Abstract
Cutaneous T-cell lymphoma is a rare cancer of skin-homing T-cells. A subgroup of patients develops large cell transformation with rapid progression to an aggressive lymphoma. Here, we investigated the transformed CTCL (tCTCL) tumor ecosystem using integrative multiomics spanning whole exome sequencing (WES), single-cell RNA-seq and immune profiling in a unique cohort of 56 patients. WES of 70 skin biopsies showed high tumor mutation burden, UV signatures that are prognostic for survival, exome-based driver events and most recurrently mutated pathways in tCTCL. Single-cell profiling of 16 tCTCL skin biopsies identified a core oncogenic program with metabolic reprogramming toward oxidative phosphorylation, cellular plasticity, upregulation of MYC and E2F activities and down-regulation of MHC-I suggestive of immune escape. Pharmacologic perturbation using OXPHOS and MYC inhibitors demonstrated potent anti-tumor activities, while immune profiling provided in situ evidence of intercellular communications between malignant T-cells expressing macrophage migration inhibitory factor and macrophages and B-cells expressing CD74.
Keywords: CTCL, large cell transformation, single-cell, genomics
INTRODUCTION
Cutaneous T-cell lymphomas (CTCLs) are a group of non-Hodgkin T-cell lymphomas. It affects male more often than females, and Black/African American (AA) patients show a more aggressive clinical course and inferior survival (1,2). The most common form of CTCL, mycosis fungoides (MF), is characterized by cutaneous patch/plaque lesions (PP) or tumors, while Sezary syndrome (SS) is the leukemic variant of CTCL with circulating malignant T-cells in the peripheral blood (3–6). Large cell transformation (LCT) occurs in a subset of MF and SS patients when the lymphoma cells undergo histopathologic transition from neoplastic small-medium sized lymphocytes to large, blast-like T-cells. The primary site of detectable MF/SS transformation is the skin (7,8). Similar to Richter’s transformation in chronic lymphocytic leukemia, transformation in CtCl heralds immediate transition to aggressive clinical behavior, especially for those who transform within 2 years of MF diagnosis (8), rapid decline in survival (median survival 19–36 months) (7–10) and resistance to multiple forms of therapy.
Major advances have been made in the treatment of advanced stage CTCL with the FDA approval of brentuximab vedotin (anti-CD30) in 2017 (ALCANZA trial) (11) and mogamulizumab (anti-CCR4) as a breakthrough therapy in 2018 (MAVORIC trial (12)). While brentuximab shows superior efficacy in the skin compartment and in CD30+ MF with LCT (13) and mogamulizumab with more reliable activity in the blood compartment (14,15), the responses are not universal or durable. The median PFS was 15.9 months for brentuximab and 7.7 months for mogamulizumab, and notably, LCT was one of the exclusion criteria in the MAVORIC trial. These trials thus highlight a critical unmet need to identify novel therapeutic targets for advanced stage CTCL, particularly following disease transformation.
While cancer death rates have significantly declined for many common cancers in the past decade, there is a sobering under-representation of this success in rare cancers, and particularly in the vulnerable racial and ethnic minority groups (16,17). The lack of research tissue samples and clinical trials further compound this inequality. Several research groups have attempted to profile the genomic landscape of SS/MF by WES and WGS (18–29). While we have gained a wealth of information from these multi-institutional sequencing efforts, there is a lack of genome-level investigation of CTCL at disease transformation, with only 7 transformed MF WES reported to date (20,23). Similarly, recent advances in single-cell technologies have provided a high-resolution window into malignant and benign T-cell transcriptomics in a small number of SS and MF samples and revealed evidence of inter- and intra-tumoral heterogeneity (30–32). Malignant T-cells in the skin showed up-regulation of T cell activation, TCR ligation, and cell cycle progression transcripts compared to those in the blood compartment (32). Despite these efforts, the paucity of investigation of CTCL tumor and immune microenvironment (TIME) at LCT or its relationship to precursor PP lesions contributes to inadequate knowledge concerning potential therapeutic targets for this deadly disease state.
To address this critical unmet need, we tackled the challenging tCTCL TIME by applying integrative multi-omics and multiplex immune profiling of skin biopsies from a rare cohort of 56 tCTCL patients. We comprehensively characterized the genomic landscape of tCTCL using tissue resource of similar size to other TCGA rare cancers, and established tCTCL as a high tumor mutation burden (TMB) cancer dominated by UV signatures that are prognostic for enhanced survival. Notably, Black/AA patients in our cohort show significantly lower contribution of UV signatures compared to the White patients. We identified predicted driver genes and recurrently altered pathways in Hippo, RAS/RTK and Notch, and showed that tCTCL in skin exhibits a distinct genomic chromosomal copy number variation (CNV) profile from SS/leukemic CTCL, which has important therapeutic implications. Using a combination of scRNAseq, scV(D)Jseq and CNV inference analytic strategies, we identified a unique malignant T-cell program with enrichment for oxidative phosphorylation (OXPHOS), cellular plasticity, upregulation of MYC and E2F activities, and down-regulation of MHC-I at transformation suggestive of immune escape. Furthermore, in vitro pharmacologic studies using novel small molecule inhibitors of OXPHOS (IACS-10759 (33)) and MYC (MYCi975 (34)) demonstrated potent anti-tumor activity. Immune profiling further revealed receptor-ligand interaction between macrophage migration inhibitory factor (MIF) in malignant T-cells and CD74 in antigen-presenting cells (APCs). Notably, malignant T-cells in tCTCL demonstrated intra-tumoral genetic and transcriptional heterogeneity, with upregulation of ribosomal protein subunit gene expression in dominant subclones in patients with the poorest clinical outcomes. Collectively, our study provided the first comprehensive compendium of genomic alterations in CTCL at disease transformation, identified a tCTCL oncogenic program that exploits metabolic reprogramming, cellular plasticity and proliferation, and highlighted potential therapeutic vulnerabilities for this incurable rare cancer.
RESULTS
tCTCL is a high TMB cancer and presence of UV signatures is prognostic for survival at LCT
We collected a unique cohort of 56 patients with biopsy-proven tCTCL (53 transformed MF, 3 SS/overlap MF-SS with transformed tumors in skin) who were seen and / or treated between 2014 to 2020. Of the 56 patients, there were 38 males (68%), 18 females (32%), and 17 patients belonging to racial/ethnic minority groups (30%). Clinical re-staging was performed at the time of LCT. The median time from initial MF diagnosis to LCT was 25.3 months (range 0 – 252.5 months), median time from LCT to death or last follow-up was 20.4 months (range 1.0 – 61.3 months), and 26 patients were deceased (46.4% of the cohort) (Supplementary Table S1). Available biopsies from these patients were carefully annotated with clinical data elements and processed for multi-omics profiling, including WES, 5’ scRNAseq, scV(D)Jseq and multiplex immunofluorescence (mIF) immune profiling (Fig. 1A; Supplementary Table S2A–D).
Figure 1. The genomic landscape of transformed CTCL.
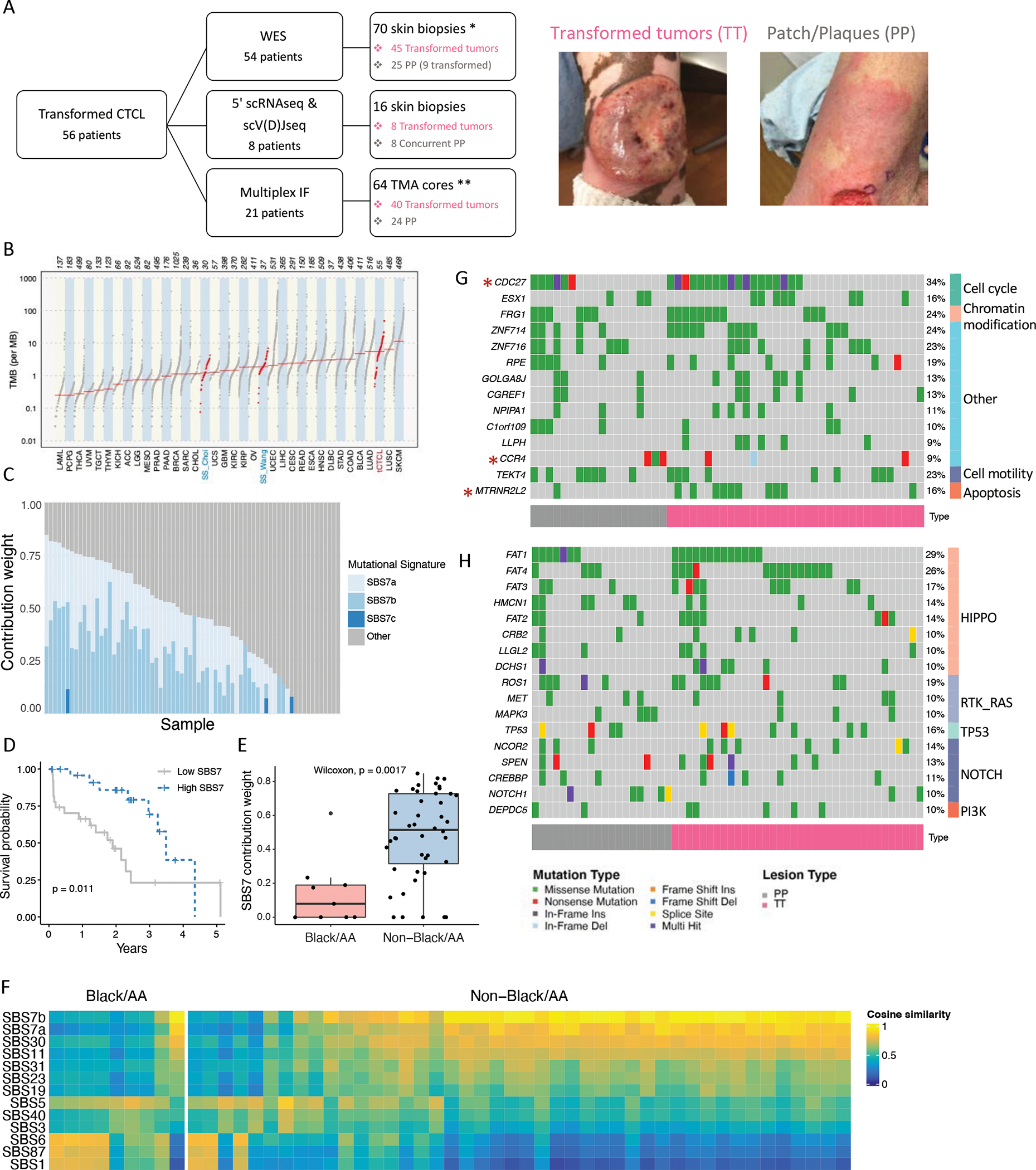
A. Flow diagram showing the tCTCL patient cohort (n= 56 patients with biopsy-proven tCTCL, including 53 transformed MF and 3 SS/overlap MF-SS with transformed tumors in skin) and correlative biospecimen studies. WES (n=54 tCTCL patients, 70 FFPE skin biopsies): 45 patients with TT, 25 patients with PP (of which 9 are transformed PP). *n=17 patients had matched transformed tumor and PP (precursor or concurrent PP; Methods). Simultaneous 5’ single cell RNAseq and single cell V(D)Jseq (n=8 tCTCL patients, each with concurrent TT and PP). Multiplex IF immune profiling by Vectra (n=21 tCTCL patients, 64 TMA cores; **n=16 patients with matched TT and PP (Methods). Clinical photographs of TT (left; PT11) and extensive patch/plaque lesions (right). B. Tumor mutation burden per MB of 33 TCGA cancer types and tCTCL, SS-Choi and SS-Wang cohorts (red). Sample size annotated at top. C. Plot represent weighted contribution of COSMIC v3.2 mutational signatures SBS7a, SBS7b, SBS7c and others in each tissue sample using deconstructSigs(38) (n=70 samples). D. Overall survival probability of tCTCL patients classified according to high versus low SBS7 mutation signature (by sum of weighted contribution from SBS7a-d) and onset of LCT to time of death or last follow up. E. Weighted contribution of SBS7a-d mutation signatures in Black/AA versus non-Black/AA patients. F. Heatmap of cosine similarities between the mutational profile of each sample and COSMIC v3.2 mutational signature in Black/AA vs non-Black/AA patients, ranked by SBS7a-d (complete COSMIC v3.2 signatures in Supplementary Fig. S3B). G, H. Oncoplot of predicted driver genes by dNdScv and MutsigCV (G) and most recurrently mutated pathway genes (H) in tCTCL (n=70 samples). Each column represents a patient tumor sample. Somatic mutations, including missense (green), nonsense (red), in frame insertion (dark grey) and deletion (light blue), frame shift insertion (orange) and deletion (blue), splice site (yellow), multi hit (purple) and genes implicated in leukemia and lymphoma by manual curation (red asterisk) are depicted. TT (magenta), PP (gray). Recurrent mutations in >10% of the samples are depicted (full mutation list in Supplemental Table S5 and S6). The predicted driver genes belong to five groups: cell cycle, chromatin modification, cell motility, apoptosis, genes implicated in leukemia/lymphomagenesis (*asterisk) and other undefined (G). Most recurrently mutated pathways are Hippo, Notch, RAS-RTK pathways and p53 (H; Supplementary Table S6).
We first sought to explore the genomic landscape of tCTCL and collected 70 skin biopsies from 54 patients with confirmed LCT in skin for WES [Fig. 1A; 45 transformed tumors (TT), 9 patch/plaque lesions (PP) with LCT, and 16 concurrent or precursor PPs to TT]. Of the 54 patients sequenced, 41 had matched germline normal samples, and 17 had paired PP and TT (Supplementary Table S2A; Methods). Somatic single nucleotide variants (SSNVs) and small indels were called by comparing tumor and PP to matched normal germline or panel of normal (n=13 patients). We identified a median of 290 mutations per sample with a dominance of missense mutations (97.1%) and C>T transitions (65.8%) that is comparable to previously described in SS and MF (Supplementary Fig. S1A–B) (19,35). We measured the tumor mutation burden (TMB) in all samples with matched germline and compared this to 33 other cancers profiled in The Cancer Genome Atlas (TCGA) and two independent cohorts of SS (Wang and Choi cohorts) (26,36). Analyses of mutation burdens of SSNVs and small indels in tCTCL demonstrated a TMB that is exceeded only by lung SCC and cutaneous melanoma and significantly higher than SS (Fig. 1B; tumor types ordered by median TMB; Supplementary Table S3A–B).
To comprehensively interrogate the operative mutational processes in tCTCL, we next catalogued the repertoire of mutational signatures with reference to the COSMIC mutational signatures (v3.2)(37). At the sample level, we used deconstructSigs (38) to analyze the weights of mutation signatures and showed that UV signatures SBS7a and SBS7b carried the maximum weight (Fig. 1C; Supplementary Table S4). Similarly, we implemented MutationalPatterns (39) and demonstrated highest cosine similarity to signatures SBS7a and SBS7b, with no significant difference between PPs and tumors (Supplementary Fig. S2A). At the cohort level, novel mutational signatures derived by non-negative matrix factorization (NMF) methods showed best match to SBS7b and SBS6 (Supplementary Fig. S2B). The predominance of UV signatures has been reported in SS/MF and other sun-protected cancers (35,40–42), and Signature 7 has been shown to be prognostic for favorable outcome in cutaneous melanoma (43). We performed survival analysis and showed that the sum of weighted signatures SBS7a, SBS7b, SBS7c and SBS7d are also prognostic for favorable survival from the time of LCT (Fig. 1D; Supplementary Table S4). Importantly, Black/AA patients in our cohort show significantly lower contribution of UV signatures compared to the white patients (Fig. 1E), with enrichment of SBS6, SBS1 and SBS87 in patients with low UV signatures (Fig. 1F, Supplementary Fig. S3A). Previous large-scale epidemiology studies showed that Black/AA patients with MF demonstrated poorer survival than white patients (1,2). We also observed a survival gap between Black/AA and white patients in this cohort (Supplementary Fig. S3B).
Exome-based sequencing identifies driver events and key oncogenic pathways in tCTCL
We next sought to detect exome-based driver events in tCTCL utilizing two mutation-based computational tools, dNdScv (44) and MutSigCV (45), to enhance the coverage of our analysis. dNdScv is a statistical model based on refined dN/dS while factoring variation of mutation rate, and MutSigCV assesses mutation significance as a function of gene size, trinucleotide context and background mutation frequency for highly recurrent mutations. When both dNdScv and MutsigCV were performed on the same dataset, a total of 20 potential driver genes were identified (Supplementary Table S5A–E), including CCR4 (chemokine receptor), FRG1 (chromatin modifying), TEKT4 (cell motility), CDC27 and ESX1 (cell cycle), MTRNR2L2 (antiapoptosis) and other novel genes previously not implicated in CTCL (Fig. 1G, Supplementary Fig. S4A–B). Notably, we observed CCR4 mutations concentrated at the C-terminus, similar to other gain-of-function CCR4 C-terminus mutations seen in adult T-cell leukemia/lymphoma (ATLL) and SS (26,46). We subsequently performed analyses of recurrently mutated pathways and showed top ranking mutations in Hippo (FAT1, FAT4), Notch (NCOR, SPEN, CREBBP), RAS (ROS1, MEK, MAPK3) pathways and p53 (Fig. 1H; Supplementary Table S6A–C). Importantly, Hippo and Notch are key signaling pathways known to be essential for organ size control in development, stemness and tumor suppression, and deletion of FAT1 has been shown to promote stemness and metastasis through epithelial to mesenchymal transition in cancer (47). We also stratified the WES samples by CD30 expression (≤1%, >1% to <10%, 10–50%, >50%) (Supplementary Methods; Supplementary Table S7) but did not detect significant differences in overall survival, TMB, UV signatures, driver or top-ranking pathway genes (Supplementary Fig. S5A–E).
Cutaneous tCTCL exhibits distinct genomic gains and losses from those of SS/leukemic CTCL
Previous work in SS reported distinct chromosomal copy number variation patterns, with characteristic recurrent alteration in genes such as ARID1A, CDKN2A, CDKN1B, ZEB1, DNMT3A, PLCG1, TP53, PDCD1 and CARD11 (21,22). We next explored the tCTCL genome for candidate somatic copy-number alterations (SCNAs). Using GISTIC 2.0, an algorithm that detects genes targeted by SCNVs that drive cancer growth (48), we mapped 42 recurrently deleted loci (33 with Q-values <0.005) and 28 recurrently amplified loci (9 with Q-value <0.005) across the genome (Q threshold 0.25; Fig. 2A, Supplementary Table S8A–C). Recurrent deletions were seen in the PRAME family of genes (cancer testis antigen/stemness), clustered Protocadherin family, Defensins (DEFB109P1 and DEFB130), RECQL4 (genomic stability and chromatin modifying), CDKN2A (cell cycle), SOC6 and SOC7 (cytokine signaling, JAK/STAT) and POLE (DNA repair), while recurrent amplifications were seen in TWIST1 (cellular plasticity/stemness), NDUFB2 and NDUFA7 (OXPHOS), RPLs and RPSs (ribosomal protein subunits), LGALS3 (Galectin3/RAS), AHR (T-cell differentiation), CARD11 (T-cell activation) and SYCP1 (cancer testis antigen). We also performed GISTIC analysis on available SS/leukemic CtCL samples (36) (Fig. 2A; Supplementary Table S8D–F). While tCTCL and SS/leukemic CTCL shared conserved alterations in genes such as CARD11 and CDKN2A, the global genomic profiles between the two entities are strikingly different, with more prominent G-scores seen in tCTCL (Fig. 2A).
Figure 2. tCTCL exhibits distinct genomic gains and losses from those of SS/leukemic CTCL.
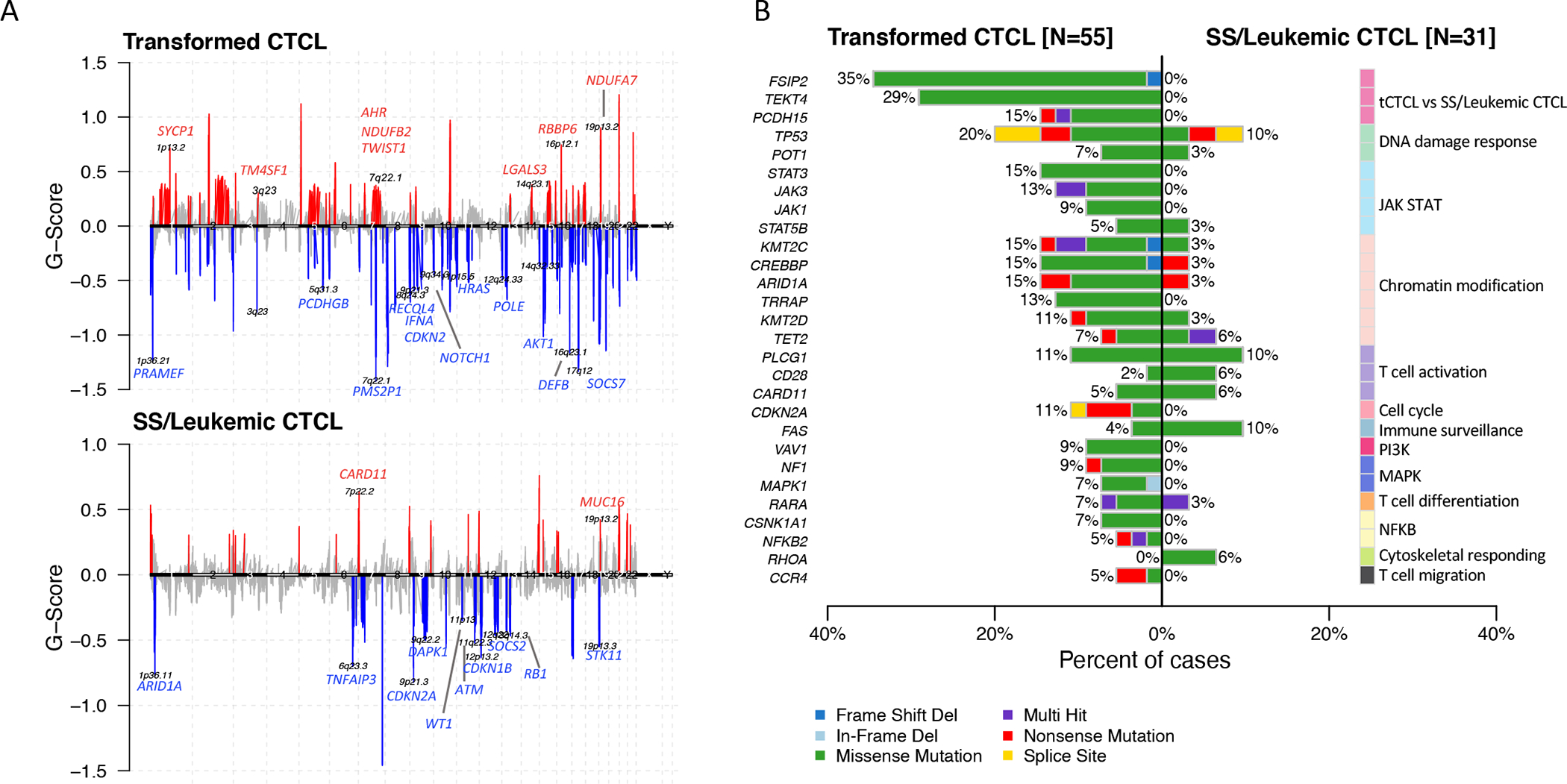
A. Composite plot of significant arm-level and focal SCNAs by WES, using GISTIC 2.0 to detect genes targeted by somatic copy-number alterations (SCNAs) that drive cancer growth. Transformed CTCL (tCTCL, top, n= 55 samples with matched germline). SS/leukemic CTCL36 (bottom, n=31 samples with matched germline). Each alteration is assigned a G-score (y-axis; frequency x amplitude). Amplifications (red, above the solid horizontal lines) and deletions (blue, below the solid horizontal lines) are plotted across the genome (x-axis). Select gene targets within the peak regions are depicted. Q-threshold= 0.25. B. Examination of 55 CTCL-associated genes reported in literature (mostly SS)19 and significantly differentially mutated genes in tCTCL (left, n=55 samples) vs SS/leukemic CTCL36 (right, n=31 samples) (p< 0.05 and q< 0.25). Depicted are select candidate genes involved in DNA damage response, JAK/STAT, chromatin modification, T-cell activation, cell cycle, immune surveillance, PI3K, MAPK, T-cell differentiation, NFKB, cytoskeletal responding, T-cell migration), and significantly differentially mutated genes from unbiased comparison between tCTCL and SS cohorts (full gene list in Supplementary Table S8).
Recent WES meta-analysis of predominantly SS patients described 55 putative CTCL driver genes involving pathways that affect chromatin remodeling, immune surveillance, MAPK, NF-κB and PI-3-kinase signaling (19). We therefore investigated these candidate genes in tCTCL and compared to a publicly available SS/leukemic CTCL dataset from this meta-analysis study (Choi cohort) (36). Interestingly, while both cohorts show similarly recurrent mutations in genes such as STAT5B, POT1, TET2, and CARD11, tCTCL demonstrated drastically more recurrent mutations in the JAK-STAT pathway genes (JAK1, JAK3, STAT3), key chromatin modifying genes (KMT2C, KMT2D, ARID1A, CREBBP), TP53 and CDKN2A (Fig 2B), suggesting more global dysregulation in genome-wide topology and gene expression in tCTCL (Supplementary Table S9A–D). Recurrent mutations previously described as hotspot mutations in T-cell and/or NK T-cell lymphomas such as CD28F511, JAK3A573V and STAT5BN642H were conserved in tCTCL, with the latter showing potential therapeutic vulnerability to JAK-STAT inhibitors (49,50). An unbiased comparison of the tCTCL and SS cohorts identified 116 significantly differentially mutated genes, including novel genes such as FSIP2, TEKT4 and Protocadherin 15 that are mutated only in tCTCL but not SS (p<0.05, Q<0.25; Supplementary Table S9E). Dissimilarity in the genomic landscape of transformed T-cells in skin versus leukemic T-cells in blood provides opportunities to exploit differential or synergistic therapeutic vulnerabilities in the two body compartments at advanced stage disease.
Dissecting the transformed CTCL TIME at single-cell resolution
To interrogate the tCTCL tumor ecosystem, we next sought to profile the tCTCL TIME at single-cell resolution. We collected 16 fresh skin biopsies from 8 tCTCL patients, each with paired PP and TT, and profiled 34,912 cells using complementary 5’ scRNAseq and scV(D)Jseq strategies (10x Chromium) (Fig. 1A, Fig. 3A). Single cell V(D)Jseq identifies the precise TCR α- and β- chain clonotype combination in individual T-cells and allows linkage of clonotype information to the whole transcriptome by complementary 5’ scRNAseq. Despite the advantage of scV(D)Jseq in identifying clonal TCR, it is still imprecise due to “dropout” read, a well-known phenomenon in both scRNAseq and scV(D)Jseq datasets. To tackle this issue, we sought to separate malignant from benign T-cells by two complementary profiling strategies. First, we defined true malignant and true benign T-cells by scV(D)J seq (Fig 3A; Supplementary Table S10). Next, we distinguished malignant cells by patterns of inferred SCNAs using the 5’ scRNAseq data (inferCNV, Fig 3B, Methods). In 7 of 8 patients, we observed a dominant TCR clonotype of paired α- and β- chain sequences, consistent with TCR monoclonality in cutaneous tCTCL (Supplementary Table S10). Concurrent PP and TT lesions from the same patient also shared the same TCR clonotype. Interestingly, even though some lesions appear to have multiple “dominant” clonotypes (e.g. PT35T, clonotypes 1–7), careful inspection reveals the same TCR α- chain (TRA: CAVGDAGGTSYGKLTF) and β-chain sequences (TRB: CASSRQGGSGNTIYF or CASSRLGGSGNTIYF) across all 7 clonotypes. Two TRB amino acid sequences that differed by single amino acid were observed in the most dominant clonotype (TRA: CAVGDAGGTSYGKLTF; TRB: CASSRQGGSGNTIYF; TRB:CASSRLGGSGNTIYF), perhaps due to somatic hypermutation. Cells exhibiting unpaired TRA and TRB (e.g. PT35 clonotypes 4, 6, 7) were present, compatible with “dropout” reads in either TCR α- or β-chain expression, respectively. Due to the observed TCR read dropout, a second malignancy detection method using inferCNV was implemented.
Figure 3. Dissecting the transformed CTCL TIME at single-cell resolution.
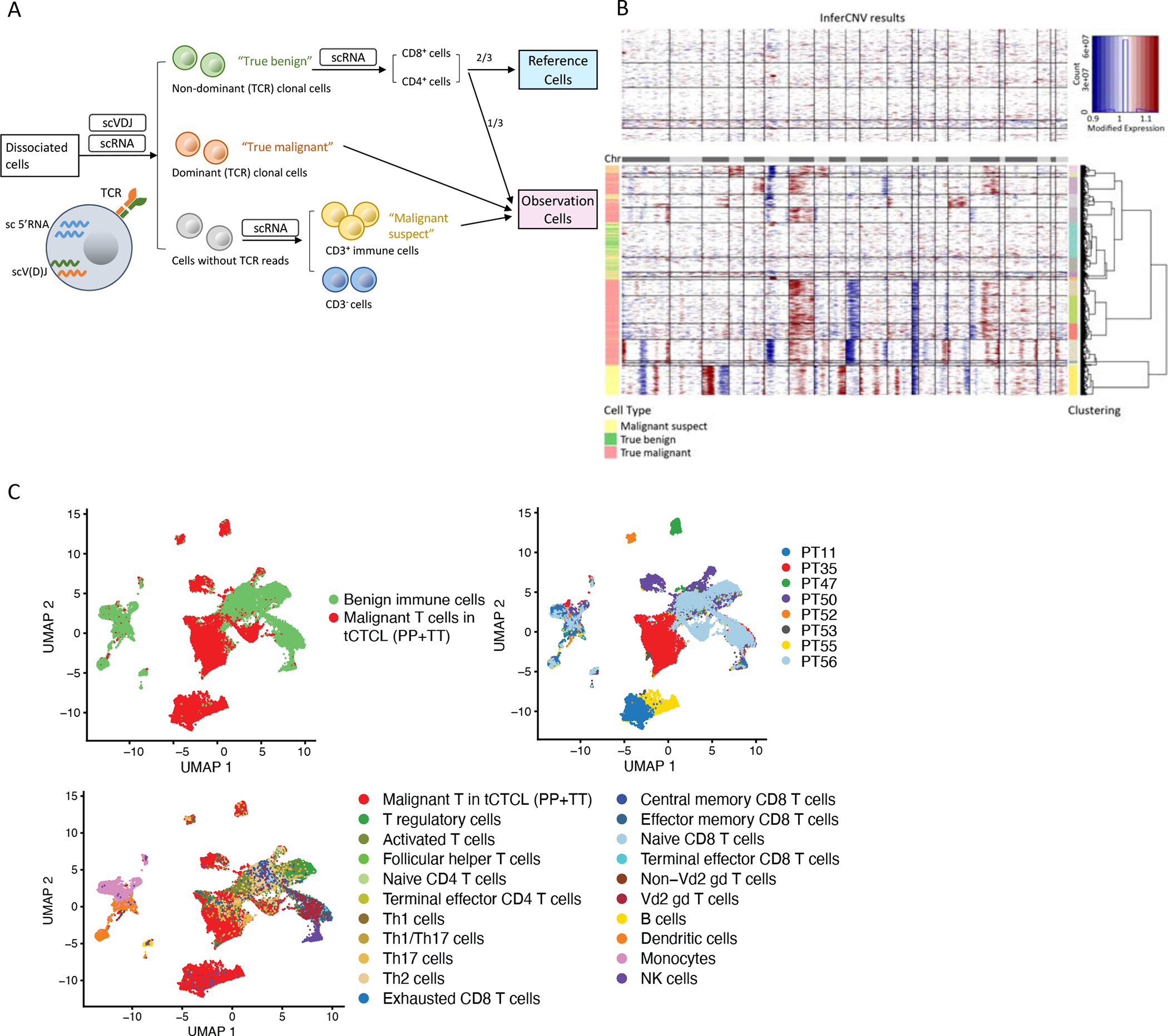
A. Single-cell profiling analytic workflow. 16 fresh skin biopsies were collected from 8 patients with CD4+ tCTCL (paired TT and PP lesions) for complementary 5’ scRNAseq and scV(D)Jseq. Single-cell V(D)J seq (10× chromium) detects the precise TCR α- and β-chain combination that defines each T-cell’s TCR clonotype. “True malignant” cells (red) = cells with dominant TCR clonotypes, while “True benign” cells (green) = cells with non-dominant/polyclonal TCR clonotypes (Supplementary Table S9). B. Malignant cells are further identified by inferred large-scale CNAs (inferCNV). The CNAs (red, amplifications; blue, deletions) are shown along the chromosomes for each cell. Two-thirds (2/3) of randomly selected non-dominant TCR/polyclonal CD4+ or CD8+ T-cells were input as the benign “Reference cells”. For the inferCNV observation group, the remaining 1/3 of “True benign” cells were “spiked in”, along with all clonal “Tue malignant” cells, and “Cells without TCR reads” (grey) that are positive for CD3 (i.e., the “Malignant suspect” cells, yellow) as the input cells. In the tCTCL TIME, “True malignant” cells by TCR clonality show malignant CNV patterns, while “True benign” cells show CNV neutral patterns. C. Uniform manifold approximation and projection (UMAP) of single-cell profile of 27,055 immune cells (dots), colored by malignant cell status (top left panel), with clear separation of malignant T-cells (red) from benign immune cell (green) clusters, patient ID (top right panel), and all malignant T-cells and benign immune cell types annotated in the TIME (bottom panel) (Supplementary Methods).
An important basis for inferCNV is the requirement for reference “normal” cells, ideally of the same cell type, and a separate group of “observation” cells for testing. Here, we selected cells harboring the non-dominant/polyclonal TCR clonotypes from scV(D)J profiling as “true normal”. We further split the “true normal” cells and input 2/3 of these cells into the inferCNV reference cell group (Fig. 3A and 3B). For the observation group, we “spiked in” the remaining “true normal” cells and input all cells with dominant TCR clonotype (i.e. “true malignant”), as well as all CD3+ T-cells with TCR dropout as “malignant suspect”. We hypothesized that malignant T-cells would demonstrate malignant CNV patterns and form distinct clusters from normal T-cells with neutral CNV patterns. Indeed, we observed a striking dichotomy in chromosomal large-scale CNV pattern between malignant versus normal cells, with increased patient-specific CNVs observed in malignant T-cells (Fig. 3B). UMAP clustering of all cells generated 17 clusters, with distinct separation between malignant T-cells harboring positive CNV patterns and benign T-cells with neutral CNV patterns (Fig. 3C). As in other cancer types, tCTCL demonstrates significant inter-patient heterogeneity by gene-expression, with malignant T-cells clustering by patient of origin, and normal T-cells from different patients clustering together, suggesting consistency of the background immune infiltrate.
Notably, of the 3 samples that lacked dominant TCR clonotypes (PT53 TT, PT55 PP, PT55 TT): PT53 TT showed CNV neutral patterns in all cells, consistent with lack of viable tumor cells in that sample, while PT55 PP and PT55 TT showed cell populations with striking malignant CNV patterns, consistent with presence of scV(D)Jseq drop out. Therefore, our approach demonstrates that the combination of scV(D)Jseq and CNV evaluation provides a more robust methodology for distinguishing malignant from benign reactive T-cells in T-cell lymphoma single-cell studies. After separating malignant T-cells from the tCTCL TIME, the remaining CD45/PTPRC+ benign immune cell populations were annotated by known marker genes (Fig. 3C; Methods, Supplementary Fig. S6; Supplementary Table S11A–C) and SingleR, a computational algorithm that performs unbiased cell type recognition by referencing transcriptomic datasets of pure cells types (Monaco immune database (51), Methods). Using these two approaches, we identified the benign immune cell clusters as B-cells, NK cells, macrophages/monocytes, dendritic cells and various T-cell subtypes. For the non-immune cells in the TIME, fibroblasts were identified by COL1A2 and endothelial cells by VWF expression.
tCTCL exploits OXPHOS metabolic reprogramming, cellular plasticity and MYC/E2F activities at transformation
To define a malignant T-cell oncogenic program in tCTCL, we first performed differentially expressed gene (DEG) and gene set enrichment pathway analyses comparing malignant T-cells in TT and benign CD4+ T-cells in the TIME (Fig. 4A). Computational overlap of genes ranked by expression fold-change with the Molecular Signatures Database hallmark gene sets (MSigDB) showed significant enrichment of genes in OXPHOS, MYC, EMT and E2F target pathways and down-regulation of IFN-α, TNF-α, and IFN-γ (Fig 4A, Supplementary Table S12A). Importantly, significant upregulation of OXPHOS, MYC, and E2F target pathways and down-regulation of IFN-α, TNF-α, and IFN-γ pathways were also observed in disease evolution from PP to TT (Fig 4B; Supplementary Table S12B). Further analysis of the DEGs between malignant T-cells in TT vs benign CD4+ T-cells highlight a 55-gene malignant T-cell signature with upregulation of genes involved in oxidative phosphorylation (SLC25A5, NDUFB2), EMT/cellular plasticity and stemness (TWIST1, EPCAM, CLDN7), MYC signaling (NME2, SRM, PIM3), chemokines (CCR7), cytokines (MIF), cell migration and motility (CD9/tetraspanin, LAIR2, RAB25), as well as down-regulation of HLA-A, HLA-B and HLA-F (MHC-I, IFN-γ pathway) that is suggestive of immune escape in tCTCL (Fig 4C, Supplementary Table S13). Of the upregulated genes, TWIST1 (cellular plasticity/stemness), NDUFB2 (OXPHOS) and LGALS3 (Galectin 3/RAS) are also predicted drivers within the GISTIC2.0 amplification loci and are therefore considered high confidence drivers in tCTCL.
Figure 4. Malignant T-cell oncogenic program in tCTCL and pharmacologic inhibition of the OXPHOS and MYC in vitro.
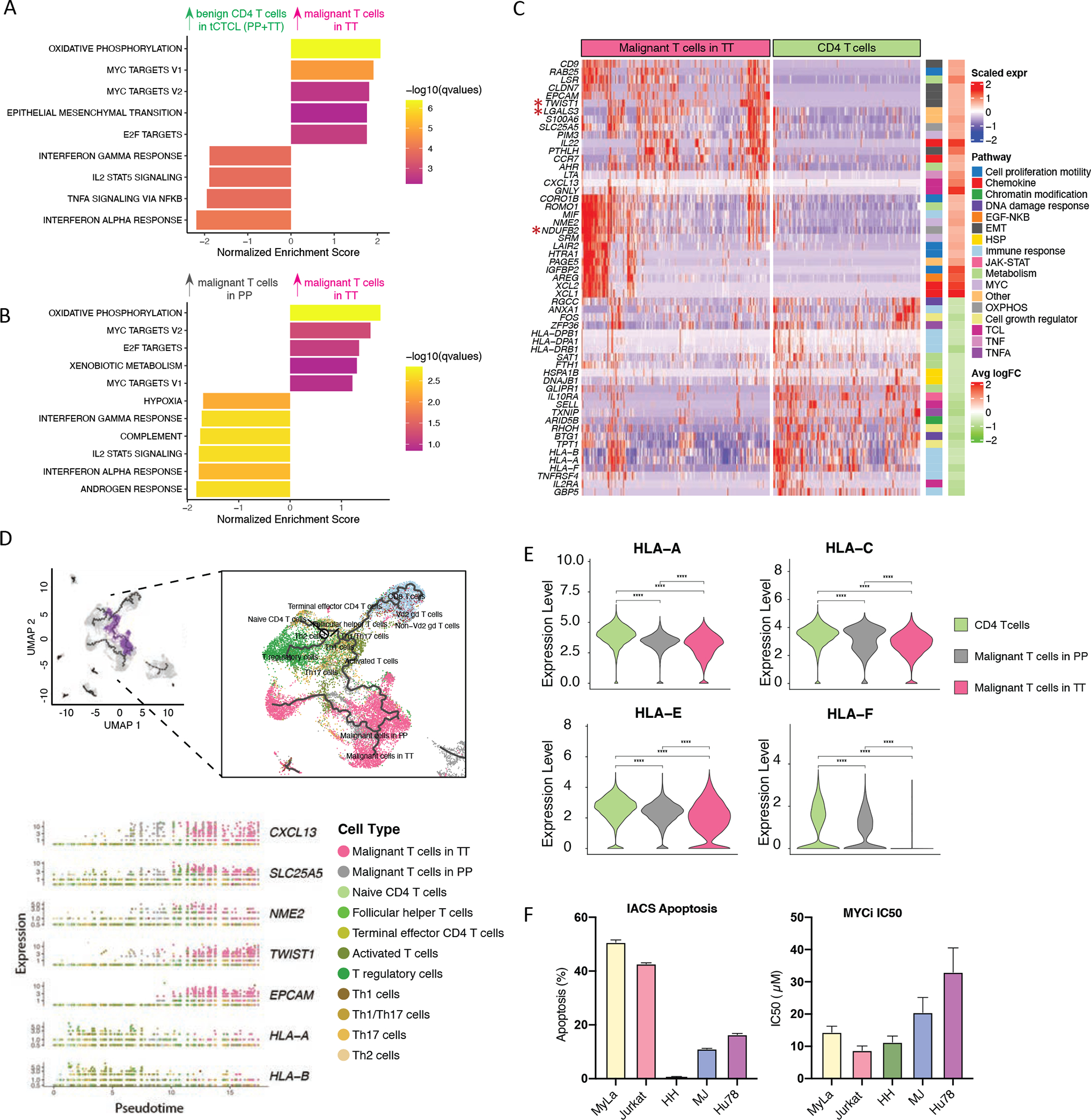
A. Gene set enrichment analysis (GSEA) comparing malignant T-cells in TT to benign CD4 T-cells showed significant enrichment of genes in OXPHOS, MYC, EMT (cellular plasticity/stemness) and E2F target pathways and down-regulation of IFN-γ, TNF-α, and IFN-α. The normalized enrichment score (NES, x-axis) reflects the extent of enrichment and allows comparison across gene sets. Listed pathways are ranked by their NES and colored by their significance. B. GSEA. Comparison of malignant T-cells in TT to malignant T-cells in PP shows upregulation of genes in OXPHOS, MYC and E2F target pathways and down-regulation of IFN-γ and IFN -α pathways from PP to TT. C. Malignant T-cell oncogenic program DEGs. Scaled expression of select genes from the 55-genes tCTCL malignant T-cell oncogenic program (y-axis, rows, full list in Supplementary Table S12) across the profiled cells (columns), malignant T-cells in TT (magenta) and benign CD4+ T-cells (green). Color bars to right denote significantly enriched pathways. Significant DEGs were filtered with a q value < 0.05 and an absolute value of fold change (FC) > 2 or <0.5 (Methods). D. Trajectories of the tCTCL TIME constituents in pseudotime by Monocle 3. UMAP for dimension reduction and visualization, including all cells previously annotated in Fig. 3C. Naïve CD4+ T cells in the graphical interface was designated as the root node, and the cellular trajectories in pseudotime were learned using the default parameters of Monocle 3 (learn_graph function). The learned trajectories reveal mono-directionality from benign T-cells to malignant T-cells in PP (grey) to malignant T-cells in TT (magenta) (right panel). A branch of the trajectory originating from naïve CD4+ T cells to malignant T cells was selected (choose_graph_segments function, left panel, purple branch), and the kinetics of the 55-gene malignant T-cell oncogenic program was plotted along pseudotime with select genes shown in the bottom panel (full list of genes in Supplementary Fig. S8; Supplementary Table S13). Cell types as annotated in the scRNAseq dataset are dotted in colors (e.g., malignant T-cells in PP – grey, malignant T-cells in TT - magenta). CXCL13 (chemokine), SLC25A5 (OXPHOS), NME2 (MYC) showed coordinated up-regulation from benign T-cells to malignant PP, while EPCAM and TWIST (EMT/cellular plasticity) showed accentuation of gene expression at the end of the trajectory in tumors. Downregulation of HLA-A and HLA-B (MHC-I) occurred early at the bifurcation from benign to malignant PP in pseudotime. E. Violin plots of distribution of HLA-A, C, E, F gene expression in malignant T-cells in TT (magenta), malignant T-cells in PP (gray) and benign CD4 T-cells (green). **** denotes p<0.001. F. T-cell lymphoma cell lines, Myla (MF), Jurkat (ATLL), HH (leukemic CTCL), MJ (ATLL) and Hu78 (SS). OXPHOS inhibitor (IACS-10759) apoptosis assay (left): indicated cell lines were seeded on 96-well plates and treated with 8 nM of IACS-010759 for 5 days. At day 5, cells were harvested and stained with Annexin V and PI following manufacture’s protocol (Biolegend Cat#640914). Annexin V+PI+ population was gated on Singlet population using FlowJo 10 software. The data was normalized to vehicle control. The error bars represent the mean ± s.e.m. n=8 (MyLa and Jurkat); n=4 (HH, MJ, Hu78). MYC inhibitor (MYCi975) cell proliferation assay (right). Indicated cell lines were seeded on 96-well plates and treated with different dose of MYCi975 for 5 days. At day 5, cells were incubated with MTS reagent following manufacture’s protocol (Promega Cat#G3580). Absorbance at OD490 nm was recorded and percentage of growth were normalized to vehicle control. Half maximal inhibitory concentration (IC50) was calculated based on curve fitting result using non-linear regression function of GraphPad Prism 8. The symbol represents the mean. The error bars represent the mean ± s.e.m. n=12 (MyLa and Jurkat); n=8 (HH, MJ, Hu78).
We next examined the benign immune cell types in the tCTCL TIME. While there was no quantitative difference in benign CD4+ T-cells, CD8+ T-cells, macrophages, NK-cells and dendritic cells between concurrent PP and TTs, B-cells were significantly enriched in TTs (Supplementary Fig. S7) and may play a pro-tumorigenic role during disease evolution.
While scRNAseq data offers a high dimensional view of cell types, cell states and malignancy at single-cell resolution, these measurements ultimately represent a snapshot in time of a complex ecosystem with heterogeneous cell substrates in a diverse range of evolutionary stages. We therefore sought to order the trajectory of tCTCL constituents in pseudotime using Monocle 3, a machine learning algorithm that constructs the trajectory of cells between one of several possible end states along geodestic distance from a root node and can learn trajectories that have loops or points of convergence (52). We first arbitrarily assigned benign T-cells as the starting root node, here CD4+ naïve T-cells, and demonstrated a mono-directional trajectory from benign to malignant CD4+ T-cells in PP and TTs (Fig. 4D top). We next tracked the kinetics of the 55-gene malignant T-cell program along pseudotime and observed coordinated upregulation of CXCL13 (chemokine), SLC25A5 (OXPHOS) and NME2 (MYC) from benign to malignant PP and accentuation of cellular plasticity genes such as EPCAM, TWIST1 and PTHLH at the end of the trajectory in TT (Fig. 4D bottom; Supplementary Fig. S8; Supplementary Table S14). Notably, downregulation of MHC-I genes (HLA-A, HLA-B and HLA-F) occurred early at the bifurcation from benign to malignant T-cells in PP (Fig 4D), suggesting immune evasion as an early event in CTCL lymphomagenesis, and is exacerbated from benign CD4 to malignant T-cells in PP to malignant T-cells in TT (Fig. 4E).
Small molecule inhibitors of OXPHOS and MYC suppress CTCL cells in vitro
Our scRNAseq analyses identified OXPHOS and MYC as the most enriched pathways in tCTCL and potential therapeutic vulnerabilities. High oxidative phosphorylation is a hallmark of multiple hematopoietic malignancies and solid tumors (33,53–55). In pre-clinical models of acute myeloid leukemia and mantle cell lymphoma, inhibition of OXPHOS by IACS-010759, a potent small-molecule inhibitor of mitochondrial electron transport chain complex I, results in inhibition of tumor cell proliferation and induction of apoptosis (33,55). Likewise, MYCi975, a small molecule MYC inhibitor, is shown to have potent preclinical in-vivo activity against MYC (34). To determine the sensitivity of CTCL and other T cell lymphomas to OXPHOS and MYC inhibition, we performed pharmacologic perturbation assays using IACS10759 and MYCi975 in MF and other TCL cell lines (Fig. 4F; Supplementary Table S15A–B). Using a group of established T-cell lymphoma cell lines, including Myla (MF), HH (MF – leukemic phase), and Hu78 (SS), we found low nanomolar sensitivity to IACS-010759 as assessed by apoptosis, particularly for Myla, and low micromolar sensitivity to MYCi975, as assessed by cell proliferation, cell viability and cytotoxicity (MTS assay). Together, these findings provide functional support for the two most significantly enriched malignant T-cell programs in tCTCL and potential use of OXPHOS and MYC inhibitors in treating aggressive T-cell lymphomas.
Analysis of cellular crosstalk between malignant T-cells and the TIME highlights MIF-CD74 interactions
To systematically disentangle the complex intercellular network within the tCTCL ecosystem, we next investigated our scRNAseq data for potential receptor-ligand interactions between malignant T-cells and the TIME. We integrated our scRNAseq data with CellPhoneDB, a computational tool that interrogates scRNAseq data for protein subunit structures and receptor-ligand interactions, and explored the crosstalk between malignant T-cells and the macrophages/monocytes, B-cells, dendritic cells, NK cells, fibroblasts and endothelial cells (Fig. 5A; Supplementary Table S16A–C). We showed top-ranking receptor-ligand pairs between MIF (ligand) in malignant T-cells and CD74, also known as HLA-DR antigens-associated invariant chain, in macrophages/monocytes, B-cells and dendritic cells (Fig. 5A). Importantly, our scRNAseq 55-gene malignant T-cell program also highlighted MIF as one of the significant differentially upregulated genes in malignant T-cells (Fig. 4C).
Figure 5. Cellular crosstalk between malignant T-cells and the tCTCL TIME highlights MIF-CD74 interactions.
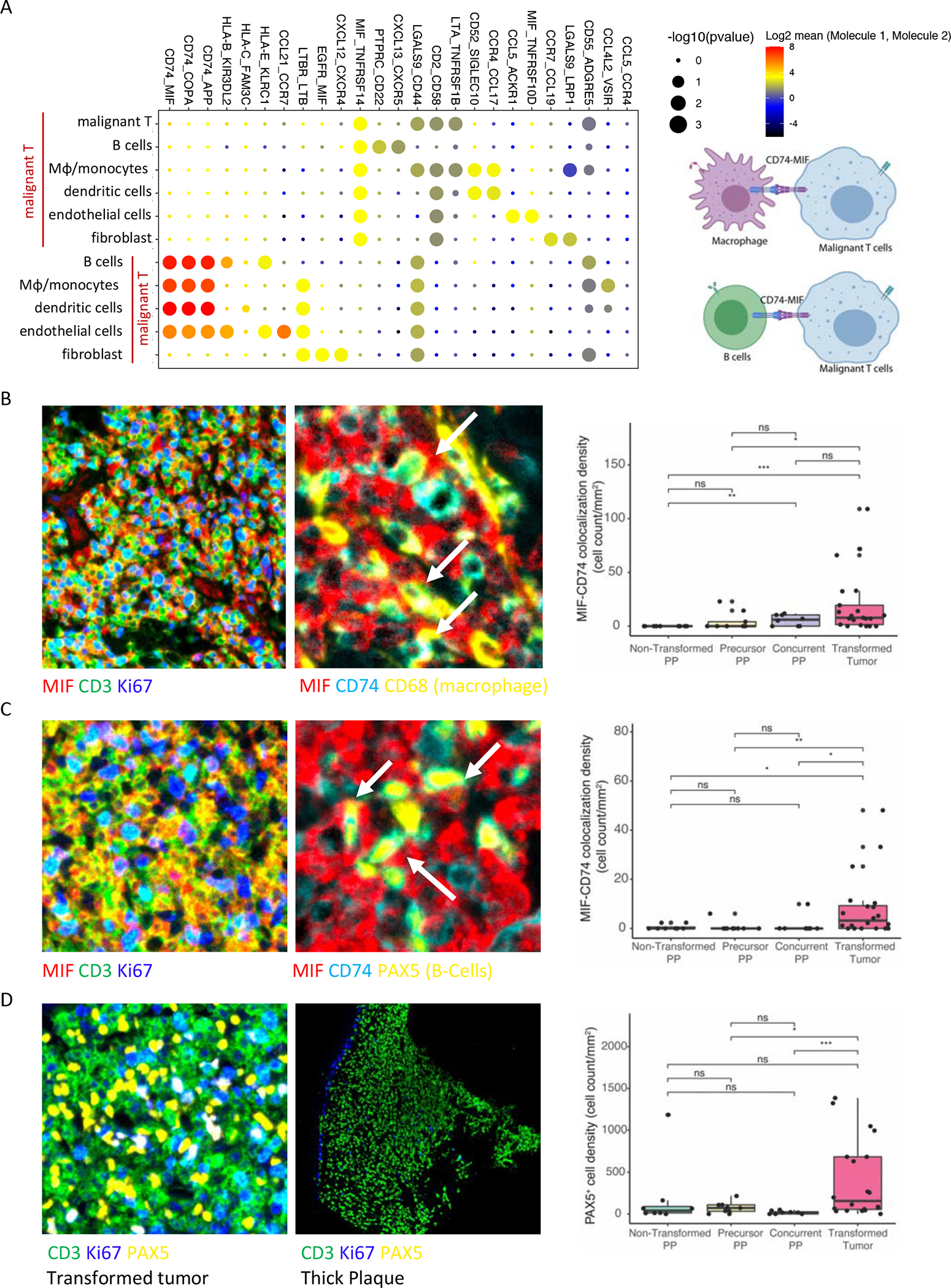
A. Overview of the statistically significant receptor-ligand interactions between malignant T-cells and macrophage/monocytes, B-cells, dendritic cells, endothelial cells and fibroblasts by integrating CellPhoneDB v2.0, a cell-cell communication informatics pipeline, with the single-cell RNAseq dataset (left; potential receptor-ligand pairs between interacting cell types denoted at top). Significance of p-values are indicated by circle size (−log10 p-value, permutation test; Methods). Color indicates the log2 means of the receptor-ligand pairs between 2 interacting cell types. Scale is shown to the right. Schematic (right) representing predicted top-ranking predicted ligand-receptor interactions between MIF in malignant T-cells and CD74 in macrophages/monocytes and B-cells in the tCTCL TIME. B-C. MIF expression and MIF-CD74 co-localization by mIF immune profiling (80 core TMA). Lesion types: PP-NT (PP from non-transformed patients, n= 16 tissue cores), PP-P (precursor PP from tCTCL patients, n= 12 cores), PP-C (concurrent PP from tCTCL patients, n= 12 cores), Transformed tumors (n= 64 cores). Multi-layer TIFF images were exported from InForm (Akoya Biosciences) into HALO Image Analysis Platform (Indica Labs) for segmentation and quantitative analysis. B. MIF (red), CD74 (sky blue), CD3 (green), Ki67 (dark blue), CD68 (yellow). Malignant Cd4+ T-cell (Cd3+ CD8− Ki67 high). Macrophages (CD68+ cells). MIF in malignant T-cells (left panel), MIF in malignant T-cells co-localizes with CD74 in macrophages (white arrow, middle panel). Boxplot depicting MIF-CD74 colocalization density (y-axis, count per mm2) against each lesion type. C. MIF (red), CD74 (sky blue), CD3 (green), Ki67 (dark blue), PAX5 (yellow). Malignant CD4+ T-cell (CD3+ CD8− Ki67 high). B-cells (PAX5+ cells). MIF in malignant T-cells (left panel), MIF in malignant T-cells co-localizes with CD74 in B-cells (white arrow, middle panel). Boxplot demonstrating MIF-CD74 colocalization density (y-axis, cell count per mm2) against each lesion type. D. TT (left panel) with dense infiltration of B-cells (PAX5+, yellow) between malignant Cd4+ T-cells (CD3+ CD8− Ki67 high). A thick PP lesion from a non-transformed patient (middle panel) showing absence of PAX5+ B-cells in the TIME. Ki67 cells (dark blue) depict basal layer of the epidermis. Boxplot depicting B-cell density (y-axis, cell count per mm2) against each lesion type.
To demonstrate in situ evidence for MIF-CD74 interactions, we built a tissue microarray (TMA, 80 tissue cores) using skin biopsies from 21 tCTCL patients (16 with matched PP-Tumor LCT; 64 cores; Fig. 1A) and 9 PP patients without any history of transformation (PP, 16 cores). We performed immune profiling by Vectra multiplex immunofluorescence (mIF), and regions of interest (ROI) in each biopsy were selected from all lymphoid-dense areas in the biopsy. We stained this TMA with 2 panels of antibodies (anti-MIF, CD74, CD3, CD8, Ki67, CD68 antibodies and DAPI; anti-MIF, CD74, CD3, CD8, Ki67, PAX5 antibodies and DAPI; Supplementary Table S17A–B) and imaged on Vectra 3. We analyzed a total of 488,399 cells by HALO software (Indica Labs) and demonstrated colocalization between MIF in malignant CD4+ T-cells (CD3+ Cd8- Ki67+ cells) and CD74 in macrophages (CD68+) (Fig. 5B) and B-cells (PAX5+) (Fig. 5C), providing in situ validation for the CellPhoneDB predicted receptor-ligand interactions. These findings together raise the possibility that MIF could be an important therapeutic target in tCTCL through inhibiting MIF-CD74 interactions between malignant T-cells and APCs. Consistent with our observation in scRNAseq dataset, we also found moderate to prominent B-cell infiltrate in transformed tumors, while the PP lesions showed either minimal or no B-cell infiltrate (Fig. 5D; Supplementary Fig. S7). This TMA was also evaluated for potential differences in the TIME by CD30 expression (≤1%, >1% to <10%, 10–50%, >50%) via immunohistochemical studies. Here, we did not detect quantitative differences in macrophage, B-cell and CD8+ T-cell infiltrates or MIF-CD74 interactions between malignant T-cells and macrophages or B-cells (Supplementary Fig. S9A–F).
Dominant subclones in tCTCL show upregulation of genes encoding ribosomal protein subunits
We have observed inter-patient tumoral heterogeneity in tCTCL (Fig. 3C), with some patients revealing more than one malignant T-cell clusters. We therefore sought to interrogate intratumoral heterogeneity (ITH) at the genetic and transcriptional level and determine if ITH correlates with patient outcomes. As CNV inference has been used to distinguish genetically distinct tumor subclones (32,56–58), we focused our attention on malignant T-cells from the scRNAseq dataset and performed CNV inference at the level of tumor subclusters using the inferCNV “subclusters mode” (qnorm method, Fig. 6A). We identified genetically distinct subclones of malignant T-cells within TTs from patients 11, 35 and 50 despite TCR monoclonality. Importantly, of the 8 patients in the scRNAseq cohort, PT 11 and 35 showed highest tumor burden (PT 11 TT clinical image Fig. 1A) and the worst clinical outcomes. UMAP clustering further demonstrated subclonal transcriptional heterogeneity in TTs from the same patients (PT11 cluster 1, 11, 12; PT 35 clusters 0, 3, 9; PT50 clusters 4, 8; Fig. 6B–C; Supplementary Table S18A–B), with dramatically elevated ribosomal gene expression (large and small ribosomal protein subunits) in the preponderant malignant T-cell subclones (PT 11 and PT35). A similar ribosomal signature has been observed in circulating tumor cells from highly aggressive breast cancer (59). Our findings suggest potential therapeutic utility in targeting the ribosomal synthesis and global eukaryotic translational machineries for patients with aggressive, transformed CTCL.
Figure 6. Dominant subclones in tCTCL show deregulation of ribosomal gene expression.
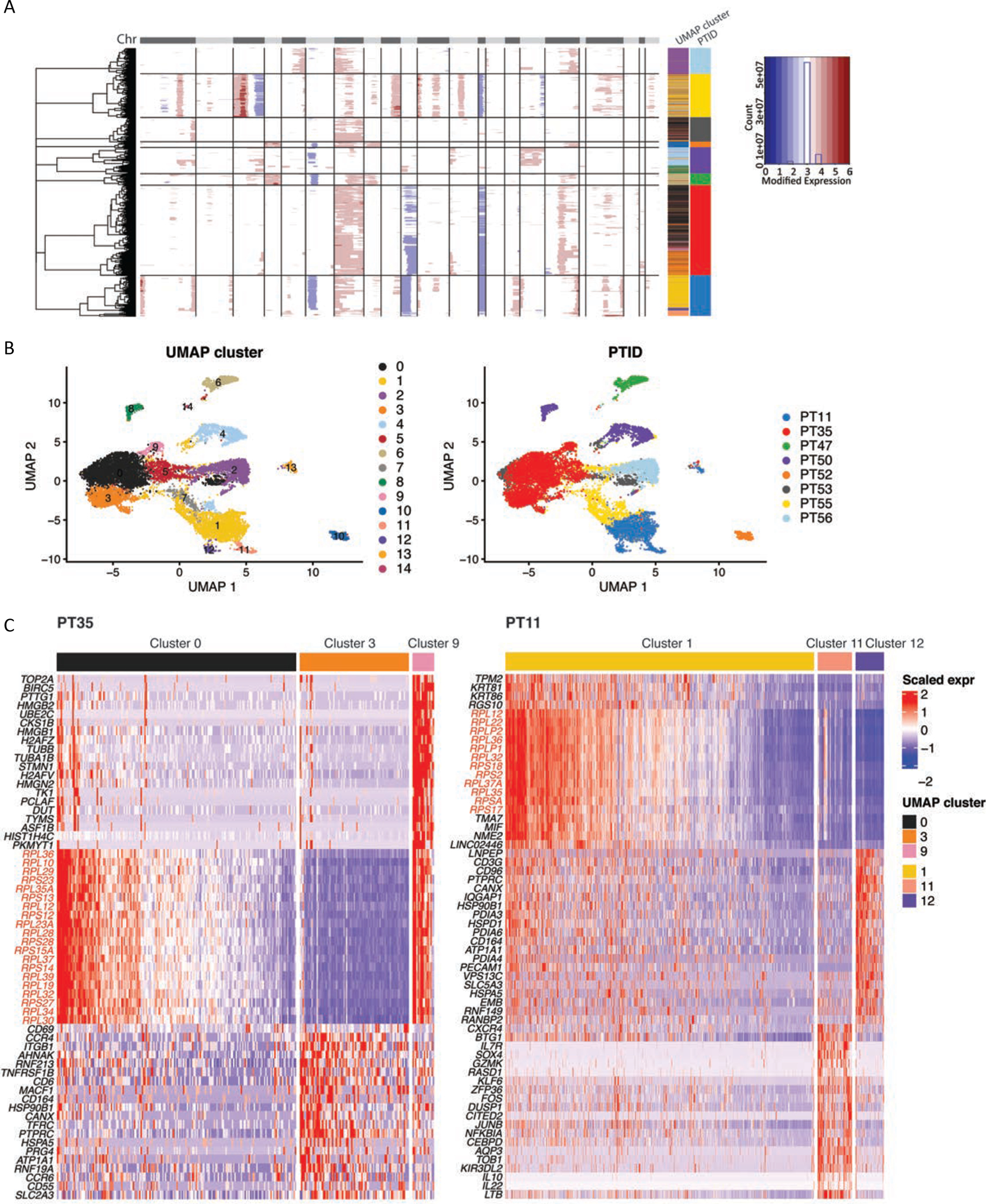
A. Identification of genetic subclones in malignant T-cells in each patient by partitioning hierarchical clustering trees (inferCNV, HMM subcluster mode, “qnorm” method). The CNAs (red, amplifications; blue, deletions) are shown along the chromosomes for each cell. Color bars to the right denote matched UMAP cluster annotation, patient ID and scaled expression.
B. UMAP plots of all malignant T-cells clustered by gene expression (left) and labeled by patient ID (right) reveal subclonal transcriptional heterogeneity in PT11 (clusters 1, 11, 12), PT 35 (clusters 0, 3, 9) and PT50 (clusters 4, 8) (Supplementary Table S17). UMAP clusters and patient IDs (PTID) are color coded to the right. C. DEG of the malignant T-cell subclusters in PT 35 (left) and PT11 (right) reveal dramatic upregulation of genes encoding ribosomal protein large and small subunits in the preponderant malignant T-cell subclones in these two patients with the worst clinical outcomes.
Cutaneous tCTCL and SS have distinct oncogenic malignant T-cell programs
Finally, as we observed distinct patterns of CNA in tCTCL and SS (Fig. 2), we sought to investigate the differences in T-cell oncogenic signatures between these two advanced stages of CTCL at single-cell resolution. A recently reported SS cohort with matched single-cell transcriptomic (expanded CRISPR-compatible cellular indexing of transcriptomes and epitopes by sequencing, ECCITE-seq) and single-cell TCR clonotype (Herrera cohort) (32) was downloaded for this analysis. Malignant T-cells from this SS cohort were identified by the dominant TCR clonotype and benign CD4+ T-cells by CD4+ polyclonal T-cells (Supplementary Methods), and UMAP clustering showed clear separation between malignant T-cells versus benign CD4 T-cells (Supplementary Fig. S10A). Interestingly, malignant T-cells in SS do not show upregulation of OXYPHOS and MYC activities (Supplementary Fig. S10B) or down-regulation of MHC-I genes compared to benign CD4 T-cells (Supplementary Fig. S10C)(Supplementary Table S19A). We next compared the gene expression profile between malignant T-cells in tCTCL (PP and TT) to malignant T-cells in SS. Here, the malignant T-cells cluster by patients of origin (Fig. 7A), with tCTCL patient clusters clearly separating from the SS patient clusters (Fig. 7B), and with benign T-cells across different patients clustering together (Supplementary Fig. S11). Importantly, malignant T-cells in tCTCL (PP and TT) show significant upregulation of OXPHOS, MYC, EMT/stemness, E2F target and TNFα activities compared to malignant T-cells in SS, as well as down-regulation of IFNα pathway genes (Fig. 7C; Supplementary Table S19B). Violin plots of MHC-I gene expression in malignant T-cells further revealed significantly lower HLA-A, B, C expression in tCTCL-TT and PP compared to SS (Fig. 7D), suggesting immunosurveillance escape through loss of MHC-I as a more prominent feature in tCTCL.
Figure 7. Cutaneous tCTCL shows distinct malignant T-cell oncogenic program from SS.
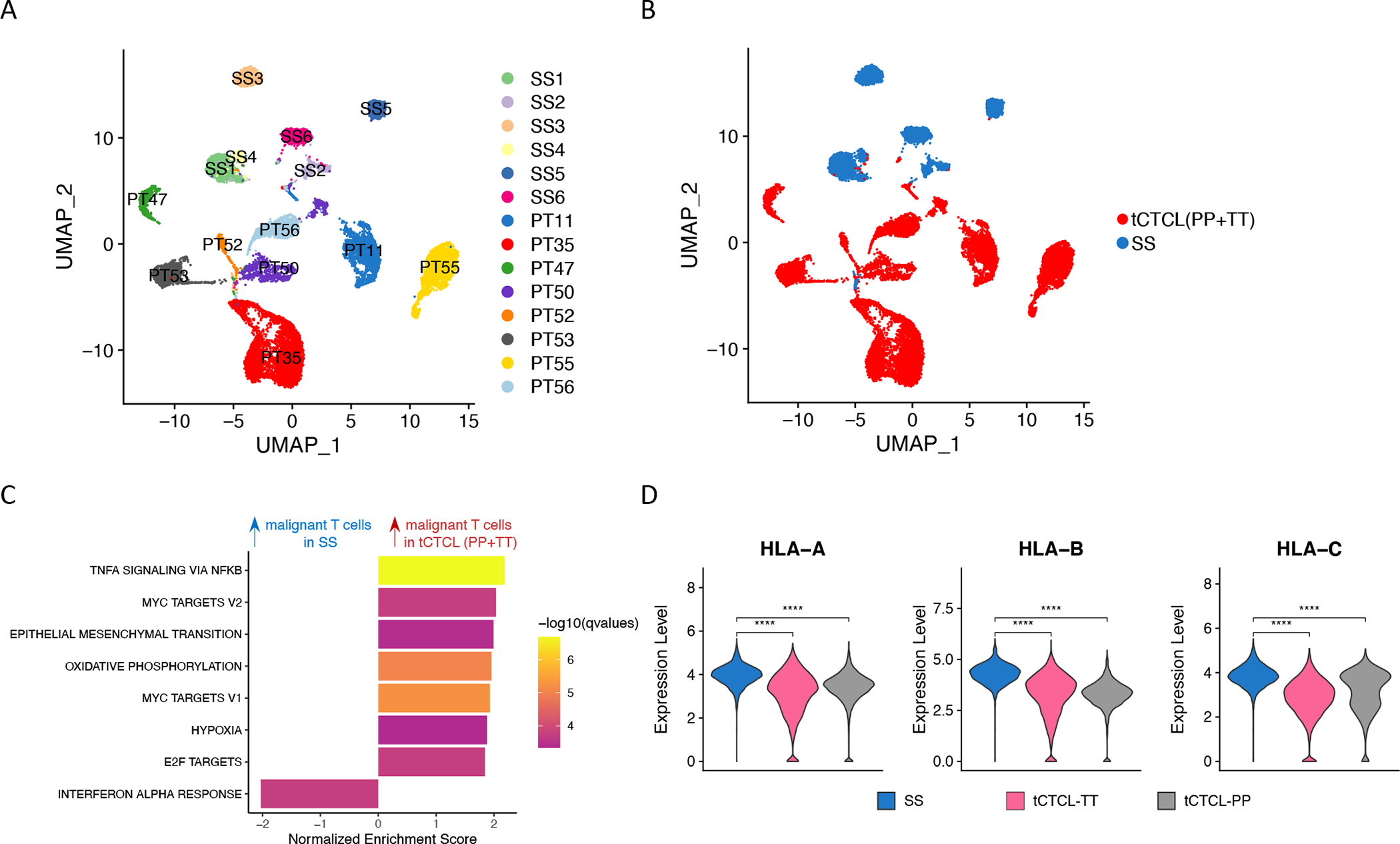
A. UMAP of malignant T-cells from 6 SS patients (Herrera cohort, patients SS1 to SS6) and malignant T-cells from 8 tCTCL patients in the current study (PT11, 35, 47, 50, 52, 53, 55, 56; each with PP and TT lesions). B. UMAP of malignant T-cells from tCTCL patients (PP+TT, red) and SS patients (blue). C. GSEA comparing malignant T-cells in tCTCL (PP+TT) vs malignant T-cells in SS shows significant upregulation of genes in TNF-a, MYC, EMT, OXPHOS and E2F target pathways and downregulation of genes in the IFN-a pathway. Normalized enrichment score (NES, x-axis). Listed pathways are ranked by their NES and colored by their significance. D. Violin plots of distribution of HLA-A, B, C gene expression (y-axis, normalized expression counts in a log scale) in malignant T-cells in SS (blue), malignant T-cells in TT (magenta) and malignant T-cells in PP (gray). **** denotes p<0.001.
DISCUSSION
Transformed CTCL is an aggressive large cell lymphoma that is resistant to multiple forms of systemic therapy. There is a critical unmet need for identifying novel therapeutic targets for this incurable cancer, yet its disease biology is poorly understood due to the multitude of challenges associated with tissue-based research in rare cancers. Genomic- and transcriptomic-level information of CTCL at disease transformation is limited, and a better understanding of tCTCL disease biology has broad implications for therapy. Our study contains the largest collection of tCTCL clinical specimens that are difficult to obtain and provides an important resource for the study of tCTCL biology and the identification of novel therapeutic vulnerabilities. We generated a WES dataset of similar size to other TCGA rare cancers, the first scRNAseq atlas of tCTCL, and immune profiled the TIME by scV(D)J seq and multiplex IF using serial biopsies from tCTCL patients. Together, this multi-omics study uncovered key driver events, prognostic mutational signatures, diverse oncogenic programs and potential receptor-ligand interactions that malignant T-cells exploit at large cell transformation.
Defining the T-cell lymphoma ecosystem at single-cell resolution is particularly challenging as the TIME is comprised of malignant CD4+ T-cells, benign T-cells, as well as other diverse immune cells. Gene expression-based clustering is imprecise in defining malignancy, and dropout read is a well-known phenomenon in scRNAseq and scVDJseq datasets. Here, we implemented a robust two-tiered algorithm using simultaneous single-cell whole transcriptome-V(D)J profiling and chromosomal copy number inference to identify malignant T-cells from the TIME, and we believe this methodology can be applied to the study of other T- and B-cell leukemia/lymphomas. While CTCL is clinically viewed as a cancer of monoclonal T-cells, a recent study showed TCR clonotypic diversity in CTCL by bulk TCR seq and suggested that T-cell tumorigenesis occurs prior to TCR gene rearrangement (60). In tCTCL, we observed TCR monoclonality, with monoclonal T-cells showing a malignant CNV patterns, and background polyclonal T-cells in the TIME showing neutral CNV patterns. The difference in these two studies could be due to differential resolution of TCR detection using scV(D)Jseq, with the ability to assign precise TCR α- and β- sequence pairs to individual T-cells. Nevertheless, our data suggest that TCR monoclonality is a key feature of transformation. Future single cell TCR studies involving CTCL lesions at different stages of disease can potentially elucidate the origin of CTCL with respect to the timing of TCR gene rearrangement.
A central finding from our single-cell study is the diverse oncogenic programs that malignant T-cells exploit to confer aggressive behavior and survival advantage at transformation, with upregulation of OXPHOS and MYC as the top enriched pathways, which are progressively upregulated in disease evolution, followed by EMT/stemness and E2F target genes (Fig. 4A and 4B). Notably, similar OXPHOS and MYC upregulation was not observed in our single-cell analysis of an independent SS cohort (Supplementary Fig. 10B, Fig. 7C, (32)). Metabolic reprogramming toward oxidative phosphorylation, and hence increased ATP production, has been reported in other high grade leukemias, lymphomas and solid tumors (33,53–55). In tCTCL, this metabolic shift can be viewed as advantageous for supplying the energy demand of rapidly growing, high-grade lymphoma cells. As MYC activation has been shown to drive cancer cell metabolism toward glutaminolysis and mitochondrial biogenesis (61,62), the shift toward OXPHOS can be related to MYC upregulation. Interestingly, quiescent T-cells (naive or memory T-cells) are known to use catabolic metabolism by oxidative phosphorylation to fuel cell survival (63), raising the possibility that the upregulation of OXPHOS in tCTCL can also be related to T-cell state at transformation. While future studies are needed to chart the mechanistic action of these pathway components, it is encouraging that novel small molecule inhibitors of OXPHOS (IACS-10759) and MYC (MYCi975) provided functional support of OXPHOS and MYC as potential therapeutic vulnerabilities. Lastly, in addition to OXPHOS and MYC, our scRNAseq dataset also demonstrated significant upregulation of MIF in malignant T-cells and interactions between malignant T-cells expressing MIF and macrophages and B-cells expressing CD74. These findings raise the possibility of MIF as potential therapeutic targets in tCTCL.
A recurrent theme observed in this study is the up-regulation of genes involved in cellular plasticity and sternness in tCTCL. In our WES dataset, GISTIC analyses aimed at detecting genomic loci targeted by SCNAs revealed recurrent amplifications in TWIST1, a master transcription factor of EMT/stemness, while the scRNAseq 55-gene malignant T-cell signature demonstrated upregulation of TWIST1, CLND7 and EPCAM, all of which are genes involved in cellular plasticity and stemness. We also observed recurrent mutations in Hippo pathway genes such FAT1 (~30%), loss of which has been implicated in driving tumor cell stemness and metastasis in cutaneous squamous cell carcinoma (47). Tumor plasticity is a key feature of aggressive cancer transition, which can certainly account for the transition of CTCL to an aggressive large cell lymphoma with blast-like morphology at transformation. In large-scale gene expression studies across 21 solid tumors, activation of stemness programs was shown to positively correlate with ITH, drive clonal evolution and limit anti-tumor immune responses (64). Indeed, we also observed ITH in tCTCL, with prominent subclonal upregulation of ribosomal subunit gene expression in patients with the worst clinical outcomes.
Besides the upregulated signatures, one intriguing observation from our single-cell dataset is the downregulation of MHC-I in malignant T-cells in tCTCL. Comparison of MHC-I gene expression levels (HLA-A, C, E, F) showed progressive and significant decreases from benign CD4 T-cells to PP to TT, and our pseudotime analysis corroborated these changes in expression across disease evolution. While high TMB cancers are theoretically more immunogenic and responsive to T-cell based immunotherapies, loss of MHC-I antigen presentation can compromise the visibility of tumor cell antigens to CD8+ T-cells and response to checkpoint blockade agents. In such MHC-I negative or low tumors with defective antigen presentation machinery, NK-cell based therapies and epigenetic drugs can be considered.
The role of B-cells in the tumor microenvironment is recently under intense investigation in various cancer types, though it is still incompletely explored compared to other benign immune substrates of the TIME. The results are also variable, with some studies showing positive correlation between B-cell infiltrate and patient outcomes or response, while other studies suggest a pro-tumorigenic role of B-cells (65). In tCTCL, our scRNAseq data shows significant enrichment of B-cell infiltrate mostly in transformed tumors, a finding that is supported by multiplex IF. Altogether, our results suggest a potential pro-tumorigenic role of B-cells in tCTCL, possibly through interactions with malignant T-cells via MIF-CD74 signaling.
Lastly, CTCL is a rare cancer with well-known racial disparity. Black/AA patients have a higher incidence rate, younger age of onset, higher disease burden and inferior survival compared to Whites, even after accounting for disease characteristics, socioeconomic factors, and treatments received (1,2). Nonetheless, potential biological factors underlying these racial disparities are poorly understood. Here, we provide the first attempt at identifying potential genomic correlates for the survival gap between Black/AA and White patients. While the sample size is admittedly small, we observed a significantly lower contribution of UV signatures that are prognostic for favorable survival and enrichment of other signatures that may drive worse outcomes in the Black/AA patients. Future studies involving larger sample size from the vulnerable population and research into their TIME, genomic correlates and potential therapeutic targets will hopefully help reduce racial disparity in CTCL.
Rare cancers represent one of the greatest inequalities in cancer research, with the lack of well-curated tissue specimens to study disease biology and resources for pharmaceutical development. Transformed CTCL exemplifies such challenge where the lack of genomic and transcriptomic level information contributes to the paucity of drug discovery efforts. In this study, we have identified potential genetic driver events and pathways in tCTCL and elucidated a malignant T-cell oncogenic program with potential novel therapeutic vulnerabilities. While further validation in larger cohorts and pre-clinical models are needed, our investigation provides a key resource with the largest collection of tCTCL samples studied to date. We anticipate results from this study can be extrapolated to other T-cell lymphomas and will help usher novel immunotherapeutic strategies to combat this currently incurable cancer.
MATERIALS AND METHODS
Note: A full description of all methods for WES (DNA extraction and library prep, data processing and somatic mutation filtering, driver gene detection and mutational signature analysis), scRNA-seq (data processing and benign cell cluster annotation), Vectra mIF studies and comparison of single-cell transcriptomic profile to SS (Herrera cohort) can be found in the Supplementary Methods.
Patient cohort and clinical annotation
A total of 56 patients with tCTCL were included in this study. The studies were conducted in accordance with the Declaration of Helsinki and approved by the Moffitt Cancer Center IRB (MCC19672). Electronic medical charts were reviewed independently by two investigators to document the clinical parameters, including age, gender, time of first rash, time of initial MF diagnosis, time of LCT, time of death / last follow up, clinical staging at time of initial MF diagnosis or presentation to MCC, clinical re-staging at time of LCT, and treatment history (Supplementary Table S1).
Whole exome sequencing
Clinical tissue samples
Of the 56 tCTCL patients, 54 had available tissues for WES (n= 70 skin formalin-fixed, paraffin-embedded (FFPE) biopsies) (Fig. 1A). For WES and immunofluorescence studies, standard of care skin biopsies (FFPE) from 54 patients with tCTCL were analyzed under Institutional Review Board (IRB) approved protocol (MCC19672) and approved waiver of consent where applicable. All samples were assessed by two pathologists for adequacy of lymphoid infiltrate, including 45 TT and 25 PP lesions (9 with LCT). 17 patients had paired PP and TT (11 precursor PP, 6 concurrent PP to the LCT lesion). Matched germline samples were available from 41 patients, while remaining patient samples used panel of normal as control. Tumor samples were micro-dissected before submitted for WES.
Cancer driver gene detection
NGS data were processed using established in-house bioinformatics pipeline (66). To enhance the coverage of our analysis, two mutation-based computational algorithms were applied, MutSigCV (version 1.41) (45) and dNdScv (RRID: SCR_017093) (44). MutSigCV was run with default parameters using the MATLAB (RRID:SCR_001622) Compiler Runtime (MCR). A global q-value threshold of 0.1 was applied for selecting driver genes as suggested by the MutSigCV developers. dNdScv was run in R (version 3.6.3) using the default parameters. A global q-value (qallsubs_cv) of < 0.05 was used as a cut-off for selecting predicted driver genes.
Copy number variation detection and analysis
CNV analyses were performed in samples with matched germline. CNVs were identified using R package ExomeLyzer (55). Copy number change was determined as the log2 ratio of tumor versus normal reads, and circular binary segmentation (CBS) algorithm was applied for CNV segmentation (67). To identify genomic regions significantly enriched with CNVs, GISTIC2.0 (RRID:SCR_000151) (48) was used with default settings. Genomic regions with FDR < 0.25 were considered as significantly enriched and highlighted in the chromosome landscape plot.
Tumor mutation burden
TMB analysis was performed in all samples with matched germline / normal tissue using Maftools (v2.6.0)(68). Results were compared to 33 TCGA cancer types (MC3 data)(69). TMB was calculated by the following formula
Results were compared to 33 TCGA cancer types (MC3 data)(69) and two SS cohorts (Wang cohort and Choi cohort) (26,36). The Choi cohort raw sequencing data was downloaded and processed using the same computational pipeline as our tCTCL cohort. The Wang cohort TMB was calculated using processed mutations from the original publication. The sequencing capture size is 39 Mb for the study cohort and 35.8 Mb for the TCGA cancer cohorts after data harmonization.
Mutational signature analysis
The repertoire of mutational signatures was catalogued at both the sample- and cohort-levels. At the sample level, contributions of known signatures (COSMIC mutation signatures version 3) in each tissue were independently calculated using two algorithms, MutationalPatterns(39) and deconstructSigs(38). Both tools were run using the default setting. To decipher the dominant mutagenic processes at the cohort-level, de novo representative signatures of the cohort was extracted using Maftools(68). A mutation context matrix was first built, and the top three mutational signatures that optimally represent the mutation profile were extracted using the non-negative matrix factorization (NMF) method. Finally, the constructed signatures were compared against COSMIC version 3 signatures (37).
Survival analysis
For Kaplan–Meier survival analysis, survival R package (version 3.2–7) was used. Survival analysis was performed from time of initial LCT to the date of death (event time) or the date of last follow up (censoring time). The patients were grouped into two sub-groups according to the median of genomic features, such as the weighted contribution of SBS7 signature. For patients with paired PP and TT tissues, data from the TT sample was used for survival analysis. Survival plot was generated using the survminer R package.
Analysis of highly mutated pathways
A variety of cancer-associated pathways were assessed for mutational enrichment (Supplementary Table S6), including eight oncogenic signaling pathways summarized by the TCGa project and implemented in Maftools (70). Genes with non-synonymous mutations observed in ≥ 3 tissues and recurrent in >10% of the samples were plotted in oncoplot using Maftools.
Single-cell RNA-seq and V(D)J-seq
Patient sample collection
Fresh tissue specimens were collected from 8 patients with tCTCL treated and managed at the H. Lee Moffitt Cancer Center Cutaneous Oncology and Malignant Hematology clinics following written informed consent under the IRB-approved Total Cancer Care Protocol (MCC14690) and processed under MCC19672. Two samples were collected per patient (concurrent PP and TT lesions).
Single-cell V(D)J-seq for TCR clonotype
TCR annotation was performed using Cell Ranger “vdj” function for sequence assembly and paired clonotype calling. Paired TCR α/β reads attributed to each cell barcode were grouped and assembled into a single contig to determine the combined TCR α/β clonotype, and contigs that were predicted as productive were selected for the downstream analyses.
Single-cell CNV Inference
InferCNV v1.3.5 (https://github.com/broadinstitute/inferCNV, Trinity CTAT Project) was used to extract chromosomal CNV patterns. Two-thirds of non-dominant TCR clonotype T-cells (CD4+, CD8+) were used as reference normal cell group for de-noise control. For the observation cell group, 1) the remaining non-dominant TCR clonotype cells, 2) all dominant TCR clonotype (“true malignant”) cells and 3) all CD3+ T-cells with TCR dropout (“malignant suspect”) were included. InferCNV was run using “denoise” mode, and “cluster by group” parameter was turned off such that observation cells can cluster regardless of tissue of origin.
Differential expression testing and gene set enrichment analysis
Differential expression testing was performed using the Wilcoxon rank sum method with two Seurat functions: FindMakers for pairwise comparisons and FindAllMarkers for comparisons across multiple groups. Both functions were run on the ‘RNA’ data assay with no pre-filters. Significant differentially expressed genes (DEGs) were filtered with a q-value < 0.05 and an absolute value of fold change (FC) > 2 or < 0.5. To reveal pathways in which DEGs are enriched, GSEA was performed on all of the detected genes ranked by average logFC. The analysis was performed using the clusterProfiler R package (RRID:SCR_016884) (71) against pre-defined hallmark gene sets of the MSigDB database.
Pseudotime trajectory analysis
T cells, both malignant and benign, were first constructed in a Seurat object and processed and clustered as described above. Trajectory analysis was performed using Monocle 3 (52,72–75). To meet the format requirement of Monocle 3, Seurat object and clustering information were converted as a cell data set using SeuratWrappers R package. Trajectory was then learned using the default Monocle 3 parameter “learn_graph” function. Naïve CD4+ T cells in the graphical interface was manually selected as the root node and cells were ordered in pseudotime. Next, a branch of the trajectory graph originating from naïve CD4+ T cells to malignant T cells was selected using the “choose_graph_segments” function. The kinetics of 55-gene malignant T-cell program were plotted along the pseudotime of the selected branch using the “plot_genes_in_pseudotime” function of Monocle 3.
CellPhoneDB intercellular communication analysis
We integrated our scRNAseq dataset with CellPhoneDB (RRID:SCR_017054) (76) and explored intercellular receptor-ligand interactions between malignant T-cells and macrophages/monocytes, B-cells, dendritic cells, NK cells, fibroblasts and endothelial cells. The raw count matrix of single-cell expression profile was normalized, and the count value of each gene in each cell was divided by the total counts of corresponding cell and multiplied by 10000. CellPhoneDB was run with the ‘statistical’ method in the virtualenv environment using default parameters. Receptor-ligand pairs with p-value < 0.05 were filtered as significantly enriched pair. To visualize the results, we selected the top 3 ligand-receptor pairs for each cell type pairs (in log2mean expression value) and summarized the results in a dot plot using R package ggplot2 (RRID:SCR_014601).
Analysis of intra-tumoral heterogeneity
InferCVN subcluster analysis for ITH at the genetic-level was performed on malignant T-cells (identified based on scV(D)J seq and copy number inference method as described above). Patient-specific clustering was restricted by setting the parameter “cluster_by_groups” as “True”. ‘Subclusters’ mode was enabled with the qnorm partition method to reveal ITH patterns. Malignant cells were clustered based on similarity of the extracted CNV patterns. Gene expression-based clustering of malignant T-cells by UMAP was performed to evaluate for ITH at the transcriptional-level.
Independent SS cohort analysis (Herrera cohort)
To perform single-cell transcriptomic analysis of SS and malignant T-cell comparison between tCTCL and SS, ECCITE-seq scRNAseq gene expression counts and TCR α/β clonotype profiles from 6 SS blood samples (SS1-SS6) were downloaded from the NCBI GEO database (GSE171811) (32)(Supplementary Methods).
Pharmacologic perturbation assays
Established T-cell lymphoma lines were used for drug assays, including Myla (MF; Sigma Aldrich Cat# 95051032), HH (MF - leukemic phase; ATCC Cat# CRL-2105), and Hu78 (SS; ATCC Cat# TIB-161), Jurkat (ATLL) and MJ (ATLL; ATCC Cat# 8294). For the apoptosis assay, indicated cell lines were seeded with the density of 1×105 cells/well in 96-well plate and treated with 8 nM of IACS-010759 (SelleckChem# S8731). At day 5, cells were harvested and stained with Annexin V and PI following manufacture’s protocol (Biolegend Cat# 640914). Annexin V+PI+ population was gated on Singlet population using FlowJo 10 software (RRID:SCR_008520). For cell proliferation assay, indicated cell lines were seeded with the density of 1×104 cells/well in 96-well plate and treated with different dose of MYCi975 (MedChemExpress Cat# HY-129601) for 5 days. At day 5, cells were incubated with MTS reagent following manufacture’s protocol (Promega Cat#G3580). Absorbance at OD490 nm was recorded and percentage of growth were normalized to vehicle control. IC50 was calculated based on curve fitting result using non-linear regression function of GraphPad Prism 8 (RRID:SCR_002798).
Vectra multiplex immunofluorescence
An 80-core tissue microarray (TMA) was built using skin biopsies from 21 tCTCL patients (64 tissue cores, including 16 patients with matched PP and TT) and additional 9 patients with PP lesions and no history of LCT (16 tissue cores). ROIs were selected based on the most lymphoid-dense/representative area in the tissue. Quantitative Image Analysis was performed using the HALO Image Analysis Platform (Indica Labs, Albuquerque, NM).
Statistical Analysis
Statistical analysis was performed using R programming language (version > 3.6). In survival test, a two-tailed log-rank test was used to determine the difference between patient subgroups. The statistical significance between category groups was determined using Wilcoxon rank sum test. ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. For comparison of survival between patient groups (Fig. 1D, Supplementary Fig. S3B), data from the TT sample was used when a patient had paired PP and TT available.
Data Availability Statement
The sequencing data, including WES, scRNA-seq and scVDJ-seq, have been submitted to NCBI BioProject database PRJNA754592. The Leukemic/SS WES data analyzed in this study were obtained from the NCBI BioProject database PRJNA285408.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Our study contributes a key resource to the community with the largest collection of tCTCL biopsies that are difficult to obtain. The multiomics data herein provide the first comprehensive compendium of genomic alterations in tCTCL and identify potential prognostic signatures and novel therapeutic targets for an incurable T-cell lymphoma.
ACKNOWLEDGEMENTS
We are grateful to the patients and their families for contributing to this work. This research was made possible through the Total Cancer Care® Research Protocol at the H. Lee Moffitt Cancer Center & Research Institute and supported by the Tissue Core, Molecular Genomics Core, and Advanced Analytical and Digital Laboratory. We thank Dr. Jennifer Wargo for her very helpful reading of the manuscript, Christophe Georgescu for his suggestions on implementing the inferCNV analytics, and Dr. Whijae Roh for his thoughtful comments on the genomics data.
Financial support:
This research was supported by generous philanthropic contributions to H. Lee Moffitt Cancer Center Moffitt Foundation, Moffitt Clinical Science Fund, Miles for Moffitt fund, Department of Pathology and Department of Cutaneous Oncology research support fund to P.L.C. Support for shared resources was provided by a Cancer Center Support Grant (CCSG) P30-CA076292 to H. Lee Moffitt Cancer Center. K.Y.T acknowledge support of Donald A. Adam Melanoma & Skin Cancer Center of Excellence and Mr. & Mrs. Winston Weber. J.R.C.G. and L.S. acknowledge support of this work by NIH R01CA240434.
Footnotes
Conflicts of interest disclosures
L.S.V. or her institution received research support from Helsinn, Eisai Co., Soligenix, Kyowa Kirin Inc., Innate Pharma, and Elorac Inc. for CTCL studies. L.S.V. is a consultant and speaker for Kyowa Kirin Inc. and Helsinn. L.S. is advisory board member of Kyowa Kirin and consultant at Dren-Bio. S.W. is advisory board member and KOL for Kyowa Kirin.
REFERENCES
- 1.Nath SK, Yu JB, Wilson LD. Poorer prognosis of African-American patients with mycosis fungoides: an analysis of the SEER dataset, 1988 to 2008. Clin Lymphoma Myeloma Leuk 2014;14(5):419–23 doi 10.1016/j.clml.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su C, Nguyen KA, Bai HX, Cao Y, Tao Y, Xiao R, et al. Racial disparity in mycosis fungoides: An analysis of 4495 cases from the US National Cancer Database. J Am Acad Dermatol 2017;77(3):497–502 e2 doi 10.1016/j.jaad.2017.04.1137. [DOI] [PubMed] [Google Scholar]
- 3.Willemze R, Cerroni L, Kempf W, Berti E, Facchetti F, Swerdlow SH, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019;133(16):1703–14 doi 10.1182/blood-2018-11-881268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scarisbrick JJ, Prince HM, Vermeer MH, Quaglino P, Horwitz S, Porcu P, et al. Cutaneous Lymphoma International Consortium Study of Outcome in Advanced Stages of Mycosis Fungoides and Sezary Syndrome: Effect of Specific Prognostic Markers on Survival and Development of a Prognostic Model. J Clin Oncol 2015;33(32):3766–73 doi 10.1200/JCO.2015.61.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol 2014;70(2):205 e1–16; quiz 21–2 doi 10.1016/j.jaad.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Yumeen S, Girardi M. Insights Into the Molecular and Cellular Underpinnings of Cutaneous T Cell Lymphoma. Yale J Biol Med 2020;93(1):111–21. [PMC free article] [PubMed] [Google Scholar]
- 7.Diamandidou E, Colome-Grimmer M, Fayad L, Duvic M, Kurzrock R. Transformation of mycosis fungoides/Sezary syndrome: clinical characteristics and prognosis. Blood 1998;92(4):1150–9. [PubMed] [Google Scholar]
- 8.Talpur R, Sui D, Gangar P, Dabaja BS, Duvic M. Retrospective Analysis of Prognostic Factors in 187 Cases of Transformed Mycosis Fungoides. Clin Lymphoma Myeloma Leuk 2016;16(1):49–56 doi 10.1016/j.clml.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Arulogun SO, Prince HM, Ng J, Lade S, Ryan GF, Blewitt O, et al. Long-term outcomes of patients with advanced-stage cutaneous T-cell lymphoma and large cell transformation. Blood 2008;112(8):3082–7 doi 10.1182/blood-2008-05-154609. [DOI] [PubMed] [Google Scholar]
- 10.Pulitzer M, Myskowski PL, Horwitz SM, Querfeld C, Connolly B, Li J, et al. Mycosis fungoides with large cell transformation: clinicopathological features and prognostic factors. Pathology 2014;46(7):610–6 doi 10.1097/PAT.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prince HM, Kim YH, Horwitz SM, Dummer R, Scarisbrick J, Quaglino P, et al. Brentuximab vedotin or physician’s choice in CD30-positive cutaneous T-cell lymphoma (ALCANZA): an international, open-label, randomised, phase 3, multicentre trial. Lancet 2017;390(10094):555–66 doi 10.1016/S0140-6736(17)31266-7. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, Bagot M, Pinter-Brown L, Rook AH, Porcu P, Horwitz SM, et al. Mogamulizumab versus vorinostat in previously treated cutaneous T-cell lymphoma (MAVORIC): an international, open-label, randomised, controlled phase 3 trial. Lancet Oncol 2018;19(9):1192–204 doi 10.1016/S1470-2045(18)30379-6. [DOI] [PubMed] [Google Scholar]
- 13.Kim YH, Prince HM, Whittaker S, Horwitz SM, Duvic M, Bechter O, et al. Response to brentuximab vedotin versus physician’s choice by CD30 expression and large cell transformation status in patients with mycosis fungoides: An ALCANZA sub-analysis. Eur J Cancer 2021;148:411–21 doi 10.1016/j.ejca.2021.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YH, Khodadoust M, de Masson A, Moins-Teisserenc H, Ito T, Dwyer K, et al. Patient characteristics of long-term responders to mogamulizumab: results from the MAVORIC study. Eur J Cancer 2021;156 Suppl 1:S48–S9 doi 10.1016/S0959-8049(21)00715-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim YH. What factors guide treatment selection in mycosis fungoides and Sezary syndrome? Hematology Am Soc Hematol Educ Program 2021;2021(1):303–12 doi 10.1182/hematology.2021000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenlee RT, Goodman MT, Lynch CF, Platz CE, Havener LA, Howe HL. The occurrence of rare cancers in U.S. adults, 1995–2004. Public Health Rep 2010;125(1):28–43 doi 10.1177/003335491012500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeSantis CE, Kramer JL, Jemal A. The burden of rare cancers in the United States. CA Cancer J Clin 2017;67(4):261–72 doi 10.3322/caac.21400. [DOI] [PubMed] [Google Scholar]
- 18.Iyer A, Hennessey D, O’Keefe S, Patterson J, Wang W, Wong GK, et al. Skin colonization by circulating neoplastic clones in cutaneous T-cell lymphoma. Blood 2019;134(18):1517–27 doi 10.1182/blood.2019002516. [DOI] [PubMed] [Google Scholar]
- 19.Park J, Yang J, Wenzel AT, Ramachandran A, Lee WJ, Daniels JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood 2017;130(12):1430–40 doi 10.1182/blood-2017-02-768234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet 2015;47(9):1056–60 doi 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen CP, et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet 2015;47(12):1465–70 doi 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J, Daniels J, Wartewig T, Ringbloom KG, Martinez-Escala ME, Choi S, et al. Integrated genomic analyses of cutaneous T-cell lymphomas reveal the molecular bases for disease heterogeneity. Blood 2021;138(14):1225–36 doi 10.1182/blood.2020009655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood 2015;126(4):508–19 doi 10.1182/blood-2014-11-611194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaque JP, Gomez-Lopez G, Monsalvez V, Varela I, Martinez N, Perez C, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood 2014;123(13):2034–43 doi 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- 25.Kiel MJ, Sahasrabuddhe AA, Rolland DCM, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun 2015;6:8470 doi 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet 2015;47(12):1426–34 doi 10.1038/ng.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woollard WJ, Pullabhatla V, Lorenc A, Patel VM, Butler RM, Bayega A, et al. Candidate driver genes involved in genome maintenance and DNA repair in Sezary syndrome. Blood 2016;127(26):3387–97 doi 10.1182/blood-2016-02-699843. [DOI] [PubMed] [Google Scholar]
- 28.Prasad A, Rabionet R, Espinet B, Zapata L, Puiggros A, Melero C, et al. Identification of Gene Mutations and Fusion Genes in Patients with Sezary Syndrome. J Invest Dermatol 2016;136(7):1490–9 doi 10.1016/j.jid.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Mirza AS, Horna P, Teer JK, Song J, Akabari R, Hussaini M, et al. New Insights Into the Complex Mutational Landscape of Sezary Syndrome. Front Oncol 2020;10:514 doi 10.3389/fonc.2020.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borcherding N, Voigt AP, Liu V, Link BK, Zhang W, Jabbari A. Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression. Clin Cancer Res 2019;25(10):2996–3005 doi 10.1158/1078-0432.CCR-18-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaydosik AM, Tabib T, Geskin LJ, Bayan CA, Conway JF, Lafyatis R, et al. Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors. Clin Cancer Res 2019;25(14):4443–54 doi 10.1158/1078-0432.CCR-19-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera A, Cheng A, Mimitou EP, Seffens A, George D, Bar-Natan M, et al. Multimodal single-cell analysis of cutaneous T-cell lymphoma reveals distinct subclonal tissue-dependent signatures. Blood 2021;138(16):1456–64 doi 10.1182/blood.2020009346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24(7):1036–46 doi 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 34.Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 2019;36(5):483–97 e15 doi 10.1016/j.ccell.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CL, Degasperi A, Grandi V, Amarante TD, Genomics England Research C, Mitchell TJ, et al. Spectrum of mutational signatures in T-cell lymphoma reveals a key role for UV radiation in cutaneous T-cell lymphoma. Sci Rep 2021;11(1):3962 doi 10.1038/s41598-021-83352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet 2015;47(9):1011–9 doi 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578(7793):94–101 doi 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31 doi 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10(1):33 doi 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Daniels J, Wartewig T, Ringbloom KG, Martinez-Escala ME, Choi S, et al. Integrated Genomic Analyses of Cutaneous T Cell Lymphomas Reveal the Molecular Bases for Disease Heterogeneity. Blood 2021. doi 10.1182/blood.2020009655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mundra PA, Dhomen N, Rodrigues M, Mikkelsen LH, Cassoux N, Brooks K, et al. Ultraviolet radiation drives mutations in a subset of mucosal melanomas. Nat Commun 2021;12(1):259 doi 10.1038/s41467-020-20432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171(5):1042–56 e10 doi 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trucco LD, Mundra PA, Hogan K, Garcia-Martinez P, Viros A, Mandal AK, et al. Ultraviolet radiation-induced DNA damage is prognostic for outcome in melanoma. Nat Med 2019;25(2):221–4 doi 10.1038/s41591-018-0265-6. [DOI] [PubMed] [Google Scholar]
- 44.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017;171(5):1029–41 e21 doi 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499(7457):214–8 doi 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto Y, Ishida T, Masaki A, Murase T, Yonekura K, Tashiro Y, et al. CCR4 mutations associated with superior outcome of adult T-cell leukemia/lymphoma under mogamulizumab treatment. Blood 2018;132(7):758–61 doi 10.1182/blood-2018-02-835991. [DOI] [PubMed] [Google Scholar]
- 47.Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 2021;589(7842):448–55 doi 10.1038/s41586-020-03046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12(4):R41 doi 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun 2015;6:6025 doi 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham HTT, Maurer B, Prchal-Murphy M, Grausenburger R, Grundschober E, Javaheri T, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest 2018;128(1):387–401 doi 10.1172/JCI94509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carre C, et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep 2019;26(6):1627–40 e7 doi 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566(7745):496–502 doi 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012;22(4):547–60 doi 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin Cancer Res 2018;24(11):2482–90 doi 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Yao Y, Zhang S, Liu Y, Guo H, Ahmed M, et al. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci Transl Med 2019;11(491) doi 10.1126/scitranslmed.aau1167. [DOI] [PubMed] [Google Scholar]
- 56.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019;178(4):835–49 e21 doi 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2020;11(1):6322 doi 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun 2020;11(1):496 doi 10.1038/s41467-019-14256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 2020;367(6485):1468–73 doi 10.1126/science.aay0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamrouni A, Fogh H, Zak Z, Odum N, Gniadecki R. Clonotypic Diversity of the T-cell Receptor Corroborates the Immature Precursor Origin of Cutaneous T-cell Lymphoma. Clin Cancer Res 2019;25(10):3104–14 doi 10.1158/1078-0432.CCR-18-4099. [DOI] [PubMed] [Google Scholar]
- 61.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 2013;3(8) doi 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goetzman ES, Prochownik EV. The Role for Myc in Coordinating Glycolysis, Oxidative Phosphorylation, Glutaminolysis, and Fatty Acid Metabolism in Normal and Neoplastic Tissues. Front Endocrinol (Lausanne) 2018;9:129 doi 10.3389/fendo.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev 2012;249(1):27–42 doi 10.1111/.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A 2019;116(18):9020–9 doi 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuen GJ, Demissie E, Pillai S. B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2016;2(12):747–57 doi 10.1016/j.trecan.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee S, Zhao L, Rojas C, Bateman NW, Yao H, Lara OD, et al. Molecular Analysis of Clinically Defined Subsets of High-Grade Serous Ovarian Cancer. Cell Rep 2020;31(2):107502 doi 10.1016/j.celrep.2020.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5(4):557–72 doi 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 68.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28(11):1747–56 doi 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst 2018;6(3):271–81 e7 doi 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173(2):321–37 e10 doi 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16(5):284–7 doi 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32(4):381–6 doi 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14(10):979–82 doi 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 2019;9(1):5233 doi 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015;162(1):184–97 doi 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15(4):1484–506 doi 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data, including WES, scRNA-seq and scVDJ-seq, have been submitted to NCBI BioProject database PRJNA754592. The Leukemic/SS WES data analyzed in this study were obtained from the NCBI BioProject database PRJNA285408.


