Abstract
Introduction:
Acute leukemia results from a series of mutational events that alter cell growth and proliferation. Mutations result in protein changes that orchestrate growth alterations characteristic of leukemia. Proteomics is a methodology appropriate for study of protein changes found in leukemia. The high-throughput reverse phase protein array (RPPA) technology is particularly well-suited for the assessment of protein changes in samples derived from clinical trials.
Areas covered:
This review discusses the technical, methodological, and analytical issues related to the successful development of acute leukemia RPPAs.
Expert commentary:
To obtain representative protein sample lysates, samples should be prepared from freshly collected blood or bone marrow material. Variables such as sample shipment, transit time, and holding temperature only have minimal effects on protein expression. CellSave preservation tubes are preferred for cells collected after exposure to chemotherapy, and incorporation of standardized guidelines for antibody validation is recommended. A more systematic biological approach to analyze protein expression is desired, searching for recurrent patterns of protein expression that allow classification of patients into risk groups, or groups of patients that may be treated similarly. Comparing RPPA protein analysis between cell lines and primary samples shows that cell lines are not representative of patient proteomic patterns.
Keywords: acute leukemia, RPPA, proteomics, sample handling, network-based approach, classification, methodology
1. Introduction
Acute leukemia forms a group of malignant diseases characterized by a block in differentiation and an uncontrolled clonal proliferation of abnormal hematopoietic progenitor cells (“blasts”) in the bone marrow. The accumulation of blasts interferes with the production of mature blood cells, causing neutropenia, thrombocytopenia and anemia, which can be rapidly fatal if left untreated. Although acute leukemia results from a series of mutational events that take place during the complex process of hematopoiesis, the exact etiology is unknown.
In addition to genetic mutations, there are many other events that influence tumor development; alterations in RNA expression and composition, post-translation protein modifications, expression and translational control of small RNA (usually non-coding RNA; e.g., microRNA, small interfering RNA), [1] environmental effects from mesenchymal stromal cells, [2] and both local and distant chemokine and cytokine production, [3] all of which affect how emerging leukemia cells develop and behave. The combined contributions from these effectors can arise from outside the leukemic cell, and these factors are independent of genetic events that drive the leukemic cell (e.g., microenvironment alternations). A means to understand the integrated effect of internal genetic events and external biology influences on the leukemic cell, therefore, is required to optimally understand how to perturb malignant cell physiology. As proteins are the central effectors that drive cell function and are the combined consequences of genetic, epigenetic and bone marrow microenvironment effects, measuring protein expression and activation could provide a summation of effects modifying upstream mutations.
One frequently used high-throughput proteomic methodology in acute leukemia research is reverse phase protein array (RPPA) technology [4,5]. As blood is easy to collect and leukemic cells are easy to isolate, acute leukemias are well suited to RPPA analysis. However, in order to obtain representative leukemia cells, correct cell purification and sample handling (i.e., processes occurring between sample collection and assay processing) must be performed to achieve accurate and representative results. In addition, while most proteomic studies have focused on individual proteins, we believe that a more systematic biological approach simultaneously analyzing hundreds of proteins is superior, as it is the net consequence of the combined influences of all proteins that determines net cell effects. This review discusses the technical and methodological issues related to the successful development of acute leukemia RPPAs, as well as solutions to investigative and analytic challenges. We also discuss potential clinical utility.
2. RPPA technology
2.1. Methodology
RPPA is an antibody-based proteomic approach [6-9]. The name “reverse phase” indicates that the antigens (protein lysates) are printed on the array and subsequently probed with a primary and a secondary antibody specific for target proteins (with or without post-translational modifications) in the solid phase lysates. This is the “reverse” of traditional “forward” immunoassays, where specific antibodies are immobilized on the solid to capture the antigen of interest.
Proteins are extracted from patient tissues or cultured cells followed by lysate preparation, which denatures the proteins. Lysates are diluted to define antigen-antibody reactions in a linear range for accurate quantification. Serially diluted cellular proteins are arrayed on nitrocellulose-coated slides (> 1000 samples can be printed on a single slide) and probed with validated antibodies that recognize signaling molecules in their functional state (e.g., total protein, post-translational modifications such as phosphorylation, cleavage, etc.). Signals are captured by a tyramide dye and a 3,3′-diaminobenzidine colorimetric reaction or by infrared fluorescence labeling. Data are collected and quantitative analysis is performed using custom software. Features of these software include automated spot identification, background correction (i.e., spatial normalization [10], topographical normalization[11]), serial dilution-signal intensity curve construction (e.g., “SuperCurve” [12,13], “modified SuperCurve” [14], “NormaCurve” [15], “serial dilution curve”[16]), and concentration determination [17]. The values derived from the slope and intercept are expressed relative to standard control cell lysates or control peptides on the array. These values indicate the levels of protein expression for either total protein or modifications (e.g., phosphorylation, cleavage, etc.). If samples are printed on different slides, replicate-based normalization [7] is used to align samples from two different slides using replicate samples printed on both slides.
2.2. Advantages of the RPPA in acute leukemia research
As RPPA is a high-throughput methodology it has several advantages for analysis of clinical trial samples. First, it can measure the expression levels of thousands of samples simultaneously, in contrast to methodologies in which samples are analyzed individually (e.g., mass spectrometry). This enables assessment of protein expression from multiple patients with varying disease characteristics and outcomes, making it suitable approach for clinical applications [18]. In addition, RPPA is a cost-effective and sample sparing, as it requires only a minimal amount of protein sample (approximately 3x105 cells for 400 proteins) [18,19]. It has a high sensitivity (pico-to-femtogram range), excellent precision, a high inter- and intra-slide reproducibility, precision of sample spotting, high throughput, and excellent reliability [20-24]. As the technology is based on an approach where samples are first printed on a large number of slides that are subsequently analyzed for protein expression using validated antibodies, it is also possible to generate and store additional slides (sample arrays), so that further analysis can be performed when new affinity reagents become available or when new hypotheses need to be tested using the same samples. While RPPA requires the availability of highly-specific antibodies, and therefore cannot be used as a de novo protein discovery platform, it can be used for screening of patients for treatments based on protein expression known to be deregulated in leukemia or other cancers types, with specific interest based on a biological rational, or proteins (in)directly targeted by drug compounds. Table 1 highlights the advantages and disadvantages of the RPPA methodology in comparison to mass spectrometry (MS), another frequently used high-throughput proteomic methodology.
Table 1.
Advantages and disadvantages of mass spectrometry and reverse phase protein arrays.
| MS | RPPA | |
|---|---|---|
| Number of proteins | Thousands, limited by sequence coverage | Hundreds, limited by number of validated antibodies |
| Amount of protein | Milligrams | Nanograms |
| Processing workflow | Single sample | > 1000 samples |
| De novo protein discovery platform | Yes | No |
| Quantitative analysis | Yes; quantitation based on size of MS peaks | No; relative expression between samples |
| Able to assess post-translational modifications (PTM) | Yes | Yes |
3. Methodological challenges of RPPA in acute leukemia
3.1. Protein sample quality
3.1.1. Ficoll separation to separate the mononuclear leukemic cell fraction
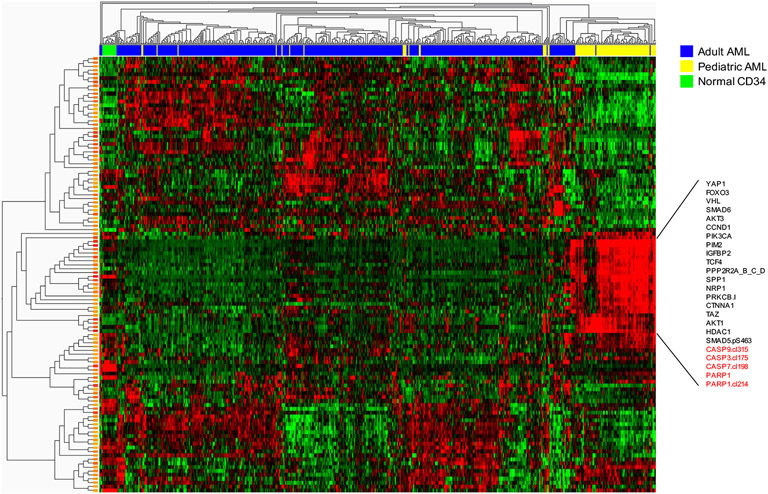
To acquire a representative protein lysate of the leukemic blasts, Ficoll separation is applied to remove dead cells, neutrophils and red blood cells from acute leukemic samples, either from blood or from frozen specimens where cell death may have occurred. However, in one of the first RPPAs that our group generated, failure to re-ficoll cryopreserved cells resulted with a very strong signal of ongoing apoptosis (high expression of cleaved caspases and cleaved PARP, Figure 1), presumably induced by the freeze-thaw process. This demonstrates the need to remove dead and dying cells from cryopreserved samples prior to making the protein preparation.
Figure 1. Heatmap showing apoptotic changes induced by freeze-thaw cycle.
Pediatric samples (annotation bar, yellow) were not re-ficolled after freeze-thaw. Notable are a block of proteins with high expression of cleavage caspase and PARP1. Scale ranging from −1 (green) to +1 (red).
3.1.2. Freshly prepared vs. cryopreserved acute leukemic samples
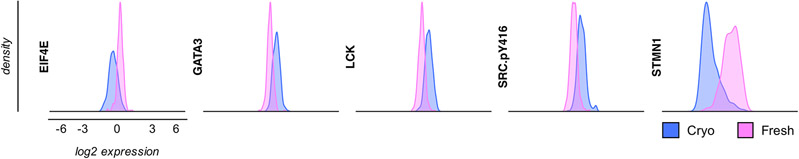
Another issue that appears when samples are not prepared from fresh material on the day of collection, but from cells that are first cryopreserved before protein lysates are made, is that several other signaling proteins can be abnormally highly expressed in the cells. Our AML719 array included over half of the samples that were prepared from cells that were first cryopreserved, and the remaining protein lysate samples from cells that were immediately processed after collection of the cells (“fresh”) and then cryopreserved for storage. Comparison of protein expression from fresh and cryopreserved samples resulted in striking differences, with changes in both directions (i.e., either higher or lower expressed) (Figure 2). At the individual protein level for these acute myeloid leukemia (AML) samples, 86% (197/228) of the proteins showed statistically significant differences in expression between lysates made from fresh cells and lysates made from cryopreserved samples [25]. Lysates were all stored frozen at −80°C until use on the RPPA. Investigation of the expression differences at the protein pathway level resulted in expression patterns in 23 of 31 protein functional groups (i.e., a group of functionally related protein based on prior literature knowledge) that were only seen in cryopreserved samples [25], but not in fresh samples. Since fresh samples were not influenced by the potential effects of cryopreservation or thawing, we assert that they represent a more accurate picture of the AML biology compared to lysates made from cryopreserved samples [26]. Similar results were observed in adult acute lymphoblastic leukemia (ALL), but not chronic lymphoblastic leukemia (CLL) (manuscript under review). Similar results were also not seen in pediatric T-cell ALL (T-ALL), suggesting that these effects may depend on the source and method of freezing. Aasebø et al. investigated changes in protein abundance in AML samples using MS. Although, the majority of proteins did not change, alterations were mainly found in mitochondrial proteins, likely secondary to oxidative stress, as well as in apoptosis-signaling proteins, cell surface interactors, ERK/MAPK targets and proteins involved in thrombin signaling [27]. Likewise, a study in Wharton’s Jelly Stem Cells comparing proteomic MS-analysis between samples prepared from fresh and frozen cells showed quantitative and qualitative changes [28]. There is also previously published data demonstrating that refrigeration enhances the stability of the transcriptome [26]. To prevent potential confounding effects, arrays in AML and ALL are now restricted to protein lysates that are prepared immediately after collected, that can then be cryopreserved and used later.
Figure 2. Selection of significantly affected proteins and post-translational protein modifications by cryopreservation in adult AML.
Expression changes in both directions (i.e., either increased expression after cryopreservation vs. fresh (GATA3, LCK, SRC.pY416) or decreased expression (EIF4E, STMN1).
3.1.3. Mycoplasma contamination affects protein expression in acute leukemia cells
Mycoplasma infection is a major challenge in cell culturing. Despite significant improvements in diagnostic and therapeutic possibilities to detect and eliminate mycoplasma contamination, it is still present in 5-30% of cell lines [29,30]. Miller et al. showed that mycoplasma infection alters gene expression of hundreds of genes in cultured human cells and Gedye at al reported poorly reproducible results from cell line aliquots that were later found to be infected with Mycoplasma hyorhinis [31,32]. As limited knowledge is available on the effects of mycoplasma on protein expression, we determined protein expression levels in mycoplasma-infected and non-infected leukemia cell lines along with post treatment mycoplasma-free cell lines. We utilized the RPPA methodology for 235 antibodies on 52 leukemic cell line samples; mycoplasma infected (n=16), uninfected post-treatment (n=19), and never-infected (n=17) [33]. Overall, unsupervised hierarchical clustering showed that protein profiles remained relatively stable. However, paired comparisons at the individual protein level identified significant changes in expression. Gene ontology analysis showed that proteins significantly altered in the mycoplasma-infected leukemia cells were enriched for apoptosis and auto-phosphorylation. While it may be expected that mycoplasma infection itself would affect protein expression, our results also showed that once mycoplasma was eliminated, their metabolism remains altered and was only partially restored. We conclude that mycoplasma infection of acute leukemic cell lines alters protein expression levels, and that mycoplasma treatment might only partially restore normal metabolism. This suggests that mycoplasma testing should be performed at regular intervals and that cells having a mycoplasma signal should be replaced with mycoplasma negative cells purchased from commercial cell line banks to avoid having altered protein expression that could confound experimental results.
3.2. Sample handling of acute leukemia samples
Because RPPA can assess protein abundance and activation states in large numbers of samples using small amounts of material, RPPA ideal for use in multi-institution clinical trials. However, in order to obtain those samples, samples often have to be shipped from one institution where cells were collected to another institution where protein lysates are prepared and analyzed. As there is concern that preanalytical handling variables can affect the integrity of protein concentrations, we sought to define the variability in preanalytical handling variables to determine how instability in protein expression could affect protein assessment [34].
3.2.1. Acute leukemia sample shipment between institutions
The effect of shipment on protein expression was evaluated in a limited set (n=7) of pediatric acute leukemia patients. Samples were split and then either held and processed at site of collection or shipped via overnight courier to another site where they were processed. To take the delay of shipment into account, the samples that were held were processed at the same time as those shipped. Median protein expression levels were compared between the held and the shipped samples for 17 protein targets. Only TP53-pSer15 had statistically lower expression after shipment, a post-translational modification that may have been more susceptible to shipment effects. Although numbers are small, the constant temperature likely protected the samples from large variations in protein expression, suggesting that shipment of samples under those conditions is not likely to adversely affect protein expression.
3.2.2. Temperature (4°C vs. room temperature) during sample storage does not adversely affect protein expression
A second pre-analytical variable that can potentially affect protein expression is temperature. Similar to the previous analysis, leukemia samples were collected, split into equal parts, and were either 1) held at collection site at ambient temperature or 2) shipped to another site without ice packs, or 3) held refrigerated at 4°C or 4) shipped with 2-3 ice packs to maintain temperature at approximately 4°C. Median protein concentrations for proteins at 4°C were compared with samples at ambient temperature. None of the 17 protein concentration distributions were statistically different between 4°C and ambient temperature, suggesting that temperature has only marginal effects on protein expression in acute leukemia for short transit times.
3.2.3. Transit time has minimal effect on protein expression
Third, transit time (i.e., time from sample collection to the start of sample processing) was assessed. Transit time was recorded for both held and shipped leukemia samples (range 12-96hr). Between samples processed immediately (< 4hr) and those processed after either 24-48hr or 48-72hr, no statistically significant differences were observed. Two proteins, PI3K (p85) and AKT1, showed significant differences between protein concentrations after 24-48hr vs. 48-72hr, with lower expression at 48-72hr in both cases. This indicates that, for most proteins, transit time (≤ 72 hours) has minimal effect on protein expression measured by RPPA [34].
3.2.4. CellSave (CS) tubes better preserve protein expression after chemotherapy
A final pre-analytical variable that was assessed by our group was collection tube type. Samples were collected in either heparin tubes (NaHeparin or LiHeparin), or CS preservative tubes (Menarini Silicon Biosystems). In acute leukemia samples (n=28) that were collected prior to the start of systemic chemotherapy, no significant differences in protein expression levels were observed between heparin and CS tubes. This suggests that there are no significant differences in CS and heparin tube type sample collection in pre-chemotherapy samples. When we repeated this experiment in samples collected 6-10hr and 24hr after the start of treatment, 10 and 14 of the 18 analyzed proteins, respectively, were significantly different between heparin and CS collection tubes. However, there was little difference in protein expression when pre-treatment samples were compared to post-treatment samples collected in CS tubes. As the CS tubes fix proteins at the time of sample collection, we hypothesized that CS tubes would better preserve protein expression after chemotherapy, preventing the continued effects of chemotherapies that could potentially change protein expression during shipment. Our data suggests that, if CS tubes are used, there are minimal changes in protein expression of leukemia cells, even in post-chemotherapy samples.
3.3. Antibody validation
Given the high dependency of RPPA on antibodies, we also reviewed how antibodies are currently validated. A review of the existing literature shows that antibodies often do not detect what they were supposed to detect and that they did not function robustly across different sample types, working in some applications but not others [35-39]. Some antibodies were not stable over time and did not always yield reproducible results between batches. There are several approaches to validate antibodies, varying from genetic strategies that knock out or overexpress the protein of interest, to demonstrating correlation with non-antibody-based methods [40-42]. Up till now, the validation procedure for RPPA typically includes the following steps. Western blot (WB) is performed using samples from a panel of cell lines along with a molecular weight calibration marker. Only those antibodies that demonstrate a single band at the correct molecular weight in the known positive controls (or other cell lines), and which do not show a signal in the knockout lines, are considered suitable for further validation. Then, quantification of signal strength is determined for later correlation with RPPA cell line control arrays. Pearson correlation coefficient between the cell lines present on both the RPPA and the WB are determined. Based on the strength of the correlation, antibodies are classified as > 0.7 (valid), 0.5–0.7 (use with caution), and < 0.5 (not reliable). For WB, antibodies that are “not usable” are those with multiple non-specific bands. “Use with caution” is intended for antibodies with one to two non-specific bands on WB and where the non-specific band is weak relative to the expected band. “Valid” or “usable” is reserved for those without non-specific bands that have a band at expected size relative to size control. The scoring could be extended by adding information about the negative control, the inducible control, or the knockdown control.
As validation of individual antibodies requires time and money, we proposed to accurately document the results once validation of an antibody has been done, and to make this validation available to other researchers. This should include the basic information (e.g., name, target, manufacturer, etc.), as well as standardized post-translational modification-convention, usage indicators (e.g., how was the antibody validated, for which sample type and application, etc.), and use of a rating metric resulting in final confidence scores (e.g., how specific was the antibody, etc.). The development of a database that collected, collated, and housed all of this information obtained from multiple laboratories would have significant utility to all doing RPPA work. However, this would require significant effort and would require significant financial support to establish and maintain.
3.4. Protein expression patterns in leukemic cell lines
3.4.1. Leukemic cell lines do not mimic acute leukemia patient samples
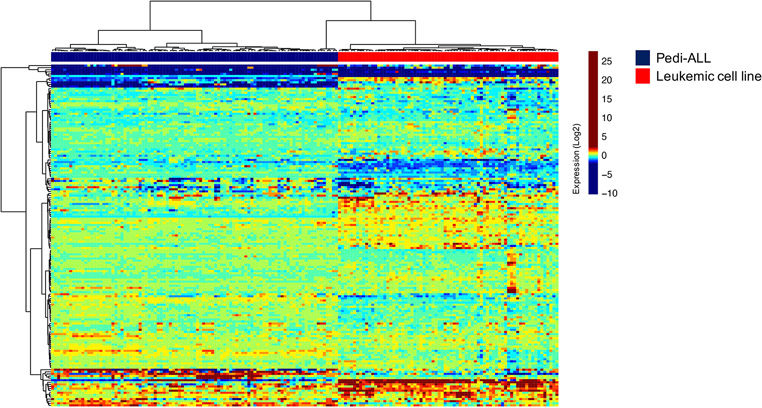
Leukemic cell lines are frequently used to investigate the pathobiology of acute leukemia, but immortalization and cryopreservation of those cells likely alter the biology from their leukemic patient cell of origin. To determine to which extent cell lines express protein expression patterns expressed in acute leukemia samples, we generated an RPPA with 95 leukemic cell line samples. When we compared cell lines against adult AML, pediatric AML and pediatric ALL patient samples, we noted that leukemia cell lines clearly had distinctive profiles that clustered separately from patient proteomic profiles; cell lines clustered completely separate from patient samples (Figure 3). This shows that inferences from cell line research should be interpreted with caution when protein analysis is done using RPPA.
Figure 3.
Heatmap showing distinct protein expression profiles between primary pediatric ALL samples (blue) and leukemic cell line samples (red).
4. Investigative challenges of RPPA protein analysis
4.1. Protein expression measurement of individual proteins in acute leukemia
Questions that can be answered with the use of proteomic data is whether individual proteins convey prognostic information and are important for leukemia initiation and progression or resistance/ relapse. For instance, protein tyrosine phosphatase 4A3 (PTP4A3 or PRL-3) was found to play a critical role in T-ALL initiation and progression by promoting leukemia cell migration [43]. Others investigated the effect of selective inhibition of choline kinase alpha 1 (ChoKa) in T-ALL, and used RPPA to evaluate the phosphorylation changes upon treatment. They found that targeting ChoKα was associated with antiproliferative activity, suggesting that targeting ChoKa may be an interesting option for treating T-ALL [44]. Irwin et al investigated additional targets for Ph+ ALL. Assessment by RPPA in 129 ALL patients showed that activity of ErbB2 was elevated in 56% of Ph+ ALL, as compared to just 5% of Ph- ALL. Inhibition of ErbB2 kinase activity with canertinib resulted in a dose-dependent decrease in the phosphorylation of an ErbB kinase signaling target P70S6-pThr389 kinase, as well as an increase of the pro-apoptotic protein BIM and activation of CASP3 associated with consequent cell death in two Ph+ ALL cell lines [45]. In a multi-resistant leukemia pediatric ALL cell line, obtained by continuous exposure to doxorubicin, RPPA probed with a panel of 90 phospho-antibodies to ERK, PCK and Akt pathways, found that whereas total AKT1 protein was higher in parental non-resistant cells, the activated isoform, AKT1-pS473, was high in the multidrug resistant cells [46]. In B-ALL, Bortolozzi et al. found that cyclin D1 levels measured by RPPA were higher in patients undergoing relapse. Based on this finding, they evaluated the effect of dual inhibition of CDK4/CDK6, part of the cyclin D1-CDK4/CDK6 complex, using ribociclib. Ribociclib induced growth inhibition of B-ALL cell lines, accompanied by strong cell cycle arrest in G1 phase, supporting the concept that pharmacologic inhibition of CDK4/ CDK6 may represent a useful therapeutic strategy to control cell proliferation in B-ALL [47].
In AML, RPPA identified mTOR pathway modulation after PIM kinase inhibition. Modulation included inhibition of protein phosphorylation of mTOR-pSer2448, P70S6K-pThr389, RPS6-pSer235_236, and 4EBP1-pSer65, which subsequently led to a reduction in protein synthesis that correlated with cell size reduction and growth inhibition [48]. Konopleva et al. explored preclinical and clinical anti-AML activity of the oral AKT inhibitor, MK-2206. They conducted a phase II trial in adults that required second salvage therapy for relapsed/ refractory AML and target inhibition was assessed with RPPA. However, when MK-2206 was used as monotherapy, they concluded that this inhibitor has insufficient clinical antileukemia activity in AML [49]. In addition, their group utilized RPPA to measure changes in multiple proteins induced by stroma in leukemic cells. The mTOR kinase inhibitor, PP242, was used to disrupt leukemia/ stroma interaction, resulting in up-regulating of multiple survival signaling pathways in primary AML cells cocultured with stroma, including PI3K/AKT and mTOR. PP242 effectively induced apoptosis in primary samples cultured with or without stroma [50].
Previously, our group has performed RPPA on 511 adult AML patient samples, and identified several protein, including Forkhead O Transcription Factor 3A (FOXO3A) [[51], Bromodomain Containing 4 (BRD4) (ASH 2017 #3794), Absent, Small, Or Homeotic Discs-like Protein (ASH2L) [[52], Tripartite Motif Containing 62 (TRIM62) [[53], Friend Leukemia Virus Integration 1 (FLI1) [[54], protein phosphatase 2A (PP2A) regulatory subunit B55α [[55], and phosphorylation of (GSKα/β) [56] as prognostic factors associated with survival in adult AML. In our pediatric AML cohort, approximately 14% of the 296 evaluated proteins were individually prognostic for OS, including both previously reported as well as novel prognostic proteins.
4.2. Analysis of protein expression in a network-based approach
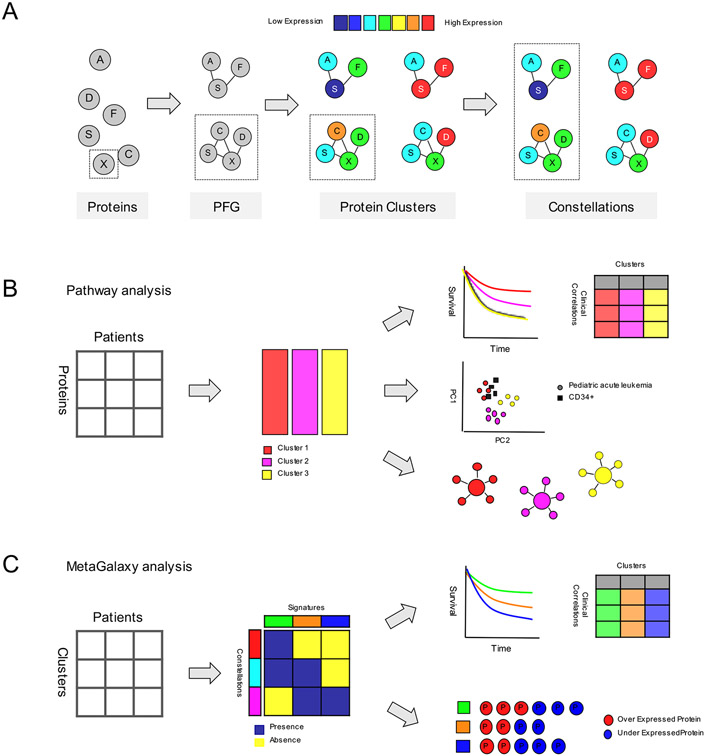
As it is the net consequence of the combined influences of all the proteins that determines the net effect on the cell, rather than of an individual protein, the focus of analyzing individual proteins has developed to a more systematic biological approach, capable of simultaneously looking at hundreds of proteins. This raises the question as to whether proteins are expressed in recurrent patterns of protein expression (i.e., expression signatures) in acute leukemia. Our group developed the novel “MetaGalaxy” approach (Figure 4), which takes into account previously known interactions between proteins, as well as proteins with functional similarities, and starts with analyzing proteins in the context of a “Protein Functional Group” (PFG). A PFG is formed by proteins that are known to be functionally related from the literature, or show a strong correlation within the RPPA data set. This method takes previously known relationships into account which traditional unsupervised hierarchical clustering does not. Within each PFG, a mathematical algorithm (Progeny Clustering [57]) is applied to identify an optimal number of “Protein Clusters”; a subset of cases with similar (correlated) expression of core protein functional group components. To obtain a cohesive understanding of the higher order structures, protein clusters from each PFG are subsequently organized in a binary matrix for each patient (i.e., “1” if a patient was a member of a given protein cluster, “0” if a patient was not a member). Using the Block Clustering algorithm, this enables recognition of the existence of recurrences between protein clusters. Those correlations define a “Protein Constellation” (CON); a group of protein clusters from protein functional groups that are strongly correlated with each other. The unique and selected combinations of CONs than enable characterization of an acute leukemia patient subpopulation into “Protein Expression Signature” (SIG).
Figure 4. Computation work flow of the MetaGalaxy analysis.
(A) Scheme showing the overall computational analysis, starting with individual proteins that are allocated into PFGs. Protein clusters are identified within a PFG and correlated to form CONs. (B) Pathway analysis: protein clusters were identified for each PFG and correlations between outcome, clinical and laboratory variables were determined. Principal component analysis was performed to visualize the distribution between the protein clusters relative to the normal CD34+ samples. Networks were generated to reveal different activation states, and to show the relative protein expression levels in a given protein cluster. (C) “Meta-galaxy” analysis: co-clustering algorithm was performed using a binary matrix indicating protein cluster membership for each patient. This identified the existence of CONs (horizontally). Patients that express similar combinations of CONs were defined as a SIG (vertically). Correlation between SIGs and outcome, clinical and laboratory variables were determined. Proteins that were significantly over or under expressed compared to CD34+ cells were identified for each CON and SIG. Figure derived from Hoff et al. Expert Review of Proteomics, 2018.
We hypothesize that the genetic complexity of the leukemic cells, ultimately results in a constrained number of protein patterns pathway utilizations, needed for the cell to become leukemic (similar to the “Hallmarks of Cancer”) [58,59]. The “Hallmarks of Cancer” are a conceptual framework of 6 (and later 10) biological capabilities acquired during the multistep development of human tumors. Though all malignancies share those same hallmarks of cancer, heterogeneities between patients complicate patients’ response to therapy and subject them to varied outcomes. If this idea is correct, then each combination of genetic events, regardless of their direct downstream consequence, must somehow meet each of the hallmarks. We hypothesize that those combinations of CONs together represent the quantitative hallmarks in acute leukemia, and can significantly accelerate the identification and development of new therapeutic targets.
4.3. Existence of SIGs across subsets of acute leukemia
Over the years the MetaGalaxy methodology was applied to several data sets, including adult and pediatric patient samples of AML, ALL, acute promyelocytic leukemia (APL), myelodysplastic syndrome, and CLL [25,60-62]. In all of these, we have seen that leukemia could indeed be defined by a limited number of recurrent patterns, typically 9-15 SIGs. While CONs were the building blocks that were expressed in unique combinations, each defining its own SIG, we also observed that several CONs existed that were shared among the SIGs, suggesting the existence of expression patterns that share mutual use of some proteins between the SIGs. Notably, this was not only observed within one disease, but also between subtypes of diseases. For instance, when protein expression patterns in pediatric T-ALL are compared to pediatric pre-B-cell ALL, or between adult ALL and adult AML or pediatric T-ALL and pediatric AML, we found that although the SIGs were largely disease-specific, overlapping CONs existed that were expressed in both diseases (manuscript in preparation). This confirms that AML, B-ALL and T-ALL are the result of different underlying biology, but that there are shared protein dysregulations in some pathways that are needed for leukemia transformation.
5. Conclusion
RPPA is a valuable tool to simultaneously quantitate protein expression in large sets of patient samples, making it ideal for use in multi-institution clinical trials. Here, we conclude that thorough assessment sample processing and handling, and universally accepted guidelines for determining the validity of antibodies, are required to ensure reliable and reproducible RPPA results. Because the combined influences of all the proteins determine the net effect on the leukemia cell, we provide support for a more systematic approach (“MetaGalaxy” approach) that takes previously known interactions between proteins into account. We found that acute leukemia can be classified into subgroups of patients based on recurrent protein expression patterns, of which some are independently prognostic for outcome and can identify patients that benefit from treatment with a proteasome inhibitor when added to standard ADE therapy (cytarabine, daunorubicin, etoposide). These results suggest the potential value for proteomics to augment the current classification systems, and to advance (individualized) therapy selection (e.g., bortezomib addition) in acute leukemia. Future studies are needed to validate our hypotheses.
6. Expert commentary
6.1. Protein expression profiles could enhance prognostication and risk stratification
Despite improvements in survival, acute leukemia remains a deadly disease with a significant mortality. In addition, treatment-related morbidity is challenging. To improve therapeutic outcome and to reduce side-effects, identification of low- and high-risk patients is crucial. Risk stratification has traditionally been based on clinical features like age, performance status and prior myelodysplasia in adults, complemented by cytogenetic abnormalities, and more recently by incorporation of prognostic information related to specific gene mutations according to the World Health Organization classification system [63]. Incorporating genetic mutation analysis into risk stratification has yielded significant prognostic information and enabled stratification of adults into “favorable”, “intermediate” and “unfavorable” risk groups. However, about half of the AML patients fall in the intermediate risk group and prognosis in this group varies widely.
In each of the analyzed leukemia data sets to which we applied the MetaGalaxy approach, SIGs were significantly associated with demographic and disease features. To simplify the usage of the protein prognostic information from the patient SIGs, they were reduced to three groups: “favorable”, “intermediate”, “unfavorable”, similar to cytogenetics. When survival analysis was restricted to existing low- or high-risk AML groups from the latest treatment protocols, SIGs retained their statistically significant prognostic grouping. Similarly, in our APL study, two SIGs were identified, one of which contained all (n = 4/4) relapse cases, suggesting that high-risk APL could be identified and stratified from low-risk APL patients based on proteomic profiles. This suggests that proteomics, in combination with cytogenetics, could enhance risk stratification by identifying high-risk patients in low-risk groups such as APL.
In order to utilize this in the clinical practice, a protein detection kit should be developed in order to facilitate the recognition of protein expression patterns present in the cell prior to chemotherapy exposure. Techniques that rapidly provide information about relative protein expressions are enzyme-linked immunosorbent assay, immunohistochemistry or a forward-phase protein array. Together with determination of cytogenetics, mutation analysis and other risk factors, these protein profiles should be used to aid prognostication.
6.2. Proteomics could help predict which leukemia patients will respond to a specific therapy
Drug initiatives that target mutations have been successful for specifically targeting FLT3[64] and mutant forms of IDH1 and 2 [65] as well as chromosomal aberration BCR-ABL1 in ALL[66]. Initiatives to target several other commonly mutated genes (NPM1, DNMT3a, TET2, ASXL1, RUNX1, etc.) in AML, as well as KMT2A (MLL) in AML and ALL, however, have been unsuccessful so far [67,68]. This makes the translation of the identification of the underlying mutations into improved clinical outcome a largely unfulfilled promise.
A potential application of proteomics may be to guide therapy selection. Given that the majority of drugs currently being tested in clinical trials target proteins, we hypothesize that abnormal protein expression could identify a critical dependency in leukemia and could pinpoint a targeted therapy across leukemia subgroups. High protein expression that results in chemotherapy resistance could result in the identification of a patient targeted for protein inhibition, while proteins with lower expression could act as target for replacement or re-activation. This would result in rational selection of therapeutic agents, either as single agent or in combination, rather than “randomly” selecting drugs in the absence of clinical features previously identified as conferring sensitivity/ resistance. Hypothetical protein targets can be identified from either a CON (and can thus also be a target across other subgroups) or a SIG. Because we are identifying aberrant protein expression patterns and not at genetic events, there is a limited number of protein expression patterns that tell us in which direction we should target.
An example is the BCL-2 inhibitor venetoclax (ABT-199), which is a promising agent in AML. Venetoclax acts as a BH3-mimetic that selectively binds and inhibits the anti-apoptotic protein BCL2, causing a subsequent release of pro-apoptotic proteins and activation of BAX/BAK, cytochrome c release and induction of apoptosis [69]. In combination with other agents, venetoclax has been shown to be very effective. Although the rationale for the combination is not completely understood, the association between azacytidine and venetoclax is effective for MDS and some AML in the elderly [70-72]. Other studies have shown that venetoclax in combination with FLT3 inhibition works synergistically in FLT3-ITD mutated patients [73]. Similarly, venetoclax is effective in combination with IDH1/2 inhibitors in IDH mutated patients. In vitro studies found that venetoclax apoptotic cell death depends on protein expression of members of the BCL-2 family; high BCL2 expression was correlated with drug efficacy, while concomitant expression of MCL1 has been shown to limit it efficacy and cause resistance [73]. This suggests that if genetics (e.g., FLT3-ITD, IDH1/2) are combined with determination of protein expression (e.g., BCL2, MCL1), this can potentially result in a selection of acute leukemia patients that are most sensitive to specific agents.
Another example are the menin-inhibitors in AML [74]. Menin is a tumor suppressor protein and critical for leukemogenesis in subsets driven by rearrangement of the lysine methyltransferase 2A (KMT2A) gene, previously known as mixed-lineage leukemia (MLL). AML and ALL with the KMT2A rearrangement are associated with high rates of resistance and relapse following conventional treatments [75-78]. However, they are predicted to respond to menin inhibition, with early clinical data validating this proof-of-concept. Recent studies have identified that mutated NPM1+ AML, most common mutation in AML, are also susceptible to menin inhibition [79,80]. We found that when pediatric AML patients were clustered based on recurrences in protein expression profiles, there is a protein expression pattern highly enriched for patients harboring the KMT2A-rearrangement (publication in press). Given that the remaining patients (without the KMT2A-rearrangement) in this protein signature express a similar protein expression profile, this may point toward the existence of MLL-like patients based on protein expression, similar to the existence of Ph+ like ALL. It might worth testing whether this proposed “KMT2A-like” group is also sensitive to menin-inhibitors, and if so, classification based on proteomics might aid in selection of these patients and guide therapy.
Thus, proteomic profiling, in combination with molecular classification, provides a means to potentially match the targeted therapies to the appropriate patient. Protein identification could facilitate the identification of effective combinations of chemotherapies to improve clinical outcome.
The Children’s Oncology Group performed a phase 3 randomized clinical trial (AAML1031) that evaluated the effect of the proteasome inhibitor bortezomib in combination with standard ADE therapy (ADEB) vs. ADE therapy alone. Interim analysis showed that 3-year OS and EFS did not improve with the addition of bortezomib and the study was closed to enrollment. We hypothesized that while the bortezomib failed to improve outcome in pediatric AML overall, there would be subgroups of patients, identified by protein expression profiles, that benefitted from bortezomib addition. We observed that some SIGs and individual protein expression patterns did identify patients that benefitted from the addition of bortezomib [81]. These results suggest a potential value for proteomics to advance (individualized) therapy selection if patients can be classified at either diagnosis or an early point in their treatment.
7. Five-year view
Many genetic “drivers” have been implicated in acute leukemia disease pathology and risk stratification. However, risk stratification is imperfect with outcome within risk groups widely varying, and only a minority of these drivers have been exploited by targeted therapeutic interventions. We believe that, if sample processing and handing are performed thoroughly, RPPA is a powerful tool to study proteomics in samples from large (multi-institutional) clinical trials. Particularly, analysis of proteins in the context of other proteins could unravel critical dependencies in leukemia, that could aid classification and could pinpoint a targeted therapy across leukemia subgroups in the near future.
Key issues.
Reverse phase protein array proteomic technology is particularly well-suited for the assessment of protein expression within the context of a clinical trial;
Variables such as sample shipment, transit time (≤ 72 hours), and holding temperature have minimal effects on protein expression in acute leukemia samples;
Incorporation of standardized guidelines for antibody validation used for RPPA is recommended;
Recurrent patterns of protein expression allow for classification of leukemia patients into risk groups independent of molecular classification. RPPA also identifies subgroups of patients that could be treated similarly;
Protein expression in acute leukemic cell lines is not representative of patient leukemia cell protein expression.
List of References
- [1].Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2016. 8(2), 3666–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kornblau SM, Ruvolo PP, Wang RY, et al. Distinct protein signatures of acute myeloid leukemia bone marrow-derived stromal cells are prognostic for patient survival. Haematologica. 2018. 103(5), 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Riether C, Schürch CM, Bührer ED, et al. CD70/CD27 signaling promotes blast stemness and is a viable therapeutic target in acute myeloid leukemia. J Exp Med. 2017. 214(2), 359–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mueller C, Liotta LA, Espina V. Reverse phase protein microarrays advance to use in clinical trials. Mol Oncol. 2010. 4(6), 461–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Akbani R, Becker KF, Carragher N, et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol Cell Proteomics. 2014. 13(7), 1625–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nishizuka S, Spurrier B, Ramalingam S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc. 2008. 3(11), 1796–1808. [DOI] [PubMed] [Google Scholar]
- [7].Akbani R, Ng PK, Werner HM, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun 2014. 5, 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc. 2008. (3), 1796–1808. [DOI] [PubMed] [Google Scholar]
- [9].O'Mahony FC, Nanda J, Laird A, et al. The use of reverse phase protein arrays (RPPA) to explore protein expression variation within individual renal cell cancers. J Vis Exp. 2013. (71), 50221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaushik P, Molinelli EJ, Miller ML, et al. Spatial normalization of reverse phase protein array data. PLoS One. 2014. 9(12), e97213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Neeley ES, Baggerly KA, Kornblau SM. Surface adjustment of reverse phase protein arrays using positive control spots. Cancer Inform. 2012. 11, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ju Z, Liu W, Roebuck PL, et al. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics. 2015. 31(6), 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007. 23(15), 1986–1994. [DOI] [PubMed] [Google Scholar]
- [14].Sun M, Lai D, Zhang L, Huang X. Modified SuperCurve method for analysis of reverse-Phase protein array data. J Comp Biol. 2015. 22(8), 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Troncale S, Barbet A, Coulibaly L, et al. NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data. PLoS One. 2012. 7(6), e38686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang L, Wei Q, Mao L, Liu W, Mills GB, Coombes K. Serial dilution curve: a new method for analysis of reverse phase protein array data. Bioinformatics. 2009. 25(5), 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Neeley ES, Kornblau SM, Coombes KR, Baggerly KA. Variable slope normalization of reverse phase protein arrays. Bioinformatics. 2019. 25(11), 1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gallagher RI, Espina V. Reverse phase protein arrays: mapping the path towards personalized medicine. Mol Diagn Ther. 2014. 18(6), 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Boellner S, Becker K. Reverse phase protein arrays-quantitative assessment of multiple biomarkers in biopsies for clinical use. Microarrays. 2015. 4(2), 98–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006. 5(10), 2512–2521. [DOI] [PubMed] [Google Scholar]
- [21].Grote T, Siwak DR, Fritsche HA, et al. Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19-9 levels in pancreatic cancer. Proteomics. 2008. 8(15), 3051–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Byron A. Reproducibility and crossplatform validation of reverse-phase protein array data. In: Reverse Phase Protein Arrays (Volume 1188). 2019. Anonymous Springer Singapore, Singapore, 181–201. [DOI] [PubMed] [Google Scholar]
- [23].Byron A, Bernhardt S, Ouine B, et al. Integrative analysis of multi-platform reverse-phase protein array data for the pharmacodynamic assessment of response to targeted therapies. Sci Rep. 2020. 10(1), 21985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dupuy L, Gauthier C, Durand G, et al. A highly sensitive near-infrared fluorescent detection method to analyze signalling pathways by reverse-phase protein array. Proteomics. 2009. 9(24), 5446–5454. [DOI] [PubMed] [Google Scholar]
- [25].Hu CW, Qiu Y, Ligeralde A, et al. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat Biomed Eng. 2019. 3(11), 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dvinge H, Ries RE, Ilagan JO, Stirewalt DL, Meshinchi S, Bradley RK. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci U S A. 2014. 124(21), 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Aasebø E, Mjaavatten O, Vaudel M, et al. Freezing effects on the acute myeloid leukemia cell proteome and phosphoproteome revealed using optimal quantitative workflows. J Proteomics. 2016. 145, 214–225. [DOI] [PubMed] [Google Scholar]
- [28].Di Giuseppe F, Pierdomenico L, Eleuterio E, et al. Cryopreservation Effects on Wharton’s Jelly Stem Cells Proteome. Stem Cell Rev and Rep. 2014. 10(3), 429–446. [DOI] [PubMed] [Google Scholar]
- [29].Drexler HG, Uphoff CC. Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology. 2002. 39(2), 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Drexler HG, Dirks WG, MacLeod RA, Uphoff CC. False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017. 140(5), 1209–1214. [DOI] [PubMed] [Google Scholar]
- [31].Miller CJ, Kassem HS, Pepper SD, Hey Y, Ward TH, Margison GP. Mycoplasma infection significantly alters microarray gene expression profiles. BioTechniques. 2003. 35(4), 812–814. [DOI] [PubMed] [Google Scholar]
- [32].Gedye C, Cardwell T, Dimopoulos N, et al. Mycoplasma infection alters cancer stem cell properties in vitro. Stem Cell Rev. 2016. 12(1), 156–161. [DOI] [PubMed] [Google Scholar]
- [33].Hoff FW, Hu CW, Qutub AA, et al. Mycoplasma contamination of leukemic cell lines alters protein expression determined by reverse phase protein arrays. Cytotechnology. 2018. 70(6), 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Horton TM, Hoff FW, van Dijk AD, et al. The effects of sample handling on proteomics assessed by reverse phase protein arrays (RPPA): functional proteomic profiling in leukemia. J Proteomics. 2021. 233(104046). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hoff FW, Lu Y, Kornblau SM. Antibody Screening (Volume 1188). Springer International Publishing AG. 2019. Singapore. [Google Scholar]
- [36].Jensen B, Swigart P, Simpson P. Ten commercial antibodies for alpha-1-adrenergic receptor subtypes are nonspecific. Naunyn Schmiemenbergs Arch Pharmacol. 2009. 379(4), 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Egelhofer TA, Minoda A, Klugman S, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011. 18(1), 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Älgenäs C, Agaton C, Fagerberg L, et al. Antibody performance in western blot applications is context-dependent. Biotechnol J. 2014. 9(3), 435–445. [DOI] [PubMed] [Google Scholar]
- [39].Berglund L, Björling E, Oksvold P, et al. A genecentric human protein atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008. 7(10), 2019–2027. [DOI] [PubMed] [Google Scholar]
- [40].Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nature Methods. 2016. 13(10), 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sjöberg R, Mattsson C, Andersson E, et al. Exploration of high-density protein microarrays for antibody validation and autoimmunity profiling. N Biotechnol. 2016. 33(5), 582–592. [DOI] [PubMed] [Google Scholar]
- [42].Treindl Fridolin, Ruprecht Benjamin, Beiter Yvonne, et al. A bead-based western for high-throughput cellular signal transduction analyses. Nat Commun. 2016. 7(1), 12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wei M, Haney MG, Rivas DR, Blackburn JS. Protein tyrosine phosphatase 4A3 (PTP4A3/PRL-3) drives migration and progression of T-cell acute lymphoblastic leukemia in vitro and in vivo. Oncogenesis. 2020. 9(1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mariotto E, Bortolozzi R, Volpin I, et al. EB-3D a novel choline kinase inhibitor induces deregulation of the AMPK-mTOR pathway and apoptosis in leukemia T-cells. Biochem Pharmacol. 2018. 155, 213–223. [DOI] [PubMed] [Google Scholar]
- [45].Irwin ME, Nelson LD, Santiago-O'Farrill JM, et al. Small-molecule ErbB inhibitors decrease proliferative signaling and promote apoptosis in Philadelphia chromosome–positive acute lymphoblastic leukemia. PLoS One. 2013. 8(8), e70608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Maraldi T, Bertacchini J, Benincasa M, et al. Reverse-phase protein microarrays (RPPA) as a diagnostic and therapeutic guide in multidrug resistant leukemia. Int J Oncol. 2011. 38(2), 427–435. *Study showing that RPPA enables recognition of overexpressed proteins associated with multidrug resistant leukemia.
- [47].Bortolozzi R, Mattiuzzo E, Trentin L, Accordi B, Basso G, Viola G. Ribociclib, a Cdk4/Cdk6 kinase inhibitor, enhances glucocorticoid sensitivity in B-acute lymphoblastic leukemia (B-All). Biochem Pharmacol. 2018. 153, 230–241. [DOI] [PubMed] [Google Scholar]
- [48].Chen LS, Yang J, Liang H, Cortes JE, Gandhi V. Protein profiling identifies mTOR pathway modulation and cytostatic effects of Pim kinase inhibitor, AZD1208, in acute myeloid leukemia. Leuk Lymphoma. 2016. 57(12), 2863–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Konopleva MY, Walter RB, Hongbo LU et al. Preclinical and early clinical evaluation of the oral AKT inhibitor, MK-2206, for the treatment of acute myelogenous leukemia. Clin Cancer Res. 2014. 20(8), 2226–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Zeng Z, Shi YX, Tsao T, et al. Targeting of mTORC1/2 by the mTOR kinase inhibitor PP242 induces apoptosis in AML cells under conditions mimicking the bone marrow microenvironment. Blood. 2012. 120(13), 2679–2689. *Authors show that RPPA can be utilized to show upregulation of survival signaling pathways were upregulated in AML cocultured with stroma cells.
- [51].Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin. Cancer Res. 2010. 16(6), 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Butler JS, Qiu YH, Zhang N, et al. Low expression of ASH2L protein correlates with a favorable outcome in acute myeloid leukemia. Leuk Lymphoma. 2017. 58(5), 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Quintas-Cardama A, Zhang N, Qiu YH, et al. Loss of TRIM62 expression is an independent adverse prognostic factor in acute myeloid leukemia. Clin. Lymphoma Myeloma Leuk 2015. 15(2), 115–127.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kornblau SM, Qiu YH, Zhang N, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011. 118(20), 5604–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ruvolo PP, Qui YH, Coombes KR, et al. Low expression of PP2A regulatory subunit B55α is associated with T308 phosphorylation of AKT and shorter complete remission duration in acute myeloid leukemia patients. Leukemia. 2011. 25(11), 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ruvolo PP, Qiu Y, Coombes KR, et al. Phosphorylation of GSK3α/β correlates with activation of AKT and is prognostic for poor overall survival in acute myeloid leukemia patients. BBA clinical. 2015. 4, 59–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hu CW, Kornblau SM, Slater JH, Qutub AA. Progeny clustering: a method to identify biological phenotypes. Sci Rep. 2015. 5, 12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000. 100(1), 57–70. [DOI] [PubMed] [Google Scholar]
- [59].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011. 144(5), 646–674. [DOI] [PubMed] [Google Scholar]
- [60].Hoff FW, Hu CW, Qiu Y, et al. Recurrent patterns of protein expression signatures in pediatric acute lymphoblastic leukemia: recognition and therapeutic guidance. Mol Cancer Res. 2018. 16(8), 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hoff FW, Hu CW, Qiu Y, et al. Recognition of recurrent protein expression patterns in pediatric acute myeloid leukemia identified new therapeutic targets. Mol Cancer Res. 2018. 16(8), 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hoff FW, Hu CW, Qutub AA, et al. Proteomic profiling of acute promyelocytic leukemia identifies two protein signatures associated with relapse. Proteomics Clin Appl. 2019. 13(4), e1800133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016. 127(20), 2391–405. [DOI] [PubMed] [Google Scholar]
- [64].Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019. 33(2), 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Golub D, Iyengar N, Dogra S, et al. Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. 2019. 9(417). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ravandi F How I treat Philadelphia chromosome–positive acute lymphoblastic leukemia. Blood. 2019. 133(2), 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pollyea DA. New drugs for acute myeloid leukemia inspired by genomics and when to use them. Hematology Am Soc Hematol Educ Progam. 2018. 2018(1), 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Estey EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol. 2018. 93(10), 1267–1291. [DOI] [PubMed] [Google Scholar]
- [69].Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013. 19(2), 202–208. [DOI] [PubMed] [Google Scholar]
- [70].Jonas BA, Pollyea DA. How we use venetoclax with hypomethylating agents for the treatment of newly diagnosed patients with acute myeloid leukemia. Leukemia. 2019. 33(12), 2795–2804. [DOI] [PubMed] [Google Scholar]
- [71].DiNardo CD, Pratz K, Pullarkat V, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019. 133(1), 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].DiNardo CD, Tiong IS, Quaglieri A, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020. 135(11), 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Singh Mali R, Zhang Q, DeFilippis RA, et al. Venetoclax combines synergistically with FLT3 inhibition to effectively target leukemic cells in FLT3-ITD+ acute myeloid leukemia models. Haematologica. 2021. 106(4), 1034–1046. **Authors report the important finding that targeting FLT3-ITD in combination with venetoclax has strong anti-leukemia activity.
- [74].Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia. 2021. 35(9), 2482–2495. [DOI] [PubMed] [Google Scholar]
- [75].Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009. 114(12), 2489–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van den Berghe H, David G, Broeckaert-Van Orshoven A, et al. A new chromosome anomaly in acute lymphoblastic leukemia (ALL). Hum Genet. 1979. 46(2), 173–180. [DOI] [PubMed] [Google Scholar]
- [77].Oshimura M, Freeman AI, Sandberg AA. Chromosomes and causation of human cancer and leukemia. XXVI. Banding studies in acute lymphoblastic leukemia (ALL). Cancer. 1977. 40(3), 1161–1172. [DOI] [PubMed] [Google Scholar]
- [78].Armstrong SA, Krivtsov AV. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007. 7(11), 823–833. [DOI] [PubMed] [Google Scholar]
- [79].Zarka J, Short NJ, Kanagal-Shamanna R, Issa GC. Nucleophosmin 1 mutations in acute myeloid leukemia. Genes. 2020. 11(6), 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood. 2020. 136(15), 1707–1721. [DOI] [PubMed] [Google Scholar]
- [81].Hoff FW, van Dijk AD, Qiu Y, et al. Heat shock factor 1 (HSF1-pSer326) predicts response to bortezomib-containing chemotherapy in pediatric AML: a COG Report. Blood. 2021. 137(8), 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]