Abstract
We investigated the in situ spatial organization of ammonia-oxidizing and nitrite-oxidizing bacteria in domestic wastewater biofilms and autotrophic nitrifying biofilms by using microsensors and fluorescent in situ hybridization (FISH) performed with 16S rRNA-targeted oligonucleotide probes. The combination of these techniques made it possible to relate in situ microbial activity directly to the occurrence of nitrifying bacterial populations. In situ hybridization revealed that bacteria belonging to the genus Nitrosomonas were the numerically dominant ammonia-oxidizing bacteria in both types of biofilms. Bacteria belonging to the genus Nitrobacter were not detected; instead, Nitrospira-like bacteria were the main nitrite-oxidizing bacteria in both types of biofilms. Nitrospira-like cells formed irregularly shaped aggregates consisting of small microcolonies, which clustered around the clusters of ammonia oxidizers. Whereas most of the ammonia-oxidizing bacteria were present throughout the biofilms, the nitrite-oxidizing bacteria were restricted to the active nitrite-oxidizing zones, which were in the inner parts of the biofilms. Microelectrode measurements showed that the active ammonia-oxidizing zone was located in the outer part of a biofilm, whereas the active nitrite-oxidizing zone was located just below the ammonia-oxidizing zone and overlapped the location of nitrite-oxidizing bacteria, as determined by FISH.
Microbial nitrification (oxidation of ammonia to nitrate via nitrite) followed by denitrification (reduction of nitrate to N2 gas) is the key process in the removal of ammonia from wastewater. Due to the slow growth rate of nitrifying bacteria and the sensitivity of these organisms to several environmental factors (e.g., pH, O2, and temperature), treatment plants frequently fail to establish stable nitrification. The process of nitrification is carried out by members of two phylogenetically unrelated groups of lithoautotrophic bacteria, the ammonia-oxidizing bacteria and the nitrite-oxidizing bacteria. A better understanding of the microbiology and ecology of nitrifying bacteria in wastewater treatment biofilms is essential for improving process performance and control. However, characterization of nitrifying bacterial populations in biofilms has been hindered by the limitations of traditional culture-dependent techniques, such as the most-probable-number and selective plating techniques, which do not allow exact localization of nitrification and nitrifying bacteria. Furthermore, these techniques often detect only a minor portion of the naturally occurring nitrifying bacteria. Biofilm spatial heterogeneity and microbial aggregation may increase the uncertainty of culture-dependent enumeration.
The recent development of fluorescence in situ hybridization (FISH) with 16S rRNA-targeted oligonucleotide probes has made it possible to analyze complex in situ microbial community structures in environmental and engineered systems. Thus, symbiotic associations of ammonia- and nitrite-oxidizing bacteria can be revealed by FISH performed with a set of fluorescently labeled 16S rRNA-targeted probes without the bias of cultivation. Previously, workers have described various combinations of numerically dominant populations of ammonia- and nitrite-oxidizing bacteria, such as Nitrosococcus mobilis and Nitrospira-like bacteria in activated sludge of an industrial wastewater treatment plant (13), Nitrosospira sp. and Nitrospira sp. in microbial aggregates of a laboratory scale nitrifying fluidized bed reactor (25), and Nitrosomonas spp. and Nitrobacter spp. in sludge flocs of a continuously stirred tank reactor (18) and in trickling filter biofilms treating aquaculture water (24). Different species of ammonia- and nitrite-oxidizing bacteria differ in their in situ growth kinetics, their substrate and oxygen affinities, and their sensitivities to environmental changes (e.g., changes in pH and NO2 and O2 contents).
Microelectrodes have been used to investigate the spatial distributions of various microbial activities in biofilms (9, 14, 30) and in microbial aggregates grown in a fluidized bed reactor (8, 9, 15). Furthermore, FISH has recently been combined successfully with microelectrode measurements to study sulfate reduction (19, 21) and nitrification (24) in trickling filter biofilms and nitrification in microbial flocs of a nitrifying fluidized bed reactor (25). The combination of the two methods provided reliable and direct information about relationships between in situ microbial activity and the occurrence of specific microorganisms in complex microbial consortia.
In this study, we investigated the ecophysiology of the numerically important ammonia and nitrite oxidizer assemblages in aerobic biofilms on a rotating disk reactor (RDR) fed with domestic wastewater. This was done by combining FISH performed with a set of fluorescently labeled 16S rRNA-targeted probes and microelectrode measurements of NH4+, NO2−, NO3−, and O2. In addition, the nitrifying community in an autotrophic nitrifying biofilm cultured with synthetic nutrient medium was also investigated and compared with the community in a wastewater biofilm in the RDR. Below we discuss the relationship between the spatial organization of nitrifying bacterial populations and the activity of these populations within biofilms.
MATERIALS AND METHODS
Biofilm samples.
Two types of biofilms, a domestic wastewater biofilm and an autotrophic nitrifying biofilm, were studied. Both biofilms were cultured in partially submerged RDR consisting of five polymethylmethacrylate disks. Eight removable slides (1 by 6 cm) were installed in each disk for biofilm sampling. The reactor volume was 1,370 cm3, and the total biofilm area was 2,830 cm2. The temperature was maintained at 20°C. The disk rotational speed was 14 rpm. The dilution rate in the reactors was 0.2 h−1. Mixed liquor from the Shoseigawa municipal wastewater treatment plant in Sapporo, Japan consisting of stage 1 (215,000 population equivalents [PE] [1 PE = 60 g of biological oxygen demand day−1]) and stage 2 (210,000 PE) was initially added to the reactor in order to add microorganisms. After this, the wastewater biofilm was cultured with the primary settling tank effluent containing 1.6 ± 0.5 mM dissolved organic carbon, 1.4 ± 0.5 mM NH4+, and less than 0.1 mM NO2− and NO3− (pH 6.8 ± 0.3). The autotrophic nitrifying biofilm was first seeded with the mixed liquor and cultured with the primary settling tank effluent for a few days like the wastewater biofilms in order to develop thin initial biofilms and then was cultured with synthetic nutrient medium without an organic carbon source. The nutrient medium contained 3.6 mM NH4Cl, 17.8 mM NaHCO3, 0.03 mM K2HPO4, 0.41 mM MgSO4 · 7H2O, and 1.25 mM NaCl, (pH 7.0 ± 0.2).
Microelectrode preparation and measurements.
Cathode type oxygen microelectrodes with tip diameters of about 10 μm were prepared and calibrated as described previously by Revsbech and Jorgensen (22). Liquid ion-exchanging membrane (LIX) microsensors for NH4+, NO2−, and NO3− were prepared as described by de Beer et al. (6, 7, 9). The microsensors were calibrated with NH4+, NO2−, and NO3− dilution series (10−3 to 10−6 M) in the medium used for the measurements. All measurements were obtained as described previously (6) by using a flow cell reactor at 20°C with an average liquid velocity of 2 to 3 cm s−1 by blowing air on the liquid surface. The composition of the medium used for microprofile measurements has been described previously by de Beer et al. (8). The biofilm samples taken from the reactor were acclimated in the medium for a few hours before the measurements were obtained to ensure that steady-state profiles were obtained. Concentration profiles in the biofilms were obtained by using a motor-driven micromanipulator (model ACV-104-HP; Chuo Precision Industrial Co., Ltd., Tokyo, Japan) at intervals of 25 to 100 μm from the bulk liquid into the biofilm. The biofilm-liquid interface was determined with a dissecting microscope (model Stemi 2000; Carl Zeiss). Microprofiles were determined three to five times at different positions in the biofilms for each species and set of conditions. The graphs below show means ± standard deviations of measurements.
Estimation of consumption and production rate profiles.
The net rates of consumption and production of NH4+, NO2−, and NO3− were estimated from the measured microprofiles by using Fick’s second law of diffusion. As a boundary condition we introduced the same concentration point below the deepest measuring point; thus, the activity below the deepest measured point was assumed to be zero. From this point, concentration profiles were calculated stepwise toward the biofilm surface by altering net activities by using Microsoft Excel to minimize the sum of squared deviations of the calculated profile from the measured profile and the sum of squared first derivatives of the guessed activities, Σ(∂A/∂z)2. The activities were assumed to be constant within a measured step. The step size of the calculation was equal to the measurement step. More details of this calculation method were described previously by Lorenzen et al. (16). Furthermore, fluxes (J) of NH4+, NO2−, NO3−, and O2 through the biofilm-liquid interface were calculated by using Fick’s first law: J = −Ds (dS/dz), where Ds is the molecular diffusion coefficient of compound S in the biofilm and dS/dz is the concentration gradient in the boundary layer at the biofilm-liquid interface. Molecular diffusion coefficients of 1.38 × 10−5 cm2 s−1 for NH4+, 1.23 × 10−5 cm2 s−1 for NO2−, and 1.23 × 10−5 cm2 s−1 for NO3− at 20°C were used for the calculations (3).
Fixation and cryosectioning of biofilm samples.
Immediately after the microelectrode measurements were obtained, the biofilms were fixed by immersing the removable slides in test tubes filled with a freshly prepared paraformaldehyde solution (4% paraformaldehyde in phosphate-buffered saline [130 mM sodium chloride, 10 mM sodium phosphate buffer; pH 7.2]) for 4 to 8 h at 4°C. Then the biofilms were rinsed twice with phosphate-buffered saline. Each fixed biofilm was placed in a small aluminum container with the biofilm side up, embedded in Tissue-Tek OCT compound (Miles, Elkhart, Ind.) overnight to infiltrate the OCT compound into the biofilm, and subsequently frozen at −20°C. The frozen biofilms were cut into 10- to 20-μm-thick vertical slices with a cryostat (Reichert-Jung Cryocut 1800; Leica) at −20°C. Each slice was placed on a gelatin (0.1% gelatin and 0.01% chromium potassium sulfate)-coated microscopic slide (Cell-line Associates) and air dried overnight. The specimen was finally dehydrated by successive 50, 80, and 98% ethanol washes (3 min each), air dried, and stored at room temperature. The ethanol dehydration procedure substantially reduced the inherent fluorescence, removed the OCT compound, and also increased probe penetration through the cell walls.
Oligonucleotide probes.
The following oligonucleotide probes were used: Eub338, Nso190, NEU, Nsm156, NmV, Nsv443, NIT2, NIT3, Ntspa454, Ntspa685, and Ntspa1026. All of the probe sequences, as well as the specificities of the probes and the hybridization conditions, are shown in Table 1. The probes were labeled with fluorescein isothiocyanate (FITC) or tetramethylrhodamine 5-isothiocyanate (TRITC). Unlabeled competitor probes CNIT3 and CTE were added with equimolar amounts of NIT3 and NEU, respectively.
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′-3′) | Target sitea | Formamide concn (%)b | NaCl concn (mM)c | Refer-ence |
|---|---|---|---|---|---|---|
| EUB338 | Domain Bacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 20 | 0.225 | 2 |
| Nso190 | Ammonia-oxidizing members of the beta subclass of the Proteobacteria | CGATCCCCTGCTTTTCTCC | 190–208 | 55 | 0.020 | 18 |
| Nsm156 | Nitrosomonas cluster | TATTAGCACATCTTTCGAT | 156–174 | 5 | 0.636 | 18 |
| NmV | Nitrosococcus mobilis lineage | TCCTCAGAGACTACGCGG | 174–191 | 35 | 0.079 | 20 |
| Nsv443 | Nitrosospira cluster | CCGTGACCGTTTCGTTCCG | 444–462 | 30 | 0.112 | 18 |
| NEU | Halophilic and halotolerant members of the genus of Nitrosomonas | CCCCTCTGCTGCACTCTA | 653–670 | 40 | 0.056 | 28 |
| CTEd | Competitor for NEU | TTCCATCCCCCTCTGCCG | 659–676 | 23 | ||
| NIT2 | Nitrobacter spp. | CGGGTTAGCGCACCGCCT | 1433–1450 | 40 | 0.056 | 26 |
| NIT3 | Nitrobacter spp. | CCTGTGCTCCATGCTCCG | 1035–1048 | 40 | 0.056 | 26 |
| CNIT3e | Competitor for NIT3 | CCTGTGCTCCAGGCTCCG | 1035–1048 | 26 | ||
| Ntspa454 | Nitrospira moscoviensis, aquarium clone 710-9 | TCCATCTTCCCTCCCGAAAA | 435–454 | 20 | 0.225 | 10 |
| Ntspa685 | Nitrospira moscoviensis, Nitrospira marina, aquarium clone 710-9 | CACCGGGAATTCCGCGCTCCTC | 664–685 | 40 | 0.056 | 10 |
| Ntspa1026 | Nitrospira moscoviensis, activated sludge clones A-4 and A-11 | AGCACGCTGGTATTGCTA | 1026–1043 | 20 | 0.225 | 13 |
16S rRNA position according to Escherichia coli numbering.
Formamide concentration in the hybridization buffer.
Sodium chloride concentration in the washing buffer.
Used as an unlabeled competitor probe together with probe NEU.
Used as an unlabeled competitor probe together with probe NIT3.
In situ hybridization.
Previously described optimal hybridization conditions were used for each probe. All in situ hybridizations were performed by using the procedure described by Manz et al. (17) and Amann (1) and hybridization buffer (0.9 M NaCl, 20 mM Tris hydrochloride [pH 7.2], 0.01% sodium dodecyl sulfate, and formamide at the concentrations shown in Table 1) at 46°C for 2 to 3 h. The final probe concentration was approximately 5 ng μ1−1. Subsequently, a stringent washing step was performed at 48°C for 20 min with 50 ml of prewarmed washing solution (NaCl at the concentrations shown in Table 1, 20 mM Tris hydrochloride [pH 7.2], 0.01% sodium dodecyl sulfate). The stringency of the washing step (at 48°C) was adjusted by lowering the sodium chloride concentration to achieve the appropriate specificity. The slides were then rinsed briefly with double-distilled H2O and air dried. Simultaneous hybridizations with probes requiring different stringency conditions were performed by using a successive hybridization procedure; hybridization with the probe requiring higher stringency was performed first, and then hybridization with the probe requiring lower stringency was performed (27). The slides were mounted with a SlowFade light antifade kit (Molecular Probes, Eugene, Oreg.).
Microscopy.
A model LSM 510 inverted confocal scanning laser microscope (CSLM) (Zeiss) supplying excitation wavelengths of 488 nm (argon laser) and 543 nm (He-Ne laser) was used to examine the biofilm specimens. Zeiss filter sets 09, 10, and 15 and ×20, ×40, and ×63 oil immersion lenses were used. All image combining, processing, and analysis were performed with the standard software package provided by Zeiss. Processed images were printed by using the software package Adobe Photoshop 3.0J (Adobe Systems Incorporated, Mountain View, Calif.).
In situ analysis of population structure of nitrifying bacteria.
The compositions of the nitrifying consortia in the wastewater biofilm and the autotrophic nitrifying biofilm were analyzed by FISH performed with a set of previously described probes. It was difficult to count the total numbers of probe-stained nitrifying bacterial cells because ammonia-oxidizing bacteria formed dense clusters and the nitrite-oxidizing bacteria were too small to identify single cells. Therefore, the surface fraction of total biomass area and the probe-stained cell (cluster) area were used to quantitatively characterize the population structure of the nitrifying bacteria. The total biomass area and the probe-stained area were determined from differential interference contrast (DIC) images and projection images, respectively, of the same microscopic filed by using image analysis software provided by Zeiss. Since the fluorescence intensity derived from probe-stained cells varied slightly for each image, the greatest fluorescence intensity of the background was first determined for each channel. This value was used as a threshold low value. The threshold values used for the 488- and 543-nm channels ranged from 20 to 50 and 30 to 60, respectively, depending on the autofluorescence intensity (colored pixels were assigned intensity levels ranging from 0 to 255). Thus, all of the pixels with fluorescence intensities greater than the threshold value were counted as part of the probe-stained area. The average surface fraction was determined by using at least 10 representative microscopic images of each cross-section of the biofilm samples.
RESULTS
Reactor performance.
The first stage of nitrification, oxidation of NH4+ to NO2−, occurred immediately after both reactors were started. However, oxidation of NO2− did not progress as rapidly as oxidation of NH4+ progressed. Thus, NO2− gradually accumulated up to a concentration of about 3.2 mM at 19 days after start-up in the purely autotrophic nitrifying biofilm reactor, and oxidation of NO2− to NO3− was completed after 1 month. In contrast, complete nitrification (NH4+ oxidation to NO3−) was achieved within 2 weeks with the wastewater biofilm due to a lower ammonium loading rate. After a steady state was reached (about 1.5 months later), biofilm samples were taken and used for in situ analyses of the spatial distributions of nitrifying bacterial populations and activity.
Biofilm structure.
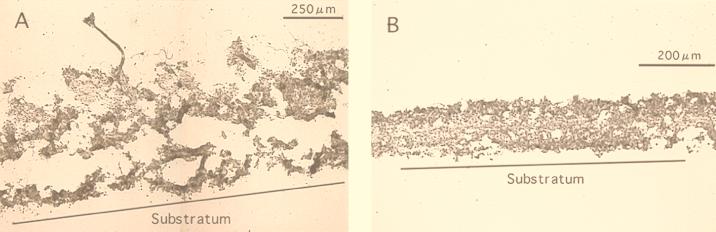
The structure of the autotrophic nitrifying biofilm differed significantly from the structure of the wastewater biofilm (Fig. 1). A cross-section of the domestic wastewater biofilm revealed a heterogeneous structure consisting of biomass and interstitial voids (Fig. 1A). The biofilm thickness was about 750 μm, and the biofilm surface was very rough and was partially covered with filamentous bacteria. The density of the biofilm was high in the upper and bottom layers, and interstitial voids occurred in the middle of the biofilm. In contrast, the autotrophic biofilm was densely packed, and the surface was smoother than the surface of the wastewater biofilm (Fig. 1B); the autotrophic biofilm thickness was about 200 μm.
FIG. 1.
Cross-sections (20 μm) obtained with a cryomicrotome of a domestic wastewater biofilm (A) and an autotrophic nitrifying biofilm (B).
In situ hybridization.
To investigate the spatial distributions of ammonia- and nitrite-oxidizing bacteria within the biofilms, simultaneous in situ hybridizations with specific probes were carried out for entire vertical sections of the domestic wastewater and autotrophic nitrifying biofilms.
(i) Wastewater biofilm.
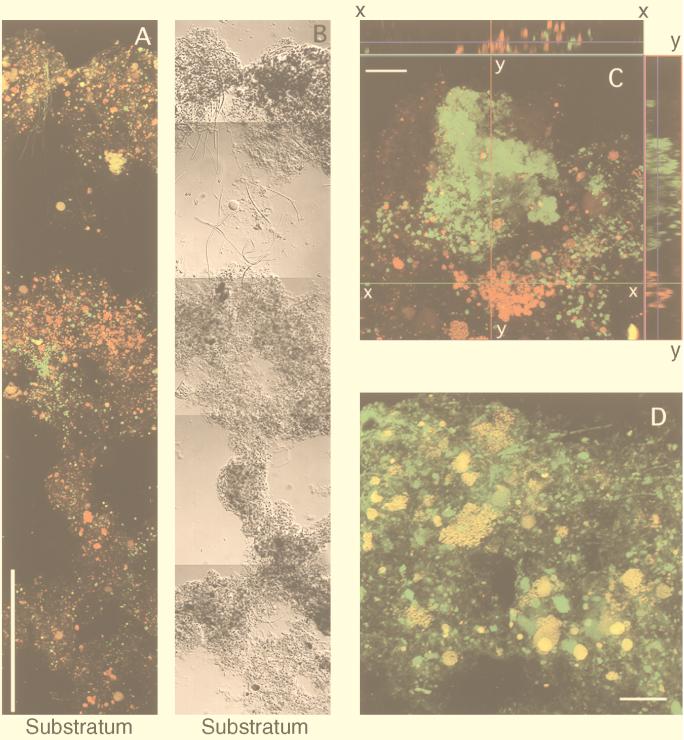
In situ hybridization clearly indicated that ammonia-oxidizing bacteria were not uniformly distributed within the wastewater biofilm (Fig. 2A), and the majority of the ammonia-oxidizing bacteria formed dense spherical microcolonies consisting of rod-shaped cells (Fig. 2C and D). These microcolonies were detected primarily in the upper and middle layers of the wastewater biofilm (Fig. 2A). In contrast, Ntspa 454 probe-stained Nitrospira-like cells formed irregularly shaped aggregates consisting of small microcolonies, which clustered around the clusters of ammonia-oxidizing bacteria (Fig. 2C). These Nitrospira clusters were found mainly in the middle part of the biofilm (Fig. 2A). Although O2 was depleted in the deeper layer, low numbers of both ammonia- and nitrite-oxidizing bacteria were present. Figure 2D shows that there was a close association of ammonia-oxidizing bacteria with heterotrophs (perhaps including nitrite-oxidizing bacteria) stained with general bacterial probe Eub338. A wide variety of heterotrophs surrounded and coexisted with ammonia-oxidizing bacteria.
FIG. 2.
Simultaneous in situ hybridization of vertical sections of a domestic wastewater biofilm with TRITC-labeled probe Nso190 and FITC-labeled probe Ntsp454. (A and B) Composite CSLM projection image (A) and corresponding DIC image (B) of the entire biofilm vertical section. The biofilm thickness was about 950 μm. Weak yellow signals were autofluorescence. Bar = 200 μm. (C) CSLM projection (xy) image (16 optical sections at 1-μm intervals) and orthogonal images (top panel, sagittal [xz] view at x-x optical cutting position; side panel, yz view at y-y optical cutting position) after hybridization with TRITC-labeled probe Nso190 and FITC-labeled probe Ntsp1026, showing the morphology and close association of ammonia- and nitrite-oxidizing bacteria. Cells of Nso190-stained ammonia-oxidizing bacteria are red; cells of Nitrospira-like bacteria are green. Bar = 20 μm. (D) CSLM projection image (thickness, 20 μm) after hybridization with TRITC-labeled probe Nso190 and FITC-labeled probe Eub338. Cells of Nso190-stained ammonia-oxidizing bacteria appear to be yellow because of binding of both probes; eubacterial cells are green. Bar = 20 μm.
(ii) Autotrophic nitrifying biofilm.
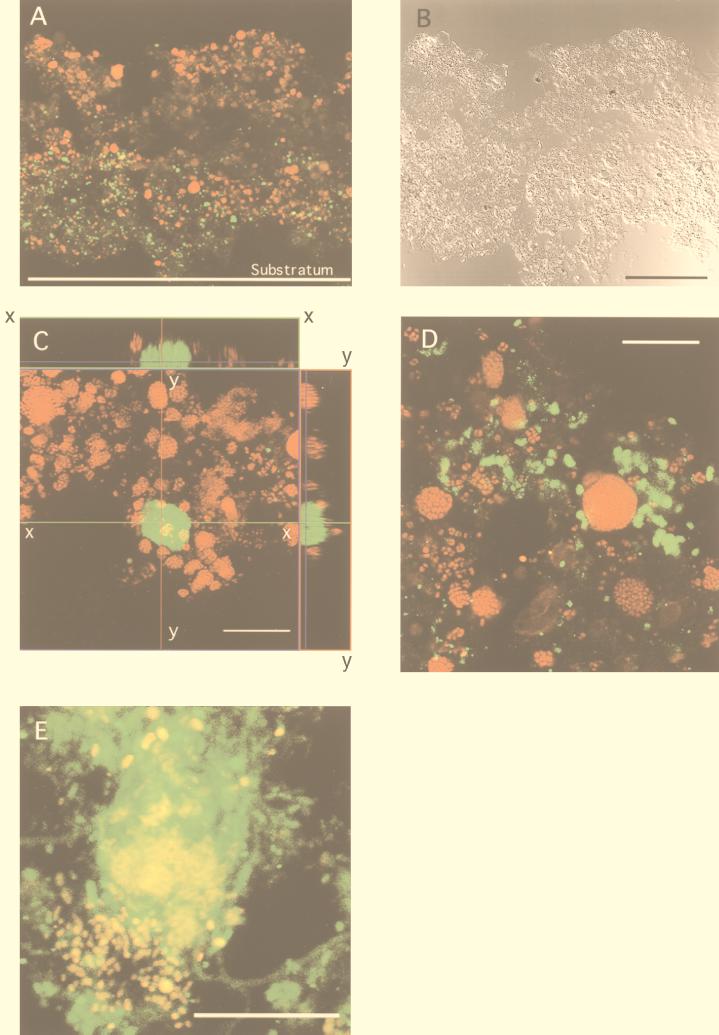
Spherical clusters of densely packed ammonia-oxidizing bacteria were present throughout the autotrophic nitrifying biofilm, indicating that the spatial distribution of the ammonia-oxidizing bacteria was homogeneous (Fig. 3A and 3B). In contrast, clusters of Ntspa 454 probe-stained Nitrospira-like cells were detected only in the deeper layer; thus, the distribution of the nitrite-oxidizing bacteria was heterogeneous. A close association between ammonia-oxidizing bacteria and nitrite-oxidizing bacteria was also observed in the autotrophic nitrifying biofilm (Fig. 3C and D). An orthogonal image showed the three-dimensional organization of the biofilm, in which relatively small ammonia-oxidizing bacterial clusters surrounded each large nitrite-oxidizing cluster (Fig. 3C). The cluster sizes of the ammonia-oxidizing bacteria were larger than the cluster sizes in the domestic wastewater biofilm, probably due to the higher ammonium loading rate. Although the biofilm was cultured in synthetic medium containing no organic carbon, a number of heterotrophs were tightly attached to the ammonia-oxidizing bacteria clusters (Fig. 3E), which may suggest that heterotrophs could utilize soluble organic compounds secreted from ammonia-oxidizing bacteria.
FIG. 3.
Simultaneous in situ hybridization of vertical sections of an autotrophic nitrifying biofilm with TRITC-labeled probe NEU and FITC-labeled probe Ntsp454. (A and B) CSLM projection image (A) and corresponding DIC image (B) of the entire biofilm vertical section. The biofilm thickness was about 350 μm. Bar = 100 μm. (C) CSLM projection (xy) image (16 optical sections at 0.85-μm intervals) and orthogonal image (top panel, sagittal [xz] view at x-x optical cutting position; side panel, yz view at y-y optical cutting position). Bar = 20 μm. (D) CSLM projection image (thickness, 16 μm) after hybridization with TRITC-labeled probe Nso190 and FITC-labeled probe Ntsp1026, showing the morphology and close association of ammonia- and nitrite-oxidizing bacteria. Cells of Nso190-stained ammonia-oxidizing bacteria are red; cells of Nitrospira-like bacteria are green. Bar = 20 μm. (E) CSLM projection image (thickness, 16 μm) after hybridization with TRITC-labeled probe Nso190 and FITC-labeled probe Eub338. Cells of Nso190-stained ammonia-oxidizing bacteria appear to be yellow because of binding of both probes; eubacterial cells are green. Bar = 20 μm.
In situ analysis of population structure of nitrifying bacteria.
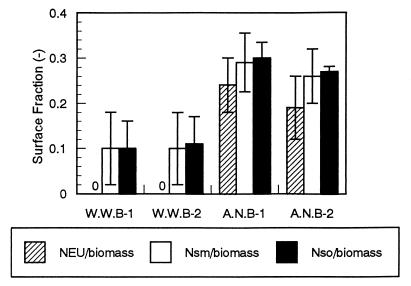
In situ diversity analyses of nitrifying bacteria in biofilm sections were performed by using FISH and different combinations of 16S rRNA-targeted oligonucleotide probes (Fig. 4). In the autotrophic nitrifying biofilm, about 30% of the total biomass area was stained with probe Nso190. In additional hybridization experiments, more than 96% of the ammonia oxidizers that were detectable with probe Nso190 were simultaneously detected with probe Nsm156. Furthermore, about 70% of the probe Nsm156-stained cells were visualized with probe NEU. No hybridization signals were observed when probes NmV and Nsv443 were used with any of the biofilm samples.
FIG. 4.
Compositions of ammonia-oxidizing bacterial consortia in the domestic wastewater biofilm and the autotrophic nitrifying biofilm, as determined by in situ hybridization with a set of fluorescent probes. W.W.B., wastewater biofilm; A.N.B., autotrophic nitrifying biofilm. The error bars indicate standard deviations (n = 10).
In contrast, the surface fraction of the Nso190-stained cells was only about 10% of the total biomass area in the wastewater biofilms. More than 98% of the cells stained with Nso190 were simultaneously detected with probe Nsm156, but no hybridization signal was detected with probes NEU, NmV, and Nsv443 in any of the biofilm samples (Fig. 4).
For nitrite-oxidizing bacteria, no hybridization signals were observed when Nitrobacter-specific probes NIT2, NIT3, and Nb1000 were used with any of the samples. Only in situ hybridization with probes Ntspa454, Ntspa686, and Ntspa1026 (specific for Nitrospira-like bacteria) revealed the presence of nitrite oxidizers in all samples, but the numbers of probe-stained cells were low compared to the number of ammonia-oxidizing bacteria.
Concentration and activity profiles. (i) Wastewater biofilm.
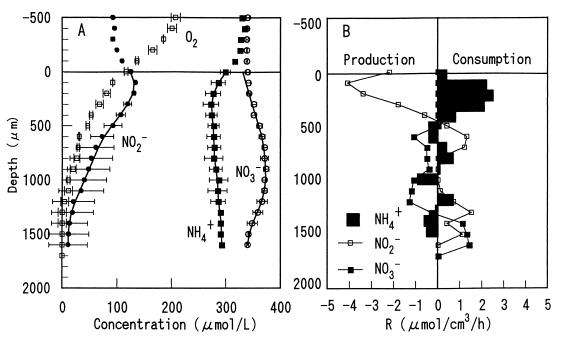
The steady-state concentration profiles of O2, NH4+, NO2−, and NO3− within the domestic wastewater biofilm were determined with microelectrodes (Fig. 5A), and the net rates of consumption and production of NH4+, NO2−, and NO3− were estimated on the basis of the measured profiles (Fig. 5B). The O2 penetration depth was about 1,200 μm in the wastewater biofilm due to the loose structure and low microbial activity. NH4+ oxidation to NO2− occurred mainly in the uppermost 500 μm, with an intermediate NO2− peak concentration of 135 μM at a depth of 200 μm, and there was no NO3− production, although both NH4+ and O2 were present. Correspondingly, NO2− production was detected in the upper 500 μm, as shown in Fig. 5B. The NO2− and NO3− profiles indicated that NO2− was oxidized to NO3− in the region that was about 500 to 1,200 μm from the surface, which was reflected by the substantially lower number of nitrite-oxidizing bacteria in the upper layer, as shown in Fig. 2A. This result clearly suggests at least that the active NH4+ oxidation zone was vertically separated from the active NO2− oxidation zone, although both NH4+ and O2 were still present. Both NO2− and NO3− concentrations decreased in the deeper microaerobic and anoxic layers (below a depth of about 1,200 μm) due to denitrification.
FIG. 5.
Steady-state concentration profiles for O2, NH4+, NO2−, and NO3− in a domestic wastewater biofilm incubated in air-saturated medium containing no acetate, 300 μM NH4+, 300 μM NO3−, and 100 μM NO2− (A) and spatial distribution of the estimated volumetric rates of consumption and production of NH4+, NO2−, and NO3− (B). The points are measured means ± standard deviations, and the lines are the calculated profiles. The biofilm surface was at a depth of zero. R, rate.
(ii) Autotrophic nitrifying biofilm.
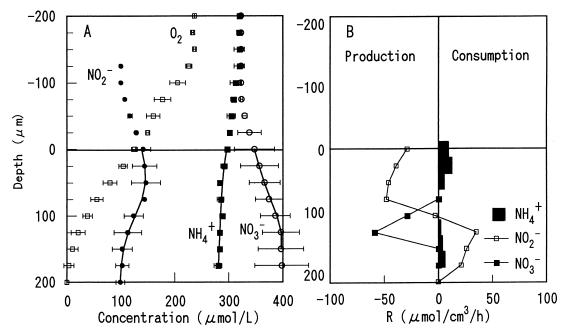
Oxygen penetrated approximately 150 μm into the autotrophic nitrifying biofilm (Fig. 6A). The NH4+ and NO2− profiles showed that the NH4+ consumed was converted to NO2− in the upper 100 μm, with a low but significant NO2− peak concentration (20 to 30 μM) at a depth of 50 to 75 μm, and there was no significant NO3− production at the surface. The NO2− produced was eventually converted to NO3− in the deeper layer (depth 100 to 150 μm) (Fig. 6B); thus, the active NO2− oxidation zone was restricted to the deeper biofilm strata, whereas the NH4+ oxidation zone was found throughout the biofilm. This result could be explained by the lower number of nitrite-oxidizing bacteria in the surface biofilm, as shown in Fig. 3A.
FIG. 6.
Steady-state concentration profiles for O2, NH4+, NO2−, and NO3− in an autotrophic nitrifying biofilm incubated in air-saturated medium containing no acetate, 300 μM NH4+, 300 μM NO3−, and 0 μM NO2− (A) and spatial distribution of the estimated volumetric rates of consumption and production of NH4+, NO2−, and NO3− (B). The points are measured means ± standard deviations, and the lines are the calculated profiles. The biofilm surface was at a depth of zero. R, rate.
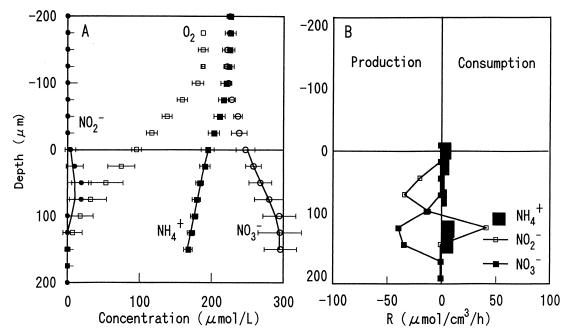
When very low levels of NO2− are encountered (Fig. 6A), the signal-to-noise ratio of the microsensors decreases and the effect of interfering substances, such as HCO3− increases. Slight shifts in the half-cell potential across the membrane result in relatively large effects on the calculated concentrations. Therefore, to verify the vertical distributions of the NH4+ oxidation zone and the NO2− oxidation zone within the biofilm, the autotrophic nitrifying biofilm was incubated in medium supplemented with 100 μM NO2− after the microprofile measurements without NO2− addition were obtained (Fig. 7A). No significant differences in the measured O2, NO3−, and NH4+ microprofiles were observed in the biofilm incubated with 100 μM NO2−. A broad NO2− peak was detected just below the surface of the biofilm, which was caused by relatively higher NH4+ oxidation activity compared with NO2− oxidation activity. The NO2− concentration decreased below a depth of about 50 μm, and the NO3− concentration increased up to a depth of about 150 μm, at which point O2 was depleted. However, no further significant NH4+ oxidation was observed below a depth of 75 μm, although both NH4+ and O2 were still present.
FIG. 7.
Steady-state concentration profiles for O2, NH4+, NO2−, and NO3− in an autotrophic nitrifying biofilm incubated in air-saturated medium containing no acetate, 300 μM NH4+, 300 μM NO3−, and 100 μM NO2− (A) and spatial distribution of the estimated volumetric rates of consumption and production of NH4+, NO2−, and NO3− (B). The points are measured means ± standard deviations, and the lines are the calculated profiles. The biofilm surface was at a depth of zero. R, rate.
Average NH4+ fluxes of 0.08 and 0.13 μmol cm−2 h−1 were found in the autotrophic nitrifying biofilms (thickness, ca. 200 μm) cultured with and without 100 μM NO2−, respectively; these values were comparable to the flux of 0.09 μmol cm−2 h−1 obtained with the wastewater biofilm, which was about 2,000 μm thick. Thus, the nitrifying activity of the autotrophic biofilm was ca. 10 times higher than the nitrifying activity of the wastewater biofilm.
DISCUSSION
Microprofiles.
Although much effort has been funneled into process engineering in order to obtain stable nitrogen removal, our knowledge of microbial community structure and the functions of the nitrifying consortia in wastewater biofilms is still very limited. Detailed in situ measurements of nitrification (oxidation of NH4+ to NO3− via NO2−) could not be obtained until LIX microsensors became available for the main reactants, intermediates, and products. Recent development of the nitrite LIX sensor by de Beer et al. (9) made it possible for us to investigate in more detail the mechanisms of the microbial nitrification process, including the production and/or consumption of the intermediate NO2− that occurs within biofilms.
In the present study, we measured the microprofiles of NH4+ (reactant), NO3− (product), and NO2− (intermediate) and could, therefore, directly correlate the activities of NH4+ and NO2− oxidation with the spatial distributions of ammonia- and nitrite-oxidizing bacterial populations within biofilms. Our data for biofilms cultured independently with different feeds showed that the active zone of oxidation of NH4+ to NO2− was vertically separated from the zone of oxidation of NO2− to NO3− within the biofilms. Microprofiles of the pH values in the biofilms showed that the pH decreased from 7.2 to 6.9 in the surface biofilm (data not shown). Therefore, the explanation for the vertical stratification of two-step nitrification processes must be the absence (or low numbers) of nitrite-oxidizing bacteria in the upper parts of the biofilms. However, Schramm et al. (25) reported that concomitant consumption of O2, NH4+, and NO2− and production of NO3− occurred in the outer 125 μm of nitrifying aggregates. This is because both numerically dominant populations of nitrifying bacteria (Nitrosospira sp. and Nitrospira sp.) were abundant in this nitrification zone.
Furthermore, the use of a set of NH4+, NO2−, and NO3− sensors showed that denitrification occurred below the nitrite oxidation zone (Fig. 5A). In this study, since no organic substrate was added to the medium, the possible electron donors for denitrification were most likely NH4+ and organic matter derived from biomass decay. The NO3− concentration never reached zero, which indicates that the denitrification rate was substantially lower than the nitrification rate.
However, there are, of course, limitations to microprofile measurements. First, it should be noted that the microprofiles presented in this paper are artificial results obtained under the conditions used for microelectrode measurements and are not profiles that actually occurred under growth conditions because, for example, the reactor hydrodynamics were different. Second, the NH4+ profile revealed that the consumption of NH4+ is always less than the production of NO2− and NO3−. A possible reason for this is that NH4+ may be liberated from biomass during measurements from biomass since biofilms have a large pool of bound NH4+ or by biomass degradation. Third, some of the profiles (e.g., the NO3− and NH4+ profiles shown in Fig. 6A) were not completely measured until the curvature ended due to thin biofilms, which led to uncertainties in the quantitative activity analyses.
In situ nitrifying activity.
In situ nitrifying activities of approximately 0.13 and 0.08 μmol of NH4+ cm−2 h−1, which gave average specific nitrifying rates of 8.7 and 4.6 μmol of NH4+ cm−3 h−1, respectively, were found in the autotrophic nitrifying biofilms fed with the synthetic medium (the thicknesses of the nitrifying layers were 150 and 175 μm, respectively [Fig. 6 and 7]); these values are lower than the values reported in previous microsensor studies of nitrifying biofilms (ca. 30 μmol of NO2− plus NO3− cm−3 h−1, measured as production of NO2− plus NO3−) (24) and nitrifying aggregates from fluidized bed reactors (ca. 12 to 50 μmol of NH4+ cm−3 h−1) (8, 9, 25). The specific autotrophic nitrifying rates obtained in this study are high compared with the rates obtained for the wastewater biofilm in this study (ca. 1.6 μmol of NH4+ cm−3 h−1) (Fig. 5) and for nitrifying sediments (11, 12). As mentioned above, since the consumption of NH4+ tends to be underestimated, the in situ nitrifying activities (measured as production of NO2− plus NO3−) were also calculated based on the NO2− and NO3− profiles. We obtained values of 0.22 and 0.30 μmol of NO2− plus NO3− cm−2 h−1, corresponding to average specific nitrifying rates of 14.7 and 17.1 μmol of NO2− plus NO3− cm−3 h−1, respectively. These values are within the lower range of the previously reported specific nitrifying rates.
Although the microprofiles presented above were averages based on three to five measurements, exact quantitative analysis of in situ nitrifying activity is hampered by biofilm heterogeneity, as shown in Fig. 1A. The influence of biofilm heterogeneity on diffusion coefficients in the biofilm was not taken into account when the volumetric consumption and production profiles were calculated (Fig. 5B, 6B, and 7B), and thus constant diffusion coefficients were used throughout the biofilm. Therefore, the profiles were prone to have relatively large errors, which limited exact quantitative analysis of the estimated in situ activity. However, this does not negate the general trend in the results presented here.
In situ identification of nitrifying bacteria.
When the phylogenetic relationships of autotrophic ammonia-oxidizing bacteria belonging to the beta subdivision of the Proteobacteria (13) were considered, the in situ hybridization results demonstrated that at least three different types of ammonia-oxidizing bacteria were present in the autotrophic nitrifying biofilm. As shown in Fig. 4, populations that hybridized with probes Nso190, Nsm156, and NEU but did not hybridize with NmV and Nsv443 (i.e., Nitrosomonas europaea, Nitrosomonas eutropha, and Nitrosomonas halophila) were the numerically dominant ammonia-oxidizing bacteria. The clusters of this group of ammonia-oxidizing bacteria strongly resembled those observed by other researchers (18, 26, 28). Other lineages that hybridized with probe Nsm156 but did not hybridize with NEU constituted the second biggest population. A third population, which hybridized with probe Nso190 but did not hybridize with Nsm156 and Nsv443, was very small, and the affiliation of these bacteria was not determined.
In contrast, the numbers of probe NEU-stained ammonia-oxidizing bacteria (i.e., N. europaea, N. eutropha, and N. halophila) were low (probably below the detection limit) in the wastewater biofilm, and other Nitrosomonas lineages which hybridized with probe Nsm156 but did not hybridize with NEU were numerically important organisms. This result indicated that after they were switched to the synthetic medium supplemented with 3.6 mM NH4+, probe NEU-stained ammonia-oxidizing bacteria (i.e., N. europaea, N. eutropha, and N. halophila) became the dominant bacteria in the autotrophic nitrifying biofilm because of the selectivity of the synthetic medium used in this study and their higher growth rates (4). This evidence indirectly supported the finding that N. europaea has been the species most commonly isolated and studied in most of the previous studies (4).
We found that Nitrosomonas spp. (excluding the probe NEU-stained ammonia-oxidizing bacteria N. europaea, N. eutropha, and N. halophila) and Nitrospira-like bacteria were numerically dominant organisms in the domestic wastewater biofilms. As previously shown, however, Nitrosomonas spp. (mainly probe NEU-stained ammonia-oxidizing bacteria) were more abundant in activated sludge (18, 26) and in trickling filter biofilms receiving an aquaculture effluent (24). The difference in the findings was probably due to the fact the NH4+ concentration in the domestic wastewater used in this study was low (0.5 to 1.1 mM) compared to the concentration used in the study of Schramm et al. (24). In the study of nitrifying aggregates from a fluidized bed reactor performed by Schramm et al. (25), Nitrosospira sp. and Nitrospira sp. populations were the numerically dominant populations of nitrifying bacteria. The dominance of Nitrosospira sp. over Nitrosomonas sp. in the system of Schramm et al. could be attributed to differences in the pH (pH 7.0 in this study versus 8.0 in the study of Schramm et al.), temperature (20°C versus 30°C), and type of inoculum. However, the details of what parameters were important to achieve this dominance are not clear at present.
Nitrospira-like bacteria were shown to be more important than Nitrobacter spp. for nitrite oxidation in wastewater treatment plants (13, 25, 26), in freshwater aquaria (10), and in a nitrite-oxidizing sequencing batch reactor (5), with exception of two reports (18, 24). Although Nitrospira spp. grow significantly more slowly in pure culture (doubling time [td], 23 to 90 h) than Nitrobacter spp. grow (td, 8 to 14 h) (29), Nitrobacter spp. could not outcompete Nitrospira spp., even in the autotrophic nitrifying biofilm. We speculate that Nitrobacter spp. compete well only if both the O2 and NO2 concentrations are high. To investigate detailed mechanisms of population dynamics of nitrite-oxidizing bacteria, isolation of Nitrospira-like bacteria followed by physiological characterization is required.
Spatial distributions.
Ammonia-oxidizing bacteria belonging to the genus Nitrosomonas were detected throughout the biofilm, whereas nitrite-oxidizing bacteria (Nitrospira-like bacteria) were found mainly in the inner parts of both domestic wastewater biofilms and autotrophic nitrifying biofilms. Nitrospira-like bacteria formed smaller and looser irregular cell clusters than Nitrosomonas spp. formed. One explanation for these findings is that since Nitrospira-like cells were present in the deeper parts of the biofilms where the O2 concentrations were low, the cluster sizes remained small to avoid oxygen depletion in their centers. We concluded that Nitrospira-like bacteria could be important members of nitrite-oxidizing bacterial populations in both biofilms, because (i) the Nitrospira-like bacteria were restricted to the active NO2− oxidation zone, (ii) the sizes of the populations of these bacteria in the biofilms increased as oxidation of NO2− to NO3− progressed, and (iii) Nitrospira-like cells were always found in vicinity of ammonia-oxidizing bacterial clusters, which may have reflected the syntrophic association between ammonia- and nitrite-oxidizing bacteria.
The distinct spatial distribution of nitrite-oxidizing bacteria could be explained in part by their slower growth rate. The growth rate of Nitrospira spp. is in general thought to be significantly slower than the growth rates of Nitrosomonas spp. and heterotrophs (29). Therefore, Nitrospira-like cells could be easily outcompeted by Nitrosomonas spp. and heterotrophs in a surface biofilm. We have frequently observed increases in NO2− peak concentrations in surface biofilms with increases in the bulk O2 concentration (data not shown). The increases in the bulk O2 concentration increase the NH4+ oxidation rates in surface biofilms but not the NO2− oxidation rates because of limitation of nitrite-oxidizing bacteria. Furthermore, active NO2− oxidation zones were found at O2 concentrations below 50 μM in all cases. This evidence may suggest that high levels of O2 inhibit the activity of Nitrospira spp. and consequently these organisms proliferate predominantly in the deeper biofilm stratum.
Concluding remarks.
Combinations of Nitrosomonas spp. and Nitrospira-like cells were identified as important populations of nitrifying bacteria in both a domestic wastewater biofilm and an autotrophic nitrifying biofilm. The active NH4+-oxidizing zone was separated from the NO2−-oxidizing zone in both biofilms, which was reflected by the spatial distributions of ammonia- and nitrite-oxidizing bacteria in the biofilms. The combination of in situ analyses of spatial distributions of nitrifying bacterial populations by FISH and activity profiles determined by using microelectrodes is a very powerful research tool and provides new insights into microbial nitrification processes that occur in environmental and engineered biofilm systems.
ACKNOWLEDGMENTS
We thank Dirk de Beer and Andreas Schramm, Max Planck Institute for Marine Microbiology, Bremen, Germany, for kind advice concerning the construction and use of all microelectrodes and for valuable comments on the manuscript. We also thank Hatsuka Naitoh for technical assistance.
This work was supported in part by CREST (Core Research for Evolutional Science and Technology), by the Japan Science and Technology Corporation (JST), and by grant-in-aid 09750627 for Developmental Scientific Research from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Amann R I. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkerman A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrussow L. Landolt-Bornstein Zahlenw, Funkt. II/5a. Berlin, Germany: Springer; 1969. Diffusion; pp. 513–727. [Google Scholar]
- 4.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 5.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Beer D, van den Heuvel J C. Gradients in immobilized biological systems. Anal Chim Acta. 1988;213:259–265. [Google Scholar]
- 7.de Beer D, van den Heuvel J C. Response of ammonium-selective microelectrodes based on the neutral carrier nonactin. Talanta. 1989;35:728–730. doi: 10.1016/0039-9140(88)80171-1. [DOI] [PubMed] [Google Scholar]
- 8.de Beer D, van den Heuvel J C, Ottengraf S P P. Microelectrode measurements of the activity distribution in nitrifying bacterial aggregates. Appl Environ Microbiol. 1993;59:573–579. doi: 10.1128/aem.59.2.573-579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Beer D, Schramm A, Santegoeds C M, Kühl M. A nitrite microsensor for profiling environmental biofilms. Appl Environ Microbiol. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovanec T A, Taylar L T, Blakis A, Delong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen K, Revsbech N P, Nielsen L P. Microscale distribution of nitrification activity in sediment determined with a shielded microsensor for nitrate. Appl Environ Microbiol. 1993;59:3287–3296. doi: 10.1128/aem.59.10.3287-3296.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen K, Sloth N P, Risgaard-Petersen N, Rysgaard S, Revsbech N P. Estimation of nitrification and denitrification from microprofiles of oxygen and nitrate in model sediment systems. Appl Environ Microbiol. 1994;60:2094–2100. doi: 10.1128/aem.60.6.2094-2100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Roser A, Koops H-P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kühl M, Jorgensen B B. Microsensor measurements of sulfate reduction and sulfide oxidation in compact microbial communities in aerobic biofilms. Appl Environ Microbiol. 1992;58:1164–1174. doi: 10.1128/aem.58.4.1164-1174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lens P L, de Beer D, Cronenberg C C H, Houwen F P, Ottengraf S P P, Verstraete W H. Heterogeneous distribution of microbial activity in methanogenic aggregates: pH and glucose microprofiles. Appl Environ Microbiol. 1993;59:3803–3815. doi: 10.1128/aem.59.11.3803-3815.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzen J, Larsen L H, Kjar T, Revsbech N P. Biosensor detection of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl Environ Microbiol. 1998;64:3264–3269. doi: 10.1128/aem.64.9.3264-3269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manz W, Amann R I, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 18.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okabe S, Mastuda T, Satoh H, Itoh T, Watanabe Y. Sulfate reduction and sulfide oxidation in aerobic mixed population biofilms. Water Sci Technol. 1998;37(4/5):131–138. [Google Scholar]
- 20.Pommerening-Roser A, Rath G, Koop H-P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 21.Ramsing N B, Kühl M, Jorgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Revsbech N P, Jorgensen B B. Microelectrodes: their use in microbial ecology. Adv Microb Ecol. 1986;9:293–352. [Google Scholar]
- 23.Schleifer K-H, Amann R, Ludwig W, Rothemund C, Springer N, Dorn S. Nucleic acid probes for the identification and in situ detection of pseudomonas. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 127–134. [Google Scholar]
- 24.Schramm A, Larsen L H, Revsbech N P, Amann R I, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner M, Rath G, Koops H-P, Flood J, Amann R I. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34(1/2):237–244. [Google Scholar]
- 27.Wagner M, Amann R I, Kampfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K-H. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst Appl Microbiol. 1994;17:405–417. [Google Scholar]
- 28.Wagner M, Rath G, Amann R I, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 29.Watson S W, Bock E, Harms H, Koops H-P, Hooper A B. Nitrifying bacteria. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins; 1989. pp. 1808–1834. [Google Scholar]
- 30.Zhang T C, Bishop P L. Evaluation of substrate and pH effects in a nitrifying biofilm. Water Environ Res. 1996;68:1107–1115. [Google Scholar]