumar L, Lal K, Kumar A, Kumar A. Res. Chem. Intermed. 2021;47:5079. doi: 10.1007/s11164-021-04588-3. [DOI] [Google Scholar]
- 10.Aarjane M, Slassi S, Amine A. J. Mol. Struct. 2021;1241:130636. doi: 10.1016/j.molstruc.2021.130636. [DOI] [Google Scholar]
- 11.Poonia N, Kumar A, Kumar V, Yadav M, Lal K. Curr. Top. Med. Chem. 2021;21:2109. doi: 10.2174/1568026621666210913122828. [DOI] [PubMed] [Google Scholar]
- 12.Rastegari A, Nadri H, Mahdavi M, Moradi A, Mirfazli SS, Edraki N, Moghadam FH, Larijani B, Akbarzadeh T, Saeedi M. Bioorg. Chem. 2019;83:391. doi: 10.1016/j.bioorg.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 13.Alam MM. Arch. Pharm. 2022;355:2100158. doi: 10.1002/ardp.202100158. [DOI] [PubMed] [Google Scholar]
- 14.Feng LS, Zheng MJ, Zhao F, Liu D. Arch. Pharm. 2021;354:2000163. doi: 10.1002/ardp.202000163. [DOI] [PubMed] [Google Scholar]
- 15.Kumar L, Lal K, Yadav P, Kumar A, Paul AK. J. Mol. Struct. 2020;1216:128253. doi: 10.1016/j.molstruc.2020.128253. [DOI] [Google Scholar]
- 16.Kaushik CP, Chahal M. Monatsh. Chem. 2021;152:1001. doi: 10.1007/s00706-021-02821-8. [DOI] [Google Scholar]
- 17.Shaikh MH, Subhedar DD, Nawale L, Sarkar D, Khan FAK, Sangshetti JN, Shingate BB. MedChemComm. 2015;6:1104. doi: 10.1039/C5MD00057B. [DOI] [Google Scholar]
- 18.Kim TW, Yong Y, Shin SY, Jung H, Park KH, Lee YH, Lim Y, Jung KY. Bioorg. Chem. 2015;59:1. doi: 10.1016/j.bioorg.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Nasli-Esfahani E, Mohammadi-Khanaposhtani M, Rezaei S, Sarrafi Y, Sharafi Z, Samadi N, Faramarzi MA, Bandarian F, Hamedifar H, Larijani B, Hajimiri M, Mahdavi M. Arch. Pharm. 2019;352:1900034. doi: 10.1002/ardp.201900034. [DOI] [PubMed] [Google Scholar]
- 20.Yadav P, Lal K, Kumar L, Kumar A, Kumar A, Paul AK, Kumar R. Eur. J. Med. Chem. 2018;155:263. doi: 10.1016/j.ejmech.2018.05.055. [DOI] [PubMed] [Google Scholar]
- 21.Xu G, Zhao J, Jiang Y, Zhang P, Li W. J. Chem. Res. 2016;40:269. doi: 10.3184/174751916X14597828245275. [DOI] [Google Scholar]
- 22.Kumar L, Lal K, Kumar A, Paul AK, Kumar A. J. Mol. Struct. 2021;1246:131154. doi: 10.1016/j.molstruc.2021.131154. [DOI] [Google Scholar]
- 23.Naveen RK, Tittal P, Yadav K, Lal VD, Ghule A. Kumar. New J. Chem. 2019;43:8052. doi: 10.1039/C9NJ00473D. [DOI] [Google Scholar]
- 24.Poonia N, Lal K, Kumar A. Res. Chem. Intermed. 2021;47:1087. doi: 10.1007/s11164-020-04318-1. [DOI] [Google Scholar]
- 25.Lal K, Kumar L, Kumar A, Kumar A. Curr. Top. Med. Chem. 2018;18:1506. doi: 10.2174/1568026618666180913110456. [DOI] [PubMed] [Google Scholar]
- 26.Verma NK, Mondal D, Bera S. Curr. Org. Chem. 2019;23:2505. [Google Scholar]
- 27.Tehrani KHME, Hashemi M, Hassan M, Kobarfard F, Mohebbi S. Chin. Chem. Lett. 2016;27:221. doi: 10.1016/j.cclet.2015.10.027. [DOI] [Google Scholar]
- 28.da Silva CM, da Silva DL, Modolo LV, Alves RB, de Resende MA, Martins CVB, de Fatima A. J. Adv. Res. 2011;2:1. doi: 10.1016/j.jare.2010.05.004. [DOI] [Google Scholar]
- 29.Patel RN, Patel PV, Desai KR, Purohit PY, Nimavat KS, Vyas KB. Heterocycl. Lett. 2012;2:99. [Google Scholar]
- 30.Sondhi SM, Arya S, Rani R, Kumar N, Roy P. Med. Chem. Res. 2012;21:3620. doi: 10.1007/s00044-011-9899-3. [DOI] [Google Scholar]
- 31.Murtaza S, Akhtar MS, Kanwal F, Abbas A, Ashiq S, Shamim S. J. Saudi Chem. Soc. 2017;21:S359. doi: 10.1016/j.jscs.2014.04.003. [DOI] [Google Scholar]
- 32.Jarrahpour A, Shirvani P, Sharghi H, Aberi M, Sinou V, Latour C, Brunel JM. Med. Chem. Res. 2015;24:4105. doi: 10.1007/s00044-015-1455-0. [DOI] [Google Scholar]
- 33.Cordeiro R, Kachroo M. Bioorganic Med. Chem. Lett. 2020;30:127655. doi: 10.1016/j.bmcl.2020.127655. [DOI] [PubMed] [Google Scholar]
- 34.Naeimi H, Safari J, Heidarnezhad A. Dyes Pigm. 2007;73:251. doi: 10.1016/j.dyepig.2005.12.009. [DOI] [Google Scholar]
- 35.Zhang Y, Fang Y, Liang H, Wang H, Hu K, Liu X, Yi X, Peng Y. Bioorganic Med. Chem. Lett. 2013;23:107. doi: 10.1016/j.bmcl.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Singh G, Singh J, Singh A, Singh J, Kumar M, Gupta K, Chhibber S. J. Organomet. Chem. 2018;871:21. doi: 10.1016/j.jorganchem.2018.06.024. [DOI] [Google Scholar]
- 37.Singh G, Arora A, Rani S, Kalra P, Aulakh D, Wriedt M. Appl. Organomet. Chem. 2017;31:e3728. doi: 10.1002/aoc.3728. [DOI] [Google Scholar]
- 38.Jiang Y, Li Y, Yang T, Shi X, Suo H, Zhang W, Xu G, Li W. J. Chin. Chem. Soc. 2020;67:165. doi: 10.1002/jccs.201800491. [DOI] [Google Scholar]
- 39.Balouiri M, Sadiki M, Ibnsouda SK. J. Pharm. Anal. 2016;6:71. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M Sketch 19.19.0 (2019) ChemAxon
- 41.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Kaushik CP, Lal K, Kumar A, Kumar S. Med. Chem. Res. 2014;23:2995. doi: 10.1007/s00044-013-0882-z. [DOI] [Google Scholar]
- 44.Systèmes D. Biovia discovery studio modeling environment. Dassault Systèmes Biovia: San Diego, CA, USA; 2016. [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bikas R, Kuncser V, Sanchiz J, Schinteie G, Siczek M, Hosseini-Monfared H, Lis T. Polyhedron. 2018;147:142. doi: 10.1016/j.poly.2018.03.019. [DOI] [Google Scholar]
- 47.Yadav P, Lal K, Rani P, Mor S, Kumar A, Kumar A. Med. Chem. Res. 2017;26:1469. doi: 10.1007/s00044-017-1845-6. [DOI] [Google Scholar]
- 48.Bikas R, Shahmoradi E, Reinoso S, Emami M, Lezama L, Sanchiz J, Noshiranzadeh N. Dalton Trans. 2019;48:13799. doi: 10.1039/C9DT01652J. [DOI] [PubMed] [Google Scholar]
- 49.Bikas R, Emami M, Ślepokura K, Noshiranzadeh N. New J. Chem. 2017;41:9710. doi: 10.1039/C7NJ01562C. [DOI] [Google Scholar]
- 50.Borisagar M, Joshi K, Ram H, Vyas K, Nimavat K. J. Chem. Pharm. Res. 2012;2:101. [Google Scholar]
- 51.Al-Ajely MS, Al-Ajely HM, Al-Naib AN. Tikrit J. Pure Sci. 2008;13:1. [Google Scholar]
- 52.Yadav M, Lal K, Kumar A, Kumar A, Kumar D. J Mol. Struct. 2022;1261:132867. doi: 10.1016/j.molstruc.2022.132867. [DOI] [Google Scholar]
- 53.Zhao YH, Abraham MH, Le J, Hersey A, Luscombe CN, Beck G, Sherborne B, Cooper I. Pharm. Res. 2002;19:1446. doi: 10.1023/A:1020444330011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.